Abstract
Cockayne syndrome (CS) is a rare inherited human genetic disorder characterized by UV sensitivity, developmental abnormalities and premature aging. The cellular and molecular phenotypes of CS include increased sensitivity to oxidative and UV-induced DNA lesions. The CSB protein is thought to play a pivotal role in transcription-coupled repair and CS-B cells are defective in the repair of the transcribed strand of active genes, both after exposure to UV and in the presence of oxidative DNA lesions. A previous study has indicated that a conserved helicase ATPase motif II residue is essential for the function of the CSB protein in responding to UV-induced DNA damage in a hamster cell line. Due to the limitations in studying a complex human disorder in another species, this study introduced the site-directed mutation of the ATPase motif II in the human CSB gene in an isogenic human cell line. The CSB mutant allele was tested for genetic complementation of UV-sensitive phenotypes in the human CS-B cell line CS1AN.S3.G2. In addition, the incision of an 8-oxoguanine lesion by extracts of the CS-B cell lines stably transfected with the wild-type or ATPase mutant CSB gene has been investigated. The ATPase motif II point mutation (E646Q) abolished the function of the CSB protein to complement the UV-sensitive phenotypes of survival, RNA synthesis recovery and apoptosis. Interestingly, whole-cell extract prepared from these mutant cells retained wild-type incision activity on an oligonucleotide containing a single 8-oxoguanine lesion, whereas the absence of the CSB gene altogether resulted in reduced incision activity relative to wild-type. These results suggest damage-specific functional requirements for CSB in the repair of UV-induced and oxidative lesions in human cells. The transfection of the mutant or wild-type CSB gene into the CS1AN.S3.G2 cells did not alter the expression of the subset of genes examined by cDNA array analysis.
INTRODUCTION
Cockayne syndrome (CS) is a rare inherited human genetic disorder categorized as a segmental progeroid disorder. Affected individuals suffer from postnatal growth failure resulting in cachectic dwarfism, photosensitivity, skeletal abnormalities, mental retardation and progressive neurological degeneration, retinopathy, cataracts and sensorineural hearing loss (1,2). Two complementation groups of CS (CS-A and CS-B) have been identified and the corresponding genes have been cloned (3,4). The cellular phenotype of CS involves an increased sensitivity to a number of DNA-damaging agents including UV radiation, ionizing radiation and H2O2 (2). Nucleotide excision repair (NER) is a complex process, which removes a broad spectrum of DNA lesions, and two subpathways have been identified: (i) the transcription-coupled repair (TCR) pathway and (ii) the global genome repair (GGR) pathway. The TCR pathway is confined to the transcribed strand of active genes, is dependent on RNA polymerase II (RNA pol II) transcription (5,6), and accelerates the repair of these regions. The GGR pathway removes lesions from both transcriptionally active and inactive genes. CS cells are defective in TCR, while maintaining a proficient GGR of UV-induced DNA damage (7–9). A characteristic feature of CS cells is the lack of RNA synthesis recovery after UV damage (10), which may be explained by the defect in TCR.
Both the CSA and CSB genes have been cloned and their products biochemically characterized. CSA is a 44 kDa protein and belongs to the ‘WD repeat’ family of proteins (4). This family has been found to exhibit structural and regulatory roles but no enzymatic activity. The CSB gene product is a 168 kDa protein (3) that belongs to the SWI/SNF family of proteins, which all contain seven sequence motifs conserved between two superfamilies of DNA and RNA helicases (11). In addition, the CSB protein contains an acidic amino acid stretch, a glycine-rich region and two putative nuclear localization signal (NLS) sequences. The CSB protein is a DNA-stimulated ATPase, but fails to exhibit DNA unwinding activity as measured by a conventional strand displacement assay (12,13). Transfection of the human CSB cDNA into the human CS-B cell line CS1AN.S3.G2 complements the UV sensitivity and the delay in RNA synthesis recovery (3). Similarly, it restores UV resistance and RNA synthesis to normal levels in the hamster CS-B homolog, UV61 (14). Gene-specific repair studies have also demonstrated complementation of the TCR defect by the human CSB gene in hamster UV61 cells (14,15).
CSB may facilitate repair of active genes by recruiting proteins involved in DNA repair. In vitro studies demonstrate that CSB exists in a quaternary complex composed of RNA pol II, CSB, DNA and the RNA transcript. ATP hydrolysis is required for formation of this complex (13). This complex recruits additional proteins, including the transcription factor IIH (TFIIH) core subunits p62 and XPB (16). TFIIH is a complex factor thought to promote local DNA unwinding in both transcription initiation by RNA pol II and promoter escape, as well as in NER (17–22). Co-immunoprecipitation studies have demonstrated an interaction between the XPG and CSB proteins (23). XPG is known to function as an endonuclease in NER and additionally interacts with multiple components of TFIIH (23). In vitro data also suggest that CSB interacts with CSA and with the NER damage recognition factor XPA (4,24). These protein interactions support a direct role of CSB protein in TCR.
While the defect in TCR of UV-induced DNA damage has been well characterized, a role for CSB in the TCR of oxidative damage has also been implicated. Treatment with H2O2 or ionizing radiation induces a large variety of altered bases, including oxidatively damaged bases, as well as single-strand breaks. 8-oxoguanine is a DNA base modification caused by oxidative stress from environmental agents and from endogenous metabolic processes and is repaired mainly via the process of base excision repair (BER) (25). Two studies have demonstrated a potential slight sensitivity in the colony forming ability of primary CS-B cells after γ-irradiation as well as defective strand-specific repair of ionizing radiation-induced damage and thymine glycols introduced by H2O2 treatment (26,27). Recently, LePage et al. (28) demonstrated that thymine glycol and 8-oxoguanine are removed by a TCR process that does not involve NER but requires the XPB and XPD components of TFIIH as well as XPG and CSB. That study further suggested that 8-oxoguanine is a block to transcription and is repaired preferentially in the transcribed strand of an active gene (29). A previous study from this laboratory has shown that in vitro incision of 8-oxoguanine lesions by extracts derived from CS-B cells was 50% of the incision level compared with extracts prepared from normal cells, whereas the incision rates of uracil and thymine glycol were similar in CS-B and normal cells (30).
CSB may indirectly stimulate TCR of UV-induced and oxidative damage by facilitating transcription. Members of the SWI2/SNF2 family are involved in transcription regulation, chromatin remodeling and DNA repair, including the disruption of protein–protein and protein–DNA interactions (reviewed in 31). In fact, it is still debated whether CS is due to a primary defect in transcription or DNA repair (2,32). A basal transcription defect has been observed in human CS-B lymphoblastoid cells and fibroblasts without any exposure to DNA-damaging agents (33). The reduced transcription in CS-B cells is complemented in chromatin by the addition of normal cell extract and in intact cells by transfection with the CSB gene (33). In a reconstituted system, purified CSB protein enhances the rate of transcription by RNA pol II (24), suggesting that CSB protein may indirectly stimulate TCR by facilitating the process of transcription. Indeed, Citterio et al. (34) recently demonstrated that CSB binding to DNA causes an alteration of the DNA conformation and CSB is able to remodel chromatin structure at the expense of ATP hydrolysis. In addition, CSB interacts directly with core histones (34). Thus, CSB may serve dual roles as a transcription elongation factor and a repair-coupling factor at the site of the RNA pol II-blocking lesions, and the CS phenotype may arise from a combined deficiency in DNA repair and transcription. Distinct functional domains of the protein may mediate the biological function of CSB in these different pathways.
To explore the molecular-genetic role of CSB in vivo, we have introduced site-directed mutations into the human CSB gene in order to investigate the functional significance of conserved and unique regions of the protein. This represents the first study of this kind in human cells, since previous studies were done in hamster cells. The mutant alleles were tested for genetic complementation of DNA damage-sensitive phenotypes in both DNA repair and transcription. In hamster cell lines, we have previously demonstrated that deletion mutations in a highly acidic region in the N-terminal portion of the protein (60% of a 39 amino acid stretch are acidic residues) did not impair the genetic function in the processing of UV-induced DNA damage (15). On the contrary, point mutation of a highly conserved glutamic acid residue in ATPase motif II abolished the ability of CSB protein to complement the UV-sensitive phenotypes of survival, RNA synthesis recovery, gene-specific repair and apoptosis following UV irradiation of the hamster CS-B homolog, UV61 cells (15,35). This ATPase mutant also failed to confer cellular resistance to 4-nitroquinoline-1-oxide (15), which introduces bulky DNA adducts repaired by GGR, suggesting CSB is involved in a TCR-independent pathway. A point mutation in ATPase motif I of CSB rendered the protein totally defective in ATP hydrolysis, but it was still able to partially rescue the delay in RNA synthesis recovery after exposure to UV light when microinjected into an immortalized human CS-B fibroblast cell line (36). This suggests a requirement for ATP hydrolysis by CSB protein in the repair of these lesions (36). The dramatic loss of RNA synthesis recovery after UV damage in the ATPase motif II mutant versus the compromised, but residual, function of the ATPase motif I mutant may be explained by a number of reasons. First, the site-directed mutations are directed at two spatially distinct sequence motifs, even though both motifs I and II are implicated in nucleotide binding and/or hydrolysis. Secondly, the two mutants were assayed in very different cellular backgrounds. The ATPase motif I mutant generated by Citterio et al. (36) was studied in a SV40-transformed human CS-B cell line, CS1AN, whereas the ATPase motif II mutant was studied in a hamster CS-B cell line, in which the CSB gene was originally identified by cross-complementation. Due to these differences, and the apparent complexity of the function of CSB, it is of benefit to study these site-directed mutations in a human cell line.
Human cells display certain phenotypical characteristics distinct from hamster cells that are likely to involve the function of the CSB protein. DNA repair (GGR and TCR) display different properties between the two organisms as hamster cells essentially only have TCR. In addition, the transcription defect observed in human CS-B cells (33) is not present in the hamster CS-B homolog cell line UV61 (our unpublished data). We recently demonstrated that the CSB apoptotic pathway triggered by DNA damage can occur independently of the transactivation potential of p53 in hamster cells (35), another distinguishing feature from human cells. It is evident that certain functions of CSB are not perfectly conserved between the two species, and that structure–function studies of CSB will be best served by the study of the in vivo functions of the human CSB protein in the human CS-B null cell line. To address the biological function of the ATPase domain of CSB protein in a human background, we have established stably transfected isogenic CS-B human lines and carefully characterized the level of mutant and wild-type CSB expression in the host cells. Having established a valid genetic system, we investigated the DNA repair/transcription status of the isogenic CS-B cell lines. This approach has enabled us to directly assess the biological importance of the ATPase domain of CSB protein in human cells. Previously, the functional importance of the ATPase domain of CSB protein in human cells was studied using microinjected purified recombinant mutant (K538R, motif I) or wild-type CSB protein (36). The present study has the advantage that the human CS-B null cell line is complemented by human CSB protein that is endogenously expressed. The isolation of isogenic clones from the transfected mass populations has enabled us to genetically characterize cell lines that have equivalent levels of expression of either the wild-type or mutant CSB protein. For these reasons, the results from this study advance our understanding of CSB function in human cells in a setting as physiological as possible.
Human CS-B fibroblasts were transfected with the wild-type CSB gene, the ATPase motif II point mutant generated by replacing a highly conserved glutamic acid residue with a neutral glutamine, or the expression vector alone. The SV40-immortalized CS1AN derivative CS1AN.S3.G2 retains one allele of the CSB gene which carries an A1088→T transversion leading to a premature stop at amino acid 337, encoding a protein of 336 amino acids lacking all postulated motifs (3). The transfected wild-type and mutant alleles were tested for their ability to function in DNA repair and their effect on gene expression. The ATPase motif II point mutation abolished the function of the CSB protein to complement the UV-sensitive phenotypes of survival, RNA synthesis recovery and apoptosis.
In addition, we wished to examine the role of this site-directed mutant in the repair of oxidative DNA lesions. The same ATPase motif II mutant complemented the incision activity of an 8-oxoguanine lesion by whole-cell extracts from transfected cell lines, whereas the absence of the CSB gene altogether resulted in reduced incision activity. These results suggest different functional requirements for CSB in the repair of UV-induced and oxidative lesions. The transfection of the mutant or wild-type CSB gene into the CS1AN cells did not alter the expression of the subset of genes examined by cDNA array analysis when compared with cells transfected with the vector alone.
MATERIALS AND METHODS
Cell lines and culture conditions
GM00637D is an SV40-transformed normal human fibroblast cell line. CS1AN.S3.G2 is an SV40-transformed human fibroblast cell line belonging to CS complementation group B and has been previously described by Mayne et al. (37). CS1AN.S3.G2 cell lines transfected with the mammalian expression vector pcDNA3.1 (pc3.1; Invitrogen, San Diego, CA) or pc3.1 containing the wild-type human CSB gene are designated CS1AN/pc3.1 and CS1AN/pc3.1-CSBwt, respectively. CS1AN cells transfected with pc3.1-CSB containing a mutation in the ATPase/helicase domain II is designated CS1AN/pc3.1-CSBE646Q. All cell lines were routinely grown in minimal essential medium supplemented with 15% fetal bovine serum and antibiotics. CS1AN.S3.G2 cell lines transfected with pc3.1, which contains the neomycin resistance gene, were grown in media containing 400 µg/ml geneticin (Life Technologies, Rockville, MD).
Site-directed mutagenesis and selection of stable transfectant cell lines
pcBLsSE6, a plasmid containing the entire human CSB cDNA (kindly provided by Dr Jan Hoeijmakers, Rotterdam University, Rotterdam, The Netherlands) was used for site-directed mutagenesis by published procedures (15). The oligonucleotide 5′-TTGTGTCCTTGGTCCAAGATC-3′ was used to replace the negatively charged Glu646 in the CSB protein to a neutral glutamine (E646Q). The CSB gene, containing mutant or wild-type sequence, was cloned into the mammalian expression vector pc3.1. The entire CSB coding sequence of pc3.1-CSBwt and pc3.1-CSBE646Q was sequenced to verify that the coding sequences of the two plasmids were identical with the exception of the site-specific mutation. CsCl-purified pc3.1-CSB plasmids were linearized as previously described by Brosh et al. (15) and transfected into CS1AN.S3.G2 cells using SuperFect transfection reagent (Qiagen, Santa Clarita, CA). Briefly, 150 000 cells were seeded in a 3-cm2 dish and allowed to grow until 35% confluent. A 10-µg aliquot of linearized plasmid DNA was preincubated with 8 µl of SuperFect reagent and added to the cells in 0.6 ml of complete media. The cells were incubated with the liposome–DNA mixture for 3 h at 37°C. The media were replaced with complete media and the cells were grown for an additional 48 h at 37°C. Cells were trypsinized and transferred to a 10-cm2 dish, and geneticin was added to a final concentration of 400 µg/ml for selection of antibiotic-resistant cells. After 14 days of selection, the surviving cells were trypsinized and seeded for isolation of clones (50 cells per 10 cm2 dish). Individual colonies were isolated and screened for expression of the CSB transcript.
Analysis of CSB expression
To evaluate the expression of intact CSB transcript in isolated transfectant clones, RNA was isolated and evaluated by reverse transcription and the polymerase chain reaction (PCR) using CSB-specific primers as previously described by Mallery et al. (38) with the following modifications. RNA was extracted using RNA STAT-60 (Tel-Test Inc., Friendswood, TX) according to the manufacturer’s protocol. cDNA synthesis was performed by incubating 5 µg of total RNA, 1.25 µg of oligo(dT)12–18 primer (Life Technologies) and 500 U Superscript II reverse transcriptase (Life Technologies) according to manufacturer’s recommendation. For PCR amplification of cDNA products, 2.5 U AmpliTaq Gold (Applied Biosystems, Foster City, CA) were used according to manufacturer’s procedures. The 4.7 kb cDNA product was amplified as six overlapping fragments ranging in size from 0.6 to 1.5 kb using primers and annealing conditions previously described by Mallery et al. (38). DNA products were verified by electrophoresis on an agarose gel with analysis by ethidium bromide staining (data not shown). For sequencing, PCR products were purified using a QIAquick PCR purification kit (Qiagen) and analyzed using an ABI automatic sequencer. Comparison of sequence analysis of the appropriate cDNA fragment obtained from CS1AN/pc3.1-CSBwt and CS1AN/pc3.1-CSBE646Q confirmed the presence of the engineered mutation at the defined position in the mutant cell line. The entire CSB cDNAs of pc3.1-CSBwt and pc3.1-CSBE646Q were sequenced to verify that the only difference between the wild-type and E646Q mutant CSB genes was the engineered mutation.
Two approaches were used to verify the relative amounts of CSB transcripts and protein expressed in the transfectant cell lines. To quantitate the relative levels of CSB expression in CS1AN/pc3.1-CSBwt and CS1AN/pc3.1-CSBE646Q, we performed relative quantitative RT–PCR experiments (QuantumRNA module; Ambion, Austin, TX). The QuantumRNA module provides a method for comparing relative transcript abundance standardized by co-amplification of a highly conserved fragment of 18S rRNA as an internal standard. The CSB transcript and 18S rRNA were analyzed during the exponential phase of amplification to determine an accurate level of relative expression. PCR cycling of the CSB fragment and 18S RNA standard was performed in a single tube. For CSB cDNA amplification, the primers 5′-GGTGTTAGGTGGCTGTGGGAATT-3′ (3F) and 5′-GTATCTCGTAAGACACACATGCACAC-3′ (3R) were used, which produce a 671-bp product from the coding sequence of CSB mRNA (38). The RNA used for cDNA synthesis was isolated from the transfectant cell lines and DNase treated as described below. cDNA synthesis was performed as described above. For PCR amplification of cDNA products, 0.5 U AmpliTaq Gold (Applied Biosystems) were used according to manufacturer’s procedures. In addition to standard reaction components, PCR mixtures contained 0.2 µM of CSB primers 3F and 3R, 0.2 µM or 18S RNA primer:competimer (4:6) in a total volume of 50 µl. A hot start for RT–PCR reactions was used; the cycling conditions were as follows: hot start 5 min at 80°C, 5 min at 49°C, 1 min at 94°C, followed by 39 cycles consisting of 30 s at 94°C, 45 s at 67°C, 1 min at 72°C; and one final extension cycle for 7 min at 72°C. The RT–PCR products were electrophoresed on 1% agarose gel stained with SybrGreen fluorescence dye and scanned with a FluoroImager and quantitated using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The results of the relative RT–PCR experiments gave ratios of the CSB transcript to 18S RNA standard (671-nt CSB fragment:488-nt 18S RNA internal standard) for CSBwt and CSBE646Q were determined to be 0.89 ± 0.12 and 0.81 ± 0.04, respectively. The results demonstrate that the wild-type and mutant CSB transcripts are expressed with nearly the same efficiency in the transfectant cell lines. There was very little to no CSB transcript from CSIAN/pc3.1 as judged by the inability to detect on a SybrGreen-stained gel a 671-bp product using the RT–PCR method and CSB site-specific primers 3F and 3R. Under identical reaction conditions, an abundant RT–PCR product (671 bp) was detected for both CSIAN/pc3.1-CSBwt and CSIAN/pc3.1-CSB-E646Q (data not shown).
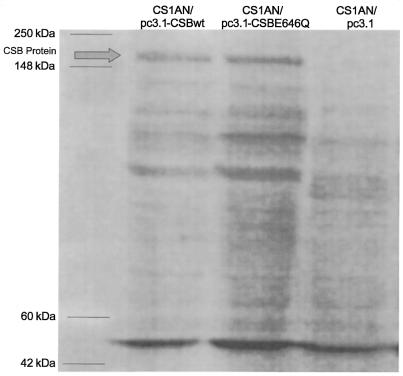
To measure the relative amounts of CSB protein in the transfected cell lines, whole-cell extracts were prepared as previously described by Manley et al. (39). A 100-µg aliquot of protein was separated on an 8–16% polyacrylamide gel and transferred to a PVDF membrane. Immunoblots were probed with human anti-ERCC6 IgG fraction monclonal antibody that recognizes the ERCC6/CSB protein (Austral Biologicals, San Ramon, CA) and developed with a chemiluminescent substrate (ECL+ kit) according to manufacturer’s directions (Amersham Pharmacia Biotech, Piscataway, NJ). The antibody detected similar levels of the CSB protein in extracts from both the CS1AN/pc3.1-CSBwt and CS1AN/pc3.1-CSBE646Q cell lines. No band corresponding to the CSB protein was detected in the CS1AN/pc3.1 cell line (Fig. 1). Purified CSB protein was included as a control.
Figure 1.
Western blot analysis of whole-cell extracts from CS1AN cell lines transfected with the vector pc3.1 or pc3.1 containing the wild-type human CSB gene (pc3.1-CSBwt) or CSB containing a mutation in the ATPase domain II (pC3.1-CSBE646Q). Lane 1, CS1AN/pc3.1-CSBwt; lane 2, CS1AN/pc3.1-CSBE646Q; lane 3, CS1AN/pc3.1. Blots were probed with a human anti-ERCC6 IgG fraction monoclonal antibody. Arrow indicates the full-length 168 kDa CSB protein. Purified CSB protein was run as a control.
UV and H2O2 survival assays
Cells were trypsinized and 500 cells were seeded per 10-cm2 dish and allowed to attach overnight. For UV treatment, cells were washed once with PBS and then irradiated at the indicated doses with a UV lamp (254 nm) and returned to complete medium. For H2O2 treatment, cells were washed and incubated with the indicated dose of H2O2 in serum-free minimal medium for 15 min in a 37°C incubator, then washed with PBS and returned to complete medium. The cells were grown for 10 days, washed once with PBS, fixed with methanol and stained with methylene blue. Blue colonies were counted to determine the clonogenic survival of cells.
RNA synthesis recovery
Transfectant cell lines were grown in the presence of [14C]thymidine (0.02 µCi/ml) for 3 days to uniformly label the DNA. The cells were washed with PBS and irradiated with UV light (254 nm) at a dose of 4 J/m2. Cells were restored to complete media and allowed to recover for the indicated times. Cells were subsequently pulse labeled with 5 µCi/ml [3H]uridine for 60 min at 37°C, washed once with PBS and lysed in 10 mM Tris, pH 8.0, 1 mM EDTA buffer containing 0.5% SDS and 100 µg/ml proteinase K for 2 h at 37°C. Trichloroacetic acid (TCA) was added to the cell lysate at a final concentration of 10%, and the samples were then spotted onto glass-fiber discs (Whatman, Maidstone, UK). The filters were sequentially washed in 5% TCA, 70% ethanol, and acetone. The TCA-precipitable radioactivity was scintillation counted.
Detection of UV-induced cell death by fluorescence staining
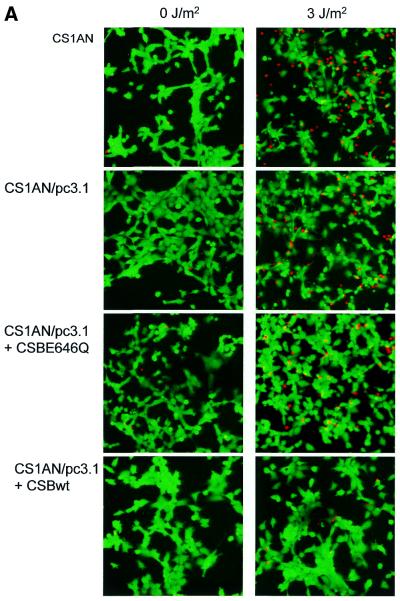
Apoptotic and necrotic cells were distinguished from viable cells by staining with fluorescein diacetate (FDA; 5 µg/ml) and propidium iodide (PI; 0.5 µg/ml). Control and UV-irradiated (254 nm, 3 J/m2) CS1AN cell lines were stained with FDA and PI 24 h post-irradiation. The proportion of apoptotic cells was determined by examining the cells in a fluorescence microscope, and the images of viable and apoptotic cells were captured using confocal microscopy. One hundred cells were counted per cell line in three independent experiments and percent apoptotic cells was determined.
Incision of 8-oxoguanine-, 5-hydroxyl-dCytosine-, thymine glycol-containing oligonucleotides by whole-cell extracts
Oligonucleotides were purchased from Midland Certified Reagent Co. (Midland, TX). The following oligonucleotides were employed: 5′-ATATACCGCG[8-oxoguanine]CCGGCCGATCAAGCTTATT-3′; 5′-ATATACCGCG[5-hydroxyl-dCytosine]CCGGCCGATCAAGCTTATT-3′; and 5′-CCAGCGCACGACGCA[Tg]GCACGACGACCGGG-3′ (Tg, thymine glycol). All oligonucleotides were 5′-end-labeled using [γ-32P]dATP and annealed to the complementary strand as previously described by Dianov and Lindahl (40).
Whole-cell extracts were prepared by the method of Manley et al. (39). Incision reactions contained 0.15 nmol of oligonucleotide duplex, 1 µg poly(dI·dC), 20 mM HEPES–KOH, pH 7.8, 100 mM KCl, 5 mM DTT, 5 mM EDTA, 2 mM MgCl2, and the indicated amount of extract protein. After incubation at 37°C for the indicated time, the reactions were terminated by the addition of SDS and proteinase K, and incubated for 10 min at 55°C. In a total 20 µl of reaction mixture, DNA precipitation was carried out by adding 4 µl of 5 mg/ml glycogen (Ambion), 8 µl of 11 M ammonium acetate and 135 µl of ice-cold ethanol. Incubation was overnight at –20°C. Centrifugation was for 1 h at 4°C. Samples were then washed with 500 µl of 70% ethanol and centrifuged for 10 min. The pellet was dried by speed-vacuum centrifugation to 500 mTorr. Samples were then resuspended in 10 µl of formamide loading dye (5% EDTA, 0.02% bromophenol blue, 0.02% xylene cyanol in 95% formamide). Samples were then electrophoresed in a 20% polyacrylamide gel containing 7 M urea, 89 mM Tris-borate pH 8.0 and 2 mM EDTA. The band images were visualized by autoradiography and quantified on a PhosphorImager (Molecular Dynamics).
Gene expression analysis using cDNA arrays
Atlas™ human and human stress/toxicology cDNA expression arrays (Clontech Laboratories, Palo Alto, CA) and a general human cDNA array containing approximately 15 000 known genes and ESTs (kindly donated by Dr Kevin Becker, NIA cDNA Array Unit, NIH) were used to examine the effect of the transfected CSB gene (either wild-type or the ATPase mutant) on gene expression in the CS1AN cells. Total RNA was isolated using RNA STAT-60 (Tel-Test Inc, Friendswood, TX) according to the manufacturer’s protocol. The RNA was treated with RNase-free DNase I (Boehringer Mannheim, Indianapolis, IN) according to the Clontech manual. A 4-µg aliquot of RNA was used for cDNA synthesis, labeling with [α-32P]dCTP (Clontech) or [α-33P]dCTP (NIA arrays) and following the procedures described in the Clontech manual and the protocol from the NIA Array Unit (http://www.grc.nia.nih.gov/branches/rrb/dna/rnalabel.htm). The labeled cDNA was hybridized to the arrays, followed by washing and exposure of the membrane on a PhosphorImager. Three independent RNA isolations were hybridized to three sets of Clontech arrays. One set of NIA arrays was probed with one RNA isolation. Image analyses and quantifications were performed by using either P-SCAN (http://abs.cit.nih.gov/pscan) (Clontech arrays) or ImageQuant and Excel (NIA arrays).
RESULTS
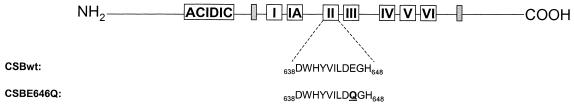
We have previously published a study detailing the generation of a site-specific mutation in the ATPase motif II of CSB, and its inability to genetically complement the UV sensitivity of the CS-B homolog, hamster UV61 cells. A mutation made to the ATPase motif I by another laboratory (36) demonstrated partial complementation of RNA synthesis by the CSB gene after UV treatment in a human CS-B cell line. This difference may be an effect of the difference in mutation location, even though both sites are involved in ATP binding and hydrolysis, or it may be a function of cellular background. In this study, we characterized the ability of the ATPase motif II mutant to complement the UV sensitivity in a human background, as well as its ability to affect gene expression and its function in an in vitro assay measuring incision of an oxidative base lesion. We have replaced the highly conserved glutamic acid found in a large number of ATPases and helicases with a neutral glutamine in ATPase motif II of CSB, designated CSBE646Q (Fig. 2). The CSB alleles were cloned behind a cytomegalovirus strong promotor in the mammalian expression vector pc3.1. The CSB expression plasmids were transfected into the human SV40-transformed CS-B fibroblast cell line CS1AN.S3.G2, and isogenic stable transfectants were obtained. Quantitation of relative amounts of CSB transcript from singular clones of the cell lines demonstrated very similar levels of expression, with CS1AN/pc3.1-CSBE646Q at 91% of CS1AN/pc3.1-CSBwt expression level. Comparison of protein levels by western blot also demonstrated similar levels of CSB protein in CS1AN/pc3.1-CSBwt and CS1AN/pc3.1-CSBE646Q, whereas none was detected in CS1AN/pc3.1 (Fig. 1). We examined repair and transcription in CS-B transfectants by genetic complementation assays following UV-induced DNA damage. We also examined the in vitro incision of an oxidative base lesion, 8-oxoguanine, by whole-cell extracts of the transfected cell lines.
Figure 2.
Schematic of the CSB protein containing the seven conserved ATPase/helicase motifs, a highly acidic region and two NLSs. A site-directed mutation was introduced in the ATPase motif II of CSB. This E646Q mutation changes a highly conserved glutamic acid to glutamine.
UV sensitivity
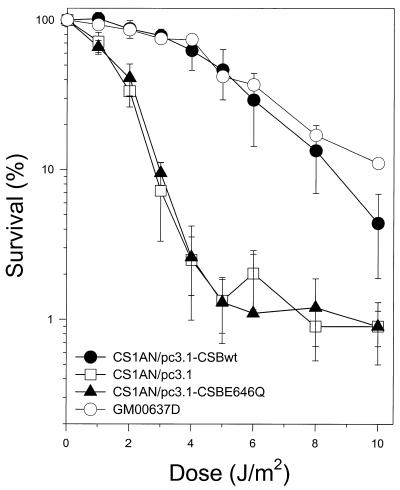
Using the classical clonogenic survival assay, we determined the effect of UV irradiation on the survival rate of the CS-B transfectant cell lines compared with a normal SV40-transformed fibroblast cell line, GM00637D. As demonstrated previously by Troelstra et al. (3), transfection of CS1AN.S3.G2 cells with the CSB wild-type allele restored UV resistance to a level comparable with a normal fibroblast cell line (Fig. 3). However, cells transfected with a CSB allele containing a point mutation in motif II of the ATPase domain, CSBE646Q, were severely compromised in their ability to complement the UV sensitivity exhibited by the vector alone (CS1AN/pc3.1). When the cells were irradiated with 4 J/m2 UV light (254 nm), 63% of cells transfected with the wild-type CSB gene were able to form colonies. Only 2.5% of cells transfected with the plasmid vector or the vector containing the ATPase mutant allele were able to form colonies following this dose. These results indicate that the integrity of the ATPase domain of CSB is critical for cellular resistance to UV light.
Figure 3.
Clonogenic UV survival of a normal fibroblast cell line, GM0637D, and isogenic clonal populations of CS1AN.S3.G2 transfectant cell lines. Cells were seeded, allowed to attach, and irradiated with the indicated doses of UV light (254 nm). After 10 days, colonies were fixed, stained and counted. Percent survival is expressed as the number of UV-irradiated cells forming colonies as a fraction of the colonies formed by unirradiated cells and represent the average of three independent experiments.
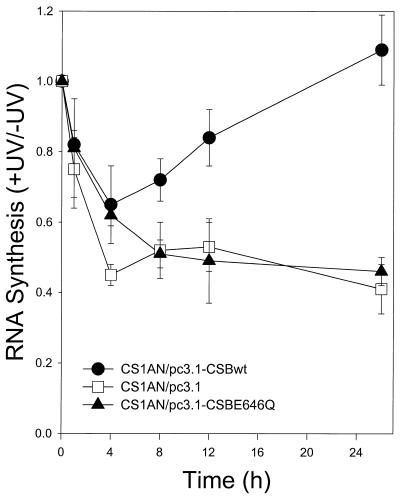
In contrast to repair proficient human cells, CS-B cells are unable to recover their RNA synthesis after UV exposure (10). To determine whether an intact ATPase motif II of CSB is necessary for recovery of RNA synthesis, cells were exposed to UV light (4 J/m2) and RNA synthesis was measured by [3H]uridine incorporation during a 60 min pulse before lysis. As shown in Figure 4, transcription initially drops during the first 4 h after irradiation in all three cell lines. As with UV survival, RNA synthesis recovers to normal levels in cells transfected with the wild-type CSB allele (CSBwt) by 24 h post-irradiation. Transcription levels were reduced to approximately one-half the level of unirradiated cells in cells transfected with the ATPase mutant or the vector alone even after 24 h of recovery. The failure of the ATPase point mutant to complement the RNA synthesis inhibition in CS-B cells strongly suggests that the integrity of the ATPase domain is instrumental for CSB protein to genetically function in the TCR pathway.
Figure 4.
RNA synthesis recovery of CS1AN.S3.G2 transfectant cell lines after UV irradiation. Cells were prelabeled with [14C]thymidine for 72 h, washed with PBS and irradiated with UV light (254 nm, 4 J/m2). Cells were returned to normal medium for the indicated recovery times and pulse labeled in medium containing [3H]uridine 1 h before lysis. Acid-insoluble radioactivity was determined.
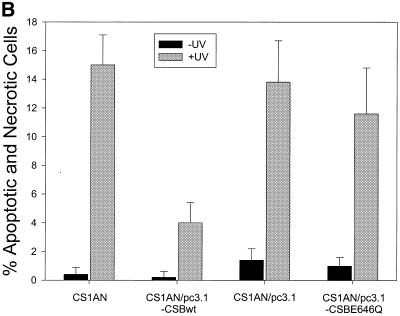
We and others have previously shown that both human and hamster CS-B cells are more susceptible to UV-induced apoptosis, and suggested that defective TCR, leading to transcription blockage, serves as the primary trigger for apoptosis in hamster CS-B cells (35,41). We examined cell death in the human CS1AN.S3.G2 transfectant cell line following treatment with 3 J/m2 UV light. Control and UV-treated cells were stained with FDA and PI to examine cell death 24 h after UV irradiation. FDA stains only the cytoplasm and nucleus of viable cells. The necrotic and apoptotic cells are stained with PI, which requires the loss of cytoplasmic and nuclear membrane integrity. Randomly chosen cells were microscopically analyzed and the images were captured using a confocal microscope (Fig. 5A). Cells stained with PI displayed the fragmented chromatin characteristic of apoptotic cells. One hundred cells per cell line were counted. There were few apoptotic and necrotic cells (∼4%) stained with PI among the unirradiated and UV-exposed CS1AN/pc3.1-CSBwt cells (Fig. 5B). In contrast, ∼15% of the untransfected and vector transfected CS1AN cells were stained with PI following treatment. The ATPase point mutant (CSBE646Q) failed to rescue the cells from death, with 12% of the cells staining with PI after irradiation. In light of our previous findings in hamster and human primary CS-B cells (35), we propose that these human CS1AN cells are undergoing an UV-induced apoptotic response that is complemented by transfection with the wild-type CSB gene, but not by the gene containing a point mutation in the ATPase motif II. Thus, the ATPase motif of CSB is essential for the inhibition of the apoptotic pathway.
Figure 5.
Analysis of UV-induced apoptosis in CS1AN.S3.G2 transfectant cell lines. (A) Cells were unirradiated or irradiated with 3 J/m2 UV light (254 nm). The cells were stained 24 h after treatment with FDA and PI to detect cell death. The viable FDA-stained cells appeared green and the necrotic and apoptotic cells appeared red. (B) One hundred cells were counted per cell line in three independent experiments and percent apoptotic and necrotic cells were determined.
A previous study has demonstrated a potential slight sensitivity to ionizing radiation in primary CS cells when compared with normal primary fibroblasts (26). Using the clonogenic survival assay, we have examined the effect of H2O2 on the survival rate of the CS1AN.S3.G2 transfectant cell lines. We saw no major differences between these cell lines in survival following this treatment (data not shown), which induces a variety of DNA lesions, including oxidatively damaged bases.
Incision of 8-oxoguanine, 5-hydroxyl-dCytosine and thymine glycol
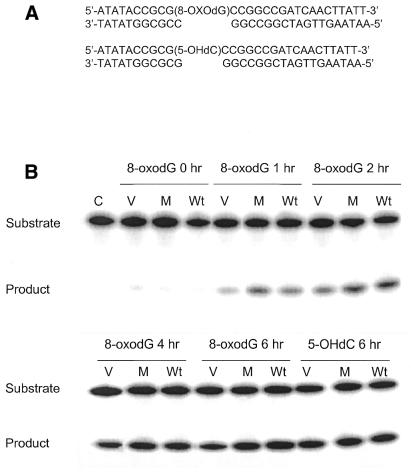
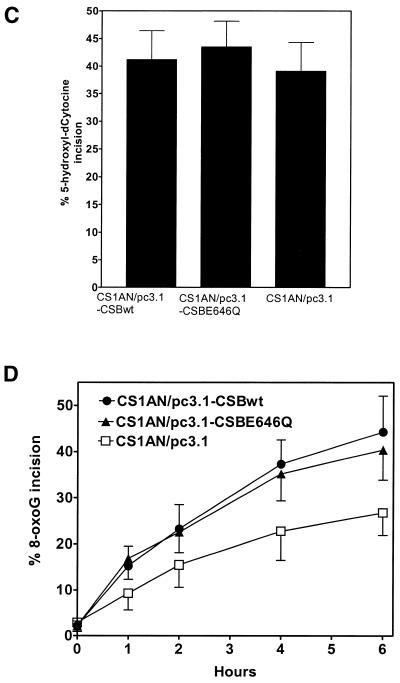
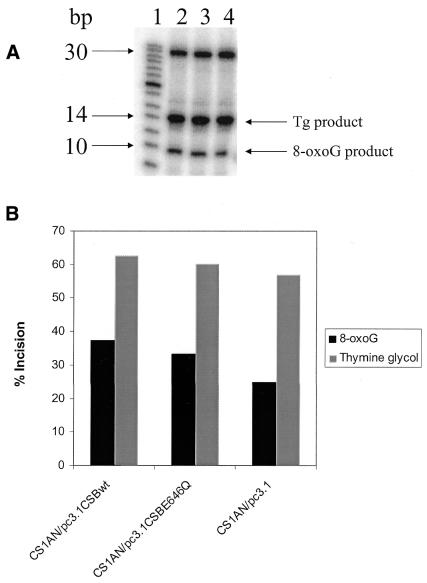
Dianov et al. (30) previously determined that both primary and transformed CS-B cells are defective in incision of 8-oxoguanine lesions by whole-cell extracts, whereas incision of thymine glycol was normal. To determine whether the wild-type CSB gene was able to complement this defect in the cell lines used in this study, and whether an intact ATPase domain is critical for this function, we examined incision activity in cell extracts prepared from these cell lines. Both 8-oxoguanine and 5-hydroxy-dCytosine are thought to be repaired by the BER pathway. In this pathway, a DNA glycosylase specific for the lesion recognizes and incises the N-glycosyl bond between the damaged base and the sugar from the oligonucleotide by a β-elimination reaction mechanism creating an apurinic/apyrimidinic site (AP site). The resulting abasic site is then recognized and cleaved by AP lyase activity of the same glycosylase or from a separate AP endonuclease. This incision results in the conversion of a 29mer 5′-end-labeled substrate into a 10mer product (Fig. 6A). We observed no differences between the three cell lines in incision of a 5-hydroxy-dCytosine containing oligonucleotide duplex (Fig. 6B and C). However, the incision of an 8-oxoguanine-containing oligonucleotide duplex was reduced in the cell line transfected with the vector alone (CS1AN/pc3.1) compared with the cell lines transfected with either the wild-type CSB gene (CS1AN/pc3.1-CSBwt) or with the ATPase point mutant (CS1AN/pc3.1-CSBE646Q) (Fig. 6B and D). The relative incision activity of 8-oxoguanine at 6 h (the time point with the highest differences in the kinetic study) in the CSBwt cells was 44%, whereas in the cells containing no CSB the relative incision was only 27%. Transfection of the CS1AN cells with the ATPase mutant CSB gene (CSBE646Q) restored the relative incision to 91% of the wild-type levels at 40% incision. Likewise, there was little difference in incision between the cell lines of a thymine glycol-containing oligonucleotide duplex (Fig. 7A and B). The reduction of incision activity of extracts on an 8-oxoguanine-containing lesion from CS1AN cells transfected with the vector alone suggests that CSB is important in the repair of 8-oxoguanine, whereas the ATPase motif II of CSB is not critical. The presence or absence of CSB protein in the extracts does not appear to affect the incision of thymine glycol and 5-hydroxy-dCytosine lesions.
Figure 6.
Incision of 8-oxoguanine (8-oxodG)-containing and 5-hydroxyl-dCytosine (5-OHdC)-containing oligonucleotides by whole-cell extracts (C, no extract control; Wt, CS1AN/pc3.1-CSBwt; M, CS1AN/pc3.1-CSBE646Q ATPase mutant; V, CS1AN/pc3.1 vector control). (A) Oligonucleotides employed. (B) The 8-oxodeoxyguanine-containing 5′-labeled oligonucleotide duplex and a 5-hydroxyl-dCytosine-containing 5′-labeled oligonucleotide duplex were incubated in the reaction mixture with 20 µg of protein from whole-cell extracts. Reactions were incubated for 6 h for 5-hydroxyl-dCytosine and the hours indicated for 8-oxodG at 37°C and terminated by addition of SDS and proteinase K. The products of incision were separated from the substrates on a 20% polyacrylamide gel. Following incision at the lesion, both oligonucleotides are cut down to a 10mer. The gels were quantified with a PhosphorImager (ImageQuant). (C) The percentage of 5-hydroxyl-dCytosine incision was calculated for the 6 h time point. (D) Incision kinetics of 8-oxoguanine were calculated, extracts were analyzed in triplicate from two biological extracts.
Figure 7.
Incision of 8-oxoguanine-containing and thymine glycol-containing oligonucleotides by whole-cell extracts. (A) An 8-oxoguanine-containing and a thymine glycol-containing 5′-end-labeled oligonucleotide duplex were incubated in the same reaction mixture with 20 µg of protein from whole-cell extracts. Reactions were incubated for 6 h and terminated by addition of SDS and proteinase K. The products of incision were separated from the substrates on a 20% polyacrylamide gel. Following incision at the lesion, the 8-oxoguanine-containing oligonucleotide is cut down to a 10mer and the thymine glycol-containing oligonucleotide is cut down to a 15mer (lane 1, marker; lane 2, CS1AN/pc3.1-CSBwt; lane 3, CS1AN/pc3.1-CSBE646Q; lane 4, CS1AN/pc3.1). (B) The gels were quantified with a PhosphorImager (ImageQuant) and the percent incision was calculated.
Analysis of differential gene expression by cDNA array
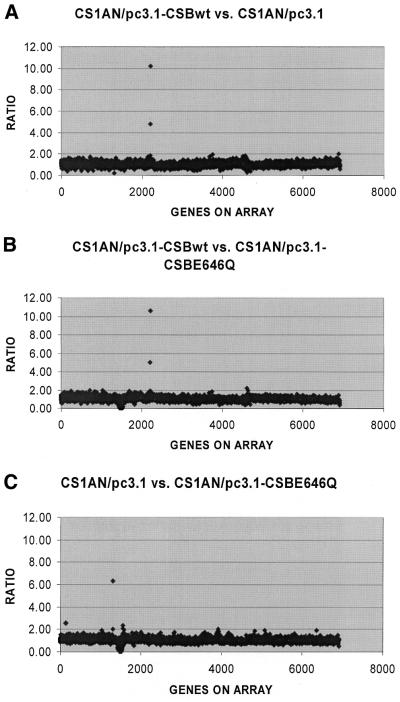
CSB belongs to the SWI/SNF family of proteins, members of which have roles in transcription regulation. CS-B cells have also been shown to have reduced basal levels of transcription compared with normal cell lines (33). To examine the role of CSB in transcription regulation and to identify gene products that might be affected by the CSB status of the cells, we used the Atlas™ cDNA expression arrays to look for differential gene expression resulting from the presence or absence of the CSB gene product. The Atlas™ human array includes 588 human cDNAs representing several different functional classes of genes, including transcription factors, cytokines, oncogenes and tumor suppressor genes. The Atlas™ human stress/toxicology arrays include 234 genes involved in stress response, xenobiotic metabolism, and DNA damage response, repair and recombination. Together, the two sets of arrays allowed us to examine 746 genes, correcting for overlap between the two membrane sets. The general human arrays from the NIA cDNA Array Unit, containing up to 15 000 ESTs, were also used. We concentrated our study to a region containing 3421 known genes and 3491 ESTs. Expression profiles were compared using total RNA for synthesis of 32P- or 33P-labeled cDNA probes. Clontech membranes were probed with three independent RNA isolations from the CS1AN/pc3.1, CS1AN/pc3.1-CSBwt and CS1AN/pc3.1-CSBE646Q cell lines (data not shown). NIA cDNA arrays were probed once (Fig. 8). No significant differences in the expression of the 746 genes on the Clontech arrays were found between the cell lines containing the transfected wild-type CSB gene, the ATPase mutant gene, or no CSB gene. Within the NIA arrays we found two genes that appeared to be overexpressed: the human guanine nucleotide exchange factor DBS and human mRNA for KIAAA232 (unknown function). These overexpressions could not be verified using either northern analysis or quantitative PCR.
Figure 8.
Ratios of gene expression from the NIA arrays comparing the following cell lines: CS1AN/pc3.1-CSBwt versus CS1AN/pc3.1 (A), CS1AN/pc3.1-CSBwt versus CS1AN/pc3.1-CSBE646Q (B) and CS1AN/pc3.1 versus CS1AN/pc3.1-CSBE646Q (C). The two outlying genes in (A) and (B) are the same and are identified as the human guanine nucleotide exchange factor DBS and human mRNA for KIAAA232 gene, whose function is unknown.
DISCUSSION
Cells from CS patients are UV sensitive and fail to recover RNA synthesis after UV irradiation (10). They demonstrate a reduced rate of repair of the transcribed strand of active genes, not only after UV exposure, but also after exposure to certain forms of oxidative stress. CS cells are unable to resume transcription after treatment with the DNA damaging agents N-acetoxy-2-acetylaminofluorene and 4-nitroquinoline-1-oxide, even though these lesions are efficiently repaired by the GGR pathway in both normal and CS cells (15,42,43). Thus, the CSB protein is involved in the processing of different classes of DNA damage and the mechanism may not be the same for all types of damage.
In this study, we demonstrated that the single amino acid substitution of glutamine for a conserved glutamic acid in motif II of the ATPase domain severely compromised the ability of the transfected CSB gene to complement the UV sensitivity of the human CS-B cell line CS1AN.S3.G2 by a colony-forming survival assay. A number of studies in which similar motif II mutations have been made to ATPases and helicases have shown that whereas the ATPase function is impaired, properties other than ATP hydrolysis or unwinding activity remained intact, such as nucleotide and DNA binding and protein interactions (44–47). Western blot analysis has demonstrated that both the CSBwt and CSBE646Q proteins are present in the transfected cell lines. The CSB ATPase point mutant failed to function in RNA synthesis recovery and rescue from apoptosis following UV damage. These results confirm in a human cell line what we have previously reported in the CS-B hamster homolog, UV61 (15,35). These results are also consistent with those seen when an invariant lysine in motif I of the ATPase domain was replaced with arginine (K538R) (36). This mutation abolished the ATPase activity of the protein and failed to fully complement RNA synthesis recovery following UV irradiation. While the ATPase motif I mutant retained partial activity in RNA synthesis recovery after UV damage and ATPase motif II mutant CSBE646Q completely abolished RNA synthesis recovery, the difference may be explained by the fact that these are mutations in two spatially distinct motifs and thus differentially impact CSB function. Finally, biochemical data demonstrate that hydrolysis of the ATP-γ phosphoanhydride bond is required for the interaction of CSB with RNA pol II stalled at a thymine dimer on a DNA template (13). These results clearly suggest that CSB functions in a process to protect cells from UV-induced damage that involves ATP hydrolysis. There is considerable evidence for interaction between CSB and transcription machinery (RNA pol II, TFIIH) in the response to UV damage, as well as NER proteins (TFIIH, XPA, XPG). Thus, CSB and its associated ATPase activity may be responsible for recruiting the repair machinery to the transcription-blocking lesion at an accelerated rate, resulting in TCR.
A previous study (30) reported that incision of 8-oxoguanine in DNA by whole-cell extracts derived from primary CS-B fibroblasts was ∼50% the level of incision in extracts prepared from normal cells. Additionally, CS1AN.S3.G2 cells transfected with a plasmid vector had reduced incision when compared with cells transfected with the same vector bearing the wild-type CSB gene (30). We have confirmed this incision defect in isogenic clonal cell lines transfected with a different expression vector. In our cell lines the incision of 8-oxoguanine in extracts from CS1AN cells transfected with the wild-type CSB gene was 2-fold greater than seen in cells transfected with the vector alone. Interestingly, CS1AN cells transfected with the CSB gene containing the single point mutation in the ATPase motif II, which were unable to complement the UV-sensitive phenotype of these cells, did complement the defect in incision of 8-oxoguanine lesions. In contrast, there was no defect in CS-B cells in the repair of the lesion 5-hydroxyl-dCytosine. This suggests clearly distinct mechanisms for facilitation of repair of various oxidative and UV-induced lesions by CSB.
In a recent study by LePage et al. (28) it was found that strand-specific removal of 8-oxoguanine and thymine glycol from an active gene was defective in CS cells, including CS-B, XP-B/CS, XP-D/CS and XP-G/CS. This defect was limited to TCR of transcribed sequence, despite its proficient repair when not transcribed. It was also found that the unrepaired 8-oxoguanine blocks transcription by RNA pol II. Our in vitro findings on an oligonucleotide substrate differed in that there was a defect in the repair of an 8-oxoguanine lesion in CS-B cells, whereas there was no defect in the repair of a thymine glycol lesion. Our previous studies utilized a plasmid containing a single 8-oxoguanine lesion, and the plasmid is non-transcribing, thus we are detecting general genome BER rather than TCR (30). Thus, we propose a general genome BER defect in CSB.
It has been suggested that CSB may either act as a recruiting factor for repair proteins at the site of RNA pol II blocking damage or that the primary defect may be in basal transcription, but both theories fail to reconcile all experimental data (reviewed in 2). Our results suggest that CSB affects the repair of at least two types of damage (UV-induced lesions and 8-oxoguanine) by different mechanisms, one requiring the ATPase function of the protein and one independent of it. The literature holds many examples of proteins and protein complexes, which are clearly multifunctional. TFIIH has proven roles in both RNA pol II transcription and NER. It has been suggested that the CSB protein is similarly endowed (2), such that mutations in this protein can result in defects in transcription, transcription-coupled NER, BER or all three.
A second example of a protein with multiple functions in DNA repair is the XPG protein. XPG is a structure-specific endonuclease that nicks damaged DNA 3′ to the lesion in an early step of NER (48–51). Another NER-defective disorder, xeroderma pigmentosum (XP), and CS are usually clinically and genetically distinct, but complementation analyses have assigned a few CS patients to the rare XP groups B, D or G. Nouspikel et al. (52) showed three XP-G/CS patients had mutations that would produce severely truncated XPG proteins. In contrast, two XPG patients without CS make full-length XPG, but with a missense mutation that inactivates its incision activity in NER. These results suggest that XP-G/CS mutations abolish interactions required for a second XPG function that leads to the CS clinical phenotype. These authors also showed that XP-G/CS cells were defective in TCR of thymine glycols whereas the lesions were repaired normally in an XPG patient, and suggested that this was the second XPG function (27). Repair of thymine glycol and 8-oxoguanine is initiated by glycosylases in a BER pathway and may require some interaction between XPG and CSB. Rather than having catalytic roles, XPG and CSB may have an assembly function in this pathway. Unlike XP, CS and XP-G/CS appear to be independent of exposure to environmentally induced DNA damage. This suggests that in CS individuals, the culpable lesions are some form of spontaneous base damage. Free radicals generated as by-products of oxidative metabolism might constitute a source of such spontaneous damage, and cells with high metabolic activity such as neurons, and those that proliferate rapidly during development, might be particularly vulnerable to such insult (53).
Experimental evidence suggests that the function of the CSB protein is associated with a defect in TCR, GGR, and removal of oxidative lesions, as well as in transcription itself, as described above. CSB belongs to the SWI2/SNF2 subfamily of proteins in which several members are ATPases involved in chromatin remodeling associated with transcriptional regulation. If CSB has such a role, a mutation in the CSB gene might affect the precise timing and/or expression of a specific subset of genes during a critical stage of postnatal development, resulting in the clinical phenotypes of CS (2). It has been suggested that the CSB protein may affect the regulation of transcription of various genes including some involved in DNA repair (30). This role of the CSB protein may be realized in one or more of the above repair pathways, and CS-B cells have been demonstrated to have reduced expression of the 8-oxoguanine-DNA glycosylase (hOGG1) gene (30). For these reasons we used cDNA expression arrays to look for differential gene expression resulting from the presence or absence of the CSB gene product. No differential expression was confirmed in the 6912 genes examined here between the three cell lines. The hOGG1 gene on the human stress/toxicology array did not display a 2-fold or greater change in expression level with this technique. If the CSB protein exerts an effect on overall basal transcription (33), it would not be detected by this technique because the analysis normalizes for overall signal, and specifically identifies differentially expressed genes. It is possible that CSB may be involved in transcription regulation, but only in response to DNA damage. Future studies of a larger subset of genes, as well as examination of mRNA levels following DNA damage, may clarify the role of CSB in transcription regulation.
In conclusion, expression of the CSB protein does not affect the differential expression of the subset of genes examined here by cDNA array analysis in undamaged cells. Our results do suggest that the CSB protein functions by different mechanisms in the repair of UV-induced and 8-oxoguanine lesions. Future studies with additional mutants will further clarify the regions of the protein involved in processing of different types of DNA damage.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. Chen and J.-P. Laine for comments. M.M. was supported by the Scientific and Technical Research Council of Turkey.
REFERENCES
- 1.Nance M. and Berry,S. (1992) Cockayne syndrome: review of 140 cases. Am. J. Med. Genet., 42, 68–84. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E.C. (1996) Cockayne syndrome—a primary defect in DNA repair, transcription, both or neither? Bioessays, 18, 731–738. [DOI] [PubMed] [Google Scholar]
- 3.Troelstra C., van Gool,A., De Wit,J., Vermeulen,W., Bootsma,D. and Hoeijmakers,H.J. (1992) ERCC6 a member of a subfamily of putative helicases is involved in Cockayne’s syndrome and preferential repair of active genes. Cell, 71, 939–953. [DOI] [PubMed] [Google Scholar]
- 4.Henning K., Li,L., Legerski,R., Iyer,N., McDaniel,L., Schultz,R., Stefanini,M., Lehmann,A., Mayne,L. and Friedberg,E. (1995) The Cockayne syndrome complementation group A gene encodes a WD-repeat protein which interacts with CSB protein and a subunit of the RNA pol II transcription factor IIH. Cell, 82, 555–566. [DOI] [PubMed] [Google Scholar]
- 5.Leadon S.A. and Lawrence,D.A. (1991) Preferential repair of DNA damage on the transcribed strand of the human metallothionein genes requires RNA polymerase II. Mutat. Res., 255, 67–78. [DOI] [PubMed] [Google Scholar]
- 6.Christians F.C. and Hanawalt,P.C. (1992) Inhibition of transcription and strand-specific DNA repair by α-amanitin in Chinese hamster ovary cells. Mutat. Res., 274, 93–101. [DOI] [PubMed] [Google Scholar]
- 7.Venema J., Mullenders,L.H., Natarajan,A.T., van Zeeland,A.A. and Mayne,L.V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hoffen A., Natarajan,A.T., Mayne,L.V., van Zeeland,A.A. and Mullenders,L.H. (1993) Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res., 21, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans M.K. and Bohr,V.A. (1994) Gene-specific DNA repair of UV-induced cyclobutane pyrimidine dimers in some cancer-prone and premature-aging human syndromes. Mutat. Res., 314, 221–231. [DOI] [PubMed] [Google Scholar]
- 10.Mayne L.V. and Lehmann,A.R. (1982) Failure of RNA synthesis to recover after UV irradiation: and early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res., 42, 1473–1478. [PubMed] [Google Scholar]
- 11.Gorbalenya A.E., Koonin,E.V., Donchenko,A.P. and Blinov,V.M. (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res., 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selby C.P. and Sancar,A. (1997) Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem., 272, 1885–1890. [DOI] [PubMed] [Google Scholar]
- 13.Tantin D., Kansal,A. and Carey,M. (1997) Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol., 17, 6803–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orren D.K., Dianov,G.L. and Bohr,V.A. (1996) The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res., 17, 3317–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brosh R.M. Jr, Balajee,A.S., Selzer,R.R., Sunesen,M., De Santis,L.P. and Bohr,V.A. (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol. Biol. Cell, 10, 3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tantin D. (1998) RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J. Biol. Chem., 273, 27794–27799. [DOI] [PubMed] [Google Scholar]
- 17.Schaeffer L., Roy,R., Humbert,S., Moncollin,V., Vermeulen,W., Hoeijmakers,J.H., Chambon,P. and Egly,J.M. (1993) DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science, 260, 58–63. [DOI] [PubMed] [Google Scholar]
- 18.Feaver W.J., Svejstrup,J.Q., Bardwell,L., Bardwell,A.J., Buratowski,S., Donahue,T.F., Friedberg,E.C. and Kornberg,R.D. (1993) Dual roles of a multiprotein complex from S.cerevisiae in transcription and DNA repair. Cell, 75, 1379–1387. [DOI] [PubMed] [Google Scholar]
- 19.van Vuuren A.J., Vermeulen,W., Ma,L., Weeda,G., Appeldoorn,E., Jaspers,N.G., Van der Eb,A.J., Bootsma,D., Hoeijmakers,J.H., Humbert,S. et al. (1994) Correction of xeroderma pigmentosum repair defect by basal transcription factor BTF2 (TFIIH). EMBO J., 13, 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaeffer L., Moncollin,V., Roy,R., Staub,A., Mezzina,M., Sarasin,A., Weeda,G., Hoeijmakers,J.H. and Egly,J.M. (1994) The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J., 13, 2388–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drapkin R., Reardon,J.T., Ansari,A., Huang,J.C., Zawel,L., Ahn,K., Sancar,A. and Reinberg,D. (1994) Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature, 368, 769–772. [DOI] [PubMed] [Google Scholar]
- 22.Douziech M., Coin,F., Chipoulet,J.M., Arai,Y., Ohkuma,Y., Egly,J.M. and Coulombe,B. (2000) Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein–DNA photo-cross-linking. Mol. Cell. Biol., 20, 8168–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer N., Reagan,M.S., Wu,K.J., Canagarajah,B. and Friedberg,E.C. (1996) Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry, 35, 2157–2167. [DOI] [PubMed] [Google Scholar]
- 24.Selby C.P. and Sancar,A. (1997) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl Acad. Sci. USA, 94, 11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dianov G., Bischoff,C., Piotrowski,J. and Bohr,V.A. (1998) Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J. Biol. Chem., 273, 33811–33816. [DOI] [PubMed] [Google Scholar]
- 26.Leadon S.A. and Cooper,P.K. (1993) Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active gene is defective in Cockayne syndrome. Proc. Natl Acad. Sci. USA, 90, 10499–10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper P.K., Nouspikel,T., Clarkson,S.G. and Leadon,S.A. (1997) Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science, 275, 990–993. [DOI] [PubMed] [Google Scholar]
- 28.Le Page F., Kwoh,E.E., Avrutskaya,A., Gentil,A., Leadon,S.A., Sarasin,A. and Cooper,P.K. (2000) Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell, 101, 159–171. [DOI] [PubMed] [Google Scholar]
- 29.Le Page F., Klungland,A., Barnes,D.E., Sarasin,A. and Boiteux,S. (2000) Transcription coupled repair of 8-oxoguanine in murine cells: the ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl Acad. Sci. USA, 97, 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dianov G., Bischoff,C., Sunesen,M. and Bohr,V.A. (1999) Repair of 8-oxoguanine in DNA is deficient inCockayne syndrome group B cells. Nucleic Acids Res., 27, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pazin M.J. and Kadonaga,J.T. (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell, 88, 737–740. [DOI] [PubMed] [Google Scholar]
- 32.vanGool A.J., vanderHorst,G., Citterio,E. and Hoeijmakers,J.J. (1997) Cockayne syndrome: defective repair of transcription? EMBO J., 16, 4155–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balajee A.S., May,A., Dianov,G.L., Friedberg,E.C. and Bohr,V.A. (1997) Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl Acad. Sci. USA, 94, 4306–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Citterio E., Van Den Boom,V., Schnitzler,G., Kanaar,R., Bonte,E., Kingston,R.E., Hoeijmakers,J.H. and Vermeulen,W. (2000) ATP-Dependent chromatin remodeling by the cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol., 20, 7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balajee A.S., DeSantis,L.P., Brosh,R.M.,Jr, Selzer,R. and Bohr,V.A. (2000) Role of the ATPase domain of the Cockayne syndrome group B protein in UV induced apoptosis. Oncogene, 19, 477–489. [DOI] [PubMed] [Google Scholar]
- 36.Citterio E., Rademakers,S., van der Horst,G.T., van Gool,A., Hoeijmakers,H.J. and Vermeulen,W. (1998) Biochemical and biological characterization of wild type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem., 273, 11844–11852. [DOI] [PubMed] [Google Scholar]
- 37.Mayne L.V., Priestley,A., James,M.R. and Burke,J.F. (1986) Efficient immortalization and morphological transformation of human fibroblasts by transfection with SV40 DNA linked to a dominant marker. Exp. Cell Res., 162, 530–538. [DOI] [PubMed] [Google Scholar]
- 38.Mallery D.L., Tanganelli,B., Colella,S., Steingrimsdottir,H., Van Gool,A.J., Troelstra,C., Stefanini,M. and Lehmann,A.R. (1998) Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet., 62, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manley J.L., Fire,A., Samuels,M. and Sharp,P. (1983) In vitro transcription: whole cell extract. Methods Enzymol., 101, 568–582. [DOI] [PubMed] [Google Scholar]
- 40.Dianov G. and Lindahl,T. (1991) Preferential recognition of 1-T base-pairs in the initiation of excision-repair by hypoxanthine-DNA glycosylase. Nucleic Acids Res., 19, 3829–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ljungman M. and Zhang,F. (1996) Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene, 13, 823–831. [PubMed] [Google Scholar]
- 42.vanOosterwijk M.F., Filon,R., Kalle,W.J., Mullenders,L.F. and vanZeeland,A.A. (1996) The sensitivity of human fibroblasts to N-acetoxy-2-acetylaminofluorene is determined by the extent of transcription-coupled repair, and/or their capability to counteract RNA synthesis inhibition. Nucleic Acids Res., 24, 4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyderwine E.G. and Bohr,V.A. (1992) Gene- and strand-specific damage and repair in Chinese hamster ovary cells treated with 4-nitroquinoline 1-oxide. Cancer Res., 52, 4183–4189. [PubMed] [Google Scholar]
- 44.Pause A. and Sonenberg,N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J., 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jindal H.K., Yong,C.B., Wilson,G.M., Tam,P. and Astell,C.R. (1994) Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem., 269, 3283–3289. [PubMed] [Google Scholar]
- 46.Brosh R.M. Jr and Matson,S.W. (1995) Mutations in motif II of E.coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J. Bacteriol., 177, 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richmond E. and Peterson,C.L. (1996) Functional analysis of the DNA stimulated ATP-ase domain of yeast SWI2/SNF2. Nucleic Acids Res., 24, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habraken Y., Sung,P., Prakash,L. and Prakash,S. (1995) Structure-specific nuclease activity in yeast nucleotide excision repair protein Rad2. J. Biol. Chem., 270, 30194–30198. [DOI] [PubMed] [Google Scholar]
- 49.O’Donovan A., Scherly,D., Clarkson,S.G. and Wood,R.D. (1994) Isolation of active recombinant XPG protein, a human DNA repair endonuclease. J. Biol. Chem., 269, 15965–15968. [PubMed] [Google Scholar]
- 50.Matsunaga T., Mu,D., Park,C.H., Reardon,J.T. and Sancar,A. (1995) Human DNA repair excision nuclease—analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem., 270, 20862–20869. [DOI] [PubMed] [Google Scholar]
- 51.Moggs J.G., Yaremar,K.J., Essigman,J.M. and Wood,R.D. (1996) Analysis of Incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of 1,3-intrastrand d(GpTpG)-Cisplatin adducts. J. Biol. Chem., 271, 7177–7186. [DOI] [PubMed] [Google Scholar]
- 52.Nouspikel T., Lalle,P., Leadon,S.A., Cooper,P.K. and Clarkson,S.G. (1997) A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc. Natl Acad. Sci. USA, 94, 3116–3121. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Hanawalt P.C. (1994) Transcription-coupled repair and human disease. Science, 266, 1957–1958. [DOI] [PubMed] [Google Scholar]