Abstract
The salamander Ambystoma mexicanum, commonly called “the axolotl” has a long, illustrious history as a model organism, perhaps with one of the longest track records as a laboratory-bred vertebrate, yet it also holds a prominent place among the emerging model organisms. Or rather it is by now an “emerged” model organism, boasting a full cohort molecular genetic tools that allows an expanding community of researchers in the field to explore the remarkable traits of this animal including regeneration, at cellular and molecular precision—which had been a dream for researchers over the years. This chapter describes the journey to this status, that could be helpful for those developing their respective animal or plant models.
1. Introduction—Reemerging the classic axolotl model into the molecular era
During the explosive growth of molecular biology in the late 20th century emerging techniques were rapidly applied to invertebrates such as drosophila, and vertebrates such as mouse, frog and zebrafish. These activities engendered the field of molecular embryology but the axolotl remained somewhat recalcitrant. Those features that had made the animal attractive for classical work such as large yolky eggs and slow cell cycle hindered direct application of molecular protocols to axolotl embryos. Additionally, regeneration of postembryonic body structures such as the limb and the tail are the axolotl’s seductive value but molecular methods were difficult to apply to adult tissues in any animal due to the complex cohorts of tissues and presence of substantial extracellular matrix. Incisive progress in the form of molecular genetics of regeneration appeared practically insurmountable. Regeneration is characterized by the changes in the adult tissue after amputation including influx of immune cells, migration of different cells layers that ultimately yield growth of new tissues. On one hand the source cells for regeneration had to be disentangled from noncontributing cells, and genetic manipulation of the regenerating cells had to be achieved, which basically required the use of genetic fate mapping and transduction of genetic material into those cells to express and downregulate molecular factors.
At the turn of this century as in situ hybridization, DNA transfection, and viral systems were further developed for mammalian systems, several brave groups among a dwindling salamander community carried the flag and reported impressive and important work characterizing gene expression during regeneration, using biolistic DNA transfection of blastema cells to study the role of retinoic acid receptors in limb segment positional identity, viral transduction to study the contribution of muscle cells to the blastema, and the role of embryonic morphogens in regeneration such as SHH and WNT (Gardiner, Blumberg, Komine, & Bryant, 1995; Kawakami et al., 2006; Kumar, Velloso, Imokawa, & Brockes, 2000; Pecorino, Lo, & Brockes, 1994; Roy, Gardiner, & Bryant, 2000). These methods were usually either cumbersome, inefficient, hard to reproduce, suffered from silencing in vivo, or damaged the transduced cells, limiting their broad use.
By the dawn of the 21st century everexpanding methodologies to transduce somatic cells and viral methods became available, followed by the next generation sequencing, and genome editing revolutions. Our account describes how we and others developed these methods for axolotls coupled with the goal to answer fundamental questions in regeneration biology which has allowed each of us and others to establish robust research programs focusing on the molecular mechanisms of axolotl regeneration. Fig. 1 provides an overview timeline as each of these methods and their associated scientific findings were published.
Fig. 1.
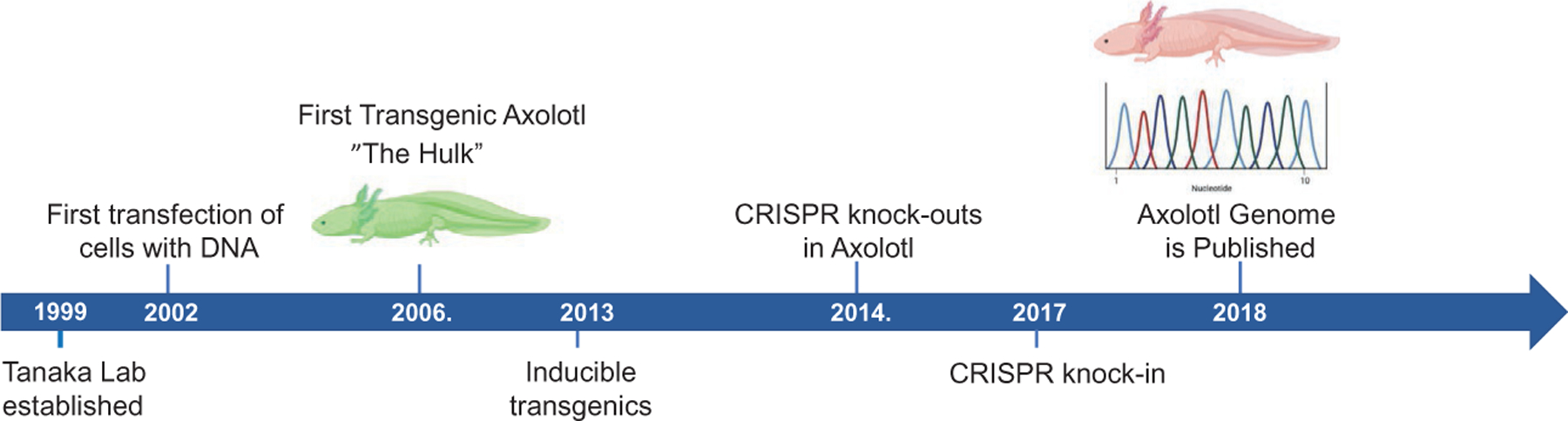
Timeline of technical developments pioneered by the Tanaka lab that have driven the advancement of the axoltol as a genetically tractable research organism.
Stepping back, it may be possible to extract a few insights for those with similar ambitions. The first might be “never say never.” This is exemplified by Karen Echeverri’s account of developing electroporation to express gene products in regenerating tissues. At the beginning of 2000s we had heard several people in the field conclude that salamander cells might not be transfectable, after they had tried the standardly available methods such as calcium phosphate precipitation and lipofection. With the broadening implementation of electroporation in chick embryos, and descriptions of ionotophoresis, Karen’s clever and dedicated effort to get this working in the axolotl tail was successful. Electroporating single spinal cord stem cells to follow their fate over time was not simple. It involved many attempts at pulling needles, engineering tilting stages to see the spinal cord lumen, and calling Jeffrey Corwin to learn that MS222 anesthetic was more toxic to the animals than benzocaine, which was critical for animal health during timelapse imaging. Having been successful, she could then apply the method not only to the spinal cord, but to testing the molecular basis of positional identity during limb regeneration (Echeverri & Tanaka, 2005). We also then applied electroporation successfully to cultured cells. In my mind, this experience addresses the oft heard anxiety of whether it is wise to potentially send PhD or postdoc careers to the graveyard developing methods. If the primary goal of the project is to answer a key question in the field by developing a method, then it is our strong belief that it will be possible to develop that method or another proxy method to address that question and even partial steps toward that goal will be publishable and appreciated if the question is important enough. Answering a question and developing a method to do so requires full commitment to analyze all the failures and to keep pushing with the attitude that something will work. This activity is not a side project.
Perhaps another insight concerns the role of emerging technologies and colleagues. As described below, transgenesis had been previously tried in salamanders, and had not been stable, but the key suggestion from wizard molecular biologist Francis Stewart that only certain promoters avoided silencing, allowed us to succeed. Similarly, we and others had consistently pursued gene sequence information for molecular studies, first in the form of expressed sequence tags (ESTs), then transcriptomes with the strong support of Bianca Haberman (Habermann et al., 2004; Putta et al., 2004). Another key turning point came when we were fortunate enough to be colleagues with Eugene Myers whose mathematical fascination to assemble genomes with newly emerging long-read technologies coupled with his generous intent to help young researchers in regeneration brought forth the planaria and axolotl genome assemblies that had been impossible to that point (Grohme et al., 2018; Nowoshilow et al., 2018). The presence of an exquisite molecular biologist, Ji-Feng Fei, in the lab was critical at this juncture, as he generated incredibly high-quality genomic DNA that showed the best output from PacBio machines that anyone had seen to that point. In parallel he was developing all the genome editing methodologies (gene mutation, gene knock-in) with remarkable care, testing many conditions, many different variants, following F0 animals into F1s and F2s with rigorous, quantitative genotyping and phenotyping that, not only filled up our axolotl colony, but lead to the breakthroughs in the field and a set of manuscripts that could withstand a very demanding review process (Fei et al., 2014; Nowoshilow et al., 2018). Many might have wondered why Ji-Feng was spending so much effort on following things through, but these quantitative observations provide a rock-solid basis for robust genome editing in our field. These anecdotes stress the importance of deep understanding and quantitative, rigorous treatment of these technology “attempts.” They also highlight the importance of connecting to colleagues in emerging technologies whether locally or by broadly by attending meetings focusing on those topics. Staying narrowly focused on the traditional scope of a given model organism may restrict one’s perspective on what is really possible.
We sincerely hope that our highly personal chapter describing the development of transient transfection, transgenesis, viral transduction, genome assembly and genome editing for the axolotl provide an amusing but also informative and inspiring account for how it is possible to address fundamental questions in regeneration biology at the molecular level in an animal previously considered as lagging in the molecular era.
2. Live cell imaging and functional studies by transient transfection
2.1. The historical starting point
A longstanding question in the field of regeneration has been, where do the cells come from to regenerate the new tissue? In the early days of the axolotl regeneration field tools to work in these animals were sparse. The first insights into the how cells reacted to injury and where the information to tell cells what must be regenerated was gained from classical transplantation experiments. Some of the first indications that blastema cells contained positional information came from studies where larval limb blastema cells were dissociated, reaggregated and then transplanted onto fin tissue, surprisingly a limb then grew from that position, suggesting that the cells contained all the information they needed to regenerate (Stocum, 1968). Other experiments using similar transplantation methods further showed positional information is set up very early during blastema formation, if a distal blastema is transplanted to a proximal stump then these cells regenerate only distal elements (Iten & Bryant, 1975; Stocum & Melton, 1977). These early grafting experiments gave important insights into regeneration and similar grafting techniques were later used to address from where the cells actually come to form the blastema. Elegant transplantation of triploid skin cells to diploid animals allowed the tracing and identification of cells population contributing to the limb blastema (Muneoka, Fox, & Bryant, 1986; Muneoka, Wise, Fox, & Bryant, 1984). These studies showed that 43% of the blastema appeared to be derived from derma skin cells, while skeletal accounted for only about 2% of the total cells in the blastema (Muneoka et al., 1986). These types of experiments gave researchers incredibly important insights into the complex regeneration process. However traditional transplantation studies give limited understanding into the cellular and molecular events that are occurring to drive the regenerative process. After reading many of the classical transplantation papers as a starting graduate student, the biggest question in my mind was where do cells come from and what do they give rise to during regeneration? I was fortunate to hear a fascinating talk from Ted Salmon at EMBL showing beautiful images of cell division in newt lung epithelial cells (Skibbens, Skeen, & Salmon, 1993; Waterman-Storer, Salmon, & Salmon, 2000; Waters, Skibbens, & Salmon, 1996) which inspired me to think about how we could possibly label cells in vivo in a live animals and image them during the course of regeneration. Thankfully Elly was completely onboard with this idea and in reading classical regeneration experiments carried out in a range of different salamander species it became clear to us that if I wanted to visualize cell behavior in vivo the white axolotl would be the most suited species for this. Beautiful TEM work from Elizabeth Hay suggested that terminally dedifferentiated muscle fibers in the salamander limb in fact dedifferentiated in response to limb amputation. From high quality serial images of the regenerating limb blastema she inferred that the muscle fibers pinched off of single cells, which then entered the blastema (Hay, 1959). The idea that terminally differentiated cells would dedifferentiate was a highly controversial idea. However given that muscle fibers are so easily visible and accessible in the axolotl tail this seemed like a logical place to start and an exciting question to revisit. And so began the adventures of getting the first white axolotls from the Stock Center in Bloomington Indiana safely to Germany and working out in vivo cell labeling and imaging.
2.2. The path to fluorescently labeling the first cells in vivo in the axolotl
The next challenge was how we would label cells in a differentiated animal, many of our colleagues were labeling cells in a developing embryo but differentiated cells remained a challenge. Elly contacted a former colleague at University College London from her days as a Postdoc there, Jon Clark. Jon had worked with labeling cells in a range of organisms including some of the very first in vivo labeling of neurons in axolotls with Nigel Holder (Clarke, 1999; Clarke, Alexander, & Holder, 1988; Clarke & Tickle, 1999; Holder, Clarke, Kamalati, & Lane, 1990; McGonnell, Clarke, & Tickle, 1998; Tawk, Bianco, & Clarke, 2009). Thankfully he didn’t think it was a crazy idea and was willing to host us for a week in his lab. Many late nights later under Jon’s patient guidance Elly and I became experts at injecting cells, perfecting the technique of gently peeling back the axolotl skin to give better access to the muscle cells and less anguish over blocked needles and broken tips. There was great excitement when we got our first muscle cells labeled and great disappointment hours later when the cells disappeared!! We discovered our conditions were too harsh and the cells gradually died, but at least we had a baseline to work from. We packed our bags and figured out the logistics of bringing axolotls back to Germany again, with some successes under our belt and a resolve from Jon to continue to help us. Early experiments with the first iterations of the lipophilic dye were ambiguous as the dye could be easily based taken up and passed to close by cells. Eventually we worked out conditions where with microinjection of rhodamine dextran we could successfully label muscle cells with no adverse effects, the labeled cells remained for days. This technique allowed us for the first time to visualize the reaction of muscle fibers to injury and live image them after amputation. This technique gave insights into muscle regeneration; however the dye was rapidly diluted and the cells could not be traced for the duration of tail regeneration (Echeverri, Clarke, & Tanaka, 2001).
2.3. Plasmid transfection: The next frontier
In Jon’s basement windowless lab at UCL he first introduced us to the labeling cells using iontophoresis (Fraser, 1996). This was a simple technique of using electrical charge to aid molecules to enter cells. This could be cheaply set up in a lab using a 6V battery and crocodile clips, it worked to label the muscle cells but they were very sensitive to the current and ultimately simple injection of dyes worked best. However we really wanted to be able to label cells for longer and image them over longer time-courses. In meantime many labs were moving beyond iontophoresis and developing electroporation as a more powerful technique for transfecting cells.
Electroporation is a technique that works by applying a short train of electrical pulses which form temporary pores in the cell membrane allowing charged molecules like DNA to enter the cell, while noncharged molecules may also enter by passive diffusion once the pores are formed (Neumann, Scaefer-Ridder, Wang, & Hofschneider, 1982; Neumann, Toensing, Kakorin, Budde, & Frey, 1998; Potter, 1988). Electroporation is used extensively by chick embryologists and more recently for in-ovo electroporation in mouse (Itasaki, Bel-Vialar, & Krumlauf, 1999; Swartz, Eberhart, Mastick, & Krull, 2001).
In many animals models bulk transfection of cells is achieved by micro-injecting plasmid DNA into the area of interest and the applying an external electrical pulse using tweezer electrodes (Harada, Omi, & Nakamura, 2017; McLennan & Kulesa, 2019; Saito & Nakaatsuji, 2001; Tawk, Tuil, Torrente, Vriz, & Paulin, 2002; Voiculescu & Stern, 2017). Advances on this standard technique have shown that a resolution of single cell electroporation can be achieved using a microelectrode technique that can achieve labeling of small groups or cells or even down to a single cell in the brain of Xenopus (Haas, Sin, Javaherian, Li, & Cline, 2001).
Luckily for us the people developing electroporation were very open and willing to help, so under guidance from Kurt Haas in Holly Cline’s lab at the time, I found myself immersed in the world of grass stimulators and electroporators, measuring voltages, tips sizes and amplitudes! What seemed like an easy straight forward technique in other systems especially the chick, proved to be more challenging in axolotls. Why axolotl cells were so much harder to transfect remains a bit of a mystery, we speculate it may be because of more mucus on skin or the cells are a surrounded by a different composition of ECM. The axolotl spinal cord is in fact a very small structure and the cells proven difficult to see, one major breakthrough for us was developing a tilted stage which allowed us to finally visualize the cells in the spinal cord and to insert an electrode that acted both as the microinjection pipette and the electrode. However through perseverance and endless testing of voltages, pulse durations, the first cells in the spinal cord were eventually successfully labeled and could be imaged over long periods of time without too much reduction in the signal due to the stability and perdurance of the GFP protein (Echeverri & Tanaka, 2003). The adaptation of this technique to the axolotl allowed cells within the spinal cord to be labeled using plasmid DNA and imaged for the first time in vivo during the time-course of regeneration could be accomplished (Echeverri & Tanaka, 2002, 2003). The cell cycle in axolotl cells is relatively long in comparison to many other species, ranging from 24 to 72h depending on the cell type and it is a state of development or regeneration, which means transfected cells are visible for much longer than in cells with rapid short cell cycle length (Rost et al., 2016).
Over time the electroporation technique itself has been further refined allowing higher efficacy of transfection to be achieved (Rodrigo Albors & Tanaka, 2015) and it has additionally shown to be an effective method for knocking down genes using morpholinos (Schnapp & Tanaka, 2005). Electroporation in axolotls has been a technique that has ultimately supported the elucidation of some key molecular insights into limb regeneration. We rapidly established that the same conditions used to label axolotl spinal cord cells could also be used to introduce plasmids and other molecules into a range of cells types, additionally other labs showed that the technique could be applied to other types of salamanders (Berg et al., 2010; Kumar, Godwin, Gates, Garza-Garcia, & Brockes, 2007).
The use of this technology in the context of limb regeneration allowed cells in different positions of the blastema to be labeled and confirmed previous transplantation data showing that the proximodistal axis is established very early in the blastema (Echeverri & Tanaka, 2005). Work from Mercader et al. elegantly illustrated how this technique could be used to overexpress proximally expressed Meis genes in the distal limb blastema leading to a proximalization of the cells and to introduce a morpholino to knock-down Meis expression, clearing illustrating that Meis genes are a target of retinoic acid (Mercader, Tanaka, & Torres, 2005). This technique has played a significant role in elucidating many of the key molecular pathways that are essential for initiating a regenerative response and patterning the regenerate. Recent work on the role of the immune system in limb regeneration has used electroporation to overexpress IL-8 in the blastema, resulting in increased recruitment of myeloid cells (Tsai, Baselga-Garriga, & Melton, 2019). The most recent example of the functionality of this system has shown how it can be used to label and examine the role of filopodia in blastemal cells, optogenic constructs could be electroporated into limb blastema cells and activated by light to change filopodia length (Zhang et al., 2021).
While the majority of uses to-date of electroporation has been to introduce plasmids to label cells or overexpress certain genes, it also has been used to knock-out genes. One caveat of studying genes in axolotl regeneration is that they can be necessary for regeneration, so in some cases knocking them out in development is not a viable option. In the early days of the development of the electroporation technology it was shown to be efficient for knocking down genes using morpholinos. Morpholinos are a very useful tool to reduce levels of proteins, however in some cases a knock-out of the gene can be more powerful. Electroporation has been shown to be an efficient technique for also introducing CRISPR guides complexed with the CAS9 protein into cells in the axolotl to specifically knock-out a gene, this has been shown to work well in the context of spinal cord regeneration (Fei et al., 2016, 2014).
The ability to label cells and knock-out or overexpress genes in the axolotl has paved the way for the axolotl to enter the stage as a genetically tractable model system. As interest waned in the axolotl due to lack of tools in comparison to systems like the zebrafish, the ability to label cells and modulate gene expression led to increased interest in the system again. However challenges remain, the axolotl can be tricky to image for long periods of time. The animal must be kept in an aqueous solution, the tissues are delicate but yet must be held down to prevent floating and going out of focus. Imaging cells closer to the lens like skin and muscle is relatively easy while cells located deep in the blastema are more challenging due to light scattering. Advances with microscopy techniques allowing gentler deeper and faster imaging over long time periods will be very important for taking in vivo imaging in the axolotl to the next level.
3. The trick to transgenics and its transformative influence on studying regenerative cells
One of the fundamental questions in the field of regenerative biology is where do the blastema cells come from? In the other words, does the blastema comprise a ubiquitous pluripotent stem cell population or tissue specific progenitors? In order to address this issue, an ideal approach is to genetically track the origin and destiny of the blastema cells. For this purpose, creation of transgenic axolotls was very critical. Transgenesis means introducing extra, generally exogenous piece of DNA into genome and creation of the stable germline transmission of integrated sequences. It allows for manipulating activities of functional genes or labeling of cells to study their roles and behaviors during tissue regeneration.
An endeavor of the Tanaka lab from the earliest days was to make transgenic axolotls to address these questions. Elly was convinced that we should adapt the REMI sperm injection method from frogs given the similar physiology of the animals but this was not very practical as axolotl sperm have giant genomes packaged into long, thin sperm that are prone to damage. Luckily, Werner Straube, a starting PhD student having learned zebrafish transgenesis through plasmid injection wanted to give it a try in axolotl and succeeded right away to produce CMV promoter-driven GFP-expressing F0 transgenics. However, as previously reported for β-gal plasmid-injected transgenic salamanders, the signal faded over time, which was interpreted as the plasmid remaining episomal rather than integrated. By then Werner, who had another project, had graciously handed over the reins to Lidia Sobkow. During her first thesis advisory committee, Francis Stewart told her that the CMV promoter is often silenced in mouse transgenics, and we should try the CAGGs promoter. Furthermore, he told us about yet unpublished work implementing the SceI meganuclease to promote integration and reduce tandem copies in medaka (Thermes et al., 2002). With this lucky advice, and many trials of finding good ways to dejelly axolotl eggs for injection, Lidia succeeded in making stable “hulk” transgenic axolotls that expressed GFP in virtually all tissues (albeit to iffering levels) and passed the transgene through the germline (Sobkow, Epperlein, Herklotz, Straube, & Tanaka, 2006). Interestingly as this work was taking place Ueda and colleagues demonstrated sperm injection-mediated transgenesis in the Japanese newt (Ueda, Kondoh, & Mizuno, 2005). Taking advantage of our axolotl CAGGS:eGFP transgenic axolotl and tissue transplantation technique (Kragl et al., 2009; Nacu, Knapp, Tanaka, & Epperlein, 2009), Kragl and colleagues have achieved the labeling of major limb tissues. Further tracing of their cell fate has revealed that each tissue tends to give rise to corresponding tissue relevant to their origin during regeneration (Kragl et al., 2009). This study has solved a long-lasting question in the field what is the potency of a limb blastema cell and reached a conclusion that the blastema is composed of lineage-restricted progenitors, instead of pluripotent stem cells.
Although combining constitutive transgenesis and tissue transplantation is very powerful using such an approach can never exclude the potential problem of grafting undesired cells during transplantation. Generation of transgenics using a tissue specific promoter could achieve genetic labeling of particular cell/tissue type. At that point Shahryar Khattak joined as a postdoc who was deeply determined to develop Cre-ERT2/loxP-mediated genetic fate mapping in axolotl, initially with the goal of fate mapping muscle vs satellite cell contribution to the limb blastema. Shahryar, with the assistance of Tobias Richter and then Maritta Schuez injecting millions of eggs, showed that well-characterized tissue-specific promoters from other animals such as mouse, drove specific expression in axolotl. He made many important tissue-specific GFP-expressing lines that are used by the field to this day (Khattak, Schuez, et al., 2013). He also systematically tried not only Cre, but also FRT and other recombinase systems (codon optimized) for axolotl, and concluded that Cre worked most efficiently and could induce recombination of a floxed GFP—Cherry loxP reporter transgenic that he created. Again, Shahryar’s success was characterized by thoroughness and care. He insisted on the transgenics always expressing a fluorescent reporter so that he could examine the animals by sections, cell by cell to confirm if the transgenic expressed the gene specifically in the correct location. This also allowed him to quantify the mosaicism of the F0 transgenic. In the search for a way to express gene × (such as Cre) together with a fluorescent protein, Shahryar, introduced the use of T2A and P2A peptides very early after their publication in our transgenics, which has been a crucial aspect of our strategy.
3.1. Inducible Cre-transgenics were not so easy…
Frustration came in trying to put together the muscle-specific and satellite-specific promoters with the Cre-ERT2 gene. After many trials, many promoters, many transgenics, we realized that somehow the ERT2 gene often induced silencing of the transgene. Although Shahryar explored many solutions such as insulator elements, it was difficult to overcome this problem with the available muscle promoters. Shahryar even developed a working virus-transduction system for this purpose (see Section 4) but the packaging limit made implementation difficult. Furthermore, the Pax7 locus and other satellite cell-specific gene loci were poorly characterized, making it impossible to make the two tissue specific Cre-ERT2 transgenics that we had envisioned at that time. Nonetheless, we were able to achieve our scientific goal, by using the CAGGS:Cre-ERT2 transgenic in a tricky way. We exploited the ability to graft blastemas from one animal to another and the fact that muscle fibers represent the product of fusion of muscle progenitors. To label limb muscle fibers, we grafted a blastema from the CAGGS:Cre-ERT2 onto the amputated stump of a CAGGs:loxP-GFP-Cherry transgenic (and vice versa) which showed muscle-specific expression, and allowed us to assess the contribution of muscle dedifferentiation to axolotl limb regeneration (Sandoval-Guzman et al., 2014).
3.2. Harvesting the power of transgenic technologies
Since the development of the first transgenic axolotl CAGGS: eGFP, this line was soon used to elucidate cell lineages of several major organs, including the teeth (Soukup, Epperlein, Horacek, & Cerny, 2008), hematopoiesis system (Lopez et al., 2014), bone tissues (McCusker, Diaz-Castillo, Sosnik, Phan, & Gardiner, 2016; Piekarski, Gross, & Hanken, 2014) during early development and regeneration in axolotls. Visualizing live cell activity has extra advantages over the static image from the fixed sections. Transplanting of lateral plate mesoderm tissue from double transgenic axolotls harboring a brainbow transgene and an inducible CAGGS: Cre-ERT2 alleles, it could be achieved the sparse, specific brainbow-labeling of the connective tissue (Currie et al., 2016; Gargioli & Slack, 2004). Connective tissue has been indicated as one of the major cell sources contributing to blastema formation (Currie et al., 2016; Dunis & Namenwirth, 1977; Kragl et al., 2009) and may carry positional information during limb regeneration (Nacu et al., 2013). Through live-imaging of the brainbow (multiple-color fluorescence) labeled connective tissue cells, Currie and colleagues has identified this cell population gives rise to only the connective tissue lineage during digit tip blastema formation and regeneration (Currie et al., 2016). It further strengthened the concept that tissue-specific progenitor cells are the major sources during axolotl tissue regeneration.
In the meanwhile, many other transgenic axolotls have been reported in the past years on solving other issues in regeneration of axolotls. Maden lab has generated a retinoic (RA) acid reporter using a characterized RA-response enhancer, which allow the monitoring of the endogenous RA activity during regeneration (Monaghan & Maden, 2012). It can be expanded to visualize the activity of other signaling molecules when particular enhancers are applied. Furthermore, although Cre-LoxP based conditional gene expression system is well established in axolotls, however, one of the drawbacks, particularly to overexpress functional genes, is that turning on the gene expression is irreversible. To overcome this obstacle, Whited and colleagues has established a more flexible E. coli Lac operon based inducible gene expression system in axolotls, which allows turning on and off of the target gene expression under the control of IPTG (Whited, Lehoczky, & Tabin, 2012).
In order to address the roles and mechanisms of how connective tissue participates in regeneration, Prayag Murawala was able to generate an inducible Prrx1:Cre-ERT2, and a Col1A2:Cre-ERT2 transgenic axolotl, in which a well characterized mouse Prrx1 and Col1A2 enhancer was employed to achieve the specific expression of gene of interest (GOI) in connective tissue (Gerber et al., 2018). When crossed with a CAGGS: floxed eGFP-Cherry reporter line followed by a pulse treatment of Tamoxifen, the Cherry-labeled PRRX1-postive connective tissue cells gives rise to only the connective tissue lineage during limb regeneration, which provides further evidence of the lineage specific origin of blastema (Gerber et al., 2018). Interestingly, analysis of the gene expression profile of these fluorescent-labeled connective tissue cells using single cell RNA sequencing technique has revealed that these cells are reprogrammed back to an embryonic like status during regeneration (Gerber et al., 2018). This study has discovered a new mechanism—the programming feature of progenitor cells prior to/during blastema formation and regeneration, that could potentially present a general mechanism of tissue regeneration.
Although applying transgenic axolotls have shown its broad application potential for answering diverse questions during axolotl tissue regeneration, and many exciting and encouraging progress have been achieved, this technique still faces many challenges. Axolotl transgenesis is established on the basis of I-Sce1 or mTol2 transposes system (Khattak et al., 2014; Khattak, Schuez, et al., 2013; Sobkow et al., 2006). In either case, the integration of the transgene-cassette into axolotl genome is random. This random integration may lead to the uneven expression of transgene in the targeted tissue, or even silencing of the transgene. Therefore, it is very necessary to characterize a safe-harbor, as the Rosa26 locus in mice (Soriano, 1999), to hold proper transgene expression in axolotls. Nonetheless, it is clear and can be easily predicted that more transgenics will be produced to facilitate the understanding of the molecular and cellular mechanisms of tissue regeneration in axolotls.
4. Viral transduction systems to study limb regeneration
Compared to other aquatic vertebrates, the axolotl boasts multiple viral transduction systems—perhaps related to the fact that axolotl is primarily used to study the postembryonic phenomenon of regeneration. Stephan Roy during his postdoc in the Bryant laboratory described the ability of the vaccinia virus to overexpress sonic hedgehog powerfully enough to generate numerous ectopic digits, providing a standard assay in the field for assessing effectiveness of viral expression in the blastema (Roy et al., 2000). He pointed out however that as vaccinia kills the infected cells, the method is appropriate mainly for testing extracellular factors.
Moving into the 21 century, two integrating retroviruses were developed for use in regeneration. Jessica Whited during her postdoc in the Tabin lab demonstrated the effectiveness of pseudotyped MMLV retrovirus for stably expressing genes in the blastema and cultured axolotl cells (Whited et al., 2013). VSV-G pseudotyped viruses were used to infect dividing blastema cells and assess the proportion of different tissue-specific progenitors in the blastema at different times of regeneration. As an integrating virus at low copy number, expression of SHH in the blastema yielded modest changes in metacarpal morphogenesis. Moving further she cleverly used transgenics to express the TVA ecotropic receptor on endothelial cells driven by the PECAM promoter, so that they could be infected with an ASLV-A glycoprotein pseudotyped MMLV virus. In parallel, Shahryar Khattak demonstrated that the axolotl blastema and cultured cells are efficiently transduced by the human foamy virus retrovirus compared to lentivirus which showed loss of expression in the limb (Khattak, Sandoval-Guzman, et al., 2013). Muscle-specific expression could be achieved by driving GFP expression with a muscle-specific actin promoter. Both viral systems which infect dividing cells have the advantage of integrated expression, but the disadvantage of strict viral genome size limits for efficient packaging, which are required for in vivo infection.
In the search for a viral system with larger packaging limits and stronger expression for driving gene expression to obtain strong phenotypes, I decided to try pseudotyped baculovirus, which, while not integrating, could accommodate inserts up to 35kb, and as a multicopy episomal system should drive strong expression, which, conveniently makes the system S1 instead of S2 genetic safety category in many situations. Knowing from the foamy and MMLV virus work that very high titers are required for in vivo transduction, my first attempts involved concentrating commercial BacMam preparations on a centricon and injecting blastemas which immediately showed strong GFP expression that persisted through regeneration and showed expression in cartilage. Based on this observation, Catarina Oliveira started her PhD thesis on developing baculovirus for axolotl regeneration studies. With this as her primary goal, she exhaustively tested viral production methods, pseudotyping methods and virus concentration eventually converging to a protocol where she pseudotyped baculovirus with a VSV-GED mutant which showed less toxicity, and a concentration through sucrose gradient to purify virus (Oliveira et al., 2018). This work allowed her not only to demonstrate efficient transduction in vivo and in vitro, a tropism of the virus to connective tissue and satellite cells, as well as strong digit duplication after SHH expression in the blastema, it allowed her to make important contributions to two functional studies in limb regeneration (Nacu, Gromberg, Oliveira, Drechsel, & Tanaka, 2016; Wagner et al., 2017). Firstly, she used her viruses encoding SHH and FGF8 to collaborate with Eugen Nacu and Elena Gromberg to demonstrate that the dependency of limb regeneration on anterior and posterior tissue represents the implementation of a SHH-FGF8 positive feedback loop by inducing limbs from anterior and posterior regions of the limb by expressing these factors respectively (Nacu et al., 2016). Secondly, she used the baculovirus to show the role of BMP-signaling in myogenic cell cycle-reentry during newt limb regeneration by expression of BMP receptor mutants (Wagner et al., 2017).
5. Overcoming a major obstacle—Assembly of the giant axolotl genome
Despite the pace of scientific and technological progress studying regeneration and developing the axolotl model system researchers in the growing axolotl field still faced a problem to be generally being perceived to be working on a viable model—and that was the genome and the ability to manipulate the genome. The axolotl has a similar gene complement to other vertebrates, and therefore, as next generation technologies including 454 sequencing and Illumina sequencing became available, the community did manage to implement reasonable transcriptome. However the unassembled genome always loomed, and this was a daunting task considering the 32gB genome. Sequencing of selected clones in bacterial artificial chromosomes had shown the presence of large intergenic and intronic sequences full of repeats, which meant that short read technologies would not suffice to obtain a high quality assembly (Smith et al., 2009). Indeed exploration of short read genome sequencing by the Voss lab even coupled to purification of specific chromosomes to ameliorate the genome size problem demonstrated the difficulty of assembling the genome (Keinath et al., 2015).
Luckily Eugene Myers who had ended up as a Director at the Max Planck Institute in Dresden decided to put his mathematical skills to the problem of genome assembly long, but error prone reads emerging from Pacific Biosciences technology. Eugene applied to the Max Planck Society to obtain the machines and consumables to sequence this giant genome along with the planaria and was fortunately successful. Working with a newly founded junior group headed by Siegfried Schloissnig, they worked on new assembly algorithms that got around unusual patterns of sequencing artifacts generated by the technology, without severing the long reads as other programs tended to do. One cannot underestimate the luck we had being a direct colleague of a leading mathematician in the genome assembly field. In 2016 we received the call that a first preliminary assembly of the axolotl data was finished. We were so astonished with the result that we decided to annotate the assembly. Then the headaches began. As the axolotl genome is 10 times the size of the human genome, none of the software used to analyze genomes was written in enough bit depth to process the data. Furthermore, as the introns were so long, ab initio gene prediction using standard software did not function. We had to find many work arounds such as splitting the genome into subparts to run software such as repeat annotation, and we had to settle with annotating genes based on mapping RNASeq data to the genome. But after many late night and weekend exchanges between Sergej Nowoshilow, Michael Hiller and I, we could annotate the genome including the genes, transposable elements (with George King). This work describing the genome assembly combined with Ji-Feng Fei’s TALEN and CRISPR generated extensive mutant analysis of the axolotl Pax7 locus showing that it substituted for a missing Pax3 generated tremendous excitement as provided the two missing elements for axolotl as a molecular genetic model organism to enable the community to work on the genetic basis of axolotl traits (Nowoshilow et al., 2018).
We were in luck again when we wanted to take the initial assembly and scaffold it to chromosome level using HiC contact data. HiC scaffolding is based on the principle when the genome is cross-linked, if one looks at any one place in the genome, the frequency of contacts to that place falls off as an exponential with distance. This is observed in HiC heat maps at the strong signal at the diagonal with falling off density from the diagonal (Burton et al., 2013; Kaplan & Dekker, 2013; Lieberman-Aiden et al., 2009). Using this method, the many contigs of a typical genome assembly can be ordered and oriented up to a contiguous chromosome scale assembly. This scaffolding effort was greatly assisted by the chromosomal scale meiotic map scaffold that had been in the mean-time produced by Jeramiah Smith and Randal Voss’s groups (Smith et al., 2019). Unfortunately the meiotic data could not orient the many of the DNA pieces along the assembly, and the resolution of the method could not allow inclusion of all contigs in the scaffolding. Akane Kawaguchi, a postdoc interested in epigenetics of regeneration, as a first step in her own aims, realized she needed this chromosome scale assembly. She adapted the protocols to perform HiC preps on axolotl cells, importantly including figuring how to synchronize AL1 cells in mitosis, since the mitotic HiC data increased the number of informative contacts for assembly tremendously. These developments took well over 1 year. In the meantime Randal Voss and Jeramiah Smith also generated HiC data. During this time, talking to HiC experts including Erez Lieberman-Aiden, Leonid Mirney, Job Dekker and Stephan Mundlos as they passed by as seminar speakers, as well as talking to local experts, Gordana Wutz, Roman Stocits and Anton Golodorodko gave us many important tips on how to go on and also how to analyze our data. At the beginning of our HiC journey, the concepts and data seemed like learning a foreign language but we persevered and became fully ensconced in this endeavor. We certainly benefited from having moved from Dresden to the Vienna Biocenter which is a hot bed of research groups examining large scale chromosome structure. In addition, we luckily could recruit computer scientist Siegfried Schloissnig to Vienna, who put his mind to the HiC scaffolding problem. Again, the available softwares could not be used because they were not written in a way to be fast enough to scaffold this giant genome, and the calculation of contact frequencies is highly disturbed in axolotl due to the presence of the many repetitive regions. Therefore expert computer programming to accelerate the processing so that many different ways of trying to calculate the contact frequencies to gain accurate scaffolding could be tried and in the end a satisfying chromosomal scale assembly (Schloissnig et al., 2021). But finally we are proud that the community can celebrate the existence of a high quality, chromosome level genome assembly that is enviable in quality for any model organism.
Transcriptome and then genome data can be overwhelming in volume so databases to easily search and access the data are essential. As we accumulated the data, we were lucky that Sergej Nowoshilow, with much input from the biologists such as Prayag Murawala, constructed a highly convenient portal called https://www.axolotl-omics.org that allows all researchers to query the genome with repeat annotation and associated gene models both in the form of BLAST searching, through a UCSC implemented genome browser, and the HiGlass browser for HiC data (Nowoshilow & Tanaka, 2020). This portal nicely complements the salamander, Sal-site (https://ambystoma.uky.edu/quick-links/sal-site) constructed by Randal Voss and colleagues (Baddar, Woodcock, Khatri, Kump, & Voss, 2015). Reflecting that axolotl is fully in the genomic age, researchers recently formalized gene nomenclature in this organism (Nowoshilow, Fei, Voss, Tanaka, & Murawala, 2021).
6. The remarkable, enabling era of genome editing
Withthe availability of gene sequence information and the tremendous progress in genome editing, making axolotl mutants, and using the genome sequence to precisely knock-in gene sequences into the genome became a reality. Editing of defined genomic locus is an important approach for creating genetic modified animals. In the past, unlike the broadly used animal models e.g. mice, it is impossible to make genetic knockout or knock-in axolotls due to the lack of proper genome editing technology. Moreover, RNA interference also does not work in axolotls. Morpholino is the only technology could be applied to axolotls to inhibit the expression of the targeted genes (Schnapp & Tanaka, 2005). Therefore, establishing an efficient genetic loss of function analysis in axolotls is in great demand in order to deeply investigate the roles of given genes during tissue regeneration. Luckily, the rapid development of genome engineering techniques provides such an opportunity to axolotls. Soon after the demonstrations of precision genome modification of the Zinc-Finger Nucleases (ZFNs) on desired genomic loci in mammalian cells and animal models (Doyon et al., 2008; Santiago et al., 2008), we started the attempt of establishing ZFN technique in axolotls, following a ZFN screening protocol published from Joung’s lab (Wright et al., 2006). We teamed up with Andrew Oates lab who is using zebrafish as a model to study the rhythmic control of embryonic body segment formation. After running over half a year between two labs on the fourth floor of MPI-CBG, we achieved the first precise modification at axolotl Oct4 genomic locus using obtained ZFN pair. However, one of the major problems we were facing at that time is the early embryonic lethality of the ZFN injected embryos, that is likely due to the unspecific toxicity of ZFN and made it very difficult to generate global gene knockout axolotls. While we were still optimizing the ZFN injection condition, a new alternative technique Transcription Activator-Like Effector Nucleases (TALENs) was developed and rapidly applied in varied species for generation of genetic modified organisms, such as mice and zebrafish (Bedell et al., 2012; Sander et al., 2011; Sung et al., 2013). After carrying out the first pilot experiment by introducing TALENs against either the endogenous Tyrosinase gene or the eGFP transgene into freshly laid eggs, we observed typical loss of function phenotypes corresponding to the relevant genes, that was truly a very exciting moment. We found that TALEN is generally more efficient and less toxic than ZFN. Importantly, the modified genomic alleles in F0 TALEN founders can be transmitted to F1 progeny when crossed with DD axolotls.
However, the progress of technology never stops. In the year of 2013, two laboratories reported successful genome engineering in eukaryotic cells using a new system Clustered Regulatory Interspaced Short Palindromic Repeat (CRISPR)/Cas9 (Cong et al., 2013; Mali et al., 2013), right after we have established TALEN approach in the lab. It took us a while to decide whether we should compare TALEN and CRISPR/Cas9 system. Amazingly, the first attempt applying CRISPR/Cas9 in axolotls to knockout Tyrosinase gene, surprisingly yield an extremely high efficiency (Fei et al., 2014; Flowers, Timberlake, Mclean, Monaghan, & Crews, 2014). Nearly all injected F0 axolotls showed massive, even complete loss of pigment, and developed properly, which were never observed from ZFN and TALEN injections. Considering it is relatively easy to synthesize gRNA, CRISPR/Cas9 system is definitely a better choice, in most of the cases, for the generation of gene knockout axolotls. Having demonstrated effective disruption of an easily scorable pigment phenotype, we moved on to mutate a gene, Sox2, involved in spinal cord stem cell function for which we had raised and antibody so we could monitor loss of expression. While Sox2 is embryonic lethal in mouse, to our great surprise, F0 axolotl Sox2 crispants with penetrant modification survived to larval stages. Careful examination of the spinal cord showed loss of SOX2 expression and lack of spinal cord regeneration. Further analysis showed that while in embryos, Sox2 and its paralog Sox3 are coexpressed in the embryo, during spinal cord regeneration Sox3 is not present and the process seems to be solely dependent on Sox2. Our observations disclosed the critical role of Sox2 gene on neural stem cell rapid amplification during spinal cord regeneration (Fei et al., 2014). The availability of TALEN and CRISPR/Cas9-mediated gene knockout provides an opportunity for deep dissection of the gene function during development and regeneration in axolotls.
6.1. Tissue-specific expression via knock-in methods
One of the major challenges is how to achieve tissue specific gene expression in transgenics. When using classical transgenic method, it generally requires a well characterized tissue specific promoter to drive the expression of GOI. However, such well-defined promoter/enhancer is normally not available. In early days, while Khattak and colleagues were trying to generate inducible Sox2:Cre-ERT2 and Pax7:Cre-ERT2 transgenic axolotls for cell fate mapping of SOX2+ neural stem cells and PAX7+ satellite cells during regeneration, it is failed to obtain any transgenics with proper GOI expression using available promoters of these two genes (Khattak, Schuez, et al., 2013). It may be due to the fact that the expression of Sox and Pax7 family genes are well known controlled by complex regulatory sequences. And it is very hard to isolate the complete promoter/enhancer sequence of such genes for transgenesis. Taking the advantages of CRISPR/Cas9 technology that could introduce a double strand DNA break (DSB) at the desired axolotl genomic loci, at the presence of exogenous DNA, this donor DNA can integrate into the DSB site (Albadri, Del Bene, & Revenu, 2017). Under this principle, either the Cherry reporter gene or inducible Cre-ERT2 cassettes were inserted into axolotl Sox2 or Pax7 genomic loci (Fei et al., 2017). In all of the knock-in lines generated, GOI expression can be faithfully detected in the expected tissues/cells. Satellite cell fate mapping using inducible Pax7: Cre-ERT2 line has shown that these cells are the major source contributing to axolotl limb regeneration (Fei et al., 2018, 2017). Extended application of the CRISPR/Cas9 mediated knock-in was also used in a recently work from Whited lab, in which they inserted the Cre recombinase into Eyes absence 2 locus to trace the limb blastema cell during development (Sousounis et al., 2020). Overall, although the CRISPR/Cas9 mediated knock-in in axolotl in a newly developed approach, it has been demonstrated to be a valuable method in studying the mechanism of tissue regeneration in axolotls.
So far, the majority achievements of using CRISPR/Cas9 system has been to generate global gene knockouts (Fei et al., 2018, 2014; Flowers, Sanor, & Crews, 2017; Flowers et al., 2014; Sanor, Flowers, & Crews, 2020). It is still challenging to establish tissue specific gene knockout in axolotls but has been achieved using delivery of CAS9-guide RNA complexes into targeted tissue by electroporation. In transgenesis, generation of a loss of function floxed allele, then combining with a tissue specific Cre line is the most conventional way to achieve tissue specific knockout of targeted gene. Until now, no floxed allele on endogenous gene has been reported in axolotls. Technically, it is not easy to insert two loxP sites separately into axolotl genome to embrace a coding exon and how to do this still needs to be explored. An alternative approach is to introduce CAS9-guide RNA complexes into targeted tissue by electroporation or viral transduction to knockout given genes. It has been demonstrated that delivery of CAS9-guide RNA complexes targeting Sox2 by electroporation led to efficient gene knock out in spinal cord ependymal cells, phenocopying the Sox2 mutant phenotype (Fei et al., 2016; Lou, Wang, Long, Liu, & Fei, 2019). So far this approach has been less efficient in the blastema, and we are actively working on alternative methods. Overall, more efforts are still needed to further optimize the CRISPR/Cas9 mediated genomic modification for diverse purposes in axolotls.
7. Parallel developments in other salamanders
The axolotl has been and continues to be a highly popular vertebrate due to the ease of breeding and rearing the animal in laboratory conditions and now with powerful molecular genetic tools. But it is important to keep in mind that regeneration has traditionally been studied in several salamander species. In parallel to developments in the axolotl, transient transfection in the eye, as well as sperm injection-mediated transgenesis were developed in the Japanese newt Pyrrhogaster (Casco-Robles, Yamada, Miura, & Chiba, 2010; Hayashi et al., 2001). However, the long generation time of this animal has made genetic analysis more difficult. Recently, Pleurodeles waltl has reemerged as a newt species for molecular regeneration studies, with gene sequence information, robust TALEN and CRISPR-mediated gene mutation (Elewa et al., 2017; Hayashi & Takeuchi, 2015; Matsunami et al., 2019). The availability of these two animal models will allow further investigation of common and divergent aspects of development and regeneration in this family.
8. Concluding remarks
Regeneration in the axolotl remains a fascinating problem to this day with the advent of understanding molecular mechanism still at its infancy. The enabling developments that we and others undertook were a labor of love for the model and the scientific problem which ameliorated worries about publications which always came in the end. We hope that this account helps others to take on this gratifying mission for their respective models and biologies.
Acknowledgments
We would like to thank all the amazing colleagues from the lab and the community who worked together on and supported over the years this wonderful model organism. K.E. is supported by NIH NICHD (R01HD092451) and the Owens Family Foundation. Work in the JFF laboratory is supported by grants from National Key R&D Program of China 2019YFE0106700, the Natural Science Foundation of China 31970782, Key-Area Research and Development Program of Guangdong Province 2018B030332001, 2019B030335001.
E.T. is an ERC Advanced Investigator (RegGeneMems 742046).
References
- Albadri S, Del Bene F, & Revenu C (2017). Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods, 121–122, 77–85. [DOI] [PubMed] [Google Scholar]
- Baddar NW, Woodcock MR, Khatri S, Kump DK, & Voss SR (2015). Sal-Site: Research resources for the Mexican axolotl. Methods in Molecular Biology, 1290, 321–336. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG 2nd, et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature, 491, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DA, Kirkham M, Beljajeva A, Knapp D, Habermann B, Ryge J, et al. (2010). Efficient regeneration by activation of neurogenesis in homeostatically quiescent regions of the adult vertebrate brain. Development, 137, 4127–4134. [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, & Shendure J (2013). Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature Biotechnology, 31, 1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casco-Robles MM, Yamada S, Miura T, & Chiba C (2010). Simple and efficient transgenesis with I-SceI meganuclease in the newt, Cynops pyrrhogaster. Developmental Dynamics, 239, 3275–3284. [DOI] [PubMed] [Google Scholar]
- Clarke JD (1999). Using fluorescent dyes for fate mapping, lineage analysis, and axon tracing in the chick embryo. Methods in Molecular Biology, 97, 319–328. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Alexander R, & Holder N (1988). Regeneration of descending axons in the spinal cord of the axolotl. Neuroscience Letters, 89, 1–6. [DOI] [PubMed] [Google Scholar]
- Clarke JD, & Tickle C (1999). Fate maps old and new. Nature Cell Biology, 1, E103–E109. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339(6121), 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, & Tanaka EM (2016). Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Developmental Cell, 39, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. (2008). Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnology, 26, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunis DA, & Namenwirth M (1977). The role of grafted skin in the regeneration of x-irradiated axolotl limbs. Developmental Biology, 56, 97–109. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Clarke JD, & Tanaka EM (2001). In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Developmental Biology, 236, 151–164. [DOI] [PubMed] [Google Scholar]
- Echeverri K, & Tanaka EM (2002). Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science, 298, 1993–1996. [DOI] [PubMed] [Google Scholar]
- Echeverri K, & Tanaka EM (2003). Electroporation as a tool to study in vivo spinal cord regeneration. Developmental Dynamics, 226, 418–425. [DOI] [PubMed] [Google Scholar]
- Echeverri K, & Tanaka EM (2005). Proximodistal patterning during limb regeneration. Developmental Biology, 279, 391–401. [DOI] [PubMed] [Google Scholar]
- Elewa A, Wang H, Talavera-Lopez C, Joven A, Brito G, Kumar A, et al. (2017). Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nature Communications, 8, 2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei JF, Knapp D, Schuez M, Murawala P, Zou Y, Singh SP, et al. (2016). Tissue- and time-directed electroporation of CAS9 protein–gRNA complexes in vivo yields efficient multigene knockout for studying gene function in regeneration. NPJ Regenerative Medicine, 1, 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei JF, Lou WP, Knapp D, Murawala P, Gerber T, Taniguchi Y, et al. (2018). Application and optimization of CRISPR-Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum). Nature Protocols, 13, 2908–2943. [DOI] [PubMed] [Google Scholar]
- Fei JF, Schuez M, Knapp D, Taniguchi Y, Drechsel DN, & Tanaka EM (2017). Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. Proceedings of the National Academy of Sciences of the United States of America, 114, 12501–12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei JF, Schuez M, Tazaki A, Taniguchi Y, Roensch K, & Tanaka EM (2014). CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Reports, 3, 444–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers GP, Sanor LD, & Crews CM (2017). Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. eLife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers GP, Timberlake AT, Mclean KC, Monaghan JR, & Crews CM (2014). Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development, 141, 2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser SE (1996). Iontophoretic dye labeling of embryonic cells. Methods in Cell Biology, 51, 147–160. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Blumberg B, Komine Y, & Bryant SV (1995). Regulation of HoxA expression in developing and regenerating axolotl limbs. Development, 121, 1731–1741. [DOI] [PubMed] [Google Scholar]
- Gargioli C, & Slack JM (2004). Cell lineage tracing during Xenopus tail regeneration. Development, 131, 2669–2679. [DOI] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, et al. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohme MA, Schloissnig S, Rozanski A, Pippel M, Young GR, Winkler S, et al. (2018). The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature, 554, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K, Sin WC, Javaherian A, Li Z, & Cline HT (2001). Single-cell electroporation for gene transfer in vivo. Neuron, 29, 583–591. [DOI] [PubMed] [Google Scholar]
- Habermann B, Bebin AG, Herklotz S, Volkmer M, Eckelt K, Pehlke K, et al. (2004). An Ambystoma mexicanum EST sequencing project: Analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biology, 5, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Omi M, & Nakamura H (2017). In ovo electroporation methods in chick embryos. Methods in Molecular Biology, 1650, 167–176. [DOI] [PubMed] [Google Scholar]
- Hay ED (1959). Electron microscopic observations of muscle dedifferentiation in regenerating Amblystoma limbs. Developmental Biology, 1, 555–585. [Google Scholar]
- Hayashi T, & Takeuchi T (2015). Gene manipulation for regenerative studies using the Iberian ribbed newt, Pleurodeles waltl. Methods in Molecular Biology, 1290, 297–305. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Yamagishi A, Kuroiwa A, Mizuno N, Kondoh H, & Okamoto M (2001). Highly efficient transfection system for functional gene analysis in adult amphibian lens regeneration. Development, Growth & Differentiation, 43, 361–370. [DOI] [PubMed] [Google Scholar]
- Holder N, Clarke JD, Kamalati T, & Lane EB (1990). Heterogeneity in spinal radial glia demonstrated by intermediate filament expression and HRP labelling. Journal of Neurocytology, 19, 915–928. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, & Krumlauf R (1999). Shocking developments in chick embryology: Electroporation and in ovo gene expression. Nature Cell Biology, 1, E203–E207. [DOI] [PubMed] [Google Scholar]
- Iten LE, & Bryant SV (1975). The interaction between the blastema and stump in the establishment of the anterior–posterior and proximal–distal organization of the limb regenerate. Developmental Biology, 44, 119–147. [DOI] [PubMed] [Google Scholar]
- Kaplan N, & Dekker J (2013). High-throughput genome scaffolding from in vivo DNA interaction frequency. Nature Biotechnology, 31, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Marti M, Dubova I, et al. (2006). Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes & Development, 20, 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath MC, Timoshevskiy VA, Timoshevskaya NY, Tsonis PA, Voss SR, & Smith JJ (2015). Initial characterization of the large genome of the salamander Ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Scientific Reports, 5, 16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak S, Murawala P, Andreas H, Kappert V, Schuez M, Sandoval-Guzman T, et al. (2014). Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nature Protocols, 9, 529–540. [DOI] [PubMed] [Google Scholar]
- Khattak S, Sandoval-Guzman T, Stanke N, Protze S, Tanaka EM, & Lindemann D (2013). Foamy virus for efficient gene transfer in regeneration studies. BMC Developmental Biology, 13, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak S, Schuez M, Richter T, Knapp D, Haigo SL, Sandoval-Guzman T, et al. (2013). Germline transgenic methods for tracking cells and testing gene function during regeneration in the axolotl. Stem Cell Reports, 1, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, et al. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature, 460, 60–65. [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, & Brockes JP (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science, 318, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Velloso CP, Imokawa Y, & Brockes JP (2000). Plasticity of retrovirus-labelled myotubes in the newt limb regeneration blastema. Developmental Biology, 218, 125–136. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Lin L, Monaghan JR, Cogle CR, Bova FJ, Maden M, et al. (2014). Mapping hematopoiesis in a fully regenerative vertebrate: The axolotl. Blood, 124, 1232–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou WP, Wang L, Long C, Liu L, & Fei JF (2019). Direct gene knock-out of axolotl spinal cord neural stem cells via electroporation of CAS9 protein-gRNA complexes. Journal of Visualized Experiments, (149). 10.3791/59850. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. (2013). RNA-guided human genome engineering via Cas9. Science, 339(6121), 823–826. 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami M, Suzuki M, Haramoto Y, Fukui A, Inoue T, Yamaguchi K, et al. (2019). A comprehensive reference transcriptome resource for the Iberian ribbed newt Pleurodeles waltl, an emerging model for developmental and regeneration biology. DNA Research, 26, 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD, Diaz-Castillo C, Sosnik J, Phan AQ, & Gardiner DM (2016). Cartilage and bone cells do not participate in skeletal regeneration in Ambystoma mexicanum limbs. Developmental Biology, 416, 26–33. [DOI] [PubMed] [Google Scholar]
- McGonnell IM, Clarke JD, & Tickle C (1998). Fate map of the developing chick face: Analysis of expansion of facial primordia and establishment of the primary palate. Developmental Dynamics, 212, 102–118. [DOI] [PubMed] [Google Scholar]
- McLennan R, & Kulesa PM (2019). In ovo electroporation of plasmid DNA and morpholinos into specific tissues during early embryogenesis. Methods in Molecular Biology, 1976, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, & Torres M (2005). Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development, 132, 4131–4142. [DOI] [PubMed] [Google Scholar]
- Monaghan JR, & Maden M (2012). Visualization of retinoic acid signaling in transgenic axolotls during limb development and regeneration. Developmental Biology, 368, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, & Bryant SV (1986). Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Developmental Biology, 116, 256–260. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Wise LD, Fox WF, & Bryant SV (1984). Improved techniques for use of the triploid cell marker in the axolotl, Ambystoma mexicanum. Developmental Biology, 105, 240–245. [DOI] [PubMed] [Google Scholar]
- Nacu E, Glausch M, Le HQ, Damanik FF, Schuez M, Knapp D, et al. (2013). Connective tissue cells, but not muscle cells, are involved in establishing the proximo-distal outcome of limb regeneration in the axolotl. Development, 140, 513–518. [DOI] [PubMed] [Google Scholar]
- Nacu E, Gromberg E, Oliveira CR, Drechsel D, & Tanaka EM (2016). FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature, 533, 407–410. [DOI] [PubMed] [Google Scholar]
- Nacu E, Knapp D, Tanaka EM, & Epperlein HH (2009). Axolotl (Ambystoma mexicanum) embryonic transplantation methods. Cold Spring Harbor Protocols, 2009. pdb.prot5265. [DOI] [PubMed] [Google Scholar]
- Neumann E, Scaefer-Ridder M, Wang Y, & Hofschneider PH (1982). Gene transfer into mouse lyoma cells by electroporation in high electric fields. The EMBO Journal, 1, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann E, Toensing K, Kakorin S, Budde P, & Frey J (1998). Mechanism of electroporative dye uptake by mouse B cells. Biophysical Journal, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowoshilow S, Fei JF, Voss RS, Tanaka EM, & Murawala P (2021). Gene and transgenics nomenclature for the laboratory axolotl—Ambystoma mexicanum. Developmental Dynamics. 10.1002/dvdy.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, et al. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature, 554, 50–55. [DOI] [PubMed] [Google Scholar]
- Nowoshilow S, & Tanaka EM (2020). Introducing www.axolotl-omics.org—An integrated -omics data portal for the axolotl research community. Experimental Cell Research, 394, 112143. [DOI] [PubMed] [Google Scholar]
- Oliveira CR, Lemaitre R, Murawala P, Tazaki A, Drechsel DN, & Tanaka EM (2018). Pseudotyped baculovirus is an effective gene expression tool for studying molecular function during axolotl limb regeneration. Developmental Biology, 433, 262–275. [DOI] [PubMed] [Google Scholar]
- Pecorino LT, Lo DC, & Brockes JP (1994). Isoform-specific induction of a retinoid-responsive antigen after biolistic transfection of chimaeric retinoic acid/thyroid hormone receptors into a regenerating limb. Development, 120, 325–333. [DOI] [PubMed] [Google Scholar]
- Piekarski N, Gross JB, & Hanken J (2014). Evolutionary innovation and conservation in the embryonic derivation of the vertebrate skull. Nature Communications, 5, 5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H (1988). Electroporation in biology: Methods, applications, and instrumentation. Analytical Biochemistry, 174, 361–373. [DOI] [PubMed] [Google Scholar]
- Putta S, Smith JJ, Walker JA, Rondet M, Weisrock DW, Monaghan J, et al. (2004). From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics, 5, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo Albors A, & Tanaka EM (2015). High-efficiency electroporation of the spinal cord in larval axolotl. Methods in Molecular Biology, 1290, 115–125. [DOI] [PubMed] [Google Scholar]
- Rost F, Rodrigo Albors A, Mazurov V, Brusch L, Deutsch A, Tanaka EM, et al. (2016). Accelerated cell divisions drive the outgrowth of the regenerating spinal cord in axolotls. eLife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Gardiner DM, & Bryant SV (2000). Vaccinia as a tool for functional analysis in regenerating limbs: Ectopic expression of Shh. Developmental Biology, 218, 199–205. [DOI] [PubMed] [Google Scholar]
- Saito T, & Nakaatsuji N (2001). Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Developmental Biology, 240, 237–246. [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, et al. (2011). Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnology, 29, 697–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Guzman T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, et al. (2014). Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell, 14, 174–187. [DOI] [PubMed] [Google Scholar]
- Sanor LD, Flowers GP, & Crews CM (2020). Multiplex CRISPR/Cas screen in regenerating haploid limbs of chimeric axolotls. eLife, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. (2008). Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America, 105, 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Kawaguchi A, Nowoshilow S, Falcon F, Otsuki L, Tardivo P, et al. (2021). The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proceedings of the National Academy of Sciences of the United States of America, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp E, & Tanaka EM (2005). Quantitative evaluation of morpholino-mediated protein knockdown of GFP, MSX1, and PAX7 during tail regeneration in Ambystoma mexicanum. Developmental Dynamics, 232, 162–170. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, & Salmon ED (1993). Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: A push-pull mechanism. The Journal of Cell Biology, 122, 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Zhu W, Pao GM, Verma IM, Hunter T, et al. (2009). Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics, 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Timoshevskaya N, Timoshevskiy VA, Keinath MC, Hardy D, & Voss SR (2019). A chromosome-scale assembly of the axolotl genome. Genome Research, 29, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobkow L, Epperlein HH, Herklotz S, Straube WL, & Tanaka EM (2006). A germline GFP transgenic axolotl and its use to track cell fate: Dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Developmental Biology, 290, 386–397. [DOI] [PubMed] [Google Scholar]
- Soriano P (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genetics, 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Soukup V, Epperlein HH, Horacek I, & Cerny R (2008). Dual epithelial origin of vertebrate oral teeth. Nature, 455, 795–798. [DOI] [PubMed] [Google Scholar]
- Sousounis K, Bryant DM, Martinez Fernandez J, Eddy SS, Tsai SL, Gundberg GC, et al. (2020). Eya2 promotes cell cycle progression by regulating DNA damage response during vertebrate limb regeneration. eLife, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocum DL (1968). The urodele limb regeneration blastema: A self-organizing system. I. Morphogenesis and differentiation of autografted whole and fractional blastemas. Developmental Biology, 18, 457–480. [DOI] [PubMed] [Google Scholar]
- Stocum DL, & Melton DA (1977). Self-organizational capacity of distally transplanted limb regeneration blastemas in larval salamanders. The Journal of Experimental Zoology, 201, 451–461. [DOI] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, et al. (2013). Knockout mice created by TALEN-mediated gene targeting. Nature Biotechnology, 31, 23–24. [DOI] [PubMed] [Google Scholar]
- Swartz M, Eberhart J, Mastick GS, & Krull CE (2001). Sparking new frontiers: Using in vivo electroporation for genetic manipulation. Developmental Biology, 233, 13–21. [DOI] [PubMed] [Google Scholar]
- Tawk M, Bianco IH, & Clarke JD (2009). Focal electroporation in zebrafish embryos and larvae. Methods in Molecular Biology, 546, 145–151. [DOI] [PubMed] [Google Scholar]
- Tawk M, Tuil D, Torrente Y, Vriz S, & Paulin D (2002). High-efficiency gene transfer into adult fish: A new tool to study fin regeneration. Genesis, 32, 27–31. [DOI] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, et al. (2002). I-SceI meganuclease mediates highly efficient transgenesis in fish. Mechanisms of Development, 118, 91–98. [DOI] [PubMed] [Google Scholar]
- Tsai SL, Baselga-Garriga C, & Melton DA (2019). Blastemal progenitors modulate immune signaling during early limb regeneration. Development, 146. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Kondoh H, & Mizuno N (2005). Generation of transgenic newt Cynops pyrrhogaster for regeneration study. Genesis, 41, 87–98. [DOI] [PubMed] [Google Scholar]
- Voiculescu O, & Stern CD (2017). Manipulating gene expression in the chick embryo. Methods in Molecular Biology, 1565, 105–114. [DOI] [PubMed] [Google Scholar]
- Wagner I, Wang H, Weissert PM, Straube WL, Shevchenko A, Gentzel M, et al. (2017). Serum proteases potentiate BMP-induced cell cycle re-entry of dedifferentiating muscle cells during newt limb regeneration. Developmental Cell, 40(608–617), e606. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon WC, & Salmon ED (2000). Feedback interactions between cell-cell adherens junctions and cytoskeletal dynamics in newt lung epithelial cells. Molecular Biology of the Cell, 11, 2471–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, & Salmon ED (1996). Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. Journal of Cell Science, 109(Pt. 12), 2823–2831. [DOI] [PubMed] [Google Scholar]
- Whited JL, Lehoczky JA, & Tabin CJ (2012). Inducible genetic system for the axolotl. Proceedings of the National Academy of Sciences of the United States of America, 109, 13662–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited JL, Tsai SL, Beier KT, White JN, Piekarski N, Hanken J, et al. (2013). Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development, 140, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, et al. (2006). Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nature Protocols, 1, 1637–1652. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Denans N, Liu Y, Zhulyn O, Rosenblatt HD, Wernig M, et al. (2021). Optogenetic manipulation of cellular communication using engineered myosin motors. Nature Cell Biology, 23, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]


