SUMMARY
Nucleotides are at the heart of the most essential biological processes in the cell, be it as key protagonists in the dogma of molecular biology or by regulating multiple metabolic pathways. The dynamic nature of nucleotides, the cross talk between them, and their constant feedback to and from the cell’s metabolic state position them as a hallmark of adaption toward environmental and growth challenges. It has become increasingly clear how the activity of RNA polymerase, the synthesis and maintenance of tRNAs, mRNA translation at all stages, and the biogenesis and assembly of ribosomes are fine-tuned by the pools of intracellular nucleotides. With all aspects composing protein synthesis involved, the ribosome emerges as the molecular hub in which many of these nucleotides encounter each other and regulate the state of the cell. In this review, we aim to highlight intracellular nucleotides in bacteria as dynamic characters permanently cross talking with each other and ultimately regulating protein synthesis at various stages in which the ribosome is mainly the principal character.
KEYWORDS: RNA, nucleotides, protein synthesis, ribosome, translation
THE DIVERSE AND DYNAMIC NATURE OF NUCLEOTIDES
Nucleotides play central roles in all domains of life, be it as major carriers for chemical energy, as triggers for protein switches, secondary messengers, regulators of transcription, translation, and protein function, and as information storage in the form of DNA or mRNA. Equally important is their role as modulators of protein activity and RNA function. Thus, nucleotides and their metabolism are at the functional heart of living cells, and the underlying mechanistic principles that enable this are universally conserved.
A nice depiction of how dynamic nucleotides are at their core is evidenced throughout the growth curve of Escherichia coli, where nucleotide pools vary as a function of the physiological state of the cell (1) (Table 1). One key difference is that purines sustain millimolar intracellular levels, while pyrimidines remain consistent around midmicromolar levels. During the exponential phase, ATP levels peak at 2 mM and then decline ~3-fold upon transitioning into the stationary phase. A sudden rebound of ATP occurs during early stationary phase, after which its levels slowly decrease as cells enter a later growth stage (1). GTP levels follow a similar trend. In the case of the pyrimidines, UTP shows the same decline while entering early stationary phase, after which it progressively increases along the rest of the growth curve, while CTP seems to keep its levels relatively constant throughout (1). An interesting observation is that despite cells dividing at a maximal rate during the mid-log phase (2), the decreased levels of ATP, GTP, and UTP tune down this rate as nutrients start to slowly become limited at this stage. In other words, the fidelity between nucleotide levels and environmental conditions is sufficiently robust to ultimately provide feedback back to the cell and regulate its metabolic state accordingly. The fundamental extent of such an interplay is so important in determining the cell’s fate that when certain bacteria recognize invading plasmids or phage DNA, short prokaryotic Argonaute (pAgo)-based complexes trigger endogenous NAD(P)+ or NAD+ depletion to promote cell death and prevent infected cells from propagating within cell cultures (3, 4). Along the same line, it was recently discovered that bacterial viperins catalyze the conversion of CTP to ddhCTP, thus favoring transcription termination and inhibiting viral transcription, offering an additional mechanism of defense against phage infections (5, 6).
TABLE 1.
Intracellular levels of nucleotides in bacteria as a function of environmental challenges
| Nucleotide | Exponential | Stationary | Stringent response | Heat shocka | Ethanol shock | Oxidative stress |
|---|---|---|---|---|---|---|
| ATP | E. coli, 1.6–2 mM (15, 127); B. sutbilis, 1.3 mM (29); S. Typhimurium, 3 mM (27) | E. coli, ~1–3.9 mM (15) | E. coli, 1.3 mM (127) | S. Typhimurium, 11 mM* (27) | S. Typhimurium, 4.5 mM (27) | |
| ADP | E. coli: 494.4 μM (127); S. Typhimurium, 0.5 mM (27); B. sutbilis, 342.3 μM (29) | E. coli, 454.2 μM (127) | S. Typhimurium, <1 mM (27) | |||
| GTP | E. coli, 0.7–1.1 mM (15, 127); S. Typhimurium, ~1.2 mM (27) | E. coli, ~0.6–0.8 μM (15) | E. coli, 278.7 μM (127) | S. Typhimurium, 4 mM* (27) | S. Typhimurium, 0.7 mM (27) | |
| GDP | E. coli, 220–301.6 μM (15, 127); S. Typhimurium, ~0.2 mM (27) | E. coli, 120 μM (15) | E. coli, 158.9 μM (127) | S. Typhimurium, 0.2 mM (27) | ||
| CTP | E. coli, 360–520.2 μM (15, 127) | E. coli, ~0.6–710 μM (15) | E. coli, 340.6 μM (127) | |||
| UTP | E. coli, 242.7–510 μM (15, 127) | E. coli, ~0.8–1.3 mM (15) | E. coli, 162 μM (127) | |||
| NAD | E. coli, 698 μM (127) | E. coli, 1,092.3 μM (127) | ||||
| FAD | E. coli, ~10 μM (128) | |||||
| dp-CoA | E. coli, ~10–100 μMb (129) | |||||
| ppGpp | E. coli, 40 μM (15); S. Typhimurium, <LOQ (27) | E. coli, 150 μM (15) | E. coli, 793.8 μM (127) | S. Typhimurium, 940 μM** (27) | ||
| pppGpp | E. coli, <40 μM (15) | E. coli, 459.8 μM (127) | S. Typhimurium, 840 μM** (27) | |||
| Ap3A | B. subtilis, 4.4 μM (29); S. Typhimurium, <5 μM (27) | S. Typhimurium, 40 μM* (27) | S. Typhimurium, 60 μM (27) | |||
| Ap4A | B. subtilis, 24.2 μM (29); S. Typhimurium, <5 μM (27) | S. Typhimurium, 30 μM* (27) | S. Typhimurium, >50 μM (27) | S. Typhimurium, 365 μM (130) | ||
| Ap3G | S. Typhimurium, ~2 μM (27) | S. Typhimurium, ~12 μM* (27) | S. Typhimurium, ~20 μM (27) | |||
| Ap4G | B. subtilis, 0.7 μM (29); S. Typhimurium, <5 μM (27) | S. Typhimurium, <10 μM (27) | ||||
| cAMP | E. coli, 140–620 μM (131) | |||||
| cGMP | E. coli, 2.5 μM (127) | E. coli, 4.4 μM (127) | ||||
| c-di-GMP | E. coli, 0.31 μM (132); V. cholerae, 0.5–2 μM (133) |
*, 28 to 50°C (severe heat shock); **, 28 to 42°C (mild heat shock).
Not a direct measurement.
Nucleotides come in all shapes and sizes (Fig. 1), and their individual levels can be finely tuned as cells respond and adapt to fluctuating environmental conditions. This highly concerted regulation in turn dictates how a cell allocates its metabolic capacities to efficiently adapt to any given circumstance. The levels of nucleotides such as GDP 3′-diphosphate (ppGpp) and GTP 3′-diphosphate (pppGpp), collectively known as (p)ppGpp, become particularly important when bacteria face growth-limiting conditions (7). When amino acids are scarce, reminiscent of the entry into stationary phase, an increase in the levels of uncharged tRNAs binding to the ribosomal A site and stalling translation triggers the stringent response (8–10). During this process, RelA/SpoT homologue (RSH) proteins catalyze the transfer of the β- and γ-phosphates from ATP to the 3′ hydroxy group of GDP or GTP to increase the levels of ppGpp or pppGpp, respectively (11). These alarmones are then able to reprogram the cellular metabolism by modulating DNA replication, transcription, translation, nucleotide metabolism, and pathogenicity, among other biological processes (12–14). Upon transitioning into the stationary phase, the levels of (p)ppGpp increase roughly up to 0.8 mM (15) (Table 1), significantly above the dissociation constants (Kds) of ribosome biogenesis and translation-associated enzymes and at sufficiently high levels at which enzymes associated with carbon metabolism and fatty acid biosynthesis can be targeted (14). As important as it is to bring the levels of (p)ppGpp up, it is equally important to bring them down once the environmental challenge is surpassed and other nucleotide pools (e.g., ATP/GTP) need to be restored. For this reason, intricate regulatory mechanisms governing the alarmone synthetases/hydrolases keep a tight control on the levels of (p)ppGpp at all times (16–20). We would also like to mention the recent discovery of (p)ppApp, a stress molecule synthesized by Pseudomonas aeruginosa via a type VI secretion effector known as Tas1 (21). This enzyme is delivered into competitor cells and pyrophosphorylates other adenosine nucleotides at a high catalytic rate, depleting ATP pools and ultimately inducing cell death. Interestingly, it has been shown that the small alarmone hydrolase (SAH) of P. aeruginosa can hydrolase (p)ppGpp and (p)ppApp, adding a layer of protection against Tas1 during biofilm formation and interbacterial competition (22).
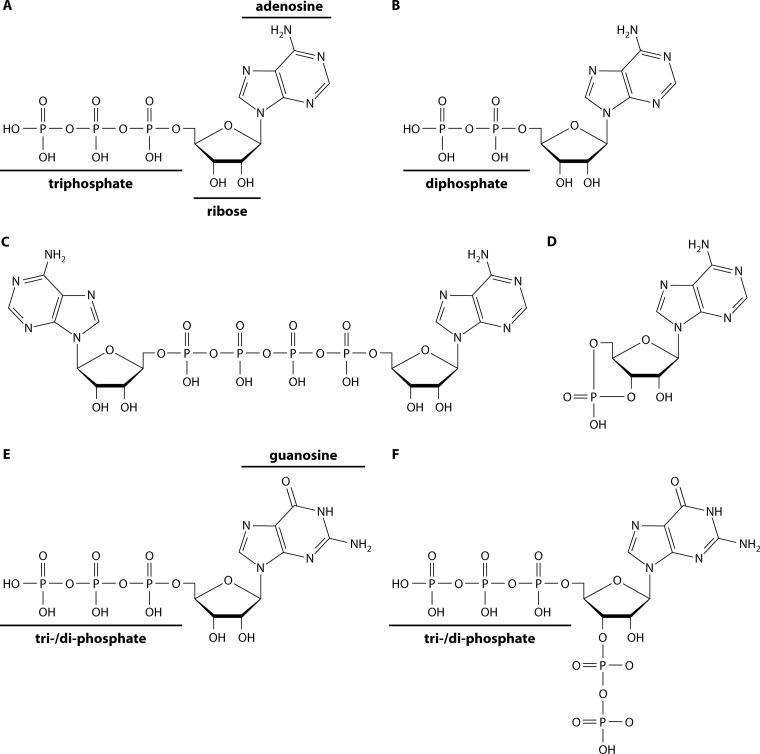
FIG 1.
Chemical structure of key nucleotides discussed in this review. (A) Adenosine triphosphate (ATP); (B) adenosine diphosphate (ADP); (C) diadenosine tetraphosphate (Ap4A); (D) cyclic adenosine monophosphate (cAMP); (E) guanosine triphosphate (GTP)/guanosine diphosphate (GDP); (F) GTP 3′-diphosphate (pppGpp)/GDP 3′-diphosphate (ppGpp).
Despite being discovered in 1966 (23), the triggering factors leading to the accumulation of the ubiquitous dinucleoside polyphosphates (NpnNs; where N represents adenine, uridine, or cytosine and n is the number of phosphates) and the physiological processes affected by them remain mostly enigmatic. Whether NpnNs are mere side products originating from the back reaction of tRNA charging or actually function as alarmones has been a topic of debate (24), albeit the latter seems to be more convincing based on previous (25–27) and recent data (28, 29). For example, when E. coli is exposed to lethal concentrations of aminoglycoside antibiotics such as kanamycin, the levels of Ap4A increase by 20-fold (28). An even bigger increase was observed when these cells were treated with hydrogen peroxide, an inducer of oxidative stress. Interestingly, when the naturally occurring hydroxyl radicals produced by the kanamycin treatment were quenched, the Ap4A levels were significantly reduced, suggesting that the levels of Ap4A functioned as a metabolic signal of aminoglycosides and/or oxidative stress in bacteria (28). A similar role was seen in the human pathogen Salmonella enterica serovar Typhimurium, in which a wide variety of heavy metal ions present in the growth media led to the accumulation of Ap3-4A and Ap3-4G to different extents (25). In addition, exposure to oxidative or osmotic stress in Myxococcus xanthus cultures increased the intracellular levels of Ap4A and Ap5A by 3.4- and 2.3-fold, respectively (26). Altogether, it seems that the levels of ApnA and perhaps other dinucleoside polyphosphates function as reporters of a wide variety of stresses that ultimately are related to the ability of bacteria to appropriately synthesize proteins. Whether these nucleotides are mere reporters of such stresses or can indeed physically interact and regulate enzymes involved in bacterial metabolism merits further investigation.
USE OF MODIFIED NUCLEOTIDES OR DINUCLEOTIDES BY RNAP
The RNA polymerase (RNAP) is the core enzyme responsible for the transcription of genetic information from DNA to RNA using canonical nucleotides (ATP, UTP, GTP, and CTP) as building blocks. Binding of the RNAP to specific sequences known as promoters on the double-stranded DNA sets the stage for the initiation of transcription. The RNAP unwinds the DNA duplex to form the transcription bubble, a process referred to as open complex formation (30, 31). Interestingly, the concentrations of the initiating nucleotide can differentially alter the transcription initiation rates of multiple promoters in bacteria. Consequently, this could potentially also alter protein synthesis rates by changing the abundance of the templates for protein synthesis. For example, short-lived open complexes, such as those found in rRNA and some tRNA and mRNA promoters, show a higher dependency on the initiating nucleotide (32–35). This regulation is nicely depicted in Bacillus subtilis, where the promoter activity from rRNA genes is highly dependent on ppGpp and GTP levels since these promoters initiate exclusively with GTP (36, 37).
Recent efforts have widened our knowledge on the vast repertoire of molecules that can be incorporated onto the nascent transcript during transcription initiation in Gram-positive and -negative bacteria and how these can uniquely influence the transcript’s processing, stability, localization, and translation efficiency (38, 39). It was demonstrated that the RNAP of E. coli can use NAD (NAD+) and dephospho-coenzyme A (dp-CoA) in vitro and in vivo as an initiating nucleotide provided that the nucleotide at the +1 position encodes ATP (40). Moreover, in vitro studies showed that RNAP is also able to use flavin adenine dinucleotide (FAD), UDP-glucose, and UDP-N-acetylglucosamine to initiate transcription, although the feasibility and relevance of this phenomena in vivo remains uncertain (41, 42). Interestingly, NAD+ and dp-CoA 5′ RNA caps were found in 10 to 15% of bacterial transcripts of ≲200 nucleotides, with the former being 2-fold higher in the stationary phase than in the exponential phase (40, 43, 44). Furthermore, the realization that most 5′ NAD-linked RNAs are associated with pathways revolving around cellular metabolism and various stress responses (45) supports the notion that the incorporation of these noncanonical molecules is not arbitrary and could prove essential when facing specific environmental challenges or growth conditions.
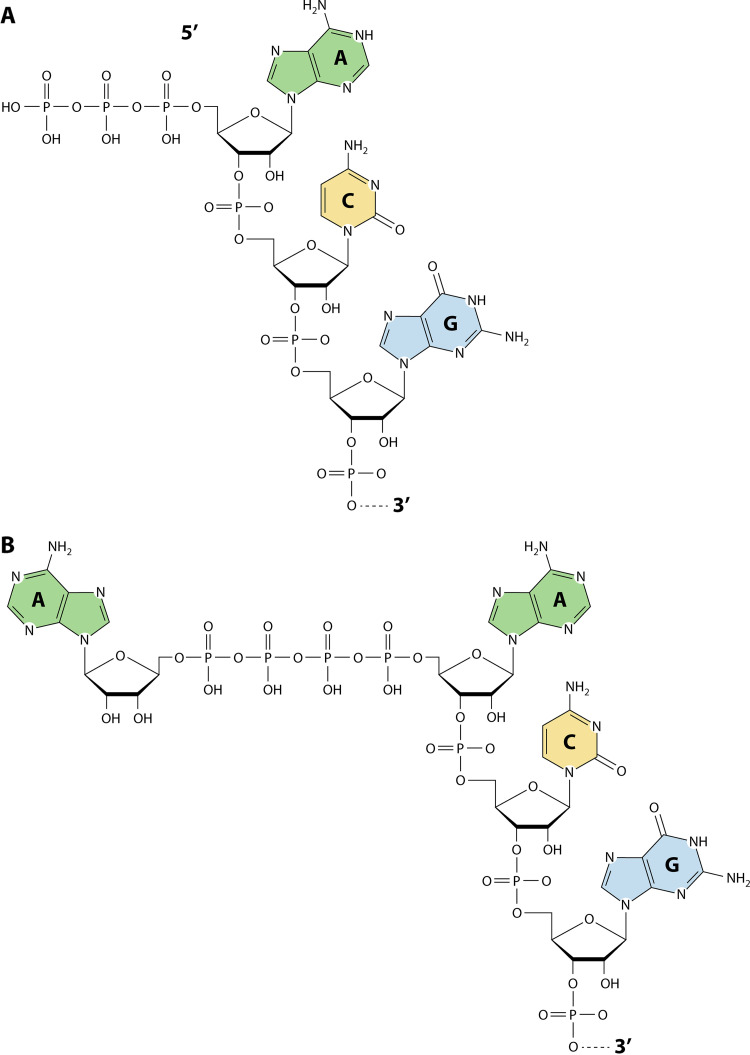
It has been reported that after treating E. coli with CdCl2, an inducer of disulfide stress, dinucleoside tetraphosphates rapidly accumulate and trigger the widespread use of ApnNs as 5′ RNA caps (Fig. 2) (46). Surprisingly, not only E. coli’s RNAP but also the lysyl-tRNA synthetase were able to catalyze this capping reaction. Additionally, it was shown that increased levels of Np4Ns inhibit ApaH, the main Np4A hydrolase in bacteria, leading to a further enrichment in Np4-capped RNAs whose stability is increased as evidenced by a lengthened half-life. The incorporation of Np4Ns into the nascent transcript seems to be considerably more efficient than ATP given that the nucleotide at position −1 of the template strand is a purine (47).
FIG 2.
Schematic of the 5′ terminal Ap4A cap. Shown are the canonical 5′ end of bacterial mRNA (A) uncapped and (B) capped by Ap4A.
Whether regulation of gene expression from Np4-capped transcripts is sufficient to explain the cellular effects observed after increased levels of dinucleoside tetraphosphates or instead is due to key direct targets of these molecules remains to be determined. Nonetheless, the implications of alternative molecules functioning as noncanonical substrates for the RNAP add an elegant, albeit still obscure, layer of regulation of gene expression that further highlights the unprecedented chemical diversity of RNA species.
SYNTHESIS AND MAINTENANCE OF TRANSFER RNAS
Since transfer RNAs (tRNAs [75 to 90 bases]) are the intermediate molecules between the nucleotide and amino acid alphabets, it is very relevant to observe how the changes in the intracellular levels of different nucleotides affect tRNA abundance and their chemical modifications. The accumulation of (p)ppGpp and consequent inhibition of the RNAP activity affect the synthesis of different families of RNAs, such as ribosomal RNAs (rRNAs), messenger RNAs (mRNAs), and tRNAs (48, 49). Regardless of RNAP regulation, increased levels of (p)ppGpp also interfere with RNA and DNA synthesis by inhibiting enzymes enabling the de novo synthesis of purines and the purine salvage pathway, such as PurF, GsK, Hpt, PpnN, and Gpt (50). Furthermore, (p)ppGpp interacts with the transcriptional factor PurR, which downregulates the transcription of several components of the aforementioned pathways (51). This is remarkable in the context of bacterial stress since different stress conditions that trigger the accumulation of (p)ppGpp also converge in the generation of oxidative stress. This redox imbalance mainly affects purines such as guanosine due to their low oxidation potential, leading to the formation of 8-hydroxyguanosine (52, 53). A similar regulation has recently been reported for the Ap4A dinucleotide, which restricts the biosynthesis of guanosine nucleotides (i.e., GMP, GDP, and GTP) in B. subtilis, by influencing the oligomerization state and activity of inosine-5′-monophosphate dehydrogenase (IMPDH) (29).
In bacteria such as E. coli, the tRNA genes are often arranged in operons that produce transcripts that contain more than one tRNA sequence, which is subject to further processing and modification steps in order to mature the functional tRNAs (54). For transcripts encoding more than one tRNA, smaller tRNA precursors are generated by the endonucleolytic activity of RNase E and RNase III downstream of the CCA terminus (55). Subsequent cleavage of the 5′ end by RNase P is a common step for all pre-tRNAs (54). Some authors have reported that the genes rnpA and rnpB, which encode the subunit C5 and the catalytic RNA subunit M1 RNA, respectively, of RNase P, are downregulated during the stringent response, which could strongly affect the global processing of tRNA (49, 56). This transcriptional regulation should significantly modulate the levels of RNase P due to the stability of the holoenzyme (half-life [t1/2] of ~60 min). However, it decreases dramatically (t1/2 of ~5 min) with the loss of the C5 subunit, becoming a potential checkpoint on the regulation of tRNA levels under some stress conditions (57). Final trimming of the 3′ end is performed by a combination of exonucleases such as RNase II, polynucleotide phosphorylase (PNPase), RNase P, RNase PH, and RNase T, depending on the size of the remaining 3′ end (58, 59). Some of these RNases also contribute to the global regulation of RNA metabolism, participating in mRNA decay. An interesting example is PNPase, which has been shown to be part of a ribonucleoprotein complex with two direct O2 sensors, DosC (direct O2-sensing cyclase) and DosP (direct O2-sensing phosphodiesterase). These two proteins adjust the levels of c-di-GMP, which ultimately targets and modulates PNPase activity (60). However, the impact of c-di-GMP on the population of tRNAs processed by PNPase has not been reported.
The different groups of tRNAs require posttranscriptional modifications of some of their nucleotides to fine tune their functionality in accordance to cellular requirements. Thus, we find more than 100 chemical modifications that generate derivatives from the nucleosides adenosine, guanosine, cytidine, and uridine. When observing the distribution of these modifications along the tRNAs in E. coli, their high frequency and diversity stand out in positions 34 and 37, corresponding to the neighborhood of the anticodon involved in codon recognition and reading frame maintenance (59, 61). One example is the lysidine modification of cytosine 34 (K2C) by the enzyme TilS, a modification that occurs in the anticodon region of tRNAIle2. K2C is required to maintain translational fidelity since, without this modification, the methionyl tRNA synthetase can recognize tRNAIle2, promoting misacylation of tRNAIle2 with a noncognate amino acid (62, 63).
Among the enzymes that carry out such modifications, many of them require nucleotide cofactors, such as ATP, GTP, NADPH, S-adenosyl-l-methionine (SAM), and/or NADP+ (Table 2). This is especially important when considering the nucleotide population as a dynamic element in terms of abundance and diversity. As an example, we can analyze the case of ATP, whose intracellular concentration can fluctuate between ~1 and 10 mM (Table 1). In addition, we must consider the concentration of ADP and changes in the ATP/ADP ratio, a nucleotide that is in constant competition for the same ATP-binding sites. Furthermore, the accumulation of nonhydrolyzable ATP analogs like Ap4A, which can also compete for some ATP-binding sites, leads to a scenario where these ATP-dependent enzymes are also dependent on all ATP analogs. Nucleotide cross talk is also important in the evolution of nucleotide-binding sites in enzymes that require cofactors. TrmD is an essential SAM-dependent methyltransferase, highly conserved in all three domains of life, that catalyzes the methylation of G37. It has been reported that in the TrmD of Firmicutes bacteria, such as Staphylococcus aureus, cAMP competes for the pocket-binding substrate and inhibits its activity. It is important to note that adenylate cyclase enzymes responsible for cAMP formation are absent in this group of bacteria. However, TrmD from cells encoding adenylate cyclases, such as E. coli, which produces cAMP as a molecule that regulates energy metabolism, has adapted the binding substrate to provide a high specificity for SAM (64).
TABLE 2.
Nucleotide modifications in E. coli tRNAs and nucleotide dependence
| Position | Modification(s) | Enzyme(s)a | Nucleotide dependence |
|---|---|---|---|
| 8 | S4U | IscS, ThiI1 | ATP (134) |
| 13 | Ψ | TruD | No |
| 16,17 | D | DusA2, DusB2, DusC2 | NADPH (135) |
| 18 | Gm | TrmH3 | SAM (136) |
| 20 | D | DusA2, DusB2, DusC2 | NADPH (135) |
| 32 | S2C, Cm, Um, Ψ | IscS, TtcA1, TrmL3, RluA | ATP (137), SAM (138) |
| 34 | I, k2C, ac4C, Um, Cm, Q, GluQ, mnm5U, cmcm5Um, cmnm5U, mnm5s2U, cmnm5s2U, mnm5se2U, mcmo5U, cmo5U | TadA, TilS1, TmcA1, TrmL3, FolE4, QueDCEF, Tgt, QueA3, QueG, YadB, MnmC13, MnmC23, MnmE4, GidA4, CmoA3, CmoB3, AroB, aroD, aroE5, aroK1, aroL1, aroA, aroC | ATP (62, 139, 140), SAM (138), GTP*, NADP+ |
| 37 | m1G, m6A, m2A, i6A, ms2i6A, t6A, m6t6A | TrmD3, TrmN6, MiaA, MiaB, IscS, IscU, TsaC, TsaD, TsaB, TsaE | SAM |
| 38 | Ψ | TruA | No |
| 39 | Ψ | TruA | No |
| 40 | Ψ | TruA | No |
| 46 | m7G | TrmB3 | SAM (141) |
| 54 | rT | TrmA3 | SAM (142) |
| 55 | Ψ | TruB | No |
| 65 | Ψ | TruC | No |
| 67 | Ψ | TruD | No |
In the “Enzymes” column, the superscript number indicates the corresponding cofactor: 1, ATP; 2, NADPH; 3, SAM; 4, GTP; 5, NADP+.
Altogether, the synthesis of mature tRNAs alone does not guarantee obtaining the necessary aminoacylated tRNAs as a functional substrate for translation. This is due to some synthetases such as E. coli isoleucyl-tRNA synthetase having their activity upregulated by ppGpp and inhibited by cAMP, which can ultimately affect the aminoacylated fraction of tRNAIle (65, 66).
HOW REGULAR AND ALTERNATIVE NUCLEOTIDES ARE INVOLVED IN THE FLOW OF THE TRANSLATION MACHINERY
Initiation
At the onset of translation, an mRNA-programmed 30S ribosomal subunit recruits three translation initiation factors (IF1 to IF3) and the initiator fMet-tRNAfMet to assemble the preinitiation complex (PIC) (for an exhaustive review of translation initiation in prokaryotes, see reference 67) (Fig. 3). IF2 controls the fidelity of translation initiation by specifically enhancing the docking of the initiator tRNA via interactions with the formyl group. This translation factor then enhances the decoding of the transcript’s start codon within the ribosomal P site of the small subunit, allowing the formation of a 30S initiation complex. The association of the 50S ribosomal subunit is then accelerated by the ribosome-bound IF2 acting as an anchor for subunit joining or by favoring an optimal ribosomal arrangement, leading to the 70S initiation complex (68). This immediately triggers the GTPase activity of IF2 which changes the environment of IF1 within the ribosome, promotes IF3 dissociation, releases and accommodates the initiator tRNA into the P site, and finally forces IF2 and IF1 out of the ribosome (69). All these highly concerted events allow the binding of elongation factor Tu (EF-Tu)-GTP-aminoacylated tRNA (aa-tRNA) (known as the ternary complex) to promote dipeptide synthesis and the formation of an elongation-competent 70S initiation complex (69).
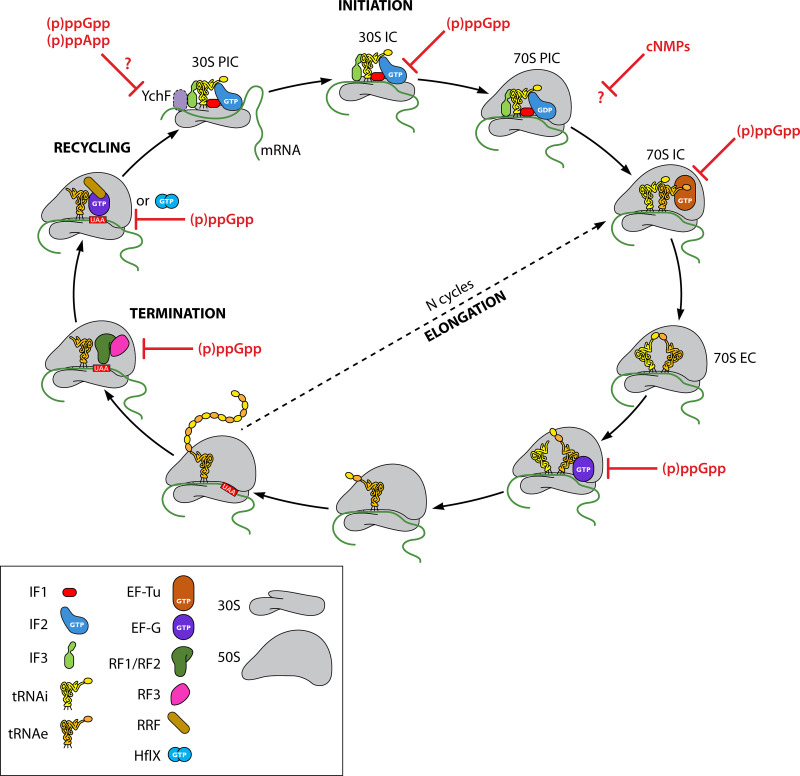
FIG 3.
Diverse involvement of nucleotides throughout the complete cycle of mRNA translation. As previously described, translation initiation starts with the formation of the 30S preinitiation complex (PIC) and the recognition of the codon-anticodon, with IF2 being regulated by (p)ppGpp levels. Upon the arrival of the next aminoacylated tRNA into the ribosomal A site with the help of elongation factor Tu, the elongation of translation begins and is also modulated by (p)ppGpp affecting EF-Tu’s function. The translocation of the P/A site tRNAs into the E/P configuration mediated by EF-G is additionally affected by the levels of (p)ppGpp. Once a stop codon is recognized in the ribosomal A site, RF1/RF2 will be recruited into the ribosome and with the aid of RF3, which will also be tuned by (p)ppGpp pools, terminates translation and releases the polypeptide chain from the ribosome. Finally, EF-G and RRF, both targets of (p)ppGpp, induces the ribosome to be recycled back into the pool of free subunits. HflX, also a (p)ppGpp binder, could potentially be aiding with the recycling of a special subset of terminated/arrested ribosomes. The precise positioning of YchF (a black dashed circle indicates its putative location) within the ribosome, its potential regulation by (p)ppGpp/(p)ppApp, and the specific translation stage or stages affected by cNMPs require further investigation.
IF2 has been defined as a metabolic sensor during translation initiation able to detect the status of the cell and either favor or preclude translation, depending on the nucleotide bound to it (68, 70). This is particularly important when we consider that during the stringent response, the levels of GTP fall from 0.7 to 1.1 mM to ~0.3 mM, while the levels of ppGpp increase up to ~0.8 mM (Table 1). Since the nucleotide-binding site of IF2 is shared by GTP and ppGpp and the dissociation constants of both ligands are comparable (1.6 and 2.8 μM, respectively), whichever is more abundant at any given point will most likely outcompete the other and hence determine the fate of translation (70–72) Fig. 3). On a mechanistic level, when ppGpp is present, the amount of fMet-tRNAfMet binding to the 30S decreases mainly due to the lower affinity of IF2-ppGpp for the ribosome compared to IF2-GTP. This in turn leads to a lower rate of dipeptide formation, by hampering the ability of IF2 to accommodate the initiator tRNA and consequently causes a decrease in the overall translation rate (70).
Nevertheless, the regulation of translation by (p)ppGpp seems to be more complex than previously expected, with recent data showing that during the stringent response, the translation of certain transcripts is affected by ppGpp levels to different extents (73). Structural features found within the 5′ untranslated region of specific mRNAs increased the affinity of GTP for IF2, increased the inhibitory concentration of ppGpp, and favor a 30S IC-like conformation, ultimately leading to a set of transcripts that can be translated at physiological concentrations of ppGpp (73). Remarkably, it was shown that pppGpp does not necessarily partake in the inhibitory nature of the stringent response as ppGpp does and allows IF2 to recruit the initiation tRNA and promote the formation of the 30S initiation complex. Whether the structural features that confer an advantage in translation during (p)ppGpp accumulation are characteristic of a certain set of mRNAs required during stress conditions, whether there are other cis- and trans-regulatory factors also affecting the translation efficiency of such mRNAs, and whether the IF2-dependent regulation of pppGpp provides another layer of translational control all remain to be answered. In addition, it was recently shown that in E. coli, the universally conserved ATPase YchF seems to act as a stress-mediated regulator of leaderless mRNA translation by interacting with the 30S small subunit and modulating the antiassociation activity of IF3 (74). What makes YchF intriguing is that it is a P-loop GTPase that preferentially hydrolyses ATP and whose levels drop during stress conditions (75). A differential regulation of YchF by GTP/ATP or its stress-induced counterparts (p)ppGpp/(p)ppApp is plausible (Fig. 3); however, whether this is true or physiologically relevant to better cope with stresses should be further examined.
Elongation
The first complete translocation of the mRNA-tRNA complex from the A and P sites to the P and E sites, respectively, catalyzed by the GTPase elongation factor G (EF-G), is defined as the start of translation elongation (translocation) (67). Now, at the posttranslocational state, the A site is vacated, and the next codon is exposed, allowing the appropriate aminoacylated tRNA to be incorporated as a ternary complex in which EF-Tu plays the major role. Once the codon-anticodon match has been successfully sampled (decoding), the EF-Tu-mediated hydrolysis of GTP is triggered, EF-Tu is released, and the cognate tRNA is accommodated into the A site, rapidly leading to the peptidyl transfer (peptide bond transfer).
As expected, the GTPases in charge of translation elongation can bind (p)ppGpp and further regulate the flow of translation (Fig. 3). It is worth mentioning that elongation factor Ts (EF-Ts), which catalyzes the exchange of GDP from EF-Tu with GTP, is indirectly inhibited by ppGpp. This is because EF-Ts is sequestered by ppGpp-bound EF-Tu, since its affinity is higher than that of GDP-bound EF-Tu (76). There seems to be, however, a differential prioritization of ppGpp over EF-Tu and EF-G as the dissociation constants (Kds) between GTP and ppGpp vary substantially between them. As such, EF-Tu and EF-G show Kds of 8 nM and 13.9 μM for ppGpp and 0.59 μM and 8.3 μM for GTP, respectively, indicating that the regulation of ppGpp on translation elongation is kinetically favored to occur through EF-Tu even at basal levels of ppGpp (72, 77–79). Interestingly, despite a previous report showing a 2-fold delay in the arrival time of EF-G to pretranslocation complexes in the presence of ppGpp, no effects on the overall elongation rates were detected. It was thought that since the pool of aminoacylated tRNAs decreases during the stringent response, fewer pretranslocation ribosomal complexes would also reduce the dependence on normal EF-G levels and, as such, inhibiting EF-G would therefore have only a marginal effect on the overall rates of elongation (76).
During stringent conditions, a limiting amino acid leads to a reduction of its corresponding cognate aa-tRNA and, consequently, a decrease of ternary complexes containing that specific aa-tRNA. This suggests that ternary complexes containing near-cognate aa-tRNAs could potentially outcompete the correct—but scarce—ternary complexes and reduce translation fidelity. Nevertheless, the accuracy of proofreading during protein elongation is preserved under high levels of (p)ppGpp (80). To explain this seemingly contradictory scenario, extensive biochemical and kinetic studies have proposed different models (80–83). The most robust of these seems to indicate that ppGpp-bound EF-Tu reduces by ~2-fold the rates for peptide bond formation and thereby widens the time frame for a higher proportion of near-cognate tRNAs to be sampled and rejected (82). This suggests that by delaying the incorporation of all aa-tRNAs and extending the time frame for proofreading, ppGpp allows the translational machinery to ensure the fidelity of aa-tRNA selection while amino acid deprivation persists. Evidently, the kinetic cost of preserving translation fidelity by delaying elongation rates would reduce overall translation rates and growth; however, misincorporation of aa-tRNAs would lead to the synthesis of aberrant proteins that could ultimately be fatal (84). Along the same line, elongation factor 4 (EF4, originally called LepA), required for 16S rRNA processing and the association of late-stage 30S ribosomal proteins, is also a target of (p)ppGpp (85, 86). Despite no clear indication of the functional basis for the alarmones targeting EF4, given its function during ribosome biogenesis, EF4 could be yet another way to control ribosome pools under conditions in which cognate aa-tRNAs are limited.
Termination
The elongation cycle will repeat itself throughout the mRNA until a stop codon is reached, which marks the start of the termination cycle. In bacteria, release factors 1 and 2 (RF1 and RF2) recognize the codons UAG/UAA and UGA/UAA, respectively, as the end of the transcript and promote the peptidyl transferase center-dependent hydrolysis of the peptidyl-tRNA, liberating the nascent polypeptide. Release factor 3 (RF3) then enhances the release of RF1/RF2 from the ribosome, setting the stage for ribosome recycling (87).
Initial estimations of the dissociation constants of GDP and GTP to RF3 (5.5 nM and 2.5 μM, respectively) pointed to a model in which RF3 mainly existed in its GDP-bound state (88). Nevertheless, these values were later revisited and the Kd of GTP to RF3 was found to be comparable to that of GDP at 76 nM (89, 90). Considering that in bacteria the levels of GTP are present in an ~5-fold excess compared to GDP throughout growth phases or under mupirocin treatment, it is believed RF3 actually favors its GTP-bound form. Furthermore, RF3 has been reported as a binding partner for the alarmones ppGpp and pppGpp, although showing higher dissociation constants at 0.8 and 15 μM, respectively (86). Interestingly, when RF1 turnover rates were probed, the efficiency of recycling was compromised in the presence of ppGpp, suggesting that under stringent conditions RF3 switches from its active GDP-bound state to an inactive ppGpp-bound form (91) Fig. 3). It was further characterized that the binding of ppGpp to RF3 would preclude the factor’s association to the ribosome due to a clash and electrostatic repulsion between the 3′ diphosphate group of ppGpp and the sarcin-ricin loop (91).
Recycling
The fourth essential step of translation ensures that the posttermination ribosomal complex releases the mRNA and tRNA and splits into functional subunits capable of binding a new transcript to start translation over again (92). This is catalyzed by the ribosome recycling factor (RRF) binding to the A site and forcing the ribosome into a fully rotated state. EF-G then binds and alters the position of RRF, forcing the tRNA out of the ribosome (93). Next, the EF-G-mediated hydrolysis of GTP pushes RRF into the intersubunit space, ultimately splitting the 70S ribosome into individual ribosomal subunits (94).
Despite not being commonly described as a player in ribosome recycling, in principle, the heat shock-induced GTPase HflX is a ribosome-splitting factor (95). It has been proposed that under stress conditions, GTP-bound HflX associates with the ribosomal E site of translationally arrested ribosomes and rescues them by inducing subunit splitting (95, 96) to rates similar to those of GTP-bound EF-G–RRF-mediated ribosome dissociation (97). Ribosomal dissociation was also observed in the presence of GDP-bound HflX, albeit at an ~20-fold lesser extent than the GTP-bound state as evaluated by the changes in light scattering (96). Since the ribosome-splitting function of HflX occurs in a nucleotide-dependent manner and it has been previously reported as a (p)ppGpp binder in bacteria, the putative regulation of translation recycling by the alarmones under specific stress conditions remains to be challenged (Fig. 3).
2′,3′-Cyclic Nucleotide Monophosphates and Translation
2′,3′-Cyclic nucleotide monophosphates (cNMPs) are a product of RNase 1-mediated RNA degradation that accumulate upon ribosome recycling, mRNA decay, and the stringent response (98). It has been shown that in E. coli, downregulated levels of cNMPs heavily modulate its transcriptome, unveiling several cNMP-dependent cellular processes, such as acid resistance, biofilm formation, motility, and other stress responses (99). However, the mechanism by which bacteria are able to sense the levels of these nucleotides is still unknown. A recent work aimed to identify cNMP binding proteins in E. coli and S. Typhimurium by employing cNMP-bound affinity chromatography resins (100). Interestingly, various ribosomal proteins were detected by a mass spectrometry-coupled protein pulldown assay, which led them to successfully probe purified 70S ribosomes as the cNMPs’ binders. Furthermore, it was shown that translation was inhibited in vitro by high levels of the cNMPs (50 to 100 mM), while at lower concentrations (5 to 10 mM), cAMP showed an increase in translation partially attributed to transcription-dependent effects (Fig. 3). All in all, it seems conceivable that intracellular levels of cNMPs can modulate translation, although the mechanistic and physiological basis for this regulation needs to be further studied.
HOW NUCLEOTIDES ARE INVOLVED IN ASSEMBLY, DEGRADATION AND BIOSYNTHESIS OF RIBOSOMES
After exploring the regulatory potential of nucleotide dynamics in the stability and abundance of mRNAs, the levels and functionality of tRNAs, and the different stages of translation, it is worth probing whether ribosome biogenesis and assembly are also influenced by nucleotides. Indeed, various screenings searching for (p)ppGpp targets have identified multiple proteins linked to the biogenesis, degradation, and assembly of ribosomes whose functions would be altered during the stringent response (86, 101–104).
Regarding ribosome biogenesis, we can distinguish between the transcriptional regulation of rRNA and the synthesis and assembly of ribosomal proteins. Irrespective of the large number of molecules involved, the rate of ribosome biogenesis is primarily determined by the rate of rRNA transcription (105). In this sense, it has been observed that during starvation in E. coli the formation of the open complex by RNAP is strongly repressed by DksA, a protein that, bound to ppGpp, inhibits the start of rRNA transcription (106). However, this regulation differs from that observed in Gram-positive bacteria such as B. subtilis, in which, as previously discussed, the regulation of rRNA transcription depends on the concentrations of its initiator NTP (36).
Due to the high number of ribosomal proteins, their synthesis must be highly regulated to obtain the precise stoichiometry of these proteins in mature ribosomes. Thus, in E. coli we can find many of them forming part of large operons whose translation and increase in free protein levels generate negative feedback on the translation of their own transcript (107). This regulation mechanism is common in eubacteria; however, the structures present in the mRNAs encoding ribosomal proteins are not highly conserved among bacteria (108). This raises the possibility of alternative RNA structures that interact with ribosomal proteins, or even with other metabolites, as is the case of Vibrio vulnificus, which contains certain riboswitches that detect adenosine and lead to the synthesis of the ribosomal S1 protein (109).
The assembly and maturation of ribosomal subunits are assisted by various factors, such as DEAD box RNA helicases, maturation factors, and ribosome-associated GTPases. Many of these GTPases have been identified as targets of (p)ppGpp, namely Era, Der (EngA), ObgE (CgtAE), RsgA, BipA (87, 101, 110, 111). Given their relevance in subunit maturation, some authors have already proposed these GTPases as regulators of ribosome biosynthesis under stress conditions (112). In addition to the regulation exerted on ribosome biosynthesis, different putative GTPases, such as RsgA, RbgA, Era, HflX, and ObgE, have been identified as (p)ppGpp targets, whose binding to (p)ppGpp negatively impacts subunit joining in Gram-positive and Gram-negative bacteria (102, 113). The accumulation of (p)ppGpp in E. coli increases the affinity of ObgE for the 50S subunit, exerting a role of antiassociation of the subunits and ultimately inhibiting the initiation of translation, although its overexpression also decreases the fraction of mature 70S ribosomes (113). It is possible that an antiassociation factor with these characteristics has a hierarchical effect on translation initiation—first on canonical translation initiation and then on alternative initiation mechanisms such as 70S-scanning or the use of leaderless mRNAs as the templates, where mechanisms that involve an interaction of the transcripts with 70S monosomes rather than to individual subunits have been proposed (113–115).
On the other hand, it has been proposed that the stringent response also involves the degradation of ribosomal proteins mediated by the Lon protease, which is highly conserved from bacteria to humans. This protease reduces the rate of protein synthesis and helps to supply back limiting amino acids during starvation (116). Lon-mediated proteolysis is activated by the accumulation of inorganic polyphosphates (PolyP), ubiquitous polymers of various lengths, depending on the species, whose intracellular levels are finely regulated by the balance between their synthesis and degradation by the enzymes polyphosphate kinase (PPK) and exopolyphosphatase (PPX), respectively (116). Agents that induce the stringent response, such as serine hydroxamate (SHX), trigger a rapid accumulation of PolyP, which is explained by a (p)ppGpp-dependent inhibition of PPX (101, 117, 118). Some of the proteins that have been identified as part of the Lon-PolyP complex are recruited very late in the formation of the subunit, such as S2, but there are also others that interact almost directly with rRNAs, such as L9 and L13 (116).
CONCLUSIONS AND PERSPECTIVES
In the present day, when high-throughput nucleic acid sequencing tools have grown vertiginously, the myriad roles of nucleotides as encoders of information are remarkable. However, before this genomic era, their study revealed them crucial elements of metabolism, as carriers of chemical energy as well as allosteric regulators of different proteins, among multiple other functions. In this review, we have tried to emphasize the relevance of changes in intracellular nucleotide levels and how their action is not isolated but depends on the entire set of nucleotides.
Throughout their study, different strategies for nucleotide characterization and quantification were performed, often sacrificing the diversity of analyzed nucleotides for improved sensitivity and reproducibility. To exemplify this, we can contrast the sensitivity of the luciferin-luciferase enzymatic assay for ATP quantification in the picogram range, while global nucleotide analysis by two-dimensional thin-layer chromatography on polyethyleneimine (PEI)-cellulose allows discrimination of around 90 nucleotide species at one time (119, 120). Additional efforts have been made in capillary electrophoresis. However, the species of nucleotides identified by this methodology depends on the electrophoresis conditions, which makes the analysis of complex samples difficult (121). Currently, chromatographic methods coupled to mass spectrometry provide us with sufficient precision for the identification and quantification of mononucleotides and dinucleotides with different degrees of phosphorylation or modifications in the bases that compose them and/or cyclic dinucleotides (122–125). One field that has advanced in parallel with the detection of nucleotides is the characterization of their targets. Different methodologies have been standardized to determine both the targets of specific nucleotides and their affinity (101, 103). An example of this is the differential radial capillary action of ligand assay (DRaCALA), which facilitates the determination of dissociation constants from different targets in a systematic way (103). These new tools force us to reanalyze dynamics at the nucleotide level, this time understanding them as a network that regulates cell metabolism and not as isolated values that change in response to some disturbance.
The ribosome is quite convincingly the molecular hub at which most of these nucleotides converge and ultimately regulate protein synthesis rates and the state and fate of the cell. In the first instance, the combination of biogenesis and assembly of ribosomes, arguably the highest energetic cost process in the cell, is constantly tuned by the pools of nucleotides, which fluctuate according to whatever challenge bacteria encounter. Then, mRNAs and tRNAs, the template-encoding substrates and the tools to build such templates for the ribosome, are also finely regulated by nucleotide levels. All stages of mRNA translation have at least one molecular reporter that senses and is affected by (p)ppGpp, meaning that the fluctuating nucleotides originating from the stringent response feedback the cell via the ribosome and its function. Furthermore, antimicrobial compounds that bind to the ribosome and preclude its function cause increased levels of Ap4A, possibly revealing a novel mechanism by which dinucleoside polyphosphates act as reporters of the state and integrity of protein synthesis in real time.
Various studies are attempting to unveil new modifications and changes in the concentrations of the nucleotide network, while others aim to characterize their targets. However, the number of processes being regulated by nucleotides is so vast that it is necessary to integrate both approaches through the generation of models, just as the field of metabolic engineering did through the development of metabolic flux analysis (126). Since many enzymes can interact to different extents with one or more nucleotides, it is difficult to assess the in vivo effect of a specific nucleotide, and hence, is why we should keep in mind the abundance, variety, and dynamics of nucleotides within the cell. An example of this is seen with regard to translation factors that show different affinities for GTP, GDP, and (p)ppGpp and where the degree of inhibition of the steps involved in translation will depend on the levels of these four nucleotides. Future research that reconnects our deep understanding of protein synthesis together with the acknowledgment of how dynamic nucleotides are will allow us to uncover the extent of how diverse nucleotide families are differentially fine-tuned when microorganisms are perturbed. This will provide a picture in which we see whether nucleotides are affecting the cell concomitantly or whether there is a prioritization of targets that changes with the fluctuating levels of nucleotides.
ACKNOWLEDGMENTS
G.B. acknowledges support from the DFG-priority program SPP1879 “Nucleotide Second Messenger Signaling in Bacteria” and the Max-Planck Society. M.I. acknowledges support from the National Science Foundation (MCB-2101998).
Contributor Information
Gert Bange, Email: gert.bange@synmikro.uni-marburg.de.
Michael Ibba, Email: ibba@chapman.edu.
REFERENCES
- 1.Buckstein MH, He J, Rubin H. 2008. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol 190:718–726. 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marr AG. 1991. Growth rate of Escherichia coli. Microbiol Rev 55:316–333. 10.1128/mr.55.2.316-333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koopal B, Potocnik A, Mutte SK, Aparicio-Maldonado C, Lindhoud S, Vervoort JJM, Brouns SJJ, Swarts DC. 2022. Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA. Cell 185:1471–1486.e19. 10.1016/j.cell.2022.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaremba M, Dakineviciene D, Golovinas E, Zagorskaitė E, Stankunas E, Lopatina A, Sorek R, Manakova E, Ruksenaite A, Silanskas A, Asmontas S, Grybauskas A, Tylenyte U, Jurgelaitis E, Grigaitis R, Timinskas K, Venclovas Č, Siksnys V. 2022. Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion. Nat Microbiol 7:1857–1869. 10.1038/s41564-022-01239-0. [DOI] [PubMed] [Google Scholar]
- 5.Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, Chandran K, Bresnick AR, Cameron CE, Almo SC. 2018. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558:610–614. 10.1038/s41586-018-0238-4. (Erratum, 562:E3, 2018; Erratum, 583:E15, 2020.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernheim A, Millman A, Ofir G, Meitav G, Avraham C, Shomar H, Rosenberg MM, Tal N, Melamed S, Amitai G, Sorek R. 2021. Prokaryotic viperins produce diverse antiviral molecules. Nature 589:120–124. 10.1038/s41586-020-2762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cashel M, Gallant J. 1969. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221:838–841. 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 8.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA 70:1564–1568. 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. 2002. Dissection of the mechanism for the stringent factor RelA. Mol Cell 10:779–788. 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 10.Kushwaha GS, Bange G, Bhavesh NS. 2019. Interaction studies on bacterial stringent response protein RelA with uncharged tRNA provide evidence for its prerequisite complex for ribosome binding. Curr Genet 65:1173–1184. 10.1007/s00294-019-00966-y. [DOI] [PubMed] [Google Scholar]
- 11.Sy J, Lipmann F. 1973. Identification of the synthesis of guanosine tetraphosphate (MS I) as insertion of a pyrophosphoryl group into the 3’-position in guanosine 5′-diphosphate. Proc Natl Acad Sci USA 70:306–309. 10.1073/pnas.70.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinchen W, Bange G. 2016. The magic dance of the alarmones (p)ppGpp. Mol Microbiol 101:531–544. 10.1111/mmi.13412. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Bittner AN, Wang JD. 2015. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 24:72–79. 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinchen W, Zegarra V, Bange G. 2020. (p)ppGpp: magic modulators of bacterial physiology and metabolism. Front Microbiol 11:2072. 10.3389/fmicb.2020.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varik V, Oliveira SRA, Hauryliuk V, Tenson T. 2017. HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci Rep 7:11022. 10.1038/s41598-017-10988-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gratani FL, Horvatek P, Geiger T, Borisova M, Mayer C, Grin I, Wagner S, Steinchen W, Bange G, Velic A, Maček B, Wolz C. 2018. Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet 14:e1007514. 10.1371/journal.pgen.1007514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J-W, Park Y-H, Seok Y-J. 2018. Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc Natl Acad Sci USA 115:E6845–E6854. 10.1073/pnas.1722514115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronneau S, Caballero-Montes J, Coppine J, Mayard A, Garcia-Pino A, Hallez R. 2019. Regulation of (p)ppGpp hydrolysis by a conserved archetypal regulatory domain. Nucleic Acids Res 47:843–854. 10.1093/nar/gky1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada H, Roghanian M, Murina V, Dzhygyr I, Murayama R, Akanuma G, Atkinson GC, Garcia-Pino A, Hauryliuk V. 2020. The C-terminal RRM/ACT domain is crucial for fine-tuning the activation of “long” RelA-SpoT homolog enzymes by ribosomal complexes. Front Microbiol 11:277. 10.3389/fmicb.2020.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roghanian M, Van Nerom K, Takada H, Caballero-Montes J, Tamman H, Kudrin P, Talavera A, Dzhygyr I, Ekström S, Atkinson GC, Garcia-Pino A, Hauryliuk V. 2021. (p)ppGpp controls stringent factors by exploiting antagonistic allosteric coupling between catalytic domains. Mol Cell 81:3310–3322.e6. 10.1016/j.molcel.2021.07.026. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad S, Wang B, Walker MD, Tran HR, Stogios PJ, Savchenko A, Grant RA, McArthur AG, Laub MT, Whitney JC. 2019. An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575:674–678. 10.1038/s41586-019-1735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinchen W, Ahmad S, Valentini M, Eilers K, Majkini M, Altegoer F, Lechner M, Filloux A, Whitney JC, Bange G. 2021. Dual role of a (p)ppGpp- and (p)ppApp-degrading enzyme in biofilm formation and interbacterial antagonism. Mol Microbiol 115:1339–1356. 10.1111/mmi.14684. [DOI] [PubMed] [Google Scholar]
- 23.Zamecnik PC, Stephenson ML, Janeway CM, Randerath K. 1966. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem Biophys Res Commun 24:91–97. 10.1016/0006-291x(66)90415-3. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson F, McLennan AG, Urbaniak MD, Jones NJ, Copeland NA. 2020. Re-evaluation of diadenosine tetraphosphate (Ap4A) from a stress metabolite to bona fide secondary messenger. Front Mol Biosci 7:606807. 10.3389/fmolb.2020.606807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pálfi Z, Surányi G, Borbély G. 1991. Alterations in the accumulation of adenylylated nucleotides in heavy-metal-ion-stressed and heat-stressed Synechococcus sp. strain PCC 6301, a cyanobacterium, in light and dark. Biochem J 276:487–491. 10.1042/bj2760487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura Y, Tanaka C, Sasaki K, Sasaki M. 2017. High concentrations of intracellular Ap4A and/or Ap5A in developing Myxococcus xanthus cells inhibit sporulation. Microbiology (Reading) 163:86–93. 10.1099/mic.0.000403. [DOI] [PubMed] [Google Scholar]
- 27.Lee PC, Bochner BR, Ames BN. 1983. AppppA, heat-shock stress, and cell oxidation. Proc Natl Acad Sci USA 80:7496–7500. 10.1073/pnas.80.24.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji X, Zou J, Peng H, Stolle A-S, Xie R, Zhang H, Peng B, Mekalanos JJ, Zheng J. 2019. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc Natl Acad Sci USA 116:9578–9585. 10.1073/pnas.1822026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giammarinaro PI, Young MKM, Steinchen W, Mais C-N, Hochberg G, Yang J, Stevenson DM, Amador-Noguez D, Paulus A, Wang JD, Bange G. 2022. Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis. Nat Microbiol 7:1442–1452. 10.1038/s41564-022-01193-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borukhov S, Severinov K. 2002. Role of the RNA polymerase sigma subunit in transcription initiation. Res Microbiol 153:557–562. 10.1016/s0923-2508(02)01368-2. [DOI] [PubMed] [Google Scholar]
- 31.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. 2015. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 4:e08504. 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sojka L, Kouba T, Barvík I, Sanderová H, Maderová Z, Jonák J, Krásny L. 2011. Rapid changes in gene expression: DNA determinants of promoter regulation by the concentration of the transcription initiating NTP in Bacillus subtilis. Nucleic Acids Res 39:4598–4611. 10.1093/nar/gkr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaal T, Bartlett MS, Ross W, Turnbough CL, Gourse RL. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092–2097. 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 34.Murray HD, Schneider DA, Gourse RL. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12:125–134. 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 35.Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. 2006. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J Bacteriol 188:5775–5782. 10.1128/JB.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krásný L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23:4473–4483. 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natori Y, Tagami K, Murakami K, Yoshida S, Tanigawa O, Moh Y, Masuda K, Wada T, Suzuki S, Nanamiya H, Tozawa Y, Kawamura F. 2009. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC. J Bacteriol 191:4555–4561. 10.1128/JB.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasilyev N, Gao A, Serganov A. 2019. Noncanonical features and modifications on the 5′-end of bacterial sRNAs and mRNAs. Wiley Interdiscip Rev RNA 10:e1509. 10.1002/wrna.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doamekpor SK, Sharma S, Kiledjian M, Tong L. 2022. Recent insights into noncanonical 5’ capping and decapping of RNA. J Biol Chem 298:102171. 10.1016/j.jbc.2022.102171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bird JG, Zhang Y, Tian Y, Panova N, Barvík I, Greene L, Liu M, Buckley B, Krásný L, Lee JK, Kaplan CD, Ebright RH, Nickels BE. 2016. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535:444–447. 10.1038/nature18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malygin AG, Shemyakin MF. 1979. Adenosine, NAD and FAD can initiate template-dependent RNA synthesis catalyzed by Escherichia coli RNA polymerase. FEBS Lett 102:51–54. 10.1016/0014-5793(79)80926-6. [DOI] [PubMed] [Google Scholar]
- 42.Julius C, Yuzenkova Y. 2017. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res 45:8282–8290. 10.1093/nar/gkx452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. 2009. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol 5:879–881. 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR. 2009. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc Natl Acad Sci USA 106:7768–7773. 10.1073/pnas.0900528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahová H, Winz M-L, Höfer K, Nübel G, Jäschke A. 2015. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519:374–377. 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 46.Luciano DJ, Levenson-Palmer R, Belasco JG. 2019. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol Cell 75:957–966.e8. 10.1016/j.molcel.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luciano DJ, Belasco JG. 2020. Np4A alarmones function in bacteria as precursors to RNA caps. Proc Natl Acad Sci USA 117:3560–3567. 10.1073/pnas.1914229117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikemura T, Dahlberg JE. 1973. Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem 248:5033–5041. 10.1016/S0021-9258(19)43667-3. [DOI] [PubMed] [Google Scholar]
- 49.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190:1084–1096. 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bange G, Brodersen DE, Liuzzi A, Steinchen W. 2021. Two P or not two P: understanding regulation by the bacterial second messengers (p)ppGpp. Annu Rev Microbiol 75:383–406. 10.1146/annurev-micro-042621-122343. [DOI] [PubMed] [Google Scholar]
- 51.Wang B, Grant RA, Laub MT. 2020. ppGpp coordinates nucleotide and amino-acid synthesis in E. coli during starvation. Mol Cell 80:29–42.e10. 10.1016/j.molcel.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jovanovic SV, Simic MG. 1986. One-electron redox potentials of purines and pyrimidines. J Phys Chem 90:974–978. 10.1021/j100277a053. [DOI] [Google Scholar]
- 53.Liu M, Gong X, Alluri RK, Wu J, Sablo T, Li Z. 2012. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol Chem 393:123–132. 10.1515/hsz-2011-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altman S. 1975. Biosynthesis of transfer RNA in Escherichia coli. Cell 4:21–29. 10.1016/0092-8674(75)90129-4. [DOI] [PubMed] [Google Scholar]
- 55.Mörl M, Marchfelder A. 2001. The final cut. The importance of tRNA 3’-processing. EMBO Rep 2:17–20. 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung YH, Lee Y. 1997. Escherichia coli rnpB promoter mutants altered in stringent response. Biochem Biophys Res Commun 230:582–586. 10.1006/bbrc.1996.6005. [DOI] [PubMed] [Google Scholar]
- 57.Kim Y, Lee Y. 2009. Novel function of C5 protein as a metabolic stabilizer of M1 RNA. FEBS Lett 583:419–424. 10.1016/j.febslet.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 58.Schedl P, Roberts J, Primakoff P. 1976. In vitro processing of E. coli tRNA precursors. Cell 8:581–594. 10.1016/0092-8674(76)90226-9. [DOI] [PubMed] [Google Scholar]
- 59.Shepherd J, Ibba M. 2015. Bacterial transfer RNAs. FEMS Microbiol Rev 39:280–300. 10.1093/femsre/fuv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol 407:633–639. 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki T. 2021. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol 22:375–392. 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 62.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. 2003. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell 12:689–698. 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 63.Nakanishi K, Bonnefond L, Kimura S, Suzuki T, Ishitani R, Nureki O. 2009. Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase. Nature 461:1144–1148. 10.1038/nature08474. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Agrebi R, Bellows LE, Collet J-F, Kaever V, Gründling A. 2017. Evolutionary adaptation of the essential tRNA methyltransferase TrmD to the signaling molecule 3’,5′-cAMP in bacteria. J Biol Chem 292:313–327. 10.1074/jbc.M116.758896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wirth R, Kohles V, Böck A. 1981. Factors modulating transcription and translation in vitro of ribosomal protein S20 and isoleucyl-tRNA synthetase from Escherichia coli. Eur J Biochem 114:429–437. 10.1111/j.1432-1033.1981.tb05164.x. [DOI] [PubMed] [Google Scholar]
- 66.Williams AL, Barnett RS. 1985. Evidence for cAMP-mediated control of isoleucyl-tRNA synthetase formation in Escherichia coli K-12. Arch Microbiol 142:190–193. 10.1007/BF00447066. [DOI] [PubMed] [Google Scholar]
- 67.Gualerzi CO, Pon CL. 2015. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell Mol Life Sci 72:4341–4367. 10.1007/s00018-015-2010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milon P, Carotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO. 2010. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep 11:312–316. 10.1038/embor.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goyal A, Belardinelli R, Maracci C, Milón P, Rodnina MV. 2015. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res 43:10700–10712. 10.1093/nar/gkv869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milon P, Tischenko E, Tomsic J, Caserta E, Folkers G, La Teana A, Rodnina MV, Pon CL, Boelens R, Gualerzi CO. 2006. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci USA 103:13962–13967. 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hauryliuk V, Mitkevich VA, Draycheva A, Tankov S, Shyp V, Ermakov A, Kulikova AA, Makarov AA, Ehrenberg M. 2009. Thermodynamics of GTP and GDP binding to bacterial initiation factor 2 suggests two types of structural transitions. J Mol Biol 394:621–626. 10.1016/j.jmb.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 72.Mitkevich VA, Ermakov A, Kulikova AA, Tankov S, Shyp V, Soosaar A, Tenson T, Makarov AA, Ehrenberg M, Hauryliuk V. 2010. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J Mol Biol 402:838–846. 10.1016/j.jmb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 73.Vinogradova DS, Zegarra V, Maksimova E, Nakamoto JA, Kasatsky P, Paleskava A, Konevega AL, Milón P. 2020. How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol 18:e3000593. 10.1371/journal.pbio.3000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landwehr V, Milanov M, Angebauer L, Hong J, Jüngert G, Hiersemenzel A, Siebler A, Schmit F, Öztürk Y, Dannenmaier S, Drepper F, Warscheid B, Koch HG. 2021. The universally conserved ATPase YchF regulates translation of leaderless mRNA in response to stress conditions. Front Mol Biosci 8:643696. 10.3389/fmolb.2021.643696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wenk M, Ba Q, Erichsen V, MacInnes K, Wiese H, Warscheid B, Koch HG. 2012. A universally conserved ATPase regulates the oxidative stress response in Escherichia coli. J Biol Chem 287:43585–43598. 10.1074/jbc.m112.413070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rojas AM, Ehrenberg M, Andersson SG, Kurland CG. 1984. ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. Mol Gen Genet 197:36–45. 10.1007/BF00327920. [DOI] [PubMed] [Google Scholar]
- 77.Anborgh PH, Okamura S, Parmeggiani A. 2004. Effects of the antibiotic pulvomycin on the elongation factor Tu-dependent reactions. Comparison with other antibiotics. Biochemistry 43:15550–15556. 10.1021/bi0487084. [DOI] [PubMed] [Google Scholar]
- 78.Miller DL, Cashel M, Weissbach H. 1973. The interaction of guanosine 5′-diphosphate, 2’ (3’)-diphosphate with the bacterial elongation factor Tu. Arch Biochem Biophys 154:675–682. 10.1016/0003-9861(73)90022-2. [DOI] [PubMed] [Google Scholar]
- 79.Hauryliuk V, Mitkevich VA, Eliseeva NA, Petrushanko IY, Ehrenberg M, Makarov AA. 2008. The pretranslocation ribosome is targeted by GTP-bound EF-G in partially activated form. Proc Natl Acad Sci USA 105:15678–15683. 10.1073/pnas.0807912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Farrell PH. 1978. The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14:545–557. 10.1016/0092-8674(78)90241-6. [DOI] [PubMed] [Google Scholar]
- 81.Wagner EG, Ehrenberg M, Kurland CG. 1982. Kinetic suppression of translational errors by (p)ppGpp. Mol Gen Genet 185:269–274. [DOI] [PubMed] [Google Scholar]
- 82.Dix DB, Thompson RC. 1986. Elongation factor Tu.guanosine 3’-diphosphate 5′-diphosphate complex increases the fidelity of proofreading in protein biosynthesis: mechanism for reducing translational errors introduced by amino acid starvation. Proc Natl Acad Sci USA 83:2027–2031. 10.1073/pnas.83.7.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner EG, Kurland CG. 1980. Translational accuracy enhanced in vitro by (p)ppGpp. Mol Gen Genet 180:139–145. 10.1007/BF00267363. [DOI] [PubMed] [Google Scholar]
- 84.Mohler K, Ibba M. 2017. Translational fidelity and mistranslation in the cellular response to stress. Nat Microbiol 2:17117. 10.1038/nmicrobiol.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gibbs MR, Moon K-M, Chen M, Balakrishnan R, Foster LJ, Fredrick K. 2017. Conserved GTPase LepA (elongation factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc Natl Acad Sci USA 114:980–985. 10.1073/pnas.1613665114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Zborníková E, Rejman D, Gerdes K. 2018. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9:e02188-17. 10.1128/mBio.02188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10:a032664. 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zavialov AV, Buckingham RH, Ehrenberg M. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107:115–124. 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 89.Koutmou KS, McDonald ME, Brunelle JL, Green R. 2014. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA 20:609–620. 10.1261/rna.042523.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. 2014. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res 42:1812–1820. 10.1093/nar/gkt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kihira K, Shimizu Y, Shomura Y, Shibata N, Kitamura M, Nakagawa A, Ueda T, Ochi K, Higuchi Y. 2012. Crystal structure analysis of the translation factor RF3 (release factor 3). FEBS Lett 586:3705–3709. 10.1016/j.febslet.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 92.Kiel MC, Kaji H, Kaji A. 2007. Ribosome recycling: an essential process of protein synthesis. Biochem Mol Biol Educ 35:40–44. 10.1002/bmb.6. [DOI] [PubMed] [Google Scholar]
- 93.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JHD. 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332:981–984. 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. 2005. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell 18:663–674. 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y, Mandava CS, Cao W, Li X, Zhang D, Li N, Zhang Y, Zhang X, Qin Y, Mi K, Lei J, Sanyal S, Gao N. 2015. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat Struct Mol Biol 22:906–913. 10.1038/nsmb.3103. [DOI] [PubMed] [Google Scholar]
- 96.Coatham ML, Brandon HE, Fischer JJ, Schümmer T, Wieden H-J. 2016. The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res 44:1952–1961. 10.1093/nar/gkv1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hirokawa G, Iwakura N, Kaji A, Kaji H. 2008. The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G). Nucleic Acids Res 36:6676–6687. 10.1093/nar/gkn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fontaine BM, Martin KS, Garcia-Rodriguez JM, Jung C, Briggs L, Southwell JE, Jia X, Weinert EE. 2018. RNase I regulates Escherichia coli 2’,3’-cyclic nucleotide monophosphate levels and biofilm formation. Biochem J 475:1491–1506. 10.1042/BCJ20170906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duggal Y, Kurasz JE, Fontaine BM, Marotta NJ, Chauhan SS, Karls AC, Weinert EE. 2022. Cellular effects of 2’,3’-cyclic nucleotide monophosphates in Gram-negative bacteria. J Bacteriol 204:e00208-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chauhan SS, Marotta NJ, Karls AC, Weinert EE. 2022. Binding of 2′,3′-cyclic nucleotide monophosphates to bacterial ribosomes inhibits translation. ACS Cent Sci 8:1518–1526. 10.1021/acscentsci.2c00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang B, Dai P, Ding D, Del Rosario A, Grant RA, Pentelute BL, Laub MT. 2019. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol 15:141–150. 10.1038/s41589-018-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Corrigan RM, Bellows LE, Wood A, Gründling A. 2016. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci USA 113:E1710–E1719. 10.1073/pnas.1522179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Orr MW, Lee VT. 2017. Differential radial capillary action of ligand assay (DRaCALA) for high-throughput detection of protein-metabolite interactions in bacteria. Methods Mol Biol 1535:25–41. 10.1007/978-1-4939-6673-8_3. [DOI] [PubMed] [Google Scholar]
- 104.Anderson BW, Schumacher MA, Yang J, Turdiev A, Turdiev H, Schroeder JW, He Q, Lee VT, Brennan RG, Wang JD. 2022. The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus. Nucleic Acids Res 50:847–866. 10.1093/nar/gkab1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nomura M. 1999. Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: diversity and common principles. J Bacteriol 181:6857–6864. 10.1128/JB.181.22.6857-6864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shin Y, Qayyum MZ, Pupov D, Esyunina D, Kulbachinskiy A, Murakami KS. 2021. Structural basis of ribosomal RNA transcription regulation. Nat Commun 12:528. 10.1038/s41467-020-20776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nomura M, Yates JL, Dean D, Post LE. 1980. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc Natl Acad Sci USA 77:7084–7088. 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu Y, Deiorio-Haggar K, Anthony J, Meyer MM. 2013. Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucleic Acids Res 41:3491–3503. 10.1093/nar/gkt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Jesus V, Qureshi NS, Warhaut S, Bains JK, Dietz MS, Heilemann M, Schwalbe H, Fürtig B. 2021. Switching at the ribosome: riboswitches need rProteins as modulators to regulate translation. Nat Commun 12:4723. 10.1038/s41467-021-25024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaczanowska M, Rydén-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71:477–494. 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gibbs MR, Moon K-M, Warner BR, Chen M, Bundschuh R, Foster LJ, Fredrick K. 2020. Functional analysis of BipA in E. coli reveals the natural plasticity of 50S subunit assembly. J Mol Biol 432:5259–5272. 10.1016/j.jmb.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zegarra V, Bedrunka P, Bange G, Czech L. 2023. How to save a bacterial ribosome in times of stress. Semin Cell Dev Biol 136:3–12. 10.1016/j.semcdb.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 113.Feng B, Mandava CS, Guo Q, Wang J, Cao W, Li N, Zhang Y, Zhang Y, Wang Z, Wu J, Sanyal S, Lei J, Gao N. 2014. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol 12:e1001866. 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamamoto H, Wittek D, Gupta R, Qin B, Ueda T, Krause R, Yamamoto K, Albrecht R, Pech M, Nierhaus KH. 2016. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc Natl Acad Sci USA 113:E1180–E1189. 10.1073/pnas.1524554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Leiva LE, Katz A. 2022. Regulation of leaderless mRNA translation in bacteria. Microorganisms 10:723. 10.3390/microorganisms10040723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705–708. 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 117.Kuroda A, Murphy H, Cashel M, Kornberg A. 1997. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem 272:21240–21243. 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 118.Rao NN, Liu S, Kornberg A. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol 180:2186–2193. 10.1128/JB.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lyman GE, DeVincenzo JP. 1967. Determination of picogram amounts of ATP using the luciferin-luciferase enzyme system. Anal Biochem 21:435–443. 10.1016/0003-2697(67)90318-1. [DOI] [PubMed] [Google Scholar]
- 120.Bochner BR, Ames BN. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem 257:9759–9769. 10.1016/S0021-9258(18)34138-3. [DOI] [PubMed] [Google Scholar]
- 121.Geldart SE, Brown PR. 1998. Analysis of nucleotides by capillary electrophoresis. J Chromatogr A 828:317–336. 10.1016/s0021-9673(98)00633-5. [DOI] [PubMed] [Google Scholar]
- 122.Strzelecka D, Chmielinski S, Bednarek S, Jemielity J, Kowalska J. 2017. Analysis of mononucleotides by tandem mass spectrometry: investigation of fragmentation pathways for phosphate- and ribose-modified nucleotide analogues. Sci Rep 7:8931. 10.1038/s41598-017-09416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen P, Liu Z, Liu S, Xie Z, Aimiuwu J, Pang J, Klisovic R, Blum W, Grever MR, Marcucci G, Chan KK. 2009. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharm Res 26:1504–1515. 10.1007/s11095-009-9863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dudley E, Bond L. 2014. Mass spectrometry analysis of nucleosides and nucleotides. Mass Spectrom Rev 33:302–331. 10.1002/mas.21388. [DOI] [PubMed] [Google Scholar]
- 125.Annibal A, Ripa R, Ballhysa E, Latza C, Hochhard N, Antebi A. 2021. Mass spectrometric characterization of cyclic dinucleotides (CDNs) in vivo. Anal Bioanal Chem 413:6457–6468. 10.1007/s00216-021-03628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Antoniewicz MR. 2015. Methods and advances in metabolic flux analysis: a mini-review. J Ind Microbiol Biotechnol 42:317–325. 10.1007/s10295-015-1585-x. [DOI] [PubMed] [Google Scholar]
- 127.Zborníková E, Knejzlík Z, Hauryliuk V, Krásný L, Rejman D. 2019. Analysis of nucleotide pools in bacteria using HPLC-MS in HILIC mode. Talanta 205:120161. 10.1016/j.talanta.2019.120161. [DOI] [PubMed] [Google Scholar]
- 128.Louie TM, Xie XS, Xun L. 2003. Coordinated production and utilization of FADH2 by NAD(P)H-flavin oxidoreductase and 4-hydroxyphenylacetate 3-monooxygenase. Biochemistry 42:7509–7517. 10.1021/bi034092r. [DOI] [PubMed] [Google Scholar]
- 129.Barvík I, Rejman D, Panova N, Šanderová H, Krásný L. 2017. Non-canonical transcription initiation: the expanding universe of transcription initiating substrates. FEMS Microbiol Rev 41:131–138. 10.1093/femsre/fuw041. [DOI] [PubMed] [Google Scholar]
- 130.Bochner BR, Lee PC, Wilson SW, Cutler CW, Ames BN. 1984. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37:225–232. 10.1016/0092-8674(84)90318-0. [DOI] [PubMed] [Google Scholar]
- 131.Saier MH, Feucht BU, McCaman MT. 1975. Regulation of intracellular adenosine cyclic 3’:5′-monophosphate levels in Escherichia coli and Salmonella typhimurium. Evidence for energy-dependent excretion of the cyclic nucleotide. J Biol Chem 250:7593–7601. 10.1016/S0021-9258(19)40859-4. [DOI] [PubMed] [Google Scholar]
- 132.Reinders A, Hee C-S, Ozaki S, Mazur A, Boehm A, Schirmer T, Jenal U. 2016. Expression and genetic activation of cyclic di-GMP-specific phosphodiesterases in Escherichia coli. J Bacteriol 198:448–462. 10.1128/JB.00604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Massie JP, Reynolds EL, Koestler BJ, Cong J-P, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci USA 109:12746–12751. 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lauhon CT, Erwin WM, Ton GN. 2004. Substrate specificity for 4-thiouridine modification in Escherichia coli. J Biol Chem 279:23022–23029. 10.1074/jbc.M401757200. [DOI] [PubMed] [Google Scholar]
- 135.Yu F, Tanaka Y, Yamashita K, Suzuki T, Nakamura A, Hirano N, Suzuki T, Yao M, Tanaka I. 2011. Molecular basis of dihydrouridine formation on tRNA. Proc Natl Acad Sci USA 108:19593–19598. 10.1073/pnas.1112352108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Persson BC, Jäger G, Gustafsson C. 1997. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2’-O-methyltransferase activity. Nucleic Acids Res 25:4093–4097. 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bouvier D, Labessan N, Clémancey M, Latour J-M, Ravanat J-L, Fontecave M, Atta M. 2014. TtcA a new tRNA-thioltransferase with an Fe-S cluster. Nucleic Acids Res 42:7960–7970. 10.1093/nar/gku508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pang P, Deng X, Wang Z, Xie W. 2017. Structural and biochemical insights into the 2’-O-methylation of pyrimidines 34 in tRNA. FEBS J 284:2251–2263. 10.1111/febs.14120. [DOI] [PubMed] [Google Scholar]
- 139.Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, Tanaka I. 2009. RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J 28:1362–1373. 10.1038/emboj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Blaise M, Becker HD, Lapointe J, Cambillau C, Giegé R, Kern D. 2005. Glu-Q-tRNA(Asp) synthetase coded by the yadB gene, a new paralog of aminoacyl-tRNA synthetase that glutamylates tRNA(Asp) anticodon. Biochimie 87:847–861. 10.1016/j.biochi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 141.De Bie LGS, Roovers M, Oudjama Y, Wattiez R, Tricot C, Stalon V, Droogmans L, Bujnicki JM. 2003. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J Bacteriol 185:3238–3243. 10.1128/JB.185.10.3238-3243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Keffer-Wilkes LC, Soon EF, Kothe U. 2020. The methyltransferase TrmA facilitates tRNA folding through interaction with its RNA-binding domain. Nucleic Acids Res 48:7981–7990. 10.1093/nar/gkaa548. [DOI] [PMC free article] [PubMed] [Google Scholar]