Abstract
The MutH protein, which is part of the Dam-directed mismatch repair system of Escherichia coli, introduces nicks in the unmethylated strand of a hemi-methylated DNA duplex. The latent endonuclease activity of MutH is activated by interaction with MutL, another member of the repair system. The crystal structure of MutH suggested that the active site residues include Asp70, Glu77 and Lys79, which are located at the bottom of a cleft where DNA binding probably occurs. We mutated these residues to alanines and found that the mutant proteins were unable to complement a chromosomal mutH deletion. The purified mutant proteins were able to bind to DNA with a hemi-methylated GATC sequence but had no detectable endonuclease activity with or without MutL. Although the data are consistent with the prediction of a catalytic role for Asp70, Glu77 and Lys79, it cannot be excluded that they are also involved in binding to MutL.
INTRODUCTION
Newly synthesized DNA of Escherichia coli contains a parental strand which is methylated at the adenine in GATC sequences and a daughter strand that is not methylated at these sequences (1). After a short time the GATC sequences in the daughter strand are methylated by DNA adenine methyltransferase (Dam) (1). This transient hemi-methylated DNA behind the replication fork is the substrate for Dam-directed DNA mismatch repair that is initiated by the action of three proteins: MutS, MutL and MutH (2–4). The MutS protein, which has weak ATPase activity, recognizes and binds to base or insertion/deletion mismatches (4,5). This is followed, in an ATP-dependent manner, by the formation of a ternary complex composed of MutL and MutH in addition to MutS. The MutL protein also binds and hydrolyzes ATP (6,7) and interacts with a variety of proteins, including MutH, whose latent endonuclease activity is activated in the ternary complex (8,9). MutH endonuclease activity is maximal on hemi-methylated DNA, 2–3-fold reduced on unmethylated DNA and minimal on fully methylated DNA. DNA cleavage on the unmethylated strand occurs 5′ to the G in GATC sequences (8,9). Genetic studies have shown that mutations in the genes encoding these three proteins results in a mutator phenotype consistent with their proposed role in preventing mutations (10).
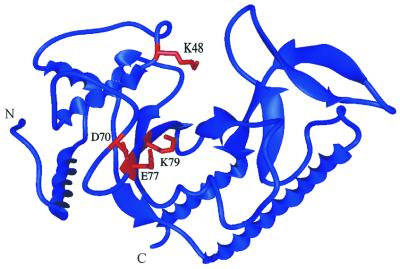
The DNA-free MutH protein structure has been determined at the atomic level (Fig. 1) (11). The MutH protein resembles a large clamp containing a cleft in which the DNA substrate is thought to bind. Movement of the clamp arms appears to be modulated by the position of the C-terminal helix, which can be likened to a lever. MutL alone, in the presence of ATP, can activate the latent single-strand endonuclease activity of MutH in vitro (12–14). An attractive feature of the structural model is that MutL binding allows movement of the lever to close the clamp holding the bound DNA. This forces the DNA into the proximity of the proposed catalytic residues at the bottom of the cleft. We have recently shown that Lys48, which is located in a flexible loop (Fig. 1), functions in catalysis, suggesting that movement of this loop occurs to position Lys48 within the catalytic core of the protein (12).
Figure 1.
Location of the mutated amino acid residues in the MutH structure. The ribbon diagram of MutH was made from the B monomer with the coordinates submitted by W. Yang (Protein Data Bank code 2AZO) using MIDAS. The residues tested from the site-directed mutagenesis, D70A, E77A and K79A, are displayed in red. An additional residue (K48) that has been shown to be involved in catalysis (12) is also displayed in red. N and C denote the N- and C-terminal ends of the protein, respectively.
The putative active site in MutH was proposed to include Asp70, Glu77 and Lys69 (Fig. 1) based on the catalytic sequence motif found in many type II restriction enzymes, including Sau3A, which also cleaves at GATC sequences (11). In the crystal structure these residues are towards the bottom of the cleft. Glu77 interacts with Glu56 in a manner analogous to that in the EcoRV active site or to Lys116, which is conserved among MutH homologs. A good fit between bent DNA and the proposed active site cleft was obtained in a molecular model if conformational changes occur upon substrate binding. In this report we confirm the prediction of Ban and Yang (11) that the Asp70, Glu77 and Lys69 residues are required for catalysis; however, we cannot exclude the possibility that these residues are also required for productive interaction with MutL.
MATERIALS AND METHODS
Strains, plasmids and media
The construction and properties of E.coli strain KM55 (ΔmutH461::Cam), a derivative of AB1157 [thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) hisG4(Oc) rfbD1 mgl-51 rpoS396(Am) rpsL31(StrR) kdgK51 xylA5 mtl-1 argE3(Oc) thi-1], have been described previously (12). Strain GM5862 [F-lacIq lacZ– proAB/ Δ mutL459::Kan ara thi Δ(gpt-lac)5 (λDE3)] containing pMQ393 (15) was used for overexpression of wild-type MutL. Plasmid pMQ402, a derivative of pBAD18 carrying the wild-type mutH gene under control of the araBAD promoter, was used to construct site-directed mutations at residues Asp70, Glu77 and Lys69 as described previously (12). The site-directed mutations were confirmed by DNA sequencing using the mutH sequence in GenBank (accession no. U16361). Both strands and the promoter region of all mutant plasmids were sequenced.
Luria–Bertani (LB) medium was made with 10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl and 1 ml 1 M sodium hydroxide. Ampicillin (Amp) and rifampicin (Rif) were added to the medium at 100 µg/ml each and chloramphenicol (Cam) at 30 µg/ml.
Mutant frequency
Cells lacking MutH protein accumulate mutations at a greater frequency than the wild-type and this can be monitored by selection on plates with Rif. The vector (pBAD18), wild-type MutH (pMQ402) and mutant (pD70A, pE77A and pK79A) plasmids were transformed into strains AB1157 and/or KM55. Ten single colonies from each strain were inoculated separately into 1 ml of LB + Amp broth, grown overnight at 37°C, diluted and spread in duplicate on LB plates with Amp or Rif + Amp. The plates were incubated at 37°C overnight and colonies counted the next day. The pMQ402 plasmid produces enough wild-type MutH protein from the araBAD promoter in the absence of inducer (arabinose) to complement the mutH strain.
Preparation of histidine-tagged MutH
The wild-type and all mutant proteins were purified in the same manner. For each strain, a 20 ml LB + Amp culture was grown overnight from a single colony. One liter of LB + Amp medium was inoculated with the 20 ml overnight culture and allowed to grow with shaking at 37°C to an OD600 of 1.0. Arabinose (Difco) was added to a final concentration of 0.2% and the culture induced for 2 h at 37°C. Cells were harvested by centrifugation for 20 min at 5000 r.p.m., washed with water and stored frozen (–80°C). The pellet was thawed, resuspended in 4 ml of reconstitution buffer (20 mM HEPES pH 7.4, 300 mM NaCl and 1 mM PMSF) and the cells lysed using a French pressure cell. The extract was then sonicated (20 pulses of a Tekmar sonifier) and centrifuged for 30 min at 15 000 r.p.m. to remove cell debris. The supernatant was applied to a 4 ml Fast Flow Chelating Sepharose (Pharmacia Biotech) column charged with 100 mM NiCl2 and equilibrated with reconstitution buffer. Step fractions of 100, 150, 250 and 400 mM imidazole (10 ml each) were collected. The 250 mM fraction contained the MutH protein and was dialyzed against two changes of 20 mM HEPES pH 7.4, 300 mM NaCl and 0.1 mM EDTA. The MutH concentration was determined by measuring the OD280 (1 OD = 0.67 mg/ml). MutH was at least 95% pure as determined in a SDS–PAGE gel stained with Coomassie Brilliant Blue.
Preparation of histidine-tagged MutL
The His-tagged MutL protein was purified essentially as described (15). Strain GM5862 was transformed with pMQ393 and the transformation mixture added to 50 ml of YT + Cam medium for overnight incubation at 37°C. Ten milliliters of this culture was inoculated into 1 l of LB + Cam broth and incubated with shaking at 37°C. At an optical density (A600) of 0.8, IPTG (Sigma) was added to 0.5 mM and the culture incubated for 90 min. The cells were harvested and clarified extracts prepared as described for MutH above. The step fractions used for elution of His-tagged MutL from the Fast Flow Chelating Sepharose column were: one 100, two 150 and one 400 mM imidazole (10 ml each). The 400 mM fraction contained the MutL protein, which was dialyzed against two changes of 20 mM HEPES pH 7.4, 300 mM NaCl and 0.1 mM EDTA. The concentration of MutL was determined by measuring the OD280 (1 OD = 1.24 mg/ml). The protein was at least 90% pure as determined in a Coomassie Brilliant Blue stained SDS–PAGE gel. Native MutL was graciously provided by Drs Paul Modrich and Claudia Spampinato (Duke University).
Endonuclease assay
Bacteriophage MR1 is a derivative of phage f1R229 that contains only one d(GATC) site (16). The covalently closed replicative form of this phage was purified as previously described from Cam-treated infected GM2807 (dam-16::Kan) cells (12). The MR1 phage was provided by Dr R. Lahue (University of Nebraska). MutH activity (amounts are indicated in the figures) was tested at 37°C in a 10 µl reaction containing 20 mM Tris–HCl pH 7.7, 5 mM MgCl2 and 25 fmol MR1 for 1 h. MutL and ATP were added at 71 pmol and 1.25 mM, respectively, to the MutH endonuclease stimulation assays. The assays were carried out with native MutL or His-tagged MutL. The reaction was stopped with 5 µl of a 50% glycerol solution containing 50 mM EDTA, 1% SDS, 0.05% Bromphenol Blue and 0.05% Xylene Cyanol. The reaction mixtures were electrophoresed in 1% agarose at 5 V/cm for 3 h. The gels were stained with Vistra Green (Amersham) following the manufacturer’s instructions (1:10 000 dilution) for 1 h with shaking. The gels were then scanned on a Storm PhosphorImager (Molecular Dynamics) and the amount of nicking was quantitated using ImageQuant (Molecular Dynamics) software. Background levels of nicked substrate were subtracted from reactions with added enzyme.
Preparation of labeled hemi-methylated DNA
A 36 bp DNA oligonucleotide, MM40 (5′-GCATACGGAAGTTAAAGTGCGGATCATCTCTAGCCA-3′; Operon Technologies), containing a single, centrally located d(GATC) site, was labeled using T4 polynucleotide kinase (PNK) (New England BioLabs). A 12 µl reaction mixture contained 2 µM oligonucleotide, 1.2 µl PNK buffer, 1 µl PNK enzyme, 1 µl [γ-32P]ATP (Amersham Pharmacia Biotech) and water to a final concentration of 0.5 µM oligonucleotide. The reaction was incubated for 1 h at 37°C and then 10 min at 75°C. Unlabeled complementary oligonucleotide [2 µM, containing N6-methyladenine in the d(GATC) site] was added to a final volume of 100 µl. This was incubated for 5 min at 95°C, 30 min at 37°C and 30 min at room temperature. The unincorporated radioactive ATP and single-stranded oligonucleotide were removed using a MicroSpin G-25 column (Pharmacia Biotech) and the labeled DNA dissolved in a final volume of 100 µl of water.
Band shift assay
MutH binding to DNA was assayed using the labeled DNA prepared as described above. A 5 µl reaction volume contained 20 mM Tris–HCl pH 7.6, 5 mM MgCl2, 0.1 mM DTT, 0.01 mM EDTA, 22.5 fmol labeled DNA and varying concentrations of MutH. MutL and ATP were added at 9 pmol and 1.25 mM, respectively, to the stimulation assays. The mixture was incubated for 30 min on ice, followed by addition of 1.6 µl of 50% sucrose and loaded on a pre-run (10 min) 6% native polyacrylamide gel in TAE buffer, pH 7.5, and electrophoresed at 50 mA for 2.5 h. The gel was scanned on a Storm PhosphorImager (Molecular Dynamics). The free DNA and shifted band intensities were measured using ImageQuant (Molecular Dynamics) software. Apparent dissociation constants (Kd) were determined by fitting the experimental data to the equation 1/r = 1 + Kd/[C]total (r = 1 – f, where f is the degree of dissociation) with the program KaleidaGraph (Synergy Software). The degree of binding or association is r and [C]total is the protein concentration in the reaction.
RESULTS
In vivo complementation assay
The MutH crystal structure and amino acid sequence comparison data obtained by Ban and Wang (7) suggested that there are three catalytic residues: Asp70, Glu77 and Lys69. To determine the role of these residues, mutations were introduced in the mutH gene on plasmid pMQ402, to convert these amino acids separately into alanines. MutH production from pMQ402 can be controlled by the amount of arabinose in the medium because mutH transcription originates from the araBAD promoter. The steady-state level of MutH, from cells grown with or without inducer, was the same for the wild-type and mutated plasmids, as determined by immunoblotting (data not shown).
A strain (KM55) with a chromosomal deletion of mutH and the vector plasmid (pBAD18) has a 175-fold increase in spontaneous mutant frequency compared to the parent strain (AB1157) with the wild-type mutH allele (Table 1). Introduction of pMQ402 into the mutH deletion strain (KM55) reduces the spontaneous mutant frequency to near background levels when these cells are grown in the absence of arabinose (Table 1). In contrast, plasmids encoding MutH variant D70A, E77A or K79A in strain KM55 show spontaneous mutant frequencies comparable to the pBAD18 vector control. The level of spontaneous mutagenesis suggests that the mutant proteins are inactive in vivo.
Table 1. In vivo complementation with wild-type mutH and mutant genes.
| Strain | Mutant frequency |
|---|---|
| pBAD18/AB1157 | 1 ± 0 |
| pBAD18/KM55 | 175 ± 25 |
| pMQ402/AB1157 | 1 ± 0 |
| pMQ402/KM55 | 3 ± 2 |
| pD70A/KM55 | 125 ± 11 |
| pE77A/KM55 | 163 ± 41 |
| pK79A/KM55 | 192 ± 105 |
Ten colonies were each inoculated into LB broth with ampicillin and grown overnight. The next day the cultures were diluted and plated on LB plates with Rif or Amp plus Rif. The mutant frequency is the number of Rif-resistant colonies per 108 Amp-resistant cells. Strain KM55 is a mutH deletion derivative of AB1157.
DNA binding assay
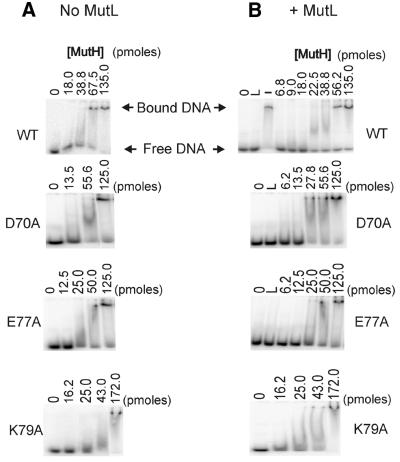
The binding of wild-type and mutant proteins to DNA was measured using a band shift assay (12). A 36mer duplex of DNA with a single central GATC sequence was used and this sequence was methylated at the adenine in one of the two strands (hemi-methylated DNA). Binding of wild-type MutH is specific because no detectable band shift was detected using a duplex with GGTC in place of GATC (data not shown). In the absence of MutL, wild-type MutH gave a complete band shift with the appearance of a broad band between the free and bound DNA (Fig. 2A). The inclusion of magnesium in the reaction mixtures or the gel did not affect this broad band. The MutH D70A, E77A and K79A mutant proteins gave results similar to the wild-type (Fig. 2A). The inclusion of MutL at a concentration that does not cause non-specific DNA binding did not significantly alter the band shift pattern of the mutant proteins compared to those without MutL (Fig. 2B). The wild-type protein showed reproducible band shifting at a lower concentration in the presence of MutL.
Figure 2.
Binding abilities of the wild-type and mutant MutH proteins. The binding ability of the wild-type and mutant MutH proteins were measured by a band shift assay using a 32P-labeled oligonucleotide. The DNA was incubated with increasing amounts of the MutH protein (indicated above each lane) in the reaction and the products separated in a 6% polyacrylamide gel. (A) All reactions contained 22 fmol DNA with a hemi-methylated GATC sequence, 1.25 mM ATP and did not contain MutL. The 0 lanes are without MutH and represent 22 fmol unbound substrate DNA. (B) All reactions contained 22 fmol DNA with a hemi-methylated GATC sequence, 1.25 mM ATP and 9 pmol MutL (except the – lane). The L lanes do not contain MutH. The – control lane contains 135 pmol MutH, 1.25 mM ATP and 22 fmol DNA.
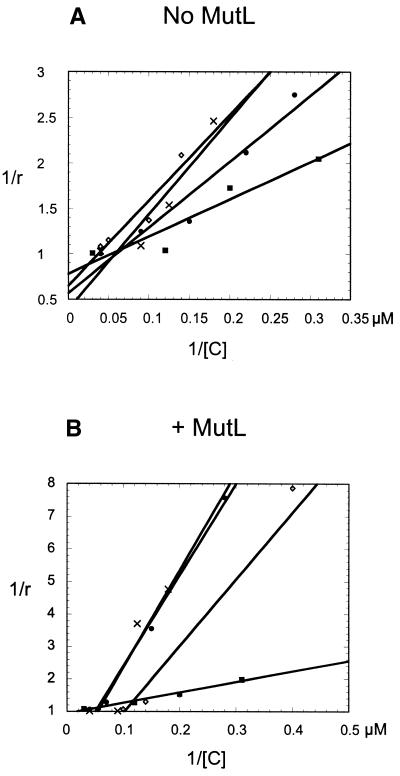
The degree of association, or band shift, was plotted versus the concentration of MutH protein using the data obtained in Figure 2. The wild-type, D70A and E77A proteins showed similar DNA binding while the K79A protein showed a higher affinity with or without MutL (Fig. 3A and B). The apparent Kd (calculated from the x-intercepts of Fig. 3A and B) for wild-type MutH increased ∼14-fold in the presence of MutL (Table 2). The K79A mutant had a Kd comparable to the wild-type protein, but the MutL stimulation was only ∼4-fold. The D70A and E77A mutant proteins had Kd values similar to the wild-type in the absence of MutL, but MutL increased these values by only ∼2- and 3-fold, respectively.
Figure 3.
Dependence of complex formation on the concentration of MutH protein. The graphs were plotted by fitting the experimental data from the band shift assay to the equation described in Materials and Methods. This allows the calculation of an apparent Kd. (A) Data plotted from the band shift assays without MutL present. The wild-type protein (circles), K79A mutant (squares), E77A mutant (diamonds) and D70A mutant (crosses) are shown. (B) Data plotted from the band shift assay containing MutL. The wild-type protein (circles), K79A mutant (squares), E77A mutant (diamonds) and D70A mutant (crosses) are shown. r is the degree of binding and [C] the MutH concentration in the reaction. Each point represents the average of at least three independent measurements.
Table 2. Apparent equilibrium dissociation constants for wild-type and mutant MutH proteins.
| MutL | Protein | Kd (µM) |
|---|---|---|
| + | Wild-type | 4.2 ± 0.7 |
| + | D70A | 48 ± 6 |
| + | E77A | 20 ± 4 |
| + | K79A | 3 ± 0.8 |
| – | Wild-type | 57 ± 0.6 |
| – | D70A | 83 ± 12 |
| – | E77A | 67 ± 10 |
| – | K79A | 14 ± 4 |
Endonuclease activity
Endonuclease activity was measured by the conversion of an unmethylated covalently closed circular (cc) DNA substrate to a nicked circular (nc) form (8). The MutH protein cleaves unmethylated substrates only 2–3-fold less efficiently than hemi-methylated DNA (8). As shown in Figure 4, 3 fmol MutH (in the presence of MutL) converts some of the substrate ccDNA to ncDNA and 31 fmol MutH converts all of the ccDNA to ncDNA. In contrast, no endonuclease activity was detected with the mutant proteins at concentrations ∼70–100-fold higher (Fig. 4). Inclusion of MutL in the endonuclease assay reduces by ∼1000-fold the amount of MutH needed to achieve the same amount of cleavage as without MutL (12; data not shown).
Figure 4.
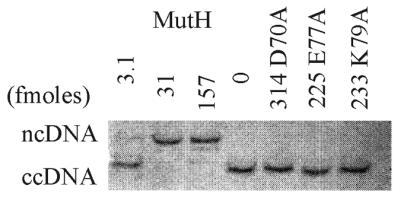
Endonuclease activity of the wild-type and mutant MutH proteins. Increasing amounts of MutH (fmol) were incubated with the MR1 ccDNA substrate which was converted to ncDNA (upper band) and linear DNA (middle band). The DNA products were separated in a 1% agarose gel before staining and scanning for quantitation. All reactions contain 71 pmol MutL, 1.25 mM ATP and 25 fmol homoduplex DNA substrate. The number of fmol of mutant protein used in the reactions is also indicated. The 0 lane represents unreacted homoduplex DNA.
DISCUSSION
Plasmids containing mutations changing Asp70, Glu77 and Lys79 to alanines were unable to complement a mutH chromosomal deletion, indicating that the mutated proteins are inactive in vivo (Table 1). The level of spontaneous mutagenesis in cells with the altered MutH proteins is comparable to that of the mutH strain with the vector alone, indicating a complete lack of function for the mutant proteins. No effect of the mutant proteins on mutation frequency in wild-type cells was detected, showing that they do not act in a dominant negative manner (data not shown). This result suggests that the mutant proteins do not interact productively with MutL to sequester it from use by wild-type MutH. It also indicates that the mutant proteins do not interfere with binding of the wild-type protein in vivo. The lack of dominant negative action cannot be due to reduced amounts of the mutant proteins as these are present at the same level as in the wild-type, as determined by immunoblotting (data not shown).
All three mutant proteins were able to bind substrate DNA (Figs 2 and 3) and MutL decreased the Kd by 2–3-fold (Table 2). However, these Kd values for the mutant proteins are in the micromolar range and therefore their relevance in vivo is questionable, as are the values for the wild-type protein, where a 14-fold effect of MutL on DNA binding is observed. The data do show, however, that the mutant proteins can bind substrate DNA in vitro under the experimental conditions used.
The interaction of MutH with MutL is probably the more important physiological effect, as a >1000-fold stimulation of MutH catalytic activity is observed (12). However, we failed to observe any endonuclease activity with the mutant proteins with or without MutL. Therefore, we cannot exclude the possibility that in vivo MutH proteins defective in catalysis are unable to interact with MutL. The lack of a dominant negative mutator phenotype with the mutated mutH plasmids is consistent with this idea. MutH protein must undergo conformational changes in order to activate its latent endonuclease activity (7). It may be that for these conformational changes to occur the protein needs to form specific contacts with MutL and hemi-methylated DNA through the proposed catalytic residues.
It is possible that the purified MutH mutant proteins have structural changes that impede efficient interactions with DNA and MutL. However, the mutant proteins are produced in the same amount as the wild-type, as determined by immunoblotting, and in addition show binding to substrate DNA. Although not conclusive, this suggests that the altered proteins do not have structural changes impeding their activity. We were not able to test the mutant proteins by circular dichroism or thermal denaturation studies.
The properties described above of the MutH Asp70, Glu77 and Lys79 mutants are consistent with the prediction of Ban and Yang (11) that these residues have a catalytic function, although they may also be defective in MutL interaction. The location of these residues near the bottom of the cleft formed by the two subdomains makes it likely that the DNA comes into close proximity to these residues for catalysis. The demonstration that Lys48 is also part of the active site suggests that upon DNA binding there is movement of MutH to position this amino acid near the catalytic core. The co-crystal structure of an active site MutH mutant protein with hemi-methylated DNA should answer this supposition.
Acknowledgments
ACKNOWLEDGEMENTS
Nyawira Gikunju, Ensar Halilovic, Mei Huan Lin and Cui Tao helped with the purification and assays of the mutant proteins. We thank Drs Claudia Spampinato and Paul Modrich (Duke University) for providing native E.coli MutL protein and Dr R. Lahue (University of Nebraska) for phage MR1. This work was supported by grant RPG-97-127-01-GMC from the American Cancer Society.
REFERENCES
- 1.Marinus M.G. (1996) Methylation of DNA. In Neidhardt,F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 782–791.
- 2.Yang W. (2000) Structure and function of mismatch repair proteins. Mutat. Res., 460, 245–256. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh P. (2001) Molecular mechanisms of DNA mismatch repair. Mutat. Res., 486, 71–87. [DOI] [PubMed] [Google Scholar]
- 4.Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 5.Parker B.O. and Marinus,M.G. (1992) Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl Acad. Sci. USA, 89, 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spampinato C. and Modrich,P. (2000) The MutL ATPase is required for mismatch repair. J. Biol. Chem., 275, 9863–9869. [DOI] [PubMed] [Google Scholar]
- 7.Ban C. and Yang,W. (1998) Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell, 95, 541–552. [DOI] [PubMed] [Google Scholar]
- 8.Welsh K.M., Lu,A.L., Clark,S. and Modrich,P. (1987) Isolation and characterization of the Escherichia coli mutH gene product. J. Biol. Chem., 262, 15624–15629. [PubMed] [Google Scholar]
- 9.Au K.G., Welsh,K. and Modrich,P. (1992) Initiation of methyl-directed mismatch repair. J. Biol. Chem., 267, 12142–12148. [PubMed] [Google Scholar]
- 10.Horst J.P., Wu,T.H. and Marinus,M.G. (1999) Escherichia coli mutator genes. Trends Microbiol., 7, 29–36. [DOI] [PubMed] [Google Scholar]
- 11.Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loh T., Murphy,K.C. and Marinus,M.G. (2001) Mutational analysis of the MutH protein from Escherichia coli. J. Biol. Chem., 276, 12113–12119. [DOI] [PubMed] [Google Scholar]
- 13.Hall M.C. and Matson,S.W. (1999) The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem., 274, 1306–1312. [DOI] [PubMed] [Google Scholar]
- 14.Hall M.C., Jordan,J.R. and Matson,S.W. (1998) Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J., 17, 1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronshtam A. and Marinus,M.G. (1996) Dominant negative mutator mutations in the mutL gene of Escherichia coli.Nucleic Acids Res., 24, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahue R.S., Su,S.S. and Modrich,P. (1987) Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc. Natl Acad. Sci. USA, 84, 1482–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]