Abstract
A secondary structure in the nascent RNA followed by a trail of U residues is believed to be necessary and sufficient to terminate transcription. Such structures represent an extremely economical mechanism of transcription termination since they function in the absence of any additional protein factors. We have developed a new algorithm, GeSTer, to identify putative terminators and analysed all available complete bacterial genomes. The algorithm classifies the structures into five classes. We find that potential secondary structure sequences are concentrated downstream of coding regions in most bacterial genomes. Interestingly, many of these structures are not followed by a discernible U-trail. However, irrespective of the nature of the trail sequence, the structures show a similar distribution, indicating that they serve the same purpose. In contrast, such a distribution is absent in archaeal genomes, indicating that they employ a distinct mechanism for transcription termination. The present algorithm represents the fastest and most accurate algorithm for identifying terminators in eubacterial genomes without being restricted by the classical Escherichia coli paradigm.
INTRODUCTION
Regulation of gene expression occurs primarily at the level of initiation. However, once the polymerase clears the promoter, at each subsequent nucleotide, in principle, it chooses whether to continue elongating or to fall off the template DNA (1). Therefore, not surprisingly, in many cases regulation also occurs at this elongation–termination decision (2,3). In Escherichia coli the decision to terminate is brought about by two mechanisms, simple and complex. Functionally, if a sequence brings about transcript release in an in vitro system with purified RNA polymerase alone, it is defined as an intrinsic, simple or factor-independent terminator. Terminators that require the presence of additional factors (including, though not restricted to, Rho) are classified as complex or factor-dependent terminators (4). These two classes of terminators are not strictly defined, as the efficiency of many intrinsic terminators is enhanced by the presence of additional factors (2).
A large body of experimental work in E.coli indicates that intrinsic terminators are characterised by a G/C-rich palindromic region followed by a trail of A residues on the template strand (5–7). The palindromic region is believed to be extruded as a hairpin in the nascent RNA, causing the polymerase to pause (8–10) and weakening its interaction with the nascent RNA and template DNA (11–12). Final release is facilitated by the U-trail (13) probably due to the unusually weak nature of the rU·dA hybrid (14). In addition, recent studies indicate that the primary role of the U-trail might be in stalling the polymerase and thereby providing time for the hairpin to form (15).
It should be noted that the requirement for the U-trail is not absolute in E.coli. For instance, Lynn et al. (13) showed that removal of up to three of nine U residues in the thr attenuator sequence had no effect on termination efficiency. However, removal of four to six U residues caused a linear decrease in the efficiency of termination and, finally, when only one or two U residues were present, termination was completely abolished. On the other hand, complete deletion of the U-trail in the crp terminator has no effect on transcription termination (16). Analysis of hybrid terminators indicates that terminators lacking a U-trail can be highly efficient, but only when joined with an appropriate sequence immediately downstream of the termination site (17). Thus, the significance of the U-trail is not completely clear in E.coli.
Despite this ambiguity, all algorithms developed so far to identify intrinsic terminators search for a stem–loop structure followed by a U-trail (18–22). Individual algorithms only differ in the manner by which they compute the stability of the stem–loop structure and the weight they attribute to the length of the U-trail. Recent attempts to extend such algorithms to other species surprisingly indicated that only a few bacterial species employ such structure-dependent terminators (22,23). In contrast, intrinsic terminators from E.coli have been shown to function in many bacteria, indicating that the underlying mechanism of transcription termination is likely to be conserved throughout the kingdom. This is not surprising since such a mechanism is independent of additional protein factors and therefore highly economical for the cell. In an attempt to resolve this paradox and formulate a general model for intrinsic transcription termination in eubacteria, the GeSTer (Genome Scanner for Terminators) algorithm has been developed to identify potential hairpin sequences in bacterial genomes. Analysis with GeSTer reveals that such sequences are concentrated in the immediate downstream region of stop codons in most bacterial genomes, a feature one would expect of intrinsic terminators. Notably, many of these structures lack a U-trail entirely. The widespread occurrence of putative terminators suggests conservation of the mechanism of intrinsic transcription termination in eubacteria.
MATERIALS AND METHODS
Definitions
A terminator is defined primarily by a double-stranded stem with a central unpaired bulb (Fig. 1). In addition, there are unpaired regions that interrupt the stem. Of these, asymmetrical regions constitute gaps while symmetrical regions constitute mismatches. Apart from these structural features, the sequence of the nucleotides trailing the structure is important, at least in some cases. The GeSTer algorithm primarily relies on identification of palindromic structures in the downstream region and analysis of their stability, distribution, nature of the trail sequences and the presence of adjacent structures. The stringency of the search is determined by both the ΔG value of the structure as well as the permitted lengths for various components of the hairpin. The default parameters shown in Figure 1 are based on the qualitative assessment of all experimentally determined terminators in different bacteria (24). All genomic sequences used in the present study were downloaded from http://www.ncbi.nlm.nih.gov:80/PMGifs/Genomes/micr.html. Wherever multiple strains of the same species have been sequenced, the strain used in the study is explicitly identified in Tables 1 and 2.
Figure 1.
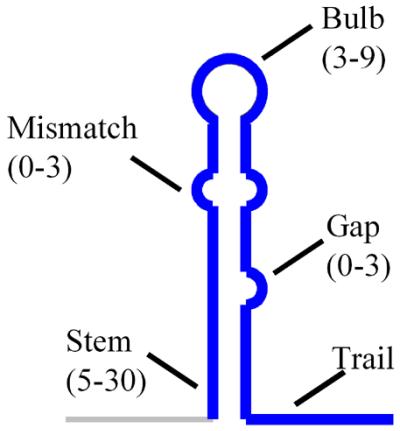
The anatomy of an imperfect palindrome. Features used to define a terminator structure by GeSTer. The numbers in parentheses denote the default number of bases (or base pairs) for each region. The trail consists of the 10 nt immediately following the structure.
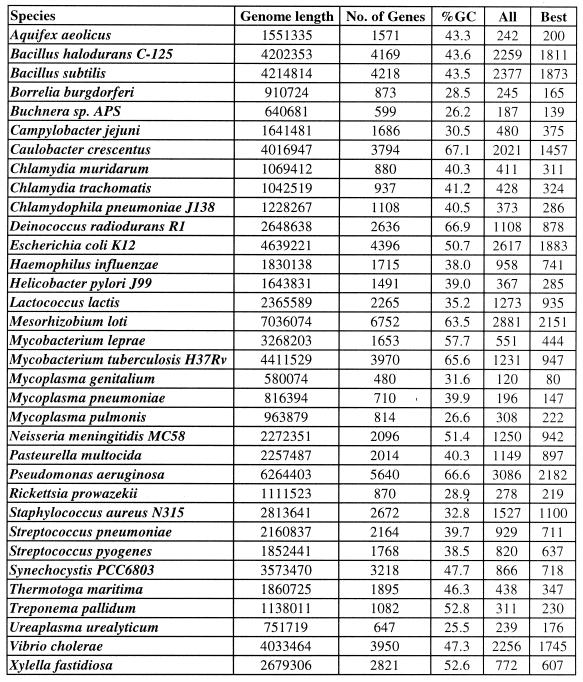
Table 1. Putative terminators in bacterial genomes.
All, total number of structures identified by GeSTer; Best, the number of structures when only the strongest structure downstream of each coding region was included (this constitutes the set of best candidate terminators, numerically equal to the number of genes with a putative terminator).
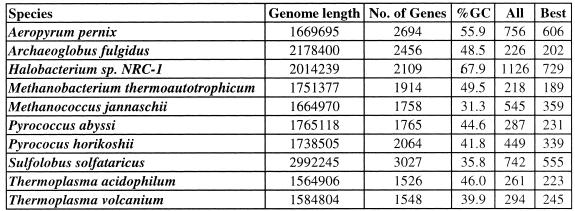
Table 2. Putative terminators in archaeal genomes.
All, total number of structures identified by GeSTer; Best, the number of structures when only the strongest structure downstream of each coding region was included (this constitutes the set of best candidate terminators, numerically equal to the number of genes with a putative terminator).
Searching for hairpins in whole genomes
The GeSTer algorithm contains three segments. The first segment accepts whole genome sequences in GenBank format and segregates the coding, upstream and downstream regions based on the feature table entries. At this stage the user has the option to tailor the various parameters used to identify the hairpin. The results presented here employ the default set of parameters indicated in Figure 1. The second segment searches for palindromic sequences in the region encompassed by –20 to +270 nt around the stop codon for each gene, without entering adjacent coding regions. The search is initiated at the first G/C-rich (>50%) tetranucleotide stretch. A reverse complementary match is sought within the next 70 nt. A match defines the base of the stem and from here the match is extended inwards until a mismatch is encountered. Next, all possible structures are computed allowing for various combinations of mismatches and gaps. All these structures constitute variants of the same structure and are mutually exclusive, therefore only the one with the lowest ΔG is retained. ΔGFormation for each of these structures was computed using the most recent parameters available (25). After the strongest structure is identified, the program searches for the next G/C-rich tetranucleotide and reinitiates the search. Again, in the case of overlapping mutually exclusive structures only the strongest structure is retained.
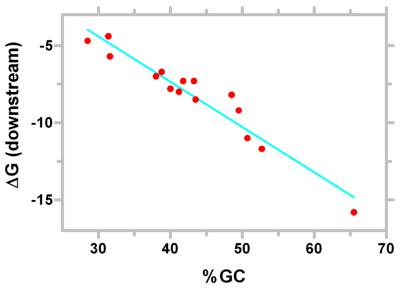
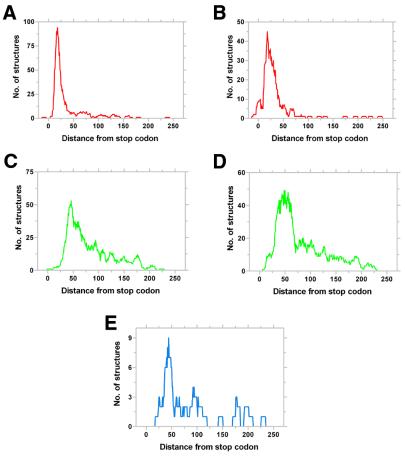
The third segment defines the final set of structures using a minimal ΔGCut-off filter. The filter is derived from two components, the G/C content of the bacteria and characteristics of structures in the upstream region. Analysis of the non-coding regions in genomes (23) reveals that the basal ΔG varies from species to species. We find a strong linear correlation between basal ΔG of the region downstream of genes and the G/C content of the genome (Fig. 2). The best linear regression fit corresponds to equation 1.
Figure 2.
Correlation of GC content with the basal ΔG of the region downstream of coding sequences. ΔG was calculated with a 60 base window using mfold as described (23). The line denoting the best linear regression fit is shown.
ΔGDownstream = –0.294 × (%GC) + 4.411 1
Since most terminator elements are expected to be present downstream of genes, the selection was optimised to minimise the identification of structures present upstream of genes. To this end, we needed to identify ‘pure upstream’ regions, i.e. regions that were not downstream with respect to any gene. In any genome, adjacent genes are arranged in four possible orientations with respect to each other (Fig. 3). When adjacent genes are transcribed in the same direction, it is difficult to theoretically distinguish between the downstream region of one and the upstream region of the next (Fig. 3A and B). On the other hand, convergently transcribed genes share a common downstream region (Fig. 3C), while divergently transcribed genes share their upstream regions (Fig. 3D). The optimised cut-off value for ΔG was derived by iteratively weighting ΔGDownstream so as to maximise the separation between structures in the pure upstream versus downstream regions. The weight parameter is composed of the optimal ΔG for E.coli divided by ΔGDownstream for E.coli. Thus, the final cut-off ΔG for any genome is computed as follows:
Figure 3.
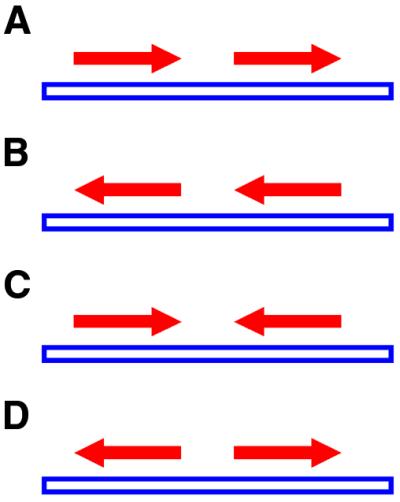
Relative orientation of adjacent genes on the genome. Adjacent genes in the genome could be both on the regular (A) or complementary strand (B). Alternatively, the genes could be oriented convergently (C) or divergently (D). The region between convergent genes constitutes the ‘pure downstream’ region while the region between divergent genes constitutes the ‘pure upstream’ region (see text).
ΔGCut-off = (12/10.5) × [–0.294 × (%GC) + 4.411] 2
With these parameters, the algorithm identified >90% of all experimentally determined terminators in different bacteria. At this stringency, false positives constitute <10% of the structures. All the putative terminators are classified based on the sequence in the trail as well as the position of adjacent structures (described below). The distribution of the structures was also analysed and is represented graphically. In the case of genes that are followed by multiple structures the best candidate is identified, again based on the lowest ΔG value. When, instead of non-coding sequences, uniform length downstream regions (–20 to +270 nt from the stop codon) were used for the analysis, very similar patterns were obtained (see Supplementary Material). It should be noted that G/U base pairing is commonly found in RNA secondary structures and therefore these have been included in the analysis, along with their energetic considerations.
Classification
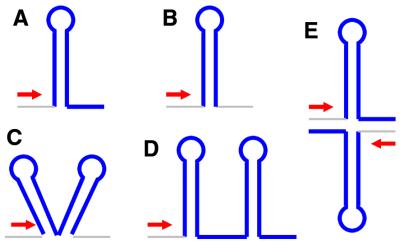
Based on the sequence content of trailing nucleotides and the presence of adjacent structures and coding sequences, structures were classified as follows. (i) E.coli type/L-shaped: those with >3 U residues present in the 10 nt trailing the structure (Fig. 4A). These represent the classical E.coli paradigm for intrinsic terminators as described in the Introduction. (ii) Mycobacterium type/I-shaped: those composed of a hairpin with 3 or fewer U residues in the trail (Fig. 4B). Such structures were first seen to predominate in M.tuberculosis (18), however, they are also present in many other species (discussed below). (iii) Streptomyces type/V-shaped: structures that are immediately followed (or preceded) by another structure (Fig. 4C). Such structures were first identified in Streptomyces sp. (26). (iv) Tandem/U-shaped: where multiple structures are present downstream of a single gene with ≤50 nt between consecutive structures (Fig. 4D). These could be composed of two or more structures. (v) Convergent/X-shaped: structures present between adjacent convergently oriented genes (Fig. 4E). It should be noted that all structures, other than the L-shaped ones, are symmetrical and could potentially work in either orientation.
Figure 4.
Classification of terminators. The terminators are denoted schematically with the relevant regions highlighted. (A) L-shaped or E.coli type; (B) I-shaped or mycobacterial type; (C) V-shaped or Streptomyces type; (D) U-shaped or tandem type; (E) X-shaped or convergent type. The arrowhead denotes the direction of transcription. The individual structures in V-, U- and X-shaped structures could be either L- or I-shaped.
Software details
The source code is in Visual Basic. The program runs in a Windows 97 environment. The program has a user friendly front end. Most parameters are set to default, however, the user has the option to change all of them, including ΔGCut-off. All outputs are generated both in the form of graphs and as tab-delimited text files. In addition, individual structures can be seen graphically. The installable version of the software and sample output files for E.coli and Pseudomonas aeruginosa are available free (for non-commercial use) at ftp.bork.embl-heidelberg.de/pub/users/suyama/GeSTer. The authors may be contacted for the source code or any commercial use of the software.
RESULTS AND DISCUSSION
The GeSTer program scans for palindromic sequences downstream of coding sequences in whole genomes. The stability of each structure is calculated using the parameters of Mathews et al. (25). All structures identified are subjected to a species-specific ΔGCut-off (derived as described in Materials and Methods). This denotes the complete (‘All’) set of putative terminators in the particular organism. Downstream of each gene, the strongest structure is judged as the ‘Best’ candidate for the primary terminator. All structures are classified based on the nature of their trail and the presence of adjacent structures (described in Materials and Methods, Fig. 4). The program also enables the user to analyse the overall distribution of terminators with respect to the stop codon for each gene in the genome. It should be noted that all classes of terminators defined by the program have been shown to function in one or other species. While L-shaped structures function in many species, including E.coli, V-shaped structures have been identified previously at least in Streptomyces sp. (26). X-shaped structures function in both E.coli (27) and Streptococcus sp. (28). In addition, we have shown that I-shaped structures, either alone or in tandem (U-shaped), function efficiently both in vivo and in vitro (24).
Bacteria, but not Archaea, employ structures as terminators
We have used the program to identify potential intrinsic terminators in all known archaeal and eubacterial genomes. Notably, in most bacterial genomes there is a preponderance of potential hairpin sequences within the first 50 nt downstream of the stop codon (Table 1 and Fig. 5), a characteristic one would expect of transcription terminators. Interestingly, the strongest terminators downstream of each gene (compare ‘Best’ with ‘All’ in Fig. 5A and C) show a more dramatic peak. Thus, the strongest structures are usually encountered first after the stop codon by the RNA polymerase and are probably responsible for most transcription termination. This is further substantiated if one looks at only the set of the strongest structures amongst the ‘Best’ candidate structures. Almost all structures apart from those that constitute the peak are eliminated in such a plot (Fig. 5B and D). In contrast, all archaeal genomes show a modest or no peak in this region, indicating that they probably employ a different mechanism of termination (Table 2 and Fig. 6A and C). Even the strongest structures are scattered throughout the entire window analysed (Fig. 6B and D). This is not surprising, since the transcription machinery in Archaea does not resemble the prokaryotic architecture.
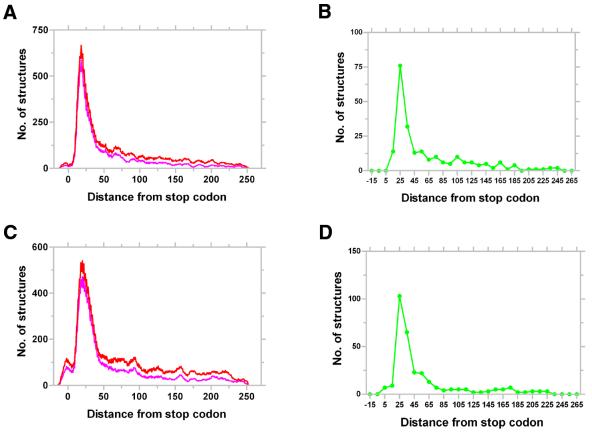
Figure 5.
Representative distribution of putative terminators in bacteria. Distribution of all (red) and the best (magenta) structures identified by GeSTer in E.coli (A) and P.aeruginosa (C) with respect to the stop codon. The number of structures were aggregated over a window of 10 bases slid one base at a time. Distribution of the strongest structures (green) amongst the best candidate terminators in E.coli (B) and P.aeruginosa (D) aggregated over 10 bases. The strongest structures were selected with a ΔG filter of mean – SD of ΔG of all the best candidate structures, i.e. those structures with a ΔG lower than the mean by at least 1 SD.
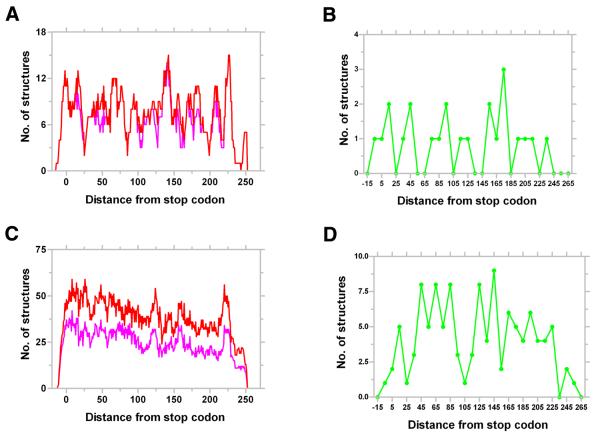
Figure 6.
Representative distribution of putative terminators in Archaea. Distribution of all (red) and the best (magenta) structures identified by GeSTer in Archaeoglobus fulgidus (A) and Halobacterium sp. (C) with respect to the stop codon. The number of structures were aggregated over a window of 10 bases slid one base at a time. Distribution of the strongest structures (green) amongst the best candidate terminators in A.fulgidus (B) and Halobacterium sp. (D). The strongest structures were selected with a ΔG filter of mean – SD of ΔG of all the best candidate structures, i.e. those structures with a ΔG lower than the mean by at least 1 SD.
Comparison with other algorithms
As discussed above, earlier theoretical analyses indicated that most bacteria use a distinct mechanism for intrinsic transcription termination. In contrast, we find that most bacterial genomes show a concentration of secondary structures downstream of coding sequences. This is probably because the majority of previous attempts did not take into account the possibility of secondary structure alone working as a terminator (18–22). As a result, they identified only the L-shaped and some X-shaped (those with L-shaped structures in both orientations) terminators identified by the present algorithm.
The only previous analysis of the distribution of secondary structures in the non-coding region similarly detected a concentration of structures downstream of the stop codon in a minority of bacterial species (23). This is probably because of the rigid 60 base window employed in the study leading to blunting of peaks. Thus, modest peaks become statistically indistinguishable from the basal species-specific ΔG. On the other hand, our algorithm varies the window size dynamically and specifically to identify individual stem–loop structures, thereby improving the sensitivity and accuracy of the prediction.
Another advantage of the GeSTer algorithm is that the cut-off parameters have been optimised to maximise the resolution between downstream structures with respect to upstream structures. In contrast, earlier workers attempted to improve their predictions by comparing structures within the coding region with those in the downstream region. It is noteworthy that structures when cloned upstream or downstream of a coding region function efficiently independent of the distance from the promoter (24). However, the same structure becomes non-functional when present within the coding region (24). This is probably because the translating ribosomes behind the polymerase prevent extrusion of the structure in the nascent RNA within the coding region.
Using the default parameters, GeSTer identified >90% of all experimentally tested intrinsic bacterial terminators. Notably, the algorithm even identified terminators that are subject to regulation. For instance, the structure present in the intergenic region between the bglG and bglF genes acts as an efficient terminator only in the absence of the anti-termination activity of bglG (29). GeSTer identified this structure correctly as an intrinsic terminator. Finally, GeSTer is the fastest algorithm for the identification of intrinsic terminators in whole genomes. On a Pentium pro(r) 450 MHz processor with 128 Mb RAM, the program took ∼19 min to scan the entire E.coli genome. This is more than twice as fast as any comparable program while allowing more liberal parameters. With identical parameters, GeSTer is nearly 4.5 times faster while detecting representatives from all classes of terminators.
Distribution of terminators in eubacteria
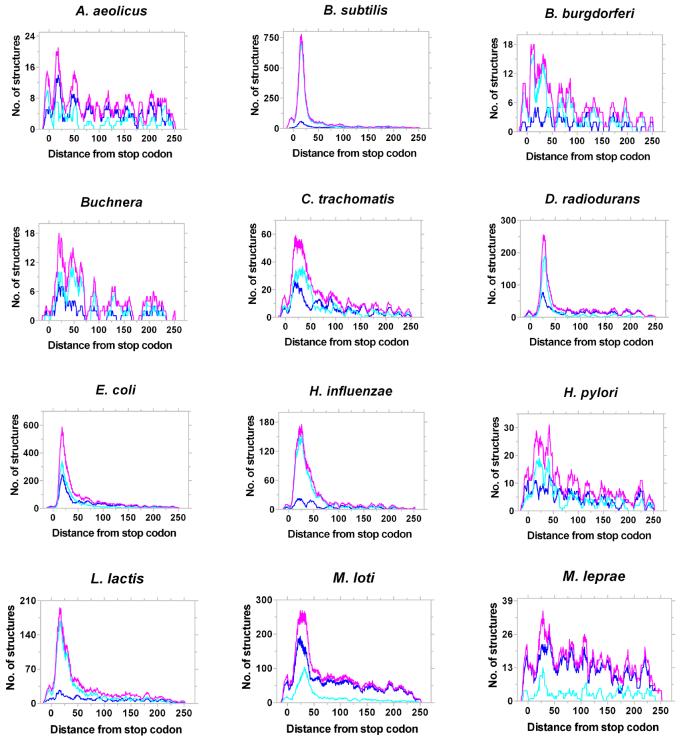
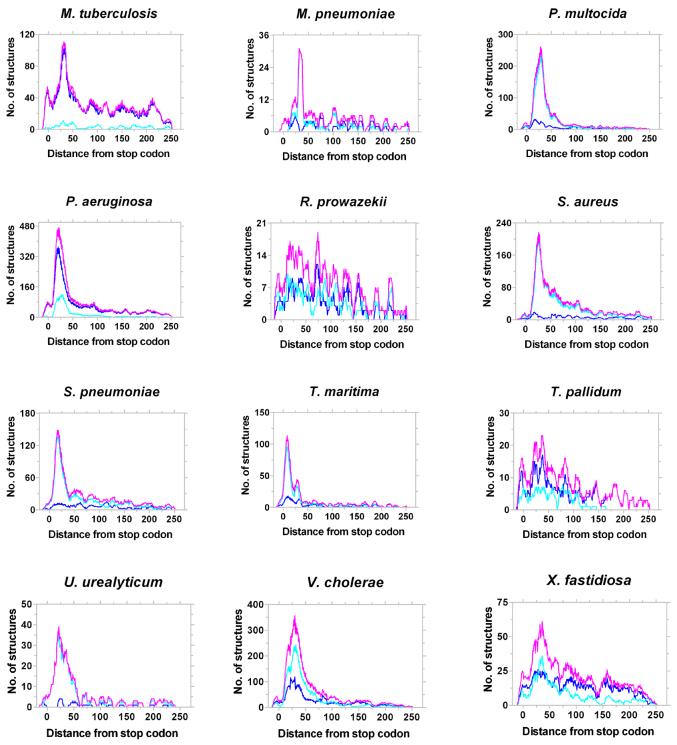
As discussed above, in most bacterial genomes the GeSTer program identified putative intrinsic terminators downstream of 12–45% of genes. Other genes are probably part of an operon or employ Rho homologues for termination. The two hyperthermophilic eubacteria, Aquifex aeolicus and Thermotoga maritima, appear to rely less on structure-based intrinsic terminators, probably because such structures would not be stable at the high temperatures at which they grow. Amongst these, the optimal growth temperature of T.maritima (80°C) is lower than that of A.aeolicus (95°C). In agreement with this, the concentration of structures in the vicinity of the stop codon in T.maritima is better than that in A.aeolicus (Fig. 7). Apart from the thermophiles, relatively few structures were identified in three species of Mycoplasma and the two spirochaetes (Borrelia burgdoferi and Treponema pallidum) and Buchnera sp. However, all these species retain the characteristic peak of structure downstream of the stop codon. Of the remaining, only Mycobacterium leprae and Rickettsia prowazekii show a broader distribution with modest peaks. All other genomes show a sharp peak within 50 nt downstream of stop codons. Another point of interest is that the majority of species that showed a lowered reliance on intrinsic terminators are obligate pathogens (Chlamydophila pneumoniae, B.burgdoferi, Helicobacter pylori, T.pallidum, M.leprae and R.prowazekii) or obligate endosymbionts (Buchnera sp.). The underlying evolutionary or molecular basis for this bias is unclear at present.
Figure 7.
Representative distribution of L- and I-shaped terminators in Bacteria. Distribution of the best candidate terminators (magenta) and their assignment as L- (light blue) or I-shaped (dark blue) structures in different bacteria is shown. The number of structures was aggregated over a window of 10 bases slid one base at a time.
Distribution and frequency of individual classes of terminators
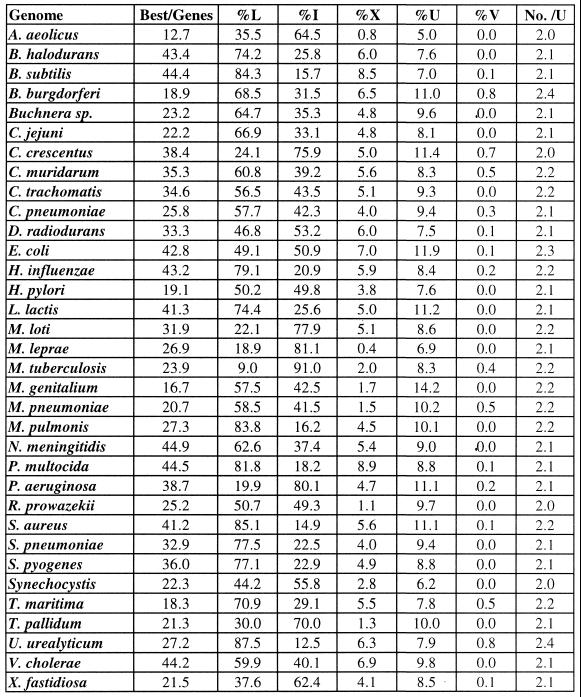
Individual bacterial species rely on various classes of terminators to different extents (Table 3). However, irrespective of their frequency, all the single structures peak at the same position (Figs 7 and 8). The peak for V- and U-shaped structures is offset by a few nucleotides downstream because they are composed of two structures and therefore their midpoint is shifted (Fig. 8). Furthermore, since the distance between the two structures in U-shaped structures varies from case to case, the peak is broader compared to that of single structures. Since V-shaped structures are rare, we have shown a single cumulative graph for all bacteria. On the other hand, for U- and X-shaped structures graphs from representative species are shown. Thus, all classes of structures appear to have a common function, as terminators.
Table 3. Frequency of occurrence of different classes of terminators in bacterial genomes.
Best/Genes, the percentage of genes that have at least one putative terminator downstream; %L, %I, percentage of best candidate structures that are L- and I-shaped, respectively; %X, %U, %V, percentage of total structures that are X-, U- and V-shaped, respectively; No. U, number of individual structures that constitute each U-shaped terminator.
Figure 8.
Representative distribution of X-, U- and V-shaped terminators in Bacteria. Distribution of X- (red) and U-shaped (green) structures in E.coli (A and C) and P.aeruginosa (B and D), respectively. (E) The overall distribution of V-shaped structures in all bacteria. The number of structures were aggregated over a window of 10 bases slid one base at a time.
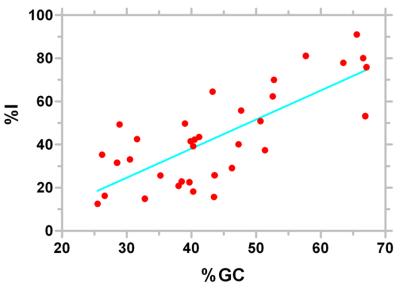
Of the more than 26 000 best candidate terminators identified in all bacterial species, approximately half (46.6%) do not have an appreciable U-trail. However, individual species show a wide range (10–90% with a standard deviation of 21.6%, Table 3) in preference for L- and I-shaped structures. Organisms like Bacillus subtilis, Staphylococcus aureus and Ureaplasma urealyticum rely largely on the L-shaped structures, while P.aeruginosa and M.tuberculosis primarily employ I-shaped structures. However, the most surprising finding was that approximately half (50.9%) of the structures even in the prototypical E.coli were I-shaped. There is a weak linear correlation between the preference for I-shaped structures and the G/C content of the organism (Fig. 9). However, this is unlikely to be the primary determinant of this preference. For instance, Caulobacter crecentus, Deinococcus radiodurans, Mycobacterium tuberculosis and P.aeruginosa have very similar G/C contents but have a highly disparate preference for I-shaped structures ranging from 53 to 91%. We believe that the primary cause for this variation may be the rate of transcription elongation in different organisms (discussed below).
Figure 9.
Correlation of GC content with a preference for I-shaped structures. The line denoting the best linear regression fit is shown.
X-shaped structures constitute up to 8.5% of the structures identified while V-shaped are the rarest. It would be interesting to test whether Streptomyces sp., where V-shaped structures have explicitly been shown to function (26), show an over-representation of this class. This test awaits a complete genome. In contrast, the tandem U-shaped structures are relatively common, constituting 5–17% of structures in different genomes. Interestingly, most of the tandem terminators are composed of two structures (No. /U in Table 3). Only rarely are more than three structures present. This is significant, since we have shown previously that two weak structures when placed in tandem can work synergistically (24). Furthermore, such structures bring about transcription termination comparable to a single strong structure (24). Therefore, additional structures would be wasteful. Thus, tandem structures provide the cell with an alternative mechanism of efficient termination using individually weak structures.
A general model for intrinsic transcription termination
The efficiency of transcription termination is believed to be determined by kinetic competition between the rates of elongation and release (1,30). In addition, recent work suggests that the primary role of the U-trail may not be to weaken the RNA–DNA hybrid, but instead it serves to stall the elongating polymerase (15), thereby allowing the hairpin to be extruded, dislodging the nascent chain from the catalytic site. In organisms, like M.tuberculosis, where the rate of RNA chain elongation is considerably slower than in E.coli (31), such a role for the U-trail would be superfluous. Therefore, an I-shaped structure, even without the U-trail, could work as efficiently as an L-shaped structure. Thus, we believe that the rate of elongation of polymerase is the primary determinant of the choice of class of structure. In agreement with this prediction, even E.coli RNA polymerase itself, when made to move slowly, terminates efficiently in the absence of a U-trail (7,32).
Conclusions and corollary
Intrinsic terminators represent a highly economical mechanism of transcription termination. Results with our algorithm imply that variants of the mechanism elucidated in E.coli are conserved in most bacteria. We have shown that a secondary structure alone is sufficient to bring about transcription termination both in vivo as well as in vitro.
A point to be noted is that two classes of highly transcribed genes, rRNA and tRNA, are known to have extensive secondary structure in their RNA. In addition, these genes are not translated; therefore, a secondary structure within the coding region of such genes could lead to premature termination since there are no translating ribosomes to protect these regions (discussed above). Significantly, the algorithm does not identify any structures in the coding regions of these genes in any of the bacterial species tested so far, further buttressing the confidence in the predictions. Thus, GeSTer represents an extremely versatile and fast algorithm for the identification and classification of terminators in bacterial genomes. Furthermore, the present algorithm is an invaluable tool that would considerably improve the accuracy of identification of intrinsic terminators in bacterial whole genome sequencing projects.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Narasimha Prakash and Monalisa Chatterji for discussions and Anirudh Goel for assistance with data analysis. R.P. is the recipient of an Indian Academy of Sciences fellowship. This research was supported by grants from the Indian Council of Medical Research, Government of India.
REFERENCES
- 1.von Hippel P.H. (1998) An integrated model of the transcription complex in elongation, termination and editing. Science, 281, 660–665. [DOI] [PubMed] [Google Scholar]
- 2.Das A. (1993) Control of transcription termination by RNA-binding proteins. Annu. Rev. Biochem., 62, 893–930. [DOI] [PubMed] [Google Scholar]
- 3.Richardson J.P. and Greenblatt,J. (1996) Control of RNA chain elongation and termination. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, DC, pp. 822–848.
- 4.Platt T. (1994) Rho and RNA: models for recognition and response. Mol. Microbiol., 11, 983–990. [DOI] [PubMed] [Google Scholar]
- 5.Platt T. (1986) Transcription termination and the regulation of gene expression. Annu. Rev. Biochem., 55, 339–372. [DOI] [PubMed] [Google Scholar]
- 6.Yager T.D. and von Hippel,P.H. (1987) Transcript elongation and termination in Escherichia coli. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, 1st Edn. ASM Press, Washington, DC, pp. 1241–1275.
- 7.Yarnell W.S. and Roberts,J.W. (1999) Mechanism of intrinsic transcription termination and antitermination. Science, 284, 611–615. [DOI] [PubMed] [Google Scholar]
- 8.Farnham P.J. and Platt,T. (1981) Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res., 9, 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D., Severinov,K. and Landick,R. (1997) Preferential interaction of the his pause RNA hairpin with RNA polymerase beta subunit residues 904–950 correlates with strong transcriptional pausing. Proc. Natl Acad. Sci. USA, 94, 8433–8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artsimovitch I. and Landick,R. (1998) Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev., 12, 3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt K.M. and Chamberlin,M.J. (1990) RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J. Mol. Biol., 213, 79–108. [DOI] [PubMed] [Google Scholar]
- 12.Wilson K.S. and von Hippel,P.H. (1995) Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl Acad. Sci. USA, 92, 8793–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynn S.P., Kasper,L.M. and Gardner,J.F. (1988) Contributions of RNA secondary structure and length of the thymidine tract to transcription termination at the thr operon attenuator. J. Biol. Chem., 263, 472–479. [PubMed] [Google Scholar]
- 14.Martin F.H. and Tinoco,I.,Jr (1980) DNA–RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res., 8, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gusarov I. and Nudler,E. (1999) The mechanism of intrinsic transcription termination. Mol. Cell, 3, 495–504. [DOI] [PubMed] [Google Scholar]
- 16.Abe H. and Aiba,H. (1996) Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie, 78, 1035–1042. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds R. and Chamberlin,M.J. (1992) Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J. Mol. Biol., 224, 53–63. [DOI] [PubMed] [Google Scholar]
- 18.Brendel V. and Trifonov,E.N. (1984) A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res., 12, 4411–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brendel V., Hamm,G.H. and Trifonov,E.N. (1986) Terminators of transcription with RNA polymerase from Escherichia coli: what they look like and how to find them. J. Biomol. Struct. Dyn., 3, 705–723. [DOI] [PubMed] [Google Scholar]
- 20.Carafa Y.d’A., Brody,E. and Thermes,C. (1990) Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol., 216, 835–858. [DOI] [PubMed] [Google Scholar]
- 21.Ermolaeva M.D., Khalak,H.G., White,O., Smith,H.O. and Salzberg,S.L. (2000) Prediction of transcription terminators in bacterial genomes. J. Mol. Biol., 301, 27–33. [DOI] [PubMed] [Google Scholar]
- 22.Lesnik E.A., Sampath,R., Levene,H.B., Henderson,T.J., McNeil,J.A. and Ecker,D.J. (2001) Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res., 29, 3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washio T., Sasayama,J. and Tomita,M. (1998) Analysis of complete genomes suggests that many prokaryotes do not rely on hairpin formation in transcription termination. Nucleic Acids Res., 26, 5456–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unniraman S., Prakash,R. and Nagaraja,V. (2001) Alternate paradigm for intrinsic transcription termination in eubacteria. J. Biol. Chem., 276, 41850–41855. [DOI] [PubMed] [Google Scholar]
- 25.Mathews D.H., Sabina,J., Zuker,M., Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 26.Ingham C.J., Hunter,I.S. and Smith,M.C. (1995) Rho-independent terminators without 3′ poly-U tails from the early region of actinophage φC31. Nucleic Acids Res., 23, 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postle K. and Good,R.F. (1985) A bidirectional rho-independent transcription terminator between the E. coli tonB gene and an opposing gene. Cell, 41, 577–585. [DOI] [PubMed] [Google Scholar]
- 28.Steiner K. and Malke,H. (1995) Transcription termination of the streptokinase gene of Streptococcus equisimilis H46A: bidirectionality and efficiency in homologous and heterologous hosts. Mol. Gen. Genet., 246, 374–380. [DOI] [PubMed] [Google Scholar]
- 29.Houman F., Diaz-Torres,M.R. and Wright,A. (1990) Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell, 62, 1153–1163. [DOI] [PubMed] [Google Scholar]
- 30.von Hippel P.H. and Yager,T.D. (1991) Transcript elongation and termination are competitive kinetic processes. Proc. Natl Acad. Sci. USA, 88, 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harshey R.M. and Ramakrishnan,T. (1977) Rate of ribonucleic acid chain growth in Mycobacterium tuberculosis H37Rv. J. Bacteriol., 129, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDowell J.C., Roberts,J.W., Jin,D.J. and Gross,C. (1994) Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science, 266, 822–825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.