Abstract
The DNA binding properties of hMutSα and hMutLα and complex formation of hMutSα with hMutLα and hMutLβ were investigated using binding experiments on magnetic bead-coupled DNA substrates with nuclear extracts as well as purified proteins. hMutSα binding to homoduplex DNA was disrupted by lower NaCl concentrations than hMutSα binding to a mismatch. ATP markedly reduced the salt resistance of hMutSα binding but hMutSα still retained affinity for heteroduplexes. hMutSα formed a complex with hMutLα and hMutLβ on DNA in the presence of ATP. This complex only formed on 81mer and not 32mer DNA substrates. Complex formation was enhanced by a mismatch in the DNA substrate, and hMutLα and hMutLβ were shown to enter the complex at different ATP concentrations. Purified hMutLα showed an intrinsic affinity for DNA, with a preference for single-stranded over double-stranded DNA.
INTRODUCTION
DNA mismatch repair is essential for the maintenance of replication fidelity and is conserved through all organisms from bacteria to humans. Its major task is to recognize mismatches as well as insertion/deletion loops which have escaped the polymerase proofreading activity in newly synthesized DNA, and to accomplish the repair of these mistakes. Bacterial mismatch repair, which has been reconstituted with purified proteins (1), is essentially carried out by the two homodimeric proteins MutS and MutL and the endonuclease MutH. MutS has been shown to recognize and bind the mismatch (2), followed by ATP-dependent recruitment of MutL (3). This complex subsequently activates the exonuclease MutH (4,5), which initiates repair of the faulty DNA area.
The homodimeric MutS protein, which exhibits an asymmetrical conformation on mismatched DNA (6,7), has evolved as heterodimers in eukaryotes and two of these (hMutSα, consisting of hMSH2 and hMSH6, and hMutSβ, consisting of hMSH2 and hMSH3) have been implicated in human mismatch repair. Human MutLα (hMLH1 paired with hPMS2) as well as hMutLβ (hMLH1 paired with hPMS1) correspond to bacterial MutL. While hMutLα alone is able to confer complete mismatch repair proficiency on hMLH1-deficient extracts (8), data for a possible contribution of hMutLβ, which is 10 times less abundant than hMutLα in HeLa nuclear extracts, to mismatch repair are conflicting (9–11).
MutS and its homologs contain a conserved ATP-binding cassette ATPase site (6,7). ATP has been observed to abolish MutS protein binding to mismatches (12–16). Further investigations indicated that MutS proteins show a translocating or sliding mode on DNA after mismatch recognition and ATP uptake, either as an ATP-driven engine (translocation model; 13) or by diffusion along DNA with the bound ATP transmitting a repair signal in a manner similar to G proteins (sliding clamp model; 17). Recently, a third model (DNA bending model; 18) was proposed, based on the findings that ATP uptake by bacterial MutS and binding to a mismatch may not be mutually exclusive (19,20) and that translocation along a DNA helix is not required in mismatch repair, since MutH activation can occur in trans (18). This model suggests that MutS remains bound to the mismatch site even after uptake of ATP, and that ATP imparts verification of the mismatch by decreasing the affinity of MutS for homoduplex DNA more than the affinity of MutS for heteroduplex DNA. In this model MutS can only activate MutH through MutL when a mismatch and ATP are bound at the same time.
MutL proteins also contain an ATPase site in each subunit. Some results indicate preferential binding of Escherichia coli MutL to single-stranded DNA (21,22) and an ATP-regulated clamp mechanism for single-stranded DNA has been suggested (22). Furthermore, MutL was shown to load DNA helicase II onto DNA (23) and to activate it in a mismatch-dependent manner (24), which promoted the development of a model of MutL inducing DNA unwinding and passing of one strand to an exonuclease (23,24). Moreover, MutL and its eukaryotic homologs have been suggested to be molecular matchmakers that signal mismatch recognition to downstream proteins responsible for excision and repair. After mismatch recognition MutS has to interact with MutL or the respective eukaryotic counterparts, and evidence for this interaction in the form of ternary complexes including MutS and MutL proteins and DNA has been provided for the bacterial, yeast and human proteins (3,13,25–32).
Conflicting results have been provided for the role of ATP in the interaction of human MutL and MutS homologs. One report showed that ATP hydrolysis is necessary for complex formation of hMSH2 and hMLH1 in HeLa extracts (28), while another found that hMutSα forms complexes with hMutLα and hMutLβ on mismatched DNA that are abolished by addition of ATP (29). Recently, an ATP- and DNA-length-dependent assembly of human MutSα and MutLα was reported (32). It was the aim of this work: (i) to establish an experimental system to examine binding of hMutSα and hMutLα to DNA; (ii) to investigate the conditions under which the interaction between hMutSα and hMutL heterodimers occurs; (iii) to clarify the role of hMutLβ in complex formation.
MATERIALS AND METHODS
Antibodies and reagents
Poly[d(I-C)] was purchased from Boehringer Mannheim (Mannheim, Germany). ADP, ATP, AMP-PNP and ATP-γ-S were from Sigma-Aldrich (Steinheim, Germany). Anti-hMLH1 (G168-728) and anti-hPMS2 (A16-4) were from Pharmingen (San Diego, CA). Anti-hMSH2 (M34520), anti-hMSH6 (G70220), anti-hMSH3 (M94120) and anti-hPCNA (P56720) were purchased from Transduction Laboratories (Lexington, KY). hPMS1 polyclonal antibody (11) and baculovirus-expressed purified wild-type hMutSα (33) and hMutLα (11) were kindly provided by Dr Josef Jiricny (University of Zürich, Switzerland).
Cell lines and nuclear extract preparation
HeLa cells were purchased from DMSZ (Braunschweig, Germany). Hec59, TK6 and MT1 cell lines were kindly supplied by Dr Josef Jiricny. HeLa, TK6, MT1 and LoVo (ATCC, Manassas, VA) cells were cultured in RPMI 1640 medium with 10% FCS. HCT-116 and HCT-116+ch3 (34) cells were kindly supplied by Dr C.Richard Boland (University of California, CA) and grown in DMEM with 10% FCS, which was supplemented with 0.4 mg/ml G-418 for HCT-116+ch3 cells. Hec59 cells were cultured in 40% DMEM F-12, 40% DMEM, 20% FCS and 1% l-glutamine.
Nuclear extraction was performed essentially as described by Dignam et al. (35). Briefly, cells were harvested and collected by centrifugation at 700 g for 5 min. All subsequent steps were performed on ice. The supernatant was removed and the cells were resuspended in three times the packed cell volume of hypotonic buffer (10 mM HEPES–KOH pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 5 mM NaF) and incubated for 10 min. Cells were lysed using a Dounce homogenizer until >90% of cells were lysed, as determined by Trypan blue staining. Nuclei were pelleted by centrifugation (3000 g, 5 min) and the supernatant was removed. The pellet was again centrifuged for 2 min at 22 000 g to remove residual cytoplasmatic extract. Nuclei were resuspended in extraction buffer (20 mM HEPES–KOH pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 5 mM NaF, 0.32 mM EDTA, 25% v/v glycerol) and kept with agitation on ice for 30 min. After centrifugation (22 000 g, 10 min), the protein concentration of the supernatant (nuclear extract) was measured and aliquots were stored at –80°C.
Oligonucleotides and preparation of DNA-coupled magnetic beads
All oligonucleotides were synthesized and, when appropriate, 5′-labeled with biotin by BioSpring (Frankfurt, Germany). The following oligonucleotides were used: biotin-5′-GCG CAC TCT TGC CCA CAC CGC CGG CGC CCA CC-3′ (29) and biotin-5′-AAA GCT GGA GCA GAA GCT TAG CTT AGG TAC ATC GAG GAT GGA CCT CGG AGC AAT TCT GCG GTA CCC TAT TCG CCC TAT AGT-3′. Bold letters indicate the position of the G-T mismatch and italic letters the area that remained single stranded in specific experiments. Duplexes were created by annealing these oligomers with their antisense oligomers as described previously (29). All oligonucleotides were coupled to Dynabeads M-280 Streptavidin (Dynal, Oslo, Norway) according to the protocol of the manufacturer. Incubations were performed with 50 pmol DNA substrate per mg Dynabeads, thus remaining below the binding capacity of the beads (200 pmol single-stranded oligomers/mg). The coupling efficiency was evaluated by comparing the DNA content before and after incubation using 15% polyacrylamide gels and silver staining. While 32mer single- and double-stranded oligomers exhibited complete binding to the beads, the binding of 81mer double-stranded DNA achieved ∼90% binding. No difference in binding efficiency was detectable between coupling of homoduplex versus heteroduplex substrates.
DNA binding assay
Before use, DNA-coupled Dynabeads were washed three times with washing buffer (20 mM Tris–HCl pH 7.9, 50 mM NaCl, 5% glycerol, 1 mM EDTA) and equal volumes were pipetted into 1.5 ml cups. After collecting the beads with a magnet the supernatant was carefully removed. Binding reactions were carried out with 50 or 100 µg nuclear extract as indicated in the individual experiments. For supplementation experiments, 185 ng purified hMutSα or 300 ng purified hMutLα were added. Nuclear extract was added to the premixed binding buffer resulting in a final composition of 20 mM Tris–HCl pH 7.9, 50 mM NaCl, 1.5 mM MgCl2, 5% glycerol, 1 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT and 1 µg poly[d(I-C)] in a total volume of 300 µl. In experiments using the same extracts or proteins, the binding mixture was prepared for all samples in one vial and then aliquoted into separate cups to avoid any variation in the incubation mixture. The mixture was incubated for 10 min at room temperature and then for 10 min on ice. Poly[d(I-C)] and the preincubation steps were omitted in experiments using only purified proteins. All subsequent steps were performed on ice. Each reaction mixture was added to aliquots of 670 µg Dynabeads coupled with substrate DNA and incubated for 25 min. In experiments including ATP, this was added now to a final concentration of 250 µM, and all samples were further incubated for 10 min if not specified otherwise. Afterwards, beads were collected with a magnet and the supernatant was removed. The cup was centrifuged, the beads collected and the remaining supernatant again taken off. For elution, the beads were resuspended in 20 µl of elution buffer (20 mM Tris–HCl pH 7.9, 1.5 mM MgCl2, 5% glycerol, 1 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT) with the NaCl concentration indicated for the individual experiments and incubated for 5 min. In experiments examining the salt resistance of the hMutSα–DNA complex in the presence of ATP, 250 µM ATP was included in the washing buffers. As shown in experiments without DNA on the beads, non-specific binding of mismatch repair proteins was negligible. Furthermore, experiments with several samples treated identically in parallel also showed identical signals on one blot, providing evidence that this procedure does not produce significant variations during sample processing.
Western blotting
The proteins eluted in the above assay were separated on 10% polyacrylamide gels, followed by western blotting on nitrocellulose membranes and antibody detection using standard procedures. Because PMS1 runs to only a slightly higher molecular weight than PMS2, both proteins were detected consecutively (first PMS1, then, after stripping of the membrane, PMS2).
RESULTS
DNA binding properties of hMutSα, hMutSβ and hMutLα
To confirm that human mismatch repair proteins bind DNA substrates coupled to magnetic beads, we incubated either nuclear extracts of the mismatch repair-proficient cell line TK6 or purified hMutLα with beads coupled with 32mer single-stranded or homoduplex DNA. Furthermore, the effect on binding of different adenine nucleotides (ADP, ATP, ATP-γ-S and AMP-PNP, each at 250 µM) was assessed. Complete elution of bound proteins was carried out with 700 mM NaCl. Beads boiled in SDS sample buffer after this elution did not show any signal (data not shown). Furthermore, the assay showed only DNA-bound mismatch repair proteins, since beads without DNA produced negligible signal (Fig. 1).
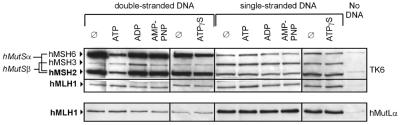
Figure 1.
DNA binding properties of hMutSα, hMutSβ and hMutLα. 32mer single-stranded or homoduplex DNA was coupled to magnetic beads and incubated with either 100 µg TK6 cell nuclear extract (upper) or 600 ng purified hMutLα (lower). After incubation for 25 min, ATP, ADP, AMP-PNP or ATP-γ-S (250 µM each) or no nucleotide (Ø) was added and bound proteins were eluted with 700 mM NaCl 10 min afterwards. The same experiment was performed in parallel with beads without DNA (No DNA). Western blots of the elution were probed for hMSH2, hMSH3, hMSH6 and hMLH1. Corresponding lanes for single- and double-stranded DNA were analyzed on the same western blot and rearranged afterwards.
As expected, hMutSα (hMSH2–hMSH6) displayed higher affinity for double-stranded DNA, which was reduced by ATP. While AMP-PNP did not affect hMutSα binding, ATP-γ-S also had a depressing effect, which is in agreement with previous findings (36). Although the effect of ATP-γ-S was much weaker than that of ATP, it was reproducible and became more intense after longer incubation times (data not shown). No hMutSβ (hMSH2–hMSH3) bound to the homoduplex substrate, while some affinity for single-stranded DNA was observed, which may reflect the preference of hMutSβ for extrahelical loops. Interestingly, binding of hMSH3 to double-stranded DNA became detectable in the presence of ADP [although the expected concomitant rise in the signal of hMSH2 could not be detected due to the significantly higher abundance of hMSH2–hMSH6 (hMutSα) in cell extracts (37)]. This may reflect an ADP-transmitted change in the hMutSβ–homoduplex interaction which did not occur with hMutSα under the assay conditions used. While hardly any hMLH1 from cell extracts bound to the substrates, hMutLα bound efficiently when used as a purified protein, which is probably attributable to the absence of competition by other DNA-binding proteins. hMutLα exhibited a three times higher affinity for single- than double-stranded DNA, as judged by densitometric evaluation of the western blot. The binding affinity of hMutLα was not affected by addition of adenine nucleotides.
hMutSα bound to a mismatch exhibits higher salt resistance than hMutSα bound to homoduplex DNA
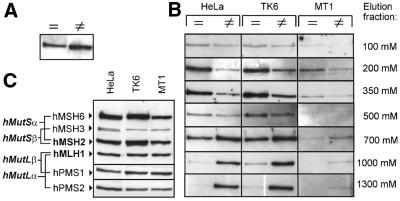
To investigate whether hMutSα binds differently to homo- and heteroduplexes in this assay, magnetic bead-coupled 32mer oligoduplex substrates, containing either a G-T mismatch or the correct base pairing, were tested for their ability to bind hMutSα by incubation with nuclear extracts from HeLa cells, which are proficient in mismatch repair. Slightly more hMutSα was retrieved from hetero- than from homoduplexes by elution with 1.5 M NaCl (Fig. 2A, hMSH2 shown; hMSH6 reacted identically, data not shown). No more mismatch repair proteins could be retrieved by boiling the beads in SDS sample buffer after the 1.5 M elution, indicating that this salt concentration was sufficient to disrupt binding of the investigated proteins to both DNA substrates. The finding that both substrates bound hMutSα efficiently is consistent with earlier reports showing that hMutSα not only binds mismatches, but also has homoduplex affinity (36). Consecutive elution with increasing salt concentrations from 100 to 1300 mM NaCl after incubation with extracts from the repair-proficient cell lines HeLa and TK6 and repair-deficient TK6 clone MT1 revealed different elution profiles of hMutSα from mismatched and correctly paired substrates. Binding of hMutSα to homoduplex DNA was disrupted at 200–700 mM NaCl, while hMutSα required 700–1300 mM NaCl to be eluted from heteroduplexes (Fig. 1B, hMSH2 shown; hMSH6 reacted identically, data not shown). hMutSα from MT1 cells, whose hMSH6 genes carry two missense mutations (38), reacted qualitatively similarly to wild-type protein, but showed reduced binding, which may be attributable either to decreased abundance of hMutSα in MT1 extracts (Fig. 1C) or to an impaired DNA binding ability of the mutant heterodimer. Similar elution profiles were obtained for 81mer duplexes (data not shown). While hMSH2 and hMSH6 reacted in parallel in these experiments, hMSH3 was again undetectable on homo- and heteroduplexes (data not shown). Although mismatch-bound hMutSα exhibited high salt resistance, the initial binding to mismatches was not efficient above 200 mM NaCl (data not shown), confirming previous studies (39).
Figure 2.
hMutSα bound to a mismatch exhibits higher salt resistance than hMutSα bound to homoduplex DNA. (A) HeLa nuclear extract (50 µg) was incubated with magnetic beads coupled with either 32mer homoduplex (=) or 32mer heteroduplex (≠) DNA as described in Materials and Methods. Bound proteins were eluted with 1500 mM NaCl. The western blot of hMSH2 in this eluate is shown. (B) HeLa, TK6 and MT1 extracts (50 µg) were incubated with beads coupled with the same DNA substrates and bound proteins were eluted consecutively with salt concentrations from 100 to 1300 mM NaCl. The hMSH2 western blots of the eluate fractions are shown. (C) Western blots of HeLa, TK6 and MT1 nuclear extracts (25 µg each).
ATP reduces the salt resistance of hMutSα binding but hMutSα retains affinity for heteroduplexes
To investigate the effect of ATP on homo- and heteroduplex binding of hMutSα in our assay, we added 250 µM ATP to the binding reaction. ATP immediately (elution 1 min after addition of ATP) disrupted the highly salt-resistant binding of HeLa hMutSα to mismatched 32mer DNA beads, and hMutSα detached at 100–350 mM NaCl from homo- and heteroduplexes (Fig. 3A, hMSH2 and eluate fractions below 500 mM shown; hMSH6 reacted identically, data not shown). To determine the ATP concentration sufficient for this effect, we performed consecutive elutions with 700 and 1000 mM NaCl. This allowed specific detection of mismatch-bound hMutSα in the 1000 mM eluate fraction in the absence of ATP (see Fig. 2B); this signal is expected to disappear when ATP takes effect (Fig. 3A). Using this approach, the ATP concentration that disrupts the specific hMutSα–mismatch complex was found to be 1–10 µM (Fig. 3B, hMSH2 shown; hMSH6 reacted identically, data not shown), which is in good agreement with the IC50 of 3 µM reported previously for human hMutSα (36).
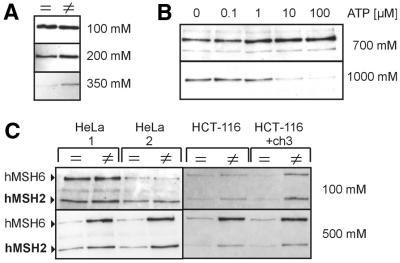
Figure 3.
ATP reduces the salt resistance of hMutSα binding but hMutSα retains affinity for heteroduplexes. (A) The binding assay was performed with HeLa nuclear extract (50 µg) as described in Figure 1B, except that ATP (250 µM final concentration) was added 1 min before elution and all elution buffers contained 250 µM ATP. Bound proteins were eluted consecutively with 100–1300 mM NaCl. hMSH2 western blots of the 100–350 mM NaCl eluate fractions are shown. (B) After incubation of HeLa nuclear extract (50 µg) with beads coupled to 32mer heteroduplexes, ATP was added to final concentrations ranging from 0 to 100 µM 10 min before elution. Bound proteins were eluted consecutively with 700 and 1000 mM NaCl. Western blots of hMSH2 in the eluate fractions are shown. (C) (Left) Incubations were performed with HeLa nuclear extract (50 µg) and homoduplex (=) or heteroduplex (≠) 32mer DNA coupled to magnetic beads. ATP (250 µM) was added to the incubation mixture either before incubation (HeLa 1) or after 25 min incubation with the beads, 10 min before elution (HeLa 2). Bound proteins were eluted consecutively with 100 and 500 mM NaCl in the continued presence of 250 µM ATP. Western blots of hMSH2 and hMSH6 in the eluate fractions are shown. (Right) Incubations were performed with HCT-116 and HCT-116+ch3 nuclear extracts. ATP was added to a final concentration of 250 µM 10 min before elution. Elution and detection were performed as above.
Interestingly, after 10 min incubation with 250 µM ATP, consecutive elution with 100 and 500 mM NaCl revealed that more protein eluted from 32mer heteroduplexes than from homoduplexes, which occurred predominantly in the 500 mM fraction. This result was similar when ATP was added before incubation with the beads and therefore was not an artefact of higher initial binding of hMutSα to heteroduplex DNA (Fig. 3C). Bacterial and yeast MutL proteins have been shown to improve the binding of MutS proteins to mismatches in the presence of ATP (30,31). However, in the present study the reaction of hMutSα in HCT-116 extracts (hMutL-deficient) was similar to that of hMutSα in HCT-116+ch3 extracts (hMutL-proficient), indicating that hMutL dimers did not affect DNA binding of hMutSα under these experimental conditions. These experiments suggest that the affinity of hMutSα to homoduplexes may be more impaired by ATP and that hMutSα is able to retain binding to heteroduplexes in the presence of ATP at salt concentrations of 50 and 100 mM NaCl.
ATP promotes binding of hMutLα and hMutLβ on 81mer DNA substrates
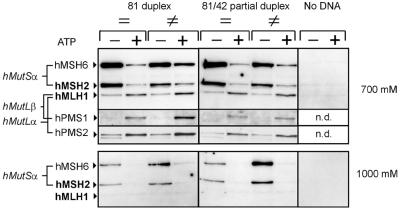
While addition of ATP did not induce hMLH1 binding to 32mer DNA substrates, hMLH1 from HeLa extracts showed enhanced binding to 81mer substrates after addition of 250 µM ATP and eluted in the 700 mM NaCl fraction. No hMutL proteins were detectable in the 1000 mM fraction or could be detached by boiling the beads in SDS sample buffer after this elution (data not shown) and no binding occurred on beads without DNA (Fig. 4), confirming that binding of hMLH1 was DNA dependent and that elution with 700 mM NaCl was quantitative. The increase in bound hMLH1 occurred on both homo- and heteroduplexes. Nuclear extracts of the mismatch repair-proficient cell lines TK6 (Fig. 5F) and HCT-116+ch3 (data not shown) reacted similarly and in all cases both hMutLα (hMLH1 with hPMS2) and hMutLβ (hMLH1 with hPMS1) were bound. Although hMutLα was shown to have a higher affinity for single-stranded DNA, the completely duplexed 81mer substrate reacted identically to one containing a duplexed area of 42 bp and a single-stranded area of 39 bp, suggesting that hMutLα binding to single-stranded DNA is not an important factor in initial complex formation. This held true whether the single-stranded area was located at the 5′- or 3′-end of the duplexed area (data not shown). In this experiment consecutive elutions with 700 and 1000 mM NaCl were performed, with the former fraction eluting hMutL proteins, homoduplex-bound hMutSα in the absence of ATP and hMutSα bound to DNA in the presence of ATP. The 1000 mM fraction was utilized to detect (ATP-induced disruption of) the mismatch-specific complex of hMutSα.
Figure 4.
ATP promotes binding of hMutLα and hMutLβ on 81mer DNA substrates. Magnetic beads were coupled with either completely duplexed (81 duplex) or partially duplexed (81/42 partial duplex) 81mer DNA substrates, which were either correctly paired (=) or contained a G-T mismatch (≠). For one experiment, beads were not coupled to DNA (No DNA). The beads were incubated with 100 µg HeLa nuclear extract for 25 min. The effect of ATP on binding of mismatch repair proteins was tested by comparing incubations without ATP (–) with incubations where ATP was added to a final concentration of 250 µM (+) as described in Materials and Methods. Following incubation, proteins were consecutively eluted with 700 and 1000 mM NaCl. The eluate fractions were probed for hMSH2, hMSH6, hMLH1, hPMS2 and hPMS1 (700 mM eluate) and hMSH2 and hMSH6 (1000 mM eluate) by western blotting. n.d., not detected.
Figure 5.
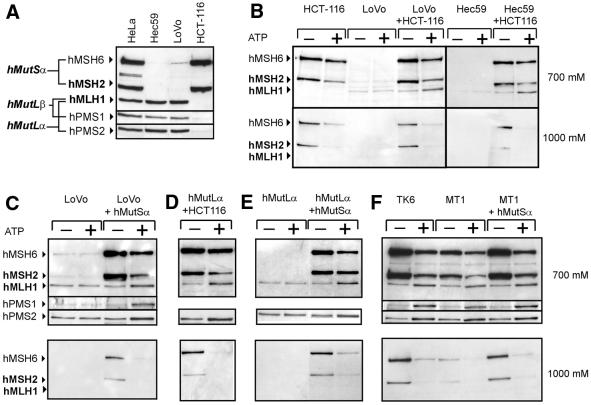
Binding of hMutLα and hMutLβ is dependent on hMutSα. (A) Western blot of nuclear extracts from HeLa, Hec59, LoVo and HCT-116 cells (25 µg each). (B–F) The effect of ATP (–, no ATP added; +, 250 µM ATP added) on binding of mismatch repair proteins was assessed as described in Figure 4 with partially duplexed (B–D and F) or duplexed (E) mismatched DNA coupled to magnetic beads. (B) (Left) DNA-coupled beads were incubated with either HCT-116 or LoVo nuclear extracts (100 µg) or a mixture of nuclear extracts from both cell lines (50 µg each). (Right) The same experiments were performed with nuclear extracts from Hec59 and HCT-116 cells. (C) Binding assays with either LoVo nuclear extract (100 µg) or LoVo nuclear extract (100 µg) supplemented with purified hMutSα (185 ng). (D) Binding assays with HCT-116 nuclear extract (100 µg) supplemented with purified hMutLα (300 ng). (E) Purified hMutLα (300 ng) was used alone or in combination with purified hMutSα (185 ng). (F) Binding assays with TK6 and MT1 nuclear extracts (100 µg) or MT1 nuclear extract (100 µg) supplemented with purified hMutSα (185 ng).
Binding of hMutLα and hMutLβ is dependent on hMutSα
LoVo and Hec59 cells are deficient in hMutS proteins (Fig. 5A) and hMLH1 in nuclear extracts of both cell lines failed to react to ATP in our binding experiments (Fig. 5B). ATP sensitivity was restored in both experiments when the extracts were supplemented with nuclear extract from hMLH1-deficient HCT-116 cells (Fig. 5B). Furthermore, purified hMutSα alone was able to restore the reaction in LoVo (Fig. 5C) and Hec59 nuclear extracts (data not shown), proving that binding of hMutL heterodimers was dependent on hMutSα. Because the reaction of hMutLα and hMutLβ with ATP was restored, hMutSα was competent for recruitment of both hMutL heterodimers. Nevertheless, hMutLβ was not essential for the interaction, because purified hMutLα alone also formed a complex with hMutSα when added to a nuclear extract of HCT-116 cells (Fig. 5D). Purified hMutLα alone also reacted with ATP when supplemented with purified hMutSα (Fig. 5E), further showing that these two heterodimers are sufficient for interaction. However, direct comparison showed that significantly more hMutSα and hMutLα bound in the supplementation experiments with cell extracts, although the same amounts of purified protein were used for both (data not shown). This effect likely reflects that additional proteins engaged in DNA manipulation and repair significantly enhance DNA binding and support complex formation of hMutSα and hMutL proteins. This concept is supported by recent findings that proliferating cell nuclear antigen (PCNA) can enhance mismatch binding of MutSα in yeast (40) and that MMR proteins seem to assemble in larger complexes with other DNA repair enzymes (41).
Interestingly, mutated hMutSα in MT1 extracts was as efficient in recruitment of hMutLα and hMutLβ as the wild-type heterodimer of the parental, mismatch repair-proficient cell line TK6 (Fig. 5F). Furthermore, addition of purified wild-type hMutSα to the MT1 extract did not increase binding of hMutL proteins, though the specific complex of mismatch-bound hMutSα in the 1000 mM NaCl fraction was restored. The mutations of hMSH6 in MT1 cells (V1260I and D1213V) are both located in the ATPase region and are likely to impair hMSH6 ATPase function. Two reasons may account for the ability of mutated MT1–hMutSα to recruit hMutL heterodimers as efficiently as wild-type protein: (i) the hMSH6 ATPase function may not be sufficiently suppressed by the mutation to abolish interaction; (ii) the interaction between hMutSα and hMutL heterodimers may be predominantly conferred by the hMSH2 ATPase.
Differential hMutSα-dependent recruitment of hMutLα and hMutLβ
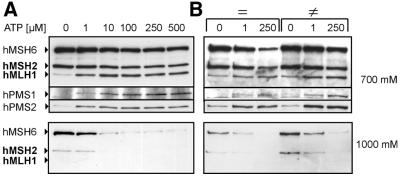
To test whether formation of complexes of hMutSα with hMutLα and hMutLβ occurs equivalently, the ATP concentration necessary for recruitment of hMutL heterodimers was assessed. Recruitment occurred at 1–10 µM ATP, but hPMS1 and hPMS2 did not react in parallel (Fig. 6A). While 1 µM ATP efficiently promoted complex formation between hMutSα and hMutLα, hMutLβ still showed a significant increase when the ATP concentration was raised from 1 to 10 µM. Direct comparison of homo- and heteroduplex DNA with 1 and 250 µM ATP showed that heteroduplex DNA was more efficient in recruitment of hMutLα and hMutLβ at both concentrations (Fig. 6B). The finding that hMutSα is able to recruit predominantly hMutLα at 1 µM ATP on homo- and heteroduplexes and the observation that in the absence of ATP hPMS2 generally exhibits higher basal binding than hPMS1 (Figs 4 and 5C–F) suggest that hMutSα may have an intrinsic preference for hMutLα.
Figure 6.
Differential hMutSα-dependent recruitment of hMutLα and hMutLβ The effect of different ATP concentrations on hMutL heterodimer recruitment was assessed with binding assays using 100 µg HeLa nuclear extracts as described in Materials and Methods with 81mer heteroduplexes (A) or 81mer homoduplexes (=) and heteroduplexes (≠) (B) coupled to magnetic beads. Elution and detection were performed as described in Figure 4. (A) ATP was added to final concentrations ranging from 0 to 500 µM as indicated. (B) The effects of 1 and 250 µM ATP were directly compared for homo- and heteroduplexes.
DISCUSSION
The binding of hMutSα to mismatches and its interaction with hMutL heterodimers on DNA was investigated by a method that enabled the use of short incubation times and cell extracts, which allowed the mimicking of biological binding conditions more closely. Binding of all proteins in this assay was confirmed to be DNA dependent, and consecutive washing steps of increasing stringency allowed the existence of mismatch-specific binding of hMutSα to be proved. Furthermore, the mismatch binding ability of hMutSα was shown to be altered at the same ATP concentration as reported previously, confirming the reliability of the method. The higher salt resistance exhibited by hMutSα when bound to heteroduplexes likely mirrors the tighter and more stable interaction of MutS proteins with mismatched DNA that has previously been visualized in the crystal structure of bacterial MutS and heteroduplex DNA (6,7). The data from this study also show that hMutLα binds single-stranded DNA, providing evidence that the function of human MutLα is similar to bacterial MutL, which also preferentially binds this substrate (21,22). Binding of hMutLα to single-stranded DNA was not, however, enhanced by the non-hydrolyzable ATP analog AMP-PNP, possibly reflecting a lower susceptibility to these nucleotides under the assay conditions used.
ATP has been shown to reduce the affinity of MutS and its eukaryotic homologs for mismatches (12–16), which was interpreted as a complete loss of mismatch binding ability. Recent studies with bacterial MutS, however, indicated that ATP binding and mismatch binding by MutS may not be mutually exclusive (18–20). The present study confirms that the affinity (in terms of salt resistance) of hMutSα for homo- and heteroduplexes is markedly reduced in the presence of ATP, but also indicates that binding to homoduplexes may be more affected than binding to heteroduplexes. With regard to the current models of mismatch repair, this observation is in accordance with the DNA bending model (18). In this case, the reduced salt resistance of the hMutSα–mismatch complex in the presence of 250 µM ATP may represent the state of verification that hMutSα would enter after binding ATP.
Furthermore, hMutSα was found to interact with hMutLα and hMutLβ on DNA in the presence of ATP. This interaction occurred only on 81mer and not on 32mer DNA substrates, reflecting a DNA length dependence of complex formation that has recently also been observed for bacterial (31) and human complexes using SPRS and gel shift assays (32). The finding that hMutLα has an intrinsic affinity for DNA supports the notion that both heterodimers may stay in contact with DNA in the complex. Because hMutSα covers ∼25 bp of DNA (36), the 32mer substrate may not be long enough for interaction.
According to recent reports, the involvement of hMutLβ in mismatch repair remains controversial. hMutLβ was suggested to participate in human mismatch repair due to a hPMS1 mutation found in a patient with hereditary non-polyposis colorectal cancer (HNPCC), a cancer predisposition syndrome associated with mutations in mismatch repair genes (9). Furthermore, PMS1-deficient mouse fibroblasts exhibit microsatellite instability, a phenotypic marker of deficient mismatch repair (10). However, hMutLβ is unable to confer mismatch repair proficiency on hMLH1-deficient extracts, which is possible with hMutLα (8,11). The present study shows that hMutLβ, although being 10 times less abundant than hMutLα in HeLa nuclear extracts (11), is efficiently recruited by hMutSα. The results also show that hMutLα is the favored partner for interaction with hMutSα at low ATP concentrations. Taken together, the results show that hMutLβ participates efficiently in mismatch repair protein complexes. It may exert a supportive function in the repair process after initial formation of the hMutSα and hMutLα complex. Further studies are necessary to elucidate the precise contribution of MutLβ to the mismatch repair process.
Under the conditions used in the present study, hMutSα interacted with hMutL heterodimers on homoduplexes and heteroduplexes, although complex formation was enhanced on heteroduplexes, which is consistent with earlier findings (28). In contrast, a recent report found complex formation to be mismatch dependent using SPRS and that different hMutSα–hMutLα complexes arose on homo- and heteroduplex DNA in gel shift assays (32), showing that the experimental procedure influences the mode of interaction. The ability to interact on homoduplex DNA may be explained by the sliding clamp model (17) as well as the translocation model (13), since both predict an interaction of hMutSα with other components of the mismatch repair machinery after ATP-induced movement from the mismatch to homoduplex DNA.
A recent report showed the dynamic nature of human MutSα–MutLα complexes on DNA, and the complexes were found to dissociate from the substrate DNA on poly[d(I-C)] challenge in gel shift assays when the DNA was not blocked at both ends (32). Although poly[d(I-C)] was present in our experiments (except for those with purified proteins) and the DNA substrates were generally blocked at only one end, hMutSα–hMutL complexes remained detectable. A possible explanation is that an equilibrium may be established between formation of hMutSα–hMutL complexes on DNA and translocation of these complexes off the oligoduplex during incubation and that only the fraction bound at the moment of elution is subsequently detected. Alternatively, under the assay conditions used, translocation off the oligoduplex may be blocked by other means than a second end block. The complex may, for example, be tethered to the substrate by additional proteins. This would also account for the weaker signals seen in experiments with purified proteins compared with cell extracts.
hMutSα contains two asymmetrical ATPase sites and it is possible that one ATPase predominantly confers interaction with hMutL proteins after mismatch recognition. The finding that the hMSH6 ATPase mutant in MT1 cells efficiently recruited hMutL heterodimers suggests that hMSH2 predominantly initiates the interaction (which has also been proposed based on considerations of the X-ray structure of bacterial MutS on a mismatch; 6), while the ATP function of hMSH6 (the subunit that directly contacts the mismatch) may promote subsequent processes, like mismatch verification in the DNA bending model. However, further studies are necessary to elucidate the different contributions of the two ATPases to these processes.
In conclusion, DNA-coupled magnetic beads provide a suitable tool for investigating protein–DNA interactions and formation of mismatch repair protein complexes. The affinity of hMutSα for homoduplexes is shown to be more reduced by ATP than the affinity for heteroduplexes, supporting the recently suggested DNA bending model of mismatch repair (18). Furthermore, hMutSα is shown to interact with hMutLα as well as hMutLβ on DNA. This interaction requires ATP and occurs only on long DNA substrates, confirming the DNA length dependence of complex formation reported earlier. Although hMutLβ is efficiently recruited by hMutSα, hMutLα seems to be the preferred partner in the initial interaction and it seems reasonable that hMutLβ exerts an auxiliary function in the mismatch repair process.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Giancarlo Marra, Markus Räschle and Josef Jiricny for helpful discussions and providing purified wild-type hMutLα and hMutSα heterodimers. This work was supported by research grant F15/99 from the University of Frankfurt to J.R.
REFERENCES
- 1.Lahue R.S., Au,K.G. and Modrich,P. (1989) DNA mismatch correction in a defined system. Science, 245, 160–164. [DOI] [PubMed] [Google Scholar]
- 2.Su S.S. and Modrich,P. (1986) Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl Acad. Sci. USA, 83, 5057–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grilley M., Welsh,K.M., Su,S.S. and Modrich,P. (1989) Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem., 264, 1000–1004. [PubMed] [Google Scholar]
- 4.Au K.G., Welsh,K. and Modrich,P. (1992) Initiation of methyl-directed mismatch repair. J. Biol. Chem., 267, 12142–12148. [PubMed] [Google Scholar]
- 5.Hall M.C. and Matson,S.W. (1999) The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem., 274, 1306–1312. [DOI] [PubMed] [Google Scholar]
- 6.Obmolova G., Ban,C., Hsieh,P. and Yang,W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature, 407, 703–710. [DOI] [PubMed] [Google Scholar]
- 7.Lamers M.H., Perrakis,A., Enzlin,J.H., Winterwerp,H.H., de Wind,N. and Sixma,T.K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature, 407, 711–717. [DOI] [PubMed] [Google Scholar]
- 8.Li G.M. and Modrich,P. (1995) Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl Acad. Sci. USA, 92, 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolaides N.C., Papadopoulos,N., Liu,B., Wei,Y.F., Carter,K.C., Ruben,S.M., Rosen,C.A., Haseltine,W.A., Fleischmann,R.D., Fraser,C.M. et al. (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature, 371, 75–80. [DOI] [PubMed] [Google Scholar]
- 10.Prolla T.A., Baker,S.M., Harris,A.C., Tsao,J.L., Yao,X., Bronner,C.E., Zheng,B., Gordon,M., Reneker,J., Arnheim,N. et al. (1998) Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nature Genet., 18, 276–279. [DOI] [PubMed] [Google Scholar]
- 11.Raschle M., Marra,G., Nystrom-Lahti,M., Schar,P. and Jiricny,J. (1999) Identification of hMutLbeta, a heterodimer of hMLH1 and hPMS1. J. Biol. Chem., 274, 32368–32375. [DOI] [PubMed] [Google Scholar]
- 12.Alani E., Sokolsky,T., Studamire,B., Miret,J.J. and Lahue,R.S. (1997) Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol. Cell. Biol., 17, 2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen D.J., Makhov,A., Grilley,M., Taylor,J., Thresher,R., Modrich,P. and Griffith,J.D. (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J., 16, 4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackwell L.J., Martik,D., Bjornson,K.P., Bjornson,E.S. and Modrich,P. (1998) Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem., 273, 32055–32062. [DOI] [PubMed] [Google Scholar]
- 15.Gradia S., Subramanian,D., Wilson,T., Acharya,S., Makhov,A., Griffith,J. and Fishel,R. (1999) hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell, 3, 255–261. [DOI] [PubMed] [Google Scholar]
- 16.Iaccarino I., Marra,G., Dufner,P. and Jiricny,J. (2000) Mutation in the magnesium binding site of hMSH6 disables the hMutSalpha sliding clamp from translocating along DNA. J. Biol. Chem., 275, 2080–2086. [DOI] [PubMed] [Google Scholar]
- 17.Fishel R. (1998) Mismatch repair, molecular switches and signal transduction. Genes Dev., 12, 2096–2101. [DOI] [PubMed] [Google Scholar]
- 18.Junop M.S., Obmolova,G., Rausch,K., Hsieh,P. and Yang,W. (2001) Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell, 7, 1–12. [DOI] [PubMed] [Google Scholar]
- 19.Joshi A., Sen,S. and Rao,B.J. (2000) ATP-hydrolysis-dependent conformational switch modulates the stability of MutS–mismatch complexes. Nucleic Acids Res., 28, 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas I. and Vijayvargia,R. (2000) Heteroduplex DNA and ATP induced conformational changes of a MutS mismatch repair protein from Thermus aquaticus. Biochem. J., 347, 881–886. [PMC free article] [PubMed] [Google Scholar]
- 21.Bende S.M. and Grafstrom,R.H. (1991) The DNA binding properties of the MutL protein isolated from Escherichia coli. Nucleic Acids Res., 19, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ban C., Junop,M. and Yang,W. (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell, 97, 85–97. [DOI] [PubMed] [Google Scholar]
- 23.Mechanic L.E., Frankel,B.A. and Matson,S.W. (2000) Escherichia coli MutL loads DNA helicase II onto DNA. J. Biol. Chem., 275, 38337–38346. [DOI] [PubMed] [Google Scholar]
- 24.Dao V. and Modrich,P. (1998) Mismatch-, MutS-, MutL- and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J. Biol. Chem., 273, 9202–9207. [DOI] [PubMed] [Google Scholar]
- 25.Galio L., Bouquet,C. and Brooks,P. (1999) ATP hydrolysis-dependent formation of a dynamic ternary nucleoprotein complex with MutS and MutL. Nucleic Acids Res., 27, 2325–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prolla T.A., Pang,Q., Alani,E., Kolodner,R.D. and Liskay,R.M. (1994) MLH1, PMS1 and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science, 265, 1091–1093. [DOI] [PubMed] [Google Scholar]
- 27.Habraken Y., Sung,P., Prakash,L. and Prakash,S. (1998) ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J. Biol. Chem., 273, 9837–9841. [DOI] [PubMed] [Google Scholar]
- 28.Gu L., Hong,Y., McCulloch,S., Watanabe,H. and Li,G.M. (1998) ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res., 26, 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matton N., Simonetti,J. and Williams,K. (2000) Identification of mismatch repair protein complexes in HeLa nuclear extracts and their interaction with heteroduplex DNA. J. Biol. Chem., 275, 17808–17813. [DOI] [PubMed] [Google Scholar]
- 30.Habraken Y., Sung,P., Prakash,L. and Prakash,S. (1997) Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr. Biol., 7, 790–793. [DOI] [PubMed] [Google Scholar]
- 31.Schofield M.J., Nayak,S., Scott,T.H., Du,C. and Hsieh,P. (2001) Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem., 276, 28291–28299. [DOI] [PubMed] [Google Scholar]
- 32.Blackwell L.J., Wang,S. and Modrich,P. (2001) DNA chain length dependence of formation and dynamics of hMutSalpha-hMutLalpha-heteroduplex complexes. J. Biol. Chem., 276, 33233–33240. [DOI] [PubMed] [Google Scholar]
- 33.Iaccarino I., Marra,G., Palombo,F. and Jiricny,J. (1998) hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J., 17, 2677–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawn M.T., Umar,A., Carethers,J.M., Marra,G., Kunkel,T.A., Boland,C.R. and Koi,M. (1995) Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res., 55, 3721–3725. [PubMed] [Google Scholar]
- 35.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gradia S., Acharya,S. and Fishel,R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell, 91, 995–1005. [DOI] [PubMed] [Google Scholar]
- 37.Genschel J., Littman,S.J., Drummond,J.T. and Modrich,P. (1998) Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem., 273, 19895–19901. [DOI] [PubMed] [Google Scholar]
- 38.Papadopoulos N., Nicolaides,N.C., Liu,B., Parsons,R., Lengauer,C., Palombo,F., D’Arrigo,A., Markowitz,S., Willson,J.K., Kinzler,K.W. et al. (1995) Mutations of GTBP in genetically unstable cells. Science, 268, 1915–1917. [DOI] [PubMed] [Google Scholar]
- 39.Blackwell L.J., Bjornson,K.P. and Modrich,P. (1998) DNA-dependent activation of the hMutSalpha ATPase. J. Biol. Chem., 273, 32049–32054. [DOI] [PubMed] [Google Scholar]
- 40.Flores-Rozas H., Clark,D. and Kolodner,R.D. (2000) Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nature Genet., 26, 375–378. [DOI] [PubMed] [Google Scholar]
- 41.Bertrand P., Tishkoff,D.X., Filosi,N., Dasgupta,R. and Kolodner,R.D. (1998) Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl Acad. Sci. USA, 24, 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]