Abstract
In higher plants, activation sequence-1 (as-1) of the cauliflower mosaic virus 35S promoter mediates both salicylic acid- and auxin-inducible transcriptional activation. Originally found in viral and T-DNA promoters, as-1-like elements are also functional elements of plant promoters activated in the course of a defence response upon pathogen attack. as-1-like elements are characterised by two imperfect palindromes with the palindromic centres being spaced by 12 bp. They are recognised by plant nuclear as-1-binding factor ASF-1, the major component of which is basic/leucine zipper (bZIP) protein TGA2.2 (∼80%) in Nicotiana tabacum. In electrophoretic mobility shift assays, ASF-1 as well as bZIP proteins TGA2.2, TGA2.1 and TGA1a showed a 3–10-fold reduced binding affinity to mutant as-1 elements encoding insertions of 2, 4, 6, 8 or 10 bp between the palindromes, respectively. This correlated with a 5–10-fold reduction in transcriptional activation from these elements in transient expression assays. Although ASF-1 and TGA factors bound efficiently to a mutant element carrying a 2 bp deletion between the palindromes [as-1/(–2)], the latter was strongly compromised with respect to mediating gene expression in vivo. A fusion protein consisting of TGA2.2 and a constitutive activation domain mediated transactivation from as-1/(–2) demonstrating binding of TGA factors in vivo. We therefore conclude that both DNA binding and transactivation require optimal positioning of TGA factors on the as-1 element.
INTRODUCTION
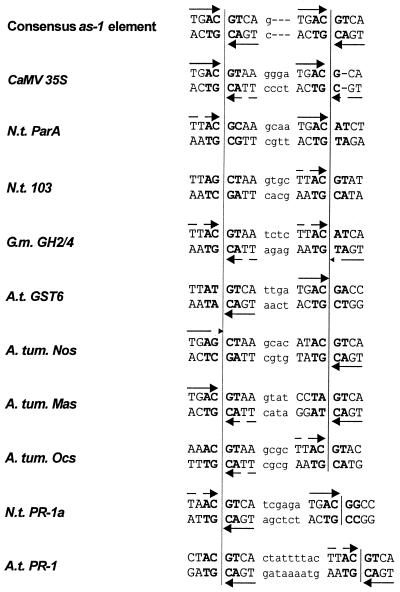
Many eukaryotic transcription factors recognise short sequence motifs with a surprisingly high tolerance for sequence deviations. Nevertheless, they efficiently recognise their binding sites in a promoter amongst a vast amount of non-specific DNA arbitrarily encoding similar sequences. Specificity can be increased by introducing the requirement for two binding sites for efficient binding (1). The cis element activation sequence-1 (as-1), which consists of two imperfect TGACGTCA palindromes (2), might be regarded as an example of this principle. The as-1 element was first identified between positions –65 and –85 of the cauliflower mosaic virus (CaMV) 35S promoter. In leaves, expression depends either on salicylic acid (SA; 3) or auxin (4), whereas expression in root tips is constitutive (5). Moreover, the element is activated in protoplasts with further stimulation by auxin and SA being possible (6,7). Since their original discovery, as-1-like elements have been identified as functional elements of other viral promoters and promoters of the Agrobacterium tumefaciens encoded T-DNA (8,9). In addition, they were found in the promoter of soybean heat shock gene Gmhsp26-A (10) and other genes that were identified either as auxin-inducible genes [Nicotiana tabacum (Nt) ParA (11,12); Nt103 (13,14); Arabidopsis thaliana (At) GST6 (15)] or as ‘immediate early’ SA-inducible genes (NtIEGT; 16). Whereas the sequence can deviate quite substantially from the consensus sequence, spacing of 12 bp between the two centres of the palindromes is conserved in all as-1-like elements (Fig. 1) that respond to auxin and SA.
Figure 1.
Alignment of different as-1-like elements from viral promoters (as-1; 2), T-DNA promoters [ocs, nos (8); mas (9)] and plant genes [ParA (12); Nt103 (14); GH2/4 (6); GST6 (15); PR-1a (18); PR-1 (17)]. Palindromes are shown in capital letters, the sequence of the spacer is shown in small letters. Positions that are not defined in the consensus sequence are indicated with n. The TGAC half sites of the 8 bp palindromes are marked by arrows. TGAC sequences carrying one mutation are marked by interrupted arrows, half sites with more than one mutations are not marked. The central 4 bp (ACGT in the consensus sequence) are indicated by bold letters. The centres of the palindromes are marked by vertical lines.
In contrast, the spacing found in as-1-like elements of the so-called ‘late’ SA-inducible promoters [AtPR-1 (17); NtPR-1a (18)] is less well conserved. Also, the PR promoters do not respond to auxin and reveal different induction kinetics upon SA treatment than the aforementioned promoters: whereas the ‘immediate early’ genes are only transiently induced after 1–2 h after SA application without requiring protein biosynthesis, the ‘late’ genes show a long lasting induction after 10–12 h and activation requires protein biosynthesis (3). This indicates that the trans-factors binding to as-1-like elements can be targets for different SA-dependent signal transduction networks.
Using electrophoretic mobility shift assays (EMSAs), a nuclear protein complex called ASF-1 (activation sequence factor-1) was identified (2), which is most likely responsible for the activation of transcription (2,3,19,20). In tobacco, four cDNAs encoding as-1-binding proteins have been described before (21–24). These ‘TGA factors’ encode a variable N-terminal domain followed by a highly conserved basic/leucine zipper (bZIP) domain. The approximately 250 amino acid C-terminal domain is moderately conserved (at least 52% identical amino acids). Based on sequence homology, TGA factors were grouped into distinct classes. TGA1a (21) and PG13 (22) belong to class I, whereas TGA2.1 (23) and TGA2.2 (24) belong to class II. Class I proteins have only been detected in root tips (25), whereas class II TGA factors have been identified in leaves as well as in roots (20,24). In tobacco leaf extracts, ASF-1 consists of TGA2.2 (∼80%) and of a TGA2.1 related activity (∼10%; 20). TGA2.1 and TGA2.2 differ in the N-terminal region preceding the bZIP region, with TGA2.1 having a 127 amino acid long extension. This extension both weakens and changes its as-1-binding activity (24), conferring the requirement of two palindromes for as-1 binding. In contrast, TGA2.2 and TGA1a are able to bind to a single palindrome. The N-terminus of TGA1a encodes a transactivation domain (26). In contrast to TGA2.2, TGA2.1 confers transcriptional activation in yeast (24). Nevertheless, overexpression of TGA2.2 in transgenic plants leads to enhanced SA and auxin inducibility of Nt103 (20), showing its function as a positive regulator despite its missing activation domain.
EMSAs have so far revealed that ASF-1 and the TGA2.2 homodimer recognise a single palindrome and bind to the second site only at higher protein concentrations (19,24). EMSAs therefore typically yield two complexes, a faster migrating ‘lower’ complex representing one TGA dimer bound to one palindrome and a slower migrating ‘upper’ complex representing two TGA dimers bound to both palindromes. In contrast, TGA1a only forms the ‘lower’ complex even at high protein concentrations, whereas the TGA2.1 homodimer requires two palindromes for binding and only forms the ‘upper’ complex. The strong conservation of the spacing in different as-1-like elements suggests that the exact arrangement of the two binding motifs is important for the activity of the element. Here we asked whether DNA binding and/or other steps leading to transcriptional activation require exact spacing of the palindromes. To address this question, we constructed a series of as-1 mutants differing in the spacing between the two palindromes, tested them in EMSAs for their relative affinities to ASF-1, TGA2.2, TGA2.1 and TGA1a, and determined their in vivo activity in transient expression assays using tobacco protoplasts.
MATERIALS AND METHODS
Plasmid constructs
pTTL-Gus, which encodes base pairs +1 to +55 of the CaMV 35S promoter upstream of gus/int was used as a starting plasmid (27). Complementary oligonucleotides encoding as-1 and EcoRI and SpeI 5′-overhangs were cloned into the respective restriction sites upstream of the minimal promoter. A synthetic linker (TAGTCTAGCTA) encoding an XbaI site was inserted into the SmaI site between the promoter and gus/int. Subsequently, the plasmid was cut with EcoRI and SpeI to replace the wild-type as-1 element by synthetic mutant as-1 elements. The wild-type spacing of 34 bp between the last base pair of the as-1 element and the first base pair of the TATA-box was maintained in all constructs. As a result of the SpeI site, the sequence was GT instead of TC at positions –51 and –52, respectively. The sequence integrity of the inserted oligonucleotides was confirmed. The TGA2.2–VP16 fusion protein was constructed by adding the amino acid sequence GGGGSGGGGS to the C-terminus of TGA2.2 using PCR. The last two amino acids (GS) were encoded by the nucleotide sequence GGATCC, thus introducing a BamHI site. The VP16 domain was amplified by PCR, adding the sequence GSGGGGS to the N-terminus. The first two amino acids (GS) were again encoded by GGATCC, introducing a BamHI site that served the in frame fusion of the two proteins. This strategy led to the introduction of a flexible amino acid linker (GGGGSGGGGSGGGGS) between the two portions of the protein. The complete coding region of the fusion protein was cloned downstream the constitutive HBT promoter (28).
Electrophoretic mobility shift assays
EMSAs using ASF-1 and in vitro translated TGA factors were done as described (20,24). As a probe, the 98 bp long as-1 encoding EcoRI/XbaI fragment from pTTL-as-1 was radiolabelled by filling in the 5′-overhangs with [α-32P]dATP and [α-32P]dCTP using the Klenow fragment and gel-purified on a 5% polyacrylamide gel. A sample (0.01 pmol) of the labelled fragment was used for each binding reaction, with 2 pmol of annealed oligonucleotides added as competitor DNA. Binding reactions (29), preparation of ASF-1 (19) and synthesis of TGA factors using a coupled in vitro transcription/translation system (24) were done as described before. The calculation for Table 2 was done according to the following simplified example: the per- cent of radioactivity per lane that was retarded by ASF-1 in the absence of competitor DNA was set 100%. If this complex was reduced to 10% by the addition of the competitor DNA encoding as-1, this value was set 1. If the complex was reduced to 50% by the addition of the competitor DNA encoding one of the mutant as-1 elements, the corresponding value would be 0.2, reflecting a 5-fold decreased affinity.
Table 2. Effect of differences in the spacing between the two palindromes of the as-1 element on binding of ASF-1, TGA2.2, TGA2.1 and TGA1a and on promoter activity.
| ASF-1 | TGA2.2 | TGA2.1 | TGA1a | GUS | |
|---|---|---|---|---|---|
| as-1 | 1.00 (± 0.00) | 1.00 (± 0.00) | 1.00 (± 0.00) | 1.00 (± 0.00) | 100.0 (± 0.0) |
| as-1/(–2)GA | 2.97 (± 0.12) | 1.09 (± 0.14) | 0.96 (± 0.32) | 1.77 (± 0.14) | 16.9 (± 1.2) |
| as-1/(–2)GG | 0.61 (± 0.03) | 0.43 (± 0.08) | 0.23 (± 0.03) | n.d. | 9.5 (± 0.5) |
| as-1/(+2) | 0.16 (± 0.02) | 0.17 (± 0.01) | 0.08 (± 0.04) | 0.29 (± 0.03) | 18.4 (± 4.4) |
| as-1/(+4) | 0.27 (± 0.01) | 0.23 (± 0.02) | 0.09 (± 0.03) | 0.46 (± 0.02) | 13.5 (± 2.8) |
| as-1/(+6) | 0.23 (± 0.01) | 0.16 (± 0.06) | 0.09 (± 0.03) | 0.44 (± 0.05) | 8.7 (± 2.8) |
| as-1/(+8) | 0.37 (± 0.02) | 0.33 (± 0.08) | 0.21 (± 0.04) | 0.62 (± 0.13) | 13.7 (± 1.2) |
| as-1/(+10) | 0.32 (± 0.06) | 0.21 (± 0.02) | 0.12 (± 0.04) | 0.61 (± 0.10) | 11.8 (± 1.2) |
To calculate the relative affinities of the different protein factors to the different as-1 elements, the percentage of radioactivity left in the presence of a 200-fold molar excess of the wild-type as-1 element was set 1 (see Materials and Methods). Relative GUS activities mediated by these elements in transiently transformed protoplasts of tobacco cell line BY-2 are given as percent of the activity of the wild-type as-1 element. Values are taken form three independent experiments.
Transient assays
Protoplasts prepared from BY-2 cells were transformed using PEG (30). After DNA transfer, protoplasts were incubated in the presence of 1 µM auxin. Protoplasts prepared from mesophyll protoplasts were transformed by electroporation (31) and incubated either in the absence or in the presence of 50 µM auxin. For fluorometric GUS assays (32), protein extracts of protoplasts were incubated with the substrate 4-methylumbelliferyl-β-glucoronide at 37°C. Quantification of the fluorescence was done using a CytoFlourII plate reader (PerSeptive).
RESULTS
The sequence between the palindromes of the as-1 element can be altered without affecting factor binding and transcriptional activation
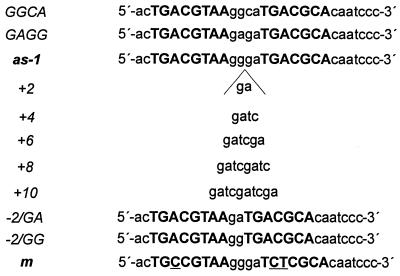
Before constructing as-1 elements with different spacings between the two palindromes, we addressed the impact of sequence alterations per se on in vitro DNA binding and in vivo transcriptional activity. Two different point mutations in the GGGA sequence located between the palindromes were introduced into the as-1 element (GAGA and GGCA). To compare the relative affinities of these as-1 mutants to ASF-1, EMSAs using the 32P-labelled wild-type as-1 element were performed in the presence of unlabelled oligonucleotides encoding the mutated as-1 sequences. Changing the sequence from GGGA to GAGA severely affected DNA binding (Table 1), indicating that the G in position 2 of the spacer sequence is important for factor binding. In order to address the question whether changing a potential Myb binding site (GGATG) overlapping the last 3 bp of the spacer and the first 2 bp of the second palindrome (33), we changed the sequence to GGCA, thus mutating the Myb binding site to GCATG. This did not alter the binding characteristics of ASF-1 (Table 1). The two point mutations were introduced within the –90 region of the truncated CaMV 35S promoter driving the β-glucuronidase (gus) reporter gene. In transient expression assays using protoplasts from tobacco cell line BY-2, the as-1 mutant encoding GAGA conferred significantly reduced transcriptional activation as compared to the wild-type element, which correlated well with reduced ASF-1 binding activity. The as-1 mutant encoding GGCA showed wild-type activity, indicating that the potential Myb site did not contribute to the activation. We thus inserted additional base pairs between the GG and the GA dinucleotide of the spacer, maintaining the sequence GGGA immediately downstream of the first palindrome as well as the GA dinucleotide immediately upstream of the second palindrome (Fig. 2). Two deletion mutants were constructed, one of them [as-1/(–2)GG] maintaining the GG dinucleotide downstream of the first palindrome and the second one [as-1/(–2)GA] maintaining the GA dinucleotide upstream of the second palindrome.
Table 1. Influence of the sequence between the palindromes on ASF-1 binding and promoter activity.
| No element | GGGA (wt) | GAGA | GGCA | |
|---|---|---|---|---|
| (a) ASF-1 complex (%) | 100.0 | 8.5 | 36.2 | 8.3 |
| (b) GUS activity (%) | 3.9 (± 0.2) | 100.0 | 19.5 (± 1.4) | 130.0 (± 1.2) |
The sequence above the columns indicates the sequence between the two palindromes of the as-1 elements, which were used as competitor oligonucleotides in EMSAs (a) or as an upstream regulatory element in transient expression analysis (b). Numbers in (a) give the relative amount of ASF-1 bound a radiolabelled as-1 element (see also Fig. 3) in the absence (100%) or presence of a 200-fold molar excess of the different competitor oligonucleotides. Numbers in (b) indicate the relative amount of GUS activity from chimeric promoters encoding different as-1 elements upstream of the –55 region of the CaMV 35S promoter. The activity conferred by the wild-type (wt) element was set 100%.
Figure 2.
Sequences of the oligonucleotides used in this study. The two palindromes are shown in capital letters, the sequence of the spacer is shown in small letters. Positions changed in the mutated oligonucleotide m, which is not recognised by ASF-1, are underlined.
Deletion of 2 bp between as-1 palindromes does not affect binding of ASF-1 and TGA factors, whereas insertions decrease factor binding in vitro
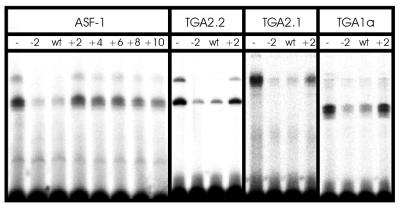
EMSAs of ASF-1 and in vitro translated TGA factors with as-1 spacing mutants are shown in Figure 3. The wild-type as-1 element competed efficiently for binding to the radioactively labelled fragment, leading to a reduction of complex formation down to 8.5%. The mutant as-1/(–2)GA element also competed efficiently for DNA binding. In contrast, increasing the distance between the palindromes by 2 or more bp decreased the efficiency of the element to compete for binding, indicating that positioning of the palindromic centres on different sides of the helix leads to reduced binding. The quantitative data of this experiment are shown in Table 2. The mutant as-1/(–2)GA competed three times more efficiently for binding than wild-type as-1 whereas the as-1/(–2)GG element competed slightly less efficiently for as-1 binding. The as-1/(+8) and the as-1/(+10) mutants, encoding the two centres of the palindromes on the same side of the helix (+8) or in a similar configuration as in the wild-type element (+10), showed somewhat increased binding to as-1 as compared with the other insertion mutants. This is presumably due to positive protein–protein interactions between the two dimers when located on the same face of the DNA helix.
Figure 3.
EMSAs of ASF-1 and in vitro translated TGA2.2, TGA2.1 and TGA1a with a radiolabelled 98 bp as-1 fragment. Numbers above the lanes indicate which oligonucleotide was used as competitor DNA (Fig. 2). For the –2 mutant, the oligonucleotide as-1(–2)GA was used. Quantitative numbers are given in Table 2. For TGA2.2, TGA2.1 and TGA1a, data obtained with oligonucleotides (+4, +6, +8 and +10) are not shown, but values are indicated in Table 2. The binding reaction contained either 5 µg nuclear extract (ASF-1) or 0.5 µl (TGA2.2), 1.0 µl (TGA2.1) or 2 µl (TGA1a) of the respective in vitro translation reactions.
In order to investigate whether the binding activity of ASF-1 represents the binding characteristics of the recombinant TGA factors, TGA1a, TGA2.1 and TGA2.2 were synthesised using a coupled in vitro transcription/translation system and subjected to the same DNA binding assay as ASF-1. As described before (24), the binding profile differed for the three factors: TGA2.2 yielded two complexes, representing single and double occupancy of the two palindromes; TGA2.1 yielded one complex representing TGA2.1 bound to both palindromes; TGA1a yielded one complex representing binding to only one half site (Fig. 3). Nevertheless, the three factors revealed the same sensitivity to alterations of the spacing (Fig. 3 and Table 2) as ASF-1, with the deletion mutant [as-1/(–2)GA] being recognised at least as well as the wild-type element, and the insertion mutants being recognised with lower efficiencies. Binding of TGA2.1, which requires two palindromes for binding, was more affected by variations of the spacing than binding of TGA2.2 and TGA1a.
Deletions and insertions between the palindromes of the as-1 element reduce its transcriptional activation capacity
The set of as-1 mutants was used to replace the wild-type as-1 element between position –85 and –65 of the –90 deletion mutant of the CaMV 35S promoter. The activities of these elements were determined in protoplasts derived from tobacco suspension line BY-2 using gus as a reporter gene. In this assay, only as-1 elements encoding 12 bp between the centres of the two palindromes conferred high transcriptional activation (Table 2). The reduced in vivo activities of the insertion mutants correlated well with their weaker binding activities. In contrast, the deletion mutant as-1/(–2)GA, which was recognised with high efficiency in the in vitro binding assay by either ASF-1 or recombinant TGA factors, conferred only 20% of the transcriptional activation mediated by the wild-type element. Deletion mutant as-1/(-2)GG, which showed 61% of ASF-1 binding activity in EMSAs, yielded only 9.5% of wild-type GUS activity.
TGA2.2–VP16 binds to as-1/(–2)GA in vivo
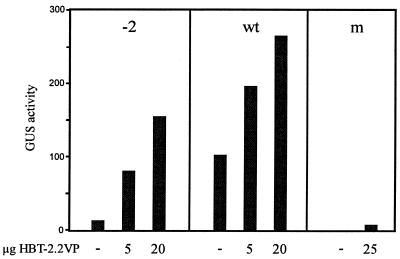
In order to test whether as-1/(–2)GA can be recognised by TGA factors in vivo, its transcriptional activity was characterised in the presence of TGA2.2 fused to the constitutive activation domain of herpes simplex virus protein 16 (VP16). TGA2.2–VP16 activated the deletion mutant as-1/(–2)GA in transiently transformed mesophyll protoplasts indicating in vivo binding (Fig. 4). However, absolute expression levels were higher when using the wild-type element, presumably due to the additive activating effect of endogenous TGA factors. Activation of both elements by TGA2.2–VP16 was observed irrespective of whether protoplasts were incubated in the presence of 50 µM auxin or whether they were incubated without the hormone. In conclusion, this experiment indicates that binding of TGA factors to the deletion mutant is possible in vivo. The inability of endogenous TGA factors to activate transcription from this element must be due the inability of a transactivation step to function if the factors are not correctly arranged on the DNA.
Figure 4.
Influence of TGA2.2–VP16 on expression from as-1 and as-1(–2)GA. Mesophyll protoplasts were electroporated with reporter plasmids (10 µg) encoding either the as-1 element (wt), the as-1(–2)GA (–2) element or a mutant as-1 element (m, unable to bind to ASF-1; see Fig. 2) within the –90 region of the CaMV 35S promoter. The amount of the co-electroporated activator plasmid encoding TGA2.2–VP16 under the control of the HBT promoter is given below the columns. The empty HBT vector was used to adjust the total amount of electroporated plasmid DNA to 30 µg. Protoplasts were incubated in the presence of 50 µM auxin. A representative experiment out of three repetitions is shown. Activation of the as-1(–2)GA (–2) element (10 µg) by TGA2.2–VP16 (20 µg) varied between 8- and 24-fold, activation of the wt element varied between 2- and 5-fold.
DISCUSSION
as-1-like elements represent a set of natural binding motifs that are recognised by ASF-1 and the TGA family of bZIP transcription factors (Fig. 1). The consensus as-1 element consists of two TGACGTCA palindromes spaced by 4 bp, which puts the palindromic centres 12 bp apart. The consensus sequence is recognised with higher affinity by ASF-1 than mutants thereof, resulting also in higher transcriptional activation in vivo (3). This indicates that ASF-1 is responsible for transcriptional activation. Here we studied the impact of the spacing between the palindromes on TGA factor binding and transcriptional activation.
As changing the spacing also affects the sequence, we asked whether the GGGA sequence between the palindromes can be disrupted without compromising its function. As seen in Figure 1, only the G in position 1 is strongly conserved among the naturally found as-1-like elements. The sequence GGCA did not change the properties of the element with respect to binding and transcriptional activation (Table 1). In contrast, the binding of ASF-1 was severely reduced in the mutant encoding the sequence GAGA, suggesting a favourable interaction of TGA factors with base G in position 2. However, this requirement was relieved in the deletion mutant as-1/(–2)GA which showed even higher binding activity than the deletion mutant as-1/(–2)GG. This indicates that location of two TGA dimers on the same side of the DNA helix alters the requirement for specific base pair contacts within the spacer.
Increasing the distance between the two palindromes decreased the binding activities of both ASF-1 and in vitro translated TGA factors TGA2.2, TGA2.1 and TGA1a. As all the insertion mutants encoded the critical GG downstream of the first palindrome and the potentially critical A immediately upstream of the second palindrome, we deduce that reduced binding activity is due to missing protein–protein contacts between two dimers. Contacts between two TGA2.1 dimers have been assumed before, as this protein requires two palindromes for efficient binding (24). Consistently, binding of this factor is more sensitive to differences in the spacing than binding of TGA2.2 and TGA1a (Table 2). As titration curves with increasing amounts of ASF-1 or TGA2.2 had not shown strong preference for double occupancy of the as-1 element (19,24) and as TGA1a prefers occupation of one palindrome even at high protein concentrations, independent binding to the two palindromes was expected. However, ASF-1, TGA1a and TGA2.2 showed decreased affinity to the insertion mutants. This observation might be explained by assuming that the on-rate of complex formation of TGA factors depends on protein–protein interactions between two dimers, while dissociation of one dimer is likely to occur subsequently, resulting in a stable complex with only one palindrome. A similar observation has been made for TGA2 from A.thaliana, which forms stable complexes with one palindrome only if a second palindrome is located in cis (34). Interestingly, the palindromes of the as-1-like elements in the promoter regions of AtPR-1 and NtPR-1a are >4 bp apart. However, in these complex promoters binding affinity might be increased by other interacting trans-factors. For instance, proteins like NPR-1 (35) and OBP1 (36) have been found to increase the DNA binding affinity of TGA factors in vitro.
The two deletion mutants of as-1, which miss 2 bp between the palindromes [as-1/(–2)GA and as-1/(–2)GG] were efficiently bound by ASF-1 and in vitro translated TGA factors. However, both elements conferred strongly reduced transcriptional activation in transiently transformed protoplasts. As a point mutation within the spacer excluded the existence of an overlapping binding site for additional trans-acting factors (Table 1), we conclude that the altered spacing of the two palindromes is responsible for the decreased transcriptional activity. A similar observation has been made by Ellis et al., who changed the spacing between the two half sites of the ocs element (10). In order to verify that the deletion mutant is recognised also in vivo by as-1-binding proteins, a TGA factor with a constitutive activation domain (TGA2.2–VP16) was co-expressed together with the reporter constructs as-1:gus and as-1/(–2)GA:gus. TGA2.2–VP16 activated transcription from as-1/(–2)GA, indicating that TGA factors are indeed able to bind to this element in vivo. The lack of transcriptional activation when driven by endogenous factors suggests that the activation step is not functioning when the two palindromes are not correctly spaced. This might be explained by assuming that the exact arrangement of two TGA dimers on the DNA is required for the recruitment of an additional protein mediating activation. In yeast, TGA2.2, which is the main component of ASF-1, does not confer transcriptional activation (24). However, its overexpression in planta has a positive effect on target gene expression (20) implying the existence of a co-activator. If the requirement for such a co-activator is relieved by fusing TGA2.2 to a constitutive activation domain, transcriptional activation can occur efficiently from as-1(–2)GA. The data imply that this co-activator associates only with two correctly spaced TGA dimers. A similar mechanism has been observed for the mammalian protein Pit1: spacing between the two Pit1 binding motifs determines whether a co-repressor is recruited by Pit1 (37). Alternatively, one might assume that as-1(–2)GA is efficiently recognised in vitro and in vivo due to favourable protein–protein contacts between the two TGA dimers when recruited to the same side of the DNA helix. As discussed above for TGA1a, one dimer might dissociate subsequently, leaving only one of the two palindromes occupied. This idea is supported by data reported by Ellis et al. (10), who found that the ratio of the lower complex to the upper complex shifts towards the lower complex (representing occupation of only one palindrome) when using a radiolabelled –2 deletion mutant of the ocs element in EMSAs with maize nuclear proteins. If this were the case, an as-1 element occupied by only one dimer would not be recognised by the postulated co-activator.
In conclusion, we report the following novel information on the interaction between TGA factors and the as-1 element. (i) The binding affinity of ASF-1 and different TGA factors to the as-1 element is highest if the spacing between the two palindromes is either 10 or 12 bp. Insertions reduce the binding activity. This result was unexpected, as titration experiments with ASF-1, TGA2.2 and TGA1a had not shown any evidence for cooperative binding. (ii) The sequence between the palindromes influences the affinity of TGA factors, but does not encode an overlapping binding site for unrelated factors. (iii) In contrast to endogenous TGA factors, a TGA factor fused to a constitutive activation domain led to transcriptional activation from a mutated as-1 element encoding 10 bp between the palindromic centres. This indicates that binding of TGA factors to this element occurs in vivo, but that the endogenous activation mechanism works only on the wild-type element encoding 12 bp between the palindromic centres. Thus, the strong conservation of the spacing in as-1-like elements is important for both the efficient recognition of the element by TGA factors and for the activation step. A challenging task for the future is the molecular characterisation of this activation mechanism.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Thomas Merkle (University of Freiburg, Germany) for his help with the transient expression assays using tobacco cell line BY-2, Dr Ingo Lenk (University of Göttingen, Germany) for providing the TGA2.2–VP16 fusion and Andreas Schiermeyer (University of Göttingen) for critically reading the manuscript. The work was supported by the Deutsche Forschungsgemeinschaft (GA330/11-1) and the Fonds der Chemischen Industrie (personal fellowships to R.N. and S.K).
REFERENCES
- 1.Berg O.G. and von Hippel,P.H. (1987) Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol., 193, 723–750. [DOI] [PubMed] [Google Scholar]
- 2.Lam E., Benfey,P.N., Gilmartin,P.M., Fang,R.-X. and Chua,N.-H. (1989) Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc. Natl Acad. Sci. USA, 86, 7890–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin X.-F., Holuigue,L. Horvath,D.M. and Chua,N.-H. (1994) Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell, 6, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X.J. and Lam,E. (1994) Two binding sites for the plant transcription factor ASF-1 can respond to auxin treatments in transgenic tobacco. J. Biol. Chem., 269, 668–675. [PubMed] [Google Scholar]
- 5.Benfey P.N., Ren,L. and Chua,N.-H. (1989) The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J., 8, 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulmasov T., Hagen,G. and Guilfoyle,T. (1994) The ocs element in the soybean GH2/4 promoter is activated by both active and inactive auxin and salicylic acid analogues. Plant Mol. Biol., 26, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 7.Pascuzzi P., Hamilton,D., Bodily,K. and Arias,J. (1998) Auxin-induced stress potentiates trans-activation by a conserved basic/leucine-zipper factor. J. Biol. Chem., 273, 26631–26637. [DOI] [PubMed] [Google Scholar]
- 8.Bouchez D., Tokuhisa,J.G., Llewellyn,D.J., Dennis,E.S. and Ellis,J.G. (1989) The ocs-element is a component of the promoters of several T-DNA and plant viral genes. EMBO J., 8, 4197–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feltkamp D., Masterson,R., Starke,J. and Rosahl,S. (1994) Analysis of the involvement of ocs-like bZip-binding elements in the differential strength of the bidirectional mas1′2′ promoter. Plant Physiol., 105, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis J.G., Tokuhisa,J.G., Llewellyn,D.J., Bouchez,D., Singh,K., Dennis,E.S. and Peacock,W.J. (1993) Does the ocs-element occur as a functional component of the promoters of plant genes? Plant J., 4, 433–443. [DOI] [PubMed] [Google Scholar]
- 11.Sakai T., Takahashi,Y. and Nagata,T. (1998) The identification of DNA binding factors specific for as-1-like sequences in auxin-responsive regions of parA, parB and parC. Plant Cell Physiol., 39, 731–739. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y., Kusaba,M., Hiraoka,Y. and Nagata,T. (1991) Characterization of the auxin-regulated par gene from tobacco mesophyll protoplasts. Plant J., 1, 327–332. [DOI] [PubMed] [Google Scholar]
- 13.Droog F.N.J., Spek,A., van der Kooy,A., der Ruyter,A., Hoge,H., Libbenga,K., Hoykaas,P. and van der Zaal,B. (1995) Promoter analysis of the auxin-regulated tobacco glutathione S-transferase genes Nt103-1 and Nt103-35. Plant Mol. Biol., 29, 413–429. [DOI] [PubMed] [Google Scholar]
- 14.Van der Zaal B.J., Droog,F.N.J., Boot,C.J., Hensgens,L.A., Hoge,J.H., Schilperoort,R.A. and Libbenga,K.R. (1991) Promoters of auxin-induced genes from tobacco can lead to auxin-inducible and root tip-specific expression. Plant Mol. Biol., 16, 983–998. [DOI] [PubMed] [Google Scholar]
- 15.Chen W., Chao,G. and Singh,K.B. (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J., 10, 955–966. [DOI] [PubMed] [Google Scholar]
- 16.Horvath D.M. and Chua,N.-H. (1996) Identification of an immediate-early salicylic acid-inducible tobacco gene and characterization of induction by other compounds. Plant Mol. Biol., 31, 1061–1072. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Fan,W., Kinkema,M., Li,X. and Dong,X. (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl Acad. Sci. USA, 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strompen G., Gruner,R. and Pfitzner,U.M. (1998) An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol., 37, 871–883. [DOI] [PubMed] [Google Scholar]
- 19.Rieping M., Fritz,M., Prat,S. and Gatz,C. (1994) A dominant negative mutant of PG13 suppresses transcription from a cauliflower mosaic virus 35S truncated promoter in transgenic tobacco plants. Plant Cell, 6, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niggeweg R., Thurow,C., Kegler,C. and Gatz,C. (2000) Tobacco transcription factor TGA2.2 is the main component of ASF-1 and is involved in salicyclic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem., 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- 21.Katagiri F., Lam,E. and Chua,N.-H. (1989) Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature, 340, 727–730. [DOI] [PubMed] [Google Scholar]
- 22.Fromm H., Katagiri,F. and Chua,N.H. (1991) The tobacco transcription activator TGA1a binds to a sequence in the 5′ upstream region of a gene encoding a TGA1a-related protein. Mol. Gen. Genet., 229, 181–188. [DOI] [PubMed] [Google Scholar]
- 23.Niggeweg R. and Gatz,C. (1997) Isolation of TGA2.1 (accession no. U90214), a member of a new subclass of the TGA family of bZIP transcription factors in Nicotiana tabacum. Plant Physiol., 113, 1464. [Google Scholar]
- 24.Niggeweg R., Thurow,C., Weigel,R., Pfitzner,U. and Gatz,C. (2000) Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol., 42, 775–788. [DOI] [PubMed] [Google Scholar]
- 25.Klinedinst S., Pascuzzi,P., Redman,J., Desai,M. and Arias,J. (2000) A xenobiotic-stress-activated transcription factor and its cognate target genes are preferentially expressed in root tip meristems. Plant Mol. Biol., 42, 679–688. [DOI] [PubMed] [Google Scholar]
- 26.Neuhaus G., Neuhaus-Url,G., Katagiri,K., Seipel,K. and Chua,N.-H. (1994) Tissue-specific expression of as-1 in transgenic tobacco. Plant Cell, 6, 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehner S., Lenk,I., Rieping,M., Herold,M. and Gatz,C. (1999) Transcriptional activator TGV mediates dexamethasone-inducible and tetracycline-inactivatable gene expression. Plant J., 19, 87–95. [DOI] [PubMed] [Google Scholar]
- 28.Sheen J. (1993) Protein phosphatase activity is required for light-inducible gene expression in maize. EMBO J., 12, 3497–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prat S., Willmitzer,L. and Sanchez-Serrano,J.J. (1989) Nuclear proteins binding to a cauliflower mosaic virus 35S truncated promoter. Mol. Gen. Genet., 17, 209–214. [DOI] [PubMed] [Google Scholar]
- 30.Haasen D., Kohler,C., Neuhaus,G. and Merkle,T. (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana.Plant J., 20, 695–705. [DOI] [PubMed] [Google Scholar]
- 31.Leborgne-Castel N., Jelitto-Van Dooren,E.P., Crofts,A.J. and Denecke,J. (1999) Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell, 11, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jefferson R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep., 5, 387–405. [Google Scholar]
- 33.Baranowskij N., Frohberg,C., Prat,S. and Willmitzer,L. (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J., 15, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam E. and Lam,Y.K.-P. (1995) Binding site requirements and differential representation of TGA factors in nuclear ASF-1 activity. Nucleic Acids Res., 23, 3778–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Despres C., DeLong,C., Glaze,S., Liu,E. and Fobert,P.R. (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell, 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B., Chen,W., Foley,R.C., Buttner,M. and Singh,K.B. (1995) Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell, 7, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scully K.M., Jacobson,E.M., Jepsen,K., Lunyak,V., Viadiu,H., Carriere,C., Rose,D.W., Hooshmand,F., Aggarwal,A.K. and Rosenfeld,M.G. (2000) Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science, 290, 1127–1131. [DOI] [PubMed] [Google Scholar]