Abstract
During cell migration and polarization, numerous signal transduction and cytoskeletal components self-organize to generate localized protrusions. Although biochemical and genetic analyses have delineated many specific interactions, how the activation and localization of so many different molecules are spatiotemporally orchestrated at the subcellular level has remained unclear. Here we show that the regulation of negative surface charge on the inner leaflet of the plasma membrane plays an integrative role in the molecular interactions. Surface charge, or zeta potential, is transiently lowered at new protrusions and within cortical waves of Ras/PI3K/TORC2/F-actin network activation. Rapid alterations of inner leaflet anionic phospholipids, such as PI(4,5)P2, PI(3,4)P2, phosphatidylserine, and phosphatidic acid, collectively contribute to the surface charge changes. Abruptly reducing the surface charge by recruiting positively charged optogenetic actuators was sufficient to trigger the entire biochemical network, initiate de novo protrusions, and abrogate pre-existing polarity. These effects were blocked by genetic or pharmacological inhibitions of key signaling components such as Akt and PI3K/TORC2. Conversely, increasing the negative surface charge deactivated the network and locally suppressed chemoattractant-induced protrusions or subverted EGF-induced ERK activation. Computational simulations involving excitable biochemical networks demonstrated that slight changes in feedback loops, induced by recruitment of the charged actuators, could lead to outsized effects on system activation. We propose that key signaling network components act on, and are in turn acted upon, by surface charge, closing feedback loops which bring about the global-scale molecular self-organization required for spontaneous protrusion formation, cell migration, and polarity establishment.
INTRODUCTION
Cell migration and polarity involve large networks of interacting signal transduction and cytoskeletal components 1-4. The spatial and temporal behavior of the molecular events are highly coordinated: For example, Ras, PI3K, and Rac activation along with actin polymerization define growing protrusions while PTEN, RhoA, and myosin II leave these regions and return to the membrane only when the protrusions are withdrawn. Where it has been examined, this complementary organization is conserved during macropinocytosis, phagocytosis, cytokinesis, within traveling cortical waves in different cells throughout phylogeny, and in apical-basal polarity of epithelial cells 5-7. Second, genetic, pharmacological, or mechanical perturbations of different individual components often result in similar phenotypic changes suggesting that the overall “setpoint” of the network is more important than any single constituent 8-18. While these emergent features could be a result of a series of stepwise protein-protein/protein-lipid interactions, it seems more likely that they are linked to some organizing principle such as a biophysical property of the plasma membrane 19. Such global organizers have been suggested 1, 10, 19, but most properties that track closely with the observed spatial and temporal organization or whose small alterations bring about large shifts in network behavior, have not been identified.
One candidate for a such a biophysical organizer is inner membrane surface charge or zeta potential. Cells maintain a negative charge on the inner leaflet of the plasma membrane, primarily by regulating the concentrations of multiple anionic phospholipids20-23. Numerous studies have focused on the signaling properties of individual lipids such as PIP324, 25. Many cytoskeletal proteins have been reported to bear PI(4,5)P2 binding motifs26. However, relatively few studies have focused on the role of surface charge itself 20, 27. These have proposed that the zeta potential, from −20 to −50 mV28-30, regulates local pH and second messenger concentrations, alters ion channel conductance, and controls peripheral membrane protein association 20-23, 31-34. Moreover, during immunological synapse and phagocytosis, the membrane surface charge changes 21, 35-37. However, whether network activation is associated with increases or decreases in surface charge or whether direct perturbations of surface charge can activate/deactivate the network has not been explored. Interestingly, it has been reported that enzymatic lowering of specific anionic lipids such as PI(4,5)P2 or PI(3,4)P2, which would be expected to decrease surface charge, can lead to the network activation and enhanced cell migration9, 38.
To assess the possible role of membrane surface charge as a biophysical organizer, we examined surface charge profile changes during cell migration and polarity and designed actuators to directly alter the charge. We discovered that PI(4,5)P2, PI(3,4)P2, phosphatidylserine (PS), and phosphatidic acid (PA) are dynamically lowered in local regions of the membrane where the Ras/PI3k/Akt/F-Actin network is activated. Consistently, we observed that the overall surface potential is high in resting membranes and is transiently lowered during network activation and protrusion formation. By recruiting positively or negatively charged optogenetic actuators to the membrane in a spatiotemporally controlled fashion, we established that the level of surface potential in local membrane domains is sufficient and necessary to control the activation of the entire signal transduction network and to direct migration and polarity phenotypes.
RESULTS
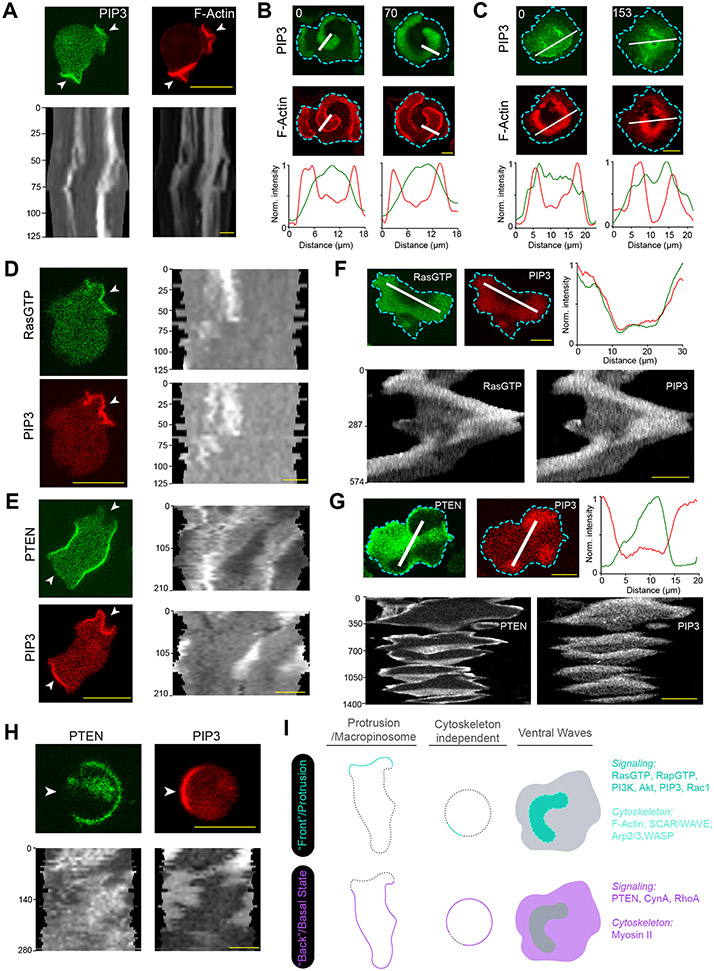
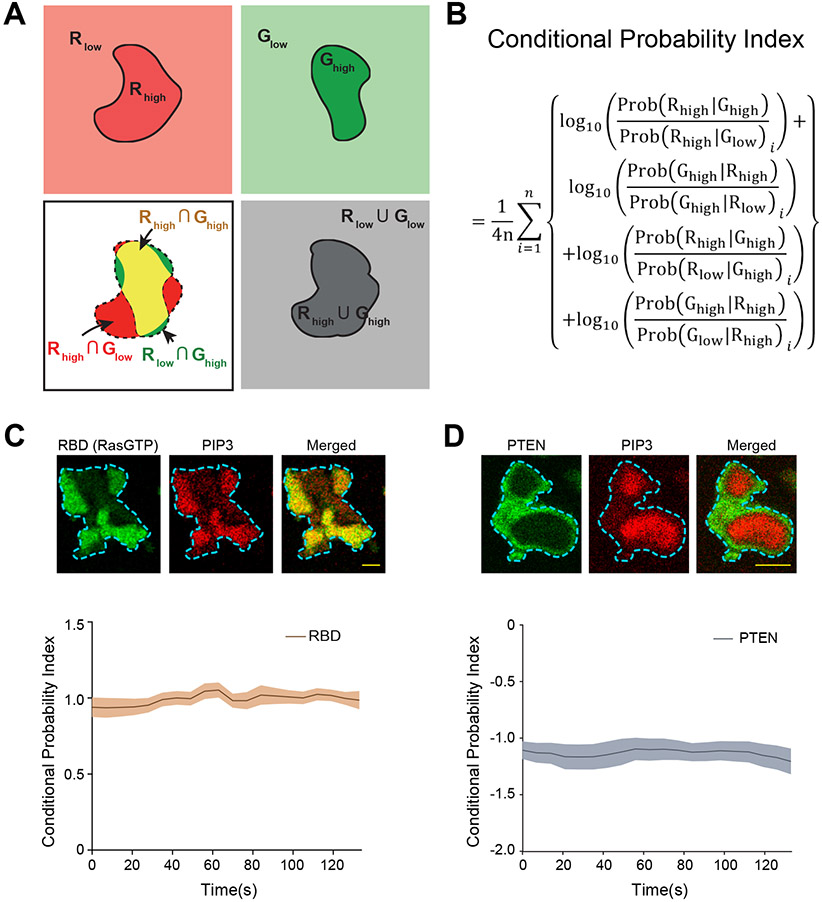
To analyze the coordinated signaling and cytoskeletal activities, we examined protrusions in migrating Dictyostelium cells as well as cortical waves in Dictyostelium and RAW 264.7 macrophages (Extended Data Figures 1A-1C). As previously noted 5, 25, 39, events that facilitate protrusion formation such as Ras/PI3K activation and actin polymerization localize to the front regions of migrating cells (Extended Data Figures 1A,1D), whereas those that suppress protrusions such as PTEN and myosin II localize to back regions (Extended Data Figures 1E). A corresponding complementarity was observed in ventral wave patterns where the molecules/events dynamically separate into broad, propagating “front-” and “back”-state regions (Extended Data Figures 1F-1I) 6, 40-42. Wave properties have been shown previously to control modes of protrusions and cell migration, and thus serve as a useful surrogate for “front/back” polarity (Extended Data Figure 1I). To quantitate the extent to which an event/component was co-localized or complementarily localized with front marker PIP3, we developed a conditional probability-based index or CP index (Methods; Extended Data Figures 2A-2D). For most of the data, we provided both normalized line-scans and CP indices to report measures of fractional changes in sensor levels and the extent of complementarity with respect to PIP3.
PI(4,5)P2, PI(3,4)P2, PS, and PA localize to back-state regions of the membrane
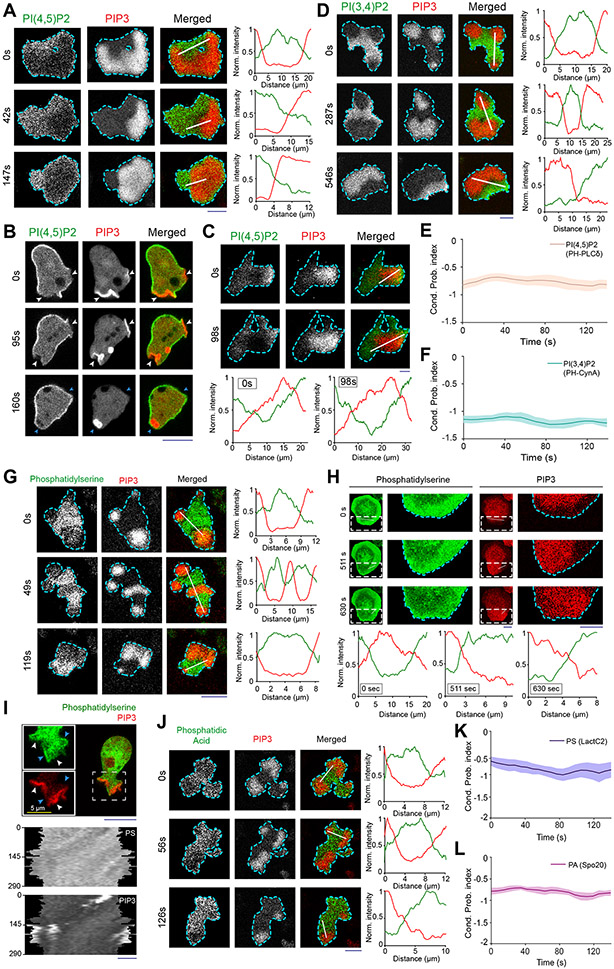
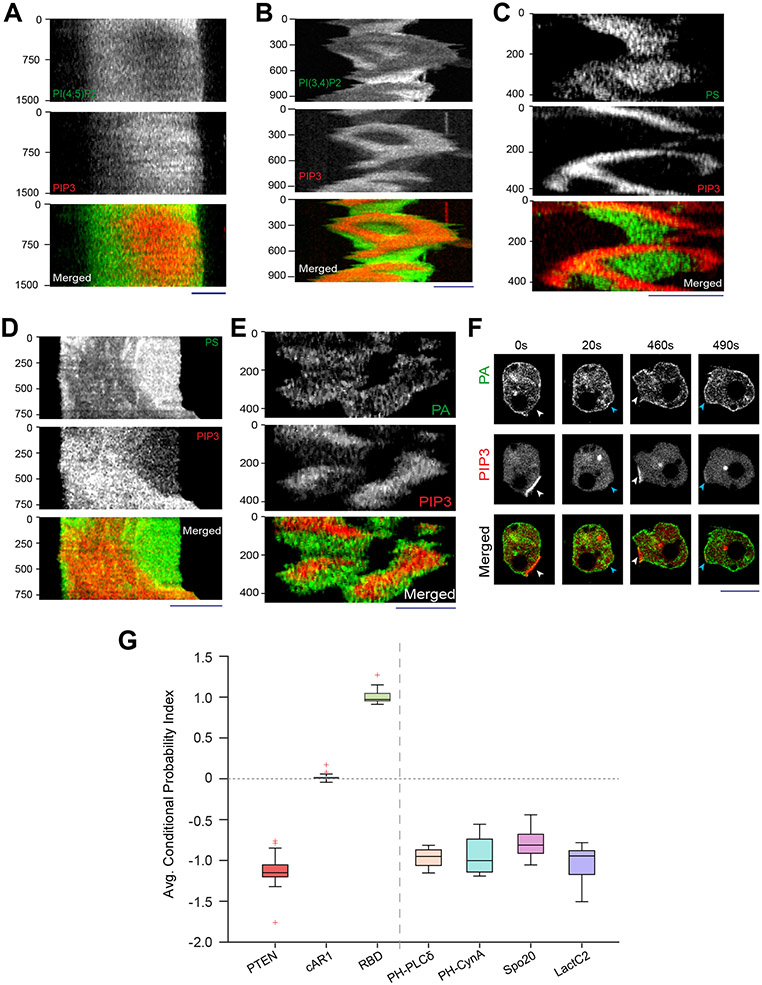
Using genetically-encoded biosensors, we discovered that several anionic phospholipids display similar dynamic patterns on the inner membrane. The distribution of biosensor PHPLCδ indicated that PI(4,5)P2 was depleted in the front-state regions of ventral waves and from protrusions in migrating Dictyostelium cells (Figures 1A,1B; Video S1). Equivalent patterns of PI(4,5)P2 were observed in RAW 264.7 macrophage ventral waves (Figure 1C; Extended Data Figure 3A; Video S1), as previously reported43, 44. PI(3,4)P2 biosensor, PHCynA, displayed a similar pattern in the Dictyostelium ventral waves (Figure 1D; Extended Data Figure 3B; Video S2), consistent with our previous reports of its depletion from the protrusions38, 45. Line-scans showed ~40-70% less PI(4,5)P2 and PI(3,4)P2 in front versus back regions of the waves. The consistently high negative CP index values for PI(4,5)P2 and PI(3,4)P2 showed that the complementarity with respect to PIP3 was maintained as waves propagated (Figures 1E,1F).
Figure 1. Multiple anionic phospholipids dynamically self-organize to the back-state regions of the membrane.
(A, B) Representative live-cell time lapse images of Dictyostelium cells co-expressing PI(4,5)P2 sensor PHPLCδ-GFP and PI(3,4,5)P3 sensor PHcrac-mCherry, during ventral wave propagation (A) or migration (B) showing PHPLCδ dynamically localizes to the back-stare regions in ventral waves (A) and analogously moved away from protrusions in migrating cells (B). White arrows showing protrusions where PIP3 is enriched and PHPLCδ is depleted whereas blue arrows showing PHPLCδ returning to the membrane domains where protrusions were retracted (B). (C) Live-cell images of RAW 264.7 macrophage co-expressing GFP-PHPLCδ and PHAkt-mCherry demonstrating dynamic complementary distribution in its ventral waves. (D) Live-cell images of Dictyostelium cells co-expressing PHcrac-mCherry and PI(3,4)P2 sensor PHCynA-KikGR, demonstrating the spatiotemporal back localization of PI(3,4)P2 in its ventral waves. (E, F) Time series plot of CP index for PI(4,5)P2 (E) and PI(3,4)P2 (F). PI(4,5)P2: nc=16 cells; PI(3,4)P2: nc=11 cells. (G-I) Dynamic back-state distribution of PS biosensor, GFP-LactC2, in ventral waves of Dictyostelium (G) and in RAW 264.7 macrophages (H). Corresponding GFP-LactC2 depletion from protrusions in migrating Dictyostelium cells (I). Protrusions and front-state regions of ventral waves are marked by PIP3 sensors. Bottom two panels of (I) show 360° membrane-kymographs around cell perimeter. In all kymographs, numbers on the left denote time in seconds, unless otherwise mentioned. (J) Complementary localization of PA sensor, GFP-Spo20, and PIP3 in Dictyostelium ventral waves. (K, L) Quantification of extent of back localization of PS and PA by time-series plot of CP index of LactC2 (K; nc=15 cells) and Spo20 (L; nc=16 cells); mean ± SEM. For all ventral wave figures, line scans are shown in right or bottom most panels. For all CP index time-plots in this paper, each of the nc cells was analyzed for nf =20 frames; all CP indices are calculated with respect to PIP3; mean ± SEM are plotted for all CP indices plots. For all figures, scale bars are 10 μm, unless otherwise mentioned.
Source numerical data are available in source data.
Since these two signaling lipids co-segregated to the back-state of the membrane, we wondered whether other major anionic phospholipids exhibit asymmetric patterns. Using biosensor LactC2, we found that phosphatidylserine , which accounts for ~20% of inner leaflet lipids46, 47, also localized to the back-states of ventral waves of Dictyostelium (Figure 1G; Extended Data Figure 3C; Video S3) and RAW 264.7 macrophages (Figure 1H; Extended Data Figure 3D; Video S3) and was depleted from protrusions in migrating Dictyostelium cells (Figure 1I). Using the biosensor Spo2048, 49, we observed that PA preferentially distributed to the back-state regions of the ventral waves as well (Figure 1J; Extended Data Figure 3E) and also away from protrusions (Extended Data Figure 3F). Line-scans showed ~30-50% less PS and PA in front versus back regions of the waves. The CP indices of PS and PA (Figures 1K,1L) were found to be consistently negative, similar to those of PI(4,5)P2, PI(3,4)P2, and PTEN (Extended Data Figure 2D,3G). Together, these observations show that four major anionic lipids decrease in the front-state region of the membrane where PIP3 increases. While PIP3 carries the most negative charge, it is a minor constituent46, 50-52.
Higher negative surface charge defines “back” regions of the membrane
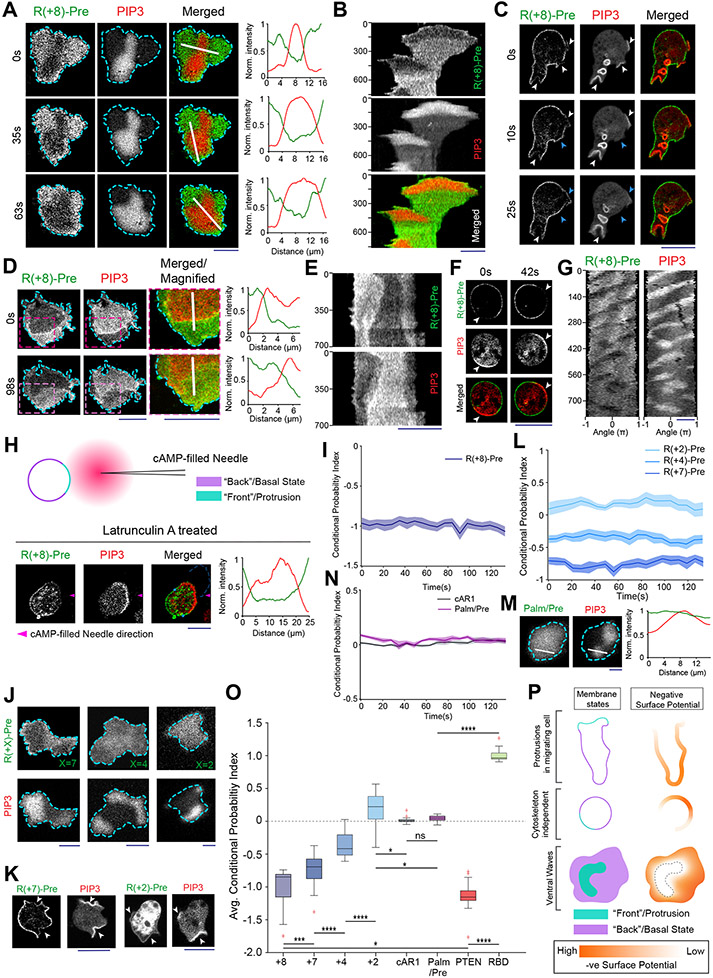
Reasoning that, with respect to charge, the decreases in the four predominant anionic phospholipids would more than offset increases in PIP3, we sought to measure spatiotemporal changes in inner membrane surface charge. We used R(+8)-Pre, a charged, prenylated peptide (Table S1), which had been characterized in vitro to detect the combination of anionic lipids 21, 28, 53. Consistent with the four anionic lipid distributions, the back-state regions of cortical waves in Dictyostelium cells are defined by higher negative surface charge versus the front-state regions (Figure 2A; Video S4). Line-scans demonstrated ~30-60% less sensor in front versus back regions of the waves (Figure 2A). Despite continuous wave propagation, as marked by PIP3, the sensor maintained its back-state localization (Figure 2B). Similarly, protrusions on migrating cells were depleted of R(+8)-Pre relative to the basal level on the membrane (Figure 2C; Video S4). An analogous spatiotemporal distribution of surface charge was also observed in the ventral waves of RAW 264.7 macrophages (Figures 2D,2E; Video S5).
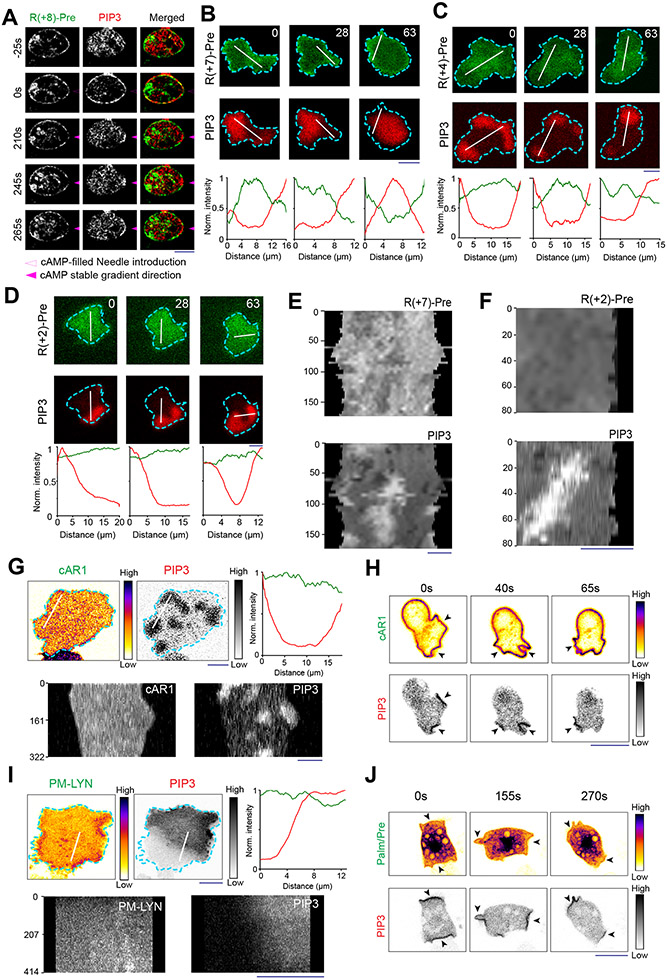
Figure 2. Back-state of the membrane maintains higher negative surface charge on the inner leaflet, compared to the front-state of the membrane.
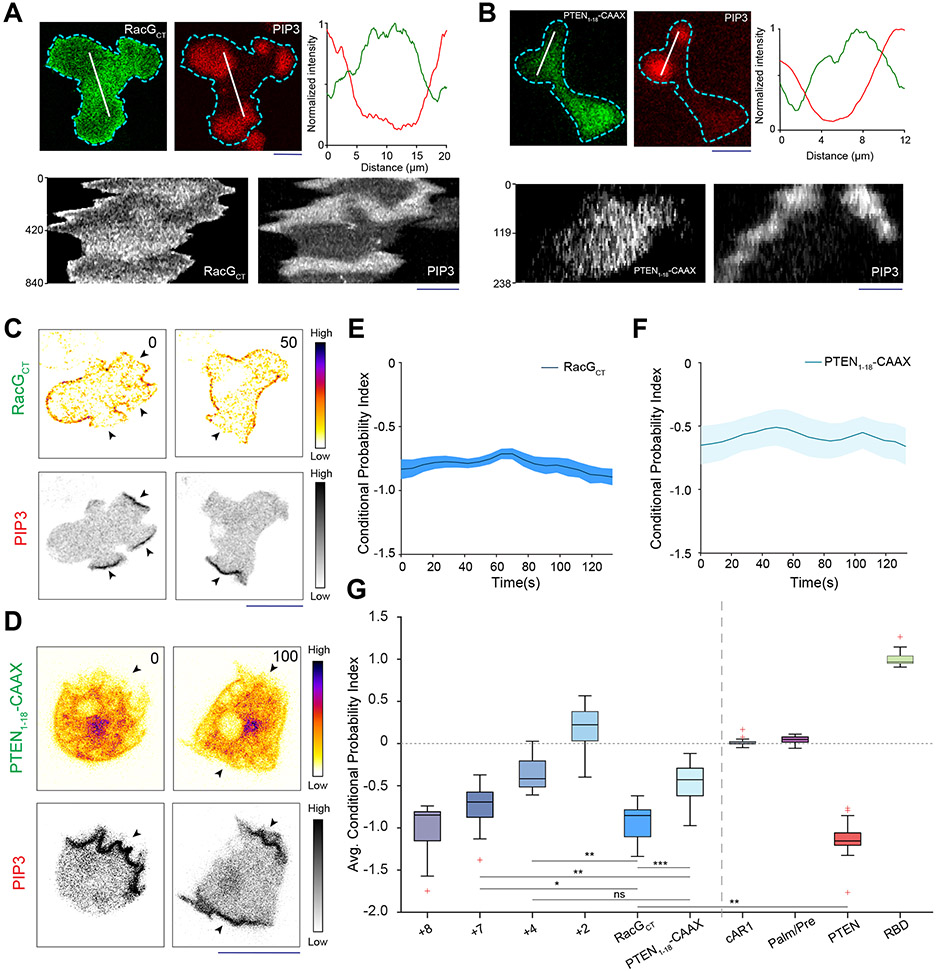
(A) Representative live-cell images of propagating ventral waves in a Dictyostelium cell co-expressing GFP-R(+8)-Pre (Surface Charge Sensor) and PHCrac-mCherry. (B) Representative line-kymograph of wave pattern shown in cell (A). (C) Live-cell images of migrating Dictyostelium cell showing GFP-R(+8)-Pre is depleted in protrusions. White arrows: PIP3 enriched protrusion; Blue arrows: Retracted protrusions. (D) Ventral waves in RAW 264.7 macrophages co-expressing GFP-R(+8)-Pre and PHAkt-mCherry, displaying analogous complementary kinetics. (E) Representative line-kymograph of wave pattern shown in cell (D). (F, G) Live-cell images (F) and 360° membrane-kymograph (G) of Actin-polymerization inhibitor Latrunculin-A treated Dictyostelium cells co-expressing GFP-R(+8)-Pre and PHCrac-mCherry. White arrows (F): Front-state, showing bright PIP3 patches and depletion of surface charge sensor. (H) R(+8)-Pre showing complementary localization with respect to front-state marker PIP3, during chemotactic gradient stimulation mediated receptor activation. Magenta arrowhead: Direction of micropipette. Cells were pre-treated with Latrunculin-A. (I) Time series plot of CP index of R(+8)-Pre, nc=29 cells. (J) Representative live-cell images of ventral waves in Dictyostelium cell membrane co-expressing PHCrac-mCherry, and GFP-R(+7)-Pre (left), GFP-R(+4)-Pre (middle), GFP-R(+2)-Pre (right). (K) Live-cell images showing R(+7)-Pre and R(+2)-Pre distribution, with respect to PIP3-rich protrusions (White arrows: protrusions), in migrating cells. (L) Time series plot of CP indices of mutated surface charge sensors R(+7)-, R(+4)-, R(+2)-Pre. +7: nc =23, +4: nc =20, +2: nc =12 cells. (M) GFP-Palm/Pre exhibiting uniform distribution in ventral waves, whereas PHCrac-mcherry was enriched in front-state regions. (N) Time series plot of CP indices of uniform membrane-markers cAR1 and Palm/Pre; cAR1: nc =20, Palm/Pre: nc =11. (O) Time averaged CP indices of surface charge sensors, uniform membrane marker controls, standard back protein PTEN (nc =17), and front sensor RBD (nc =15). To generate each datapoint in box plot, CP index values of nf =20 frames were averaged for each cell. Box and whiskers are graphed using Tukey’s method. The p-values by Mann-Whitney-Wilcoxon test. (P) Schematic of negative surface potential distribution in front- and back-state of the cell membrane during migration, cytoskeleton-independent symmetry breaking and ventral wave propagation.
Source numerical data are available in source data.
As previously reported for patterns of signaling events such as PIP3 accumulation and PTEN dissociation (Extended Data Figure 1H) 41, 42, 54, the dynamic distribution of surface charge was relatively independent of the actin cytoskeleton. In Latrunculin A-treated Dictyostelium cells where periodic circulating waves were induced, the charge sensor continuously adjusted its localization towards the back-state (Figures 2F,2G). Consistently, as shown in Figure 2H and Extended Data Figure 4A, when Latrunculin-treated cells were exposed to a chemotactic gradient, R(+8)-Pre, like standard back-protein PTEN, moved away from the typical front-region crescent of PIP3 55, 56. In all these scenarios, the CP indices indicated that the extent of complementary to PIP3 was comparable to that of established back protein, PTEN (Figure 2I; Extended Data Figure 2D).
These asymmetric distributions depended on charge and not on the specific amino acid sequences of the sensor. As the positively charged arginines of the sensor were sequentially replaced with neutral glutamines (Table S1), the distribution became increasingly uniform (Figures 2J-2L; Extended Data Figures 4B-4F). The dynamics of R(+2)-Pre (Figures 2J,2K; Extended Data Figures 4D,4F) resembled the uniform distribution of the G-protein-coupled receptor (GPCR) cAR1 (Extended Data Figure 4G,4H), the weakly charged myristoylated sequence of LYN (Extended Data Figure 4I), and the uncharged prenylated C-terminal tail of HRas (Figure 2M; Extended Data Figures 4J), as quantified by CP indices (Figures 2L,2N,2O). In contrast, two distinct polybasic sequences, PTEN1-18-CAAX and RacGCT (Table S1), carrying +6 and +7 charges respectively, displayed a complementarity to PIP3 in ventral waves and protrusions, which was roughly correlated with their net charge (Extended Data Figures 5A-5G). Thus, in protrusions of migrating cells, in activated crescents of Latrunculin-A treated cells, and in activated regions of propagating waves, the front-state of the membrane maintains a lower negative surface charge, compared to the back-state (Figure 2P).
Anionic lipids contribute to surface charge in an integrative manner
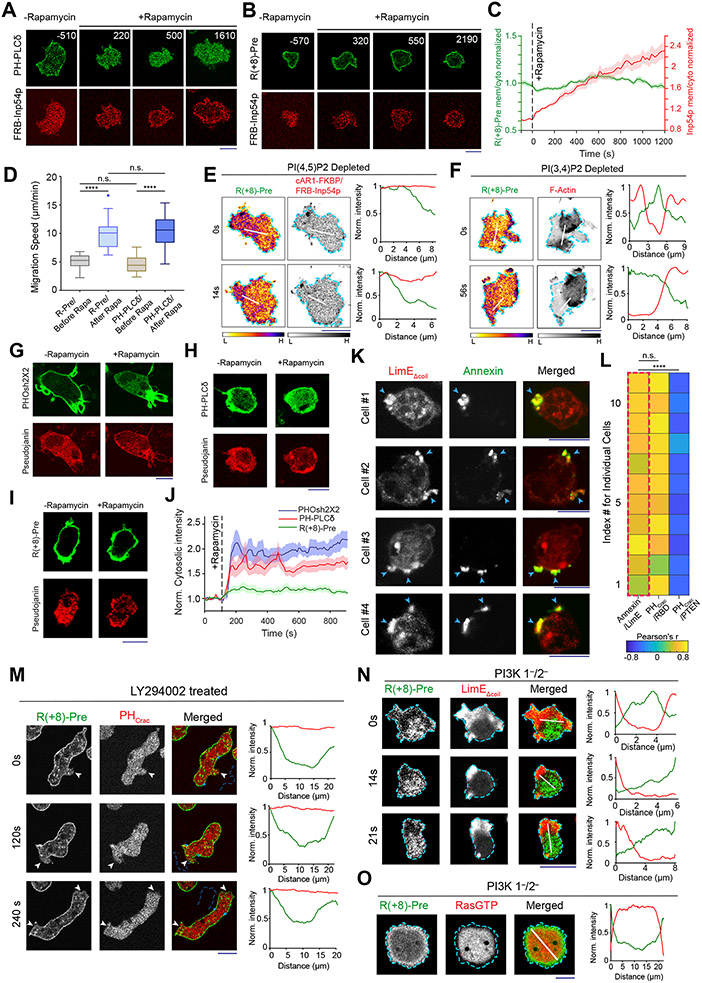
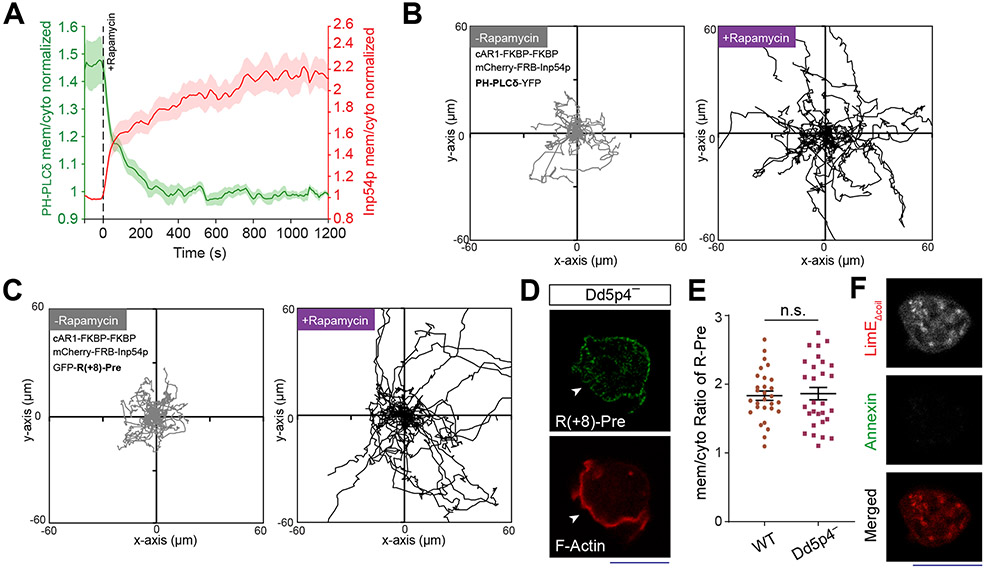
Neither PI(4,5)P2 nor PI(3,4)P2 fully accounted for the asymmetric distribution of the charge sensor. When we depleted PI(4,5)P2 by recruiting Inp54p to the membrane, PHPLCδ moved to the cytosol (Figure 3A; Extended Data Figure 6A), as expected. However, R(+8)-Pre remained membrane bound (Figures 3B,3C). As previously published38, 57, cells with low PI(4,5)P2 displayed altered migration (Figure 3D; Extended Data Figures 6B,6C) and expanded front regions. Importantly, R(+8)-Pre remained confined to diminished yet dynamically changing back regions (Figure 3E) . Similarly, in Dd5p4− Dictyostelium cells, where PI(3,4)P2 is low, R(+8)-Pre stayed on the membrane (Extended Data Figures 6D,6E), marking the smaller moving back-state regions (Figure 3F). The membrane retention of R(+8)-Pre following Inp54p recruitment was not due to its binding to the product PI4P, since recruitment of pseudojanin, which simultaneously converts PI(4,5)P2 to PI4P and PI4P to PI58, caused sensors for both PI(4,5)P2 (Figure 3G; Video S6) and PI4P (Figure 3H; Video S6) to move to the cytosol but R(+8)-Pre largely remained on the membrane (Figures 3I,3J; Video S7). Thus, dynamic regulation of the surface charge depends on PI(4,5)P2 and PI(3,4)P2, but there are additional key contributors (such as phosphatidylserine and phosphatidic acid).
Figure 3. Dynamics of inner membrane surface charge sensor R(+8)-Pre in different anionic phospholipid depleted cells.
(A, B) The dynamics of membrane localization of PHPLCδ-GFP (A) or GFP-R(+8)-Pre (B), before and after PI(4,5)P2 depletion in Dictyostelium cells, by recruiting mCherry-FRB-Inp54p to membrane-bound cAR1-FKBP. Numbers indicate time in seconds; rapamycin added at 0s. (C) Time course of membrane-to-cytosol ratio of R(+8)-Pre (green) and Inp54p (red) upon rapamycin addition (indicated by dashed vertical line); nc=20 cells; mean ± SEM. (D) Migration speed of cells, co-expressing chemically inducible dimerization system for PI(4,5)P2 depletion, along with either GFP-R(+8)-Pre or PHPLCδ-GFP, before and after rapamycin addition; nc=32 cells tracked for nf=60 frames for each case. (E) Spatiotemporal back localization of R(+8)-Pre in PI(4,5)P2 depleted cells, shown in “fire invert” colormap of Fiji/ImageJ; Inp54p recruited to uniform membrane-anchor cAR1 is symmetric in ventral waves (middle panel). (F) Complementary localization of GFP-R(+8)-Pre and LimE-mCherry in ventral waves of Dd5p4− Dictyostelium cells. (E, F): L, low; and H, high. (G-I) Live-cell images of RAW 264.7 macrophages showing the membrane localization profile of PI4P biosensor PHOsh2X2-GFP(G), PI(4,5)P2 biosensor PHPLCδ-GFP (H), or GFP-R(+8)-Pre (I), before and after recruiting Pseudojanin to membrane anchor Lyn-FRB-CFP. (J) Time course of normalized cytosolic intensity of PHOsh2X2, PHPLCδ, and R(+8)-Pre, upon rapamycin addition; time of addition is indicated by dashed vertical line; PHOsh2X2: nc=16 cells, PHPLCδ: nc=10 cells, R(+8)-Pre: nc=12 cells; mean ± SEM. (K) Representative examples of protrusion forming Dictyostelium cells expressing LimE-GFP, whose outer leaflet of membrane was allowed to transiently bind with Annexin V-Alexa Fluor 488. (L) Heatmap of Pearson correlation coefficients (PCC) between Annexin V and LimE (leftmost column, highlighted with dashed red rectangle). PCCs between (PHCrac and RBD) and (PHCrac and PTEN) are shown to demonstrate standard Co- and counter- localization profiles. Correlation coefficients were calculated along cell membrane; nc=11 cells in each case. (M) Live-cell images of LY294002-treated migrating Dictyostelium cell co-expressing GFP-R(+8)-Pre and PHCrac-mCherry. White arrows: Protrusions where PIP3 was depleted yet surface charge gradient was maintained. (N, O) Live-cell images of ventral waves in PI3K 1—/2— Dictyostelium cells co-expressing GFP-R(+8)-Pre and front-state markers LimE-mCherry (N) or RBD-mCardinal (O). All p-values by Mann-Whitney-Wilcoxon test.
Source numerical data are available in source data.
We sought a possible mechanism for the decreases of PS inside the activated regions of inner membrane since, unlike phosphoinositides, PS is not modified by kinase/phosphatases. Reversible non-apoptotic flipping of PS was reported in several physiological contexts such as cytosolic calcium release and immunological stimulation59, 60, although a connection with signaling and cytoskeleton networks has not been established21, 46. We performed a transient Annexin-V binding assay (see Methods) to label outer leaflet PS during protrusion formation. Remarkably, on the outer leaflet, PS strongly localized to the protrusions (Figure 3K), closely opposing its back-state distribution on the inner leaflet (Figure 1G-1I). In quiescent cells where protrusions were absent, Annexin V did not bind to outer leaflet (Extended Data Figures 6F). Pearson’s correlation coefficient (PCC) values between front-state/protrusion marker LimE and Annexin V were similarly positive to that of two front-state markers PHCrac and RBD (Figure 3L), while PCC for complementary pair PHCrac and PTEN was negative (Figure 3L). Obviously, more studies will be required, but our data suggest that inner leaflet PS may transiently flip to outer leaflet in the membrane domains where the networks are activated.
PI(3,4,5)P3 is present at negligible amounts in the membrane46, 50-52 and theoretically, it can perform its well-known signaling functions without significant contribution to surface charge51. Nevertheless, we directly tested the contribution of PIP3 to surface charge profile. First, cells were treated with PI3K inhibitor LY29400261-64, which resulted in a loss of PIP3 accumulation in the protrusions (Figure 3M). Yet, the typical depletion of the surface charge sensor from the protrusions was largely unchanged (Figure 3M). Second, in Dictyostelium PI3K 1−/2− cells where PIP3 production is severely impaired62, 65, R(+8)-Pre profile remained strongly complementary to front-state sensors for actin polymerization (Figure 3N) and Ras-activation (Figure 3O). These results demonstrate that PIP3 does not significantly contribute to the membrane surface charge.
Decreasing net surface charge triggers protrusions and abrogates polarity
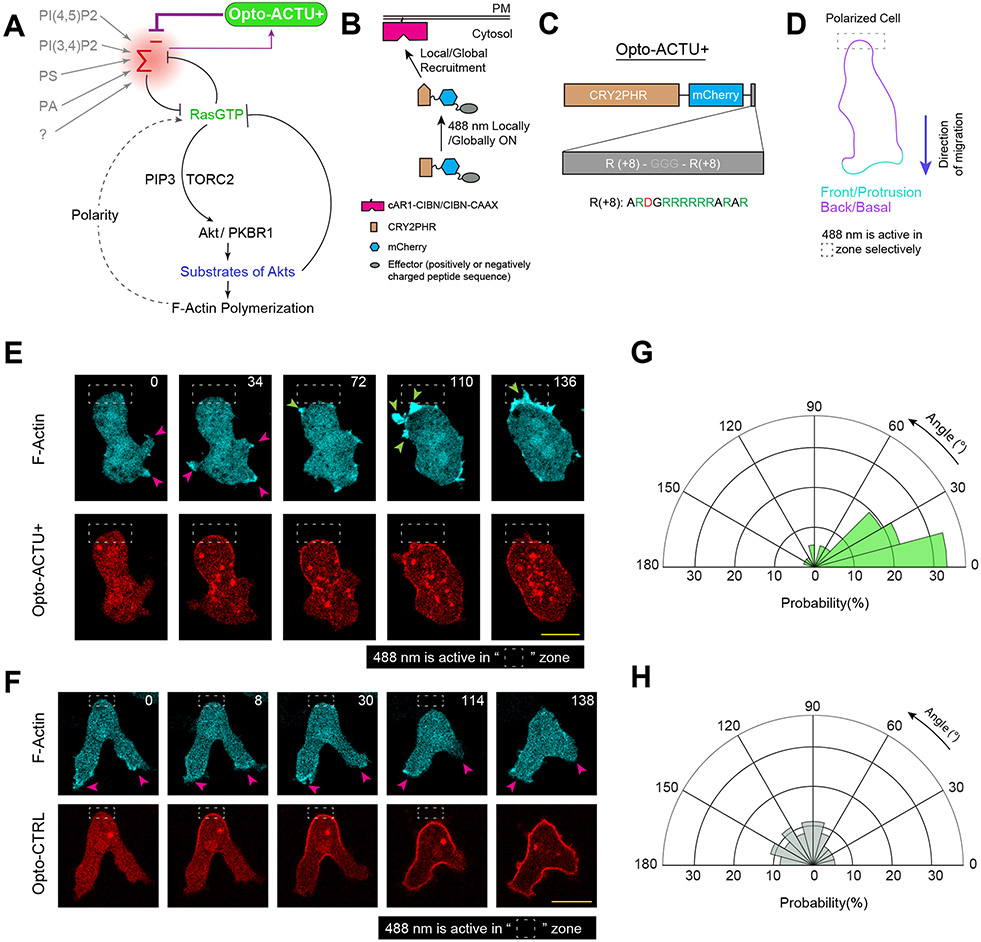
The dynamic patterns of the surface charge suggests that it could serve as a biophysical organizer of signaling and cytoskeletal networks. These networks have been previously modeled as excitable systems consisting of positive and delayed negative feedback loops 57, 66-70. Studies suggested that the delayed negative feedback loop includes the action of substrates of Akts and other refractory molecules57, 71. The positive feedback was implemented as a mutually inhibitory interaction between front- and back-states, consisting of Ras/Rap and PI(4,5)P2/PI(3,4)P2 respectively, given that induced decreases in these lipids activated the network 9, 38. However, we considered these effects might be attributable to decreases in surface charge rather than the signaling properties of these lipids (Figure 4A). In this scenario, the activated state would decrease surface charge while decreasing negative surface charge would lead to activation (Figure 4A). To test this concept, we developed two non-specific actuators to acutely decrease or increase the negative surface potential profile via optogenetic recruitment (Figure 4B) 72-76. Consisting of a short-chain of charged amino acids, these actuators (unlike conventional GEF/GAP/RGS/kinase/phosphatase-based actuators) are not directed at any particular biochemical reaction.
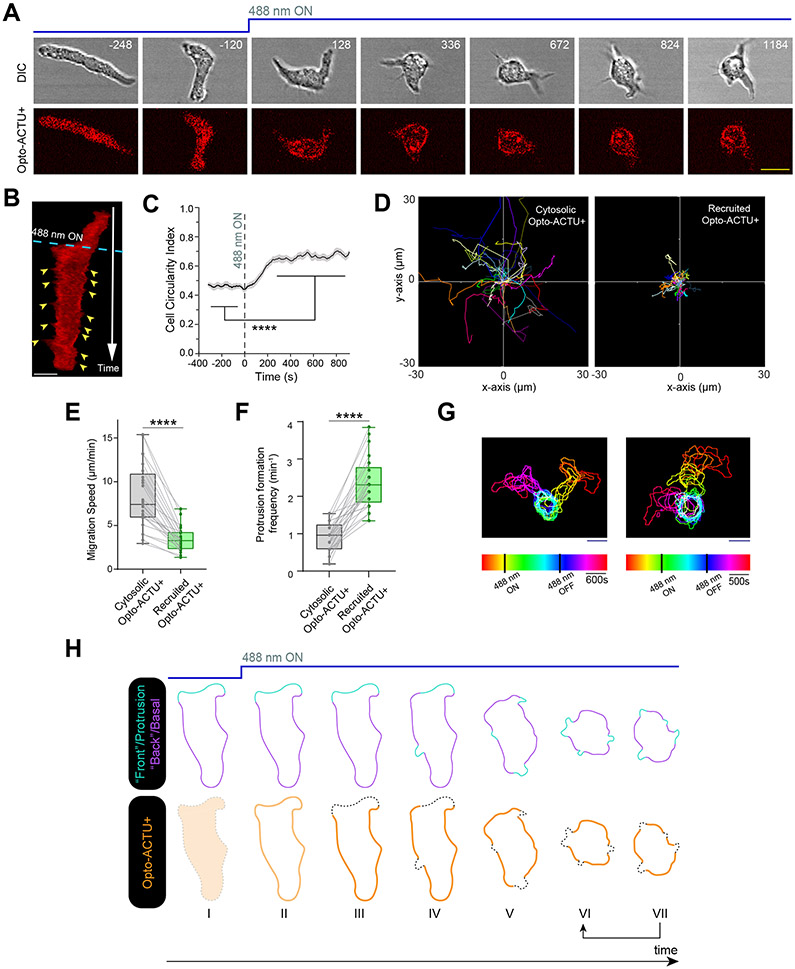
Figure 4. Localized lowering of membrane surface charge can trigger de novo protrusion generation from the area of reduced surface charge.
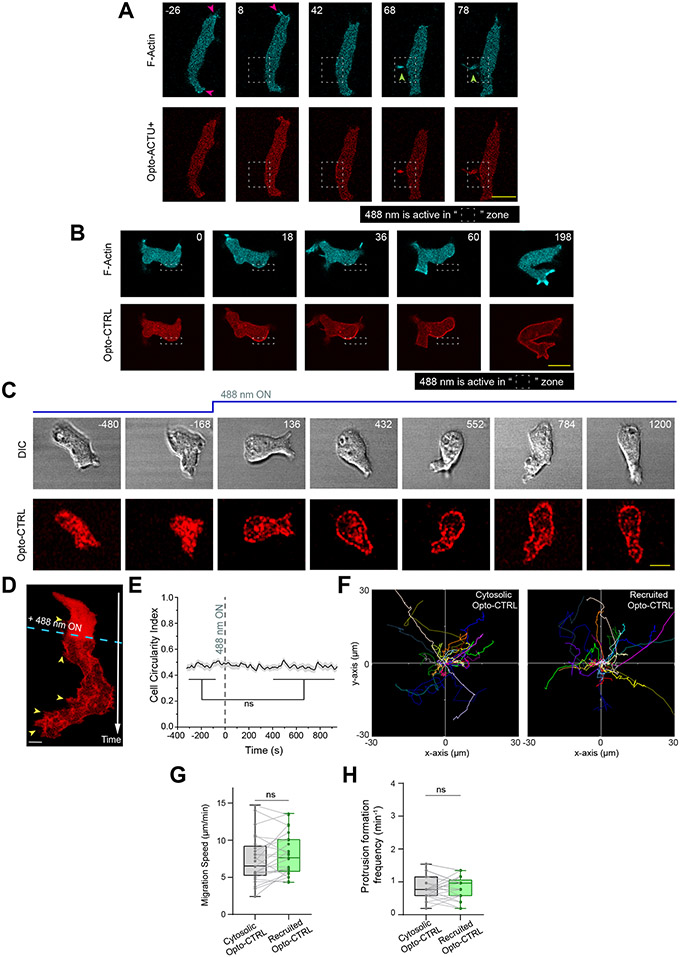
(A) Scheme for lowering negative surface charge by the recruitment of positively charged optogenetic-actuator, Opto-ACTU+, in context of overall biochemical excitable network topology. Σ—: back-state defined by overall negative surface charge. Opto-ACTU+ interfere into the topology by getting associated with Σ—, but in turn it provides a negative feedback to Σ—. (B) Scheme for optogenetic actuator recruitments. Turning on 488 nm laser changes the conformation of cytosolic cryptochrome module and as a result, CRY2PHR along with its associated positively or negatively charged peptide, gets recruited to the plasma membrane bound CIBN. The cAR1-CIBN is used in all Dictyostelium experiments and CIBN-CAAX is used in all mammalian systems as membrane anchor. (C) Design of Opto-ACTU+ with net charge +16. Positively charged amino acids are shown in green and the negatively charged amino acids are shown in red. (D) Experimental setup of selective optical recruitment at the back of polarized Dictyostelium cells. (E) Representative time-lapse images of selective de novo protrusion formation from the area of recruitment in Dictyostelium cells co-expressing Opto-ACTU+, cAR1-CIBN, and LimE-Halo. (F) Representative time-lapse images of Dictyostelium cells co-expressing Opto-CTRL, cAR1-CIBN, and LimE-Halo demonstrating locally restricted recruitment of Opto-CTRL did not generate new protrusions from the site of recruitment. In (E) and (F), dashed rectangle shows the area where 488 nm laser was selectively illuminated for recruitment; magenta and green arrows show existing and newly induced protrusions, respectively; time in seconds. (G, H) Polar histogram of angle of protrusion formation with respect to recruitment area, in case of Opto-ACTU+ (G) and Opto-CTRL (H) recruitment; Probabilities were calculated for nc=23 cells, np=36 protrusions in (G) and for nc=20 cells, np=36 protrusions in (H).
Source numerical data are available in source data.
The first actuator, designated Opto-ACTU+, had +16 net charge (Figure 4C). Upon spatially restricted recruitment from cytosol to membrane (Figure 4D), it should reduce the net negative surface charge on inner membrane and, thereby increase the positive feedback (Figure 4A). That would increase the probability of spontaneous firing and de novo protrusion generation. Indeed, when Opto-ACTU+ was recruited to a quiescent back-region of a polarized Dictyostelium cell, new protrusions started forming nearby (Figure 4E; Extended Data Figures 7A; Video S8), resembling the effects of previously reported specific perturbations of signaling molecules (such as Rac1, Cdc42, or GPCR opsins) 77-79. With recruitment of uncharged Opto-CTRL, new protrusions rarely appeared near the irradiation area (Figure 4F; Extended Data Figure 7B; Video S8), indicating that the protrusion formation was due to the surface charge reduction and not irradiation or cryptochrome recruitment. Angular histograms demonstrates that the probability of new protrusion generation was highest in the vicinity of the Opto-ACTU+ recruitment (Figure 4G) whereas Opto-CTRL recruitment did not induce a bias (Figure 4H).
We anticipated that when Opto-ACTU+ was globally recruited in polarized cells, more protrusions would be generated and polarity would be disrupted. Indeed, within few minutes of global recruitment of Opto-ACTU+, cells began to extend protrusions rapidly all along the cortex (Figures 5A,5B; Video S9), including from domains of erstwhile back-states. Consequently, polarity was abrogated (Figure 5C) and migration was impaired (Figures 5D,5E). The number of new protrusions increased about 2.5 fold (Figure 5F) which theoretically is consistent with amount of surface charge reduction (Supplementary Note 1 and 2). When the 488 nm laser was turned off, Opto-ACTU+ returned to the cytosol, cells repolarized and resumed polarized migration (Figure 5G; Video S10). No consistent changes in polarity, migration, or protrusion formations were observed upon uncharged Opto-CTRL recruitment (Extended Data Figures 7C,7D; Video S9), as quantified in terms of cell circularity index (Extended Data Figures 7E), speed (Extended Data Figures 7F,7G), and protrusion formation frequency (Extended Data Figures 7H).
Figure 5. Globally lowering negative surface charge of the membrane can abolish the pre-existing polarity by consistently generating new protrusions from the back-state regions.
(A, B) Time-lapse snapshots (A) and time-stack (B) demonstrating cell morphology and migration mode changes in a polarized Dictyostelium cell, upon recruitment of Opto-ACTU+. Cells were co-expressing Opto-ACTU+ and cAR1-CIBN. Time in seconds. 488 nm laser switched ON globally at t=0s. Yellow arrowheads in (B): newly generated protrusions. Note that Opto-ACTU+ is consistently depleted in the protrusions. (C-F) Quantification of cell morphology and migration mode changes in terms of cell circularity index (C), cell tracks (D), migration speed (E), and new protrusion formation frequency (F) upon Opto-ACTU+ recruitment (n=25 cells). Data are mean ± SEM in (C). In (D-F), for either before or after recruitment, each cell was tracked for nf=40 frames (8 sec/frame was the image acquisition frequency). Tracks were reset to the same origin in (D). For pairwise comparison, tracks are color-coded in (D) and data from same cell are connected by gray lines in (E) and (F). All p-values by Mann-Whitney-Wilcoxon test. (G) Two representative examples of temporally color-coded cell outlines showing cell morphology and migratory mode before 488nm was turned on, during 488nm kept on, and after 488nm was switched off. (H) Schematic proposing how Opto-ACTU+ recruitment changes cell morphology and migratory mode. Opto-ACTU+ is recruited globally as expected (I to II); however, presumably due to its positive charge, it quickly accumulates along the back regions of the cell (III). Consequently, new protrusions are elicited from these back regions and the cell begins to lose polarity (IV and V). And at the same time, as some areas of erstwhile back regions are converted to front, Opto-ACTU+ redistributes again to the newly formed back regions (IV to VII). This in turn generates fresh protrusions there and this entire cycle is repeated (shown in arrows between VI and VII). As a result, protrusions are generated randomly, migration becomes impaired, and pre-existing polarity is abrogated.
Source numerical data are available in source data.
Careful observation revealed that in addition to the increase in protrusion formation induced by the recruitment of the Opto-ACTU+, a series of events was occurring: As new protrusions/front-state regions established, recruited Opto-ACTU+ quickly moved away from these regions (Figures 5A,5B; Video S9) and gradually accumulated toward other back-state regions. That accumulation triggered another protrusion there and the entire cycle was repeated (Figure 5H).
Surface charge alters migration and polarity through signaling and cytoskeleton networks
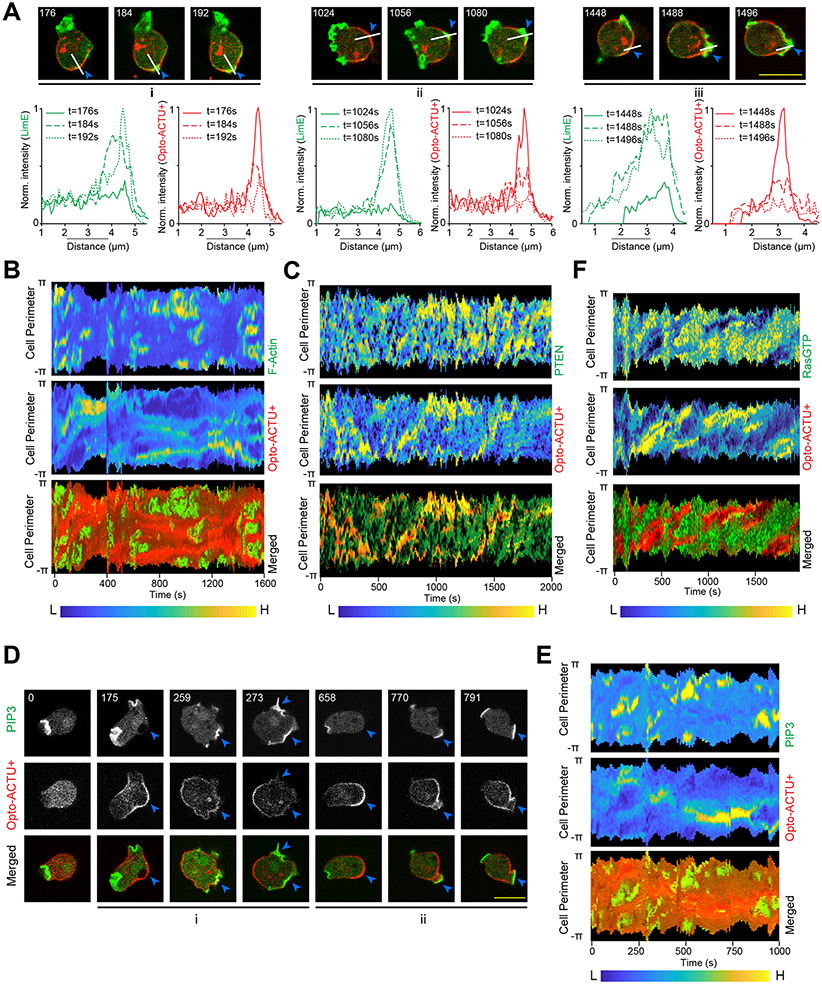
To investigate the molecular mechanisms through which membrane surface charge acts, we used a series of genetically-encoded biosensors and different pharmacological and genetic perturbations, in conjunction with our optogenetic actuators (Figure 6A). First, when Opto-ACTU+ was recruited, it accumulated to the existing back-state regions of the cortex and actin polymerization was eventually initiated there (Figure 6B). The local accumulation of Opto-ACTU+ consistently preceded actin polymerization (Figure 6B; Extended Data Figures 8A,8B). Next, we monitored the activation and localization kinetics of different upstream signaling molecules during surface charge perturbation. Localization of Opto-ACTU+ to a spatially confined domain within the back-states of the membrane consistently caused dissociation of established back protein PTEN from that domain (Figure 6C; Extended Data Figure 8C). Next, using the standard biosensors for PIP3 and GTP-bound Ras, we found that PI3K (Extended Data Figures 8D,8E) and Ras (Figure 6D; Extended Data Figure 8F) were dynamically activated in the membrane domains where surface charge was lowered.
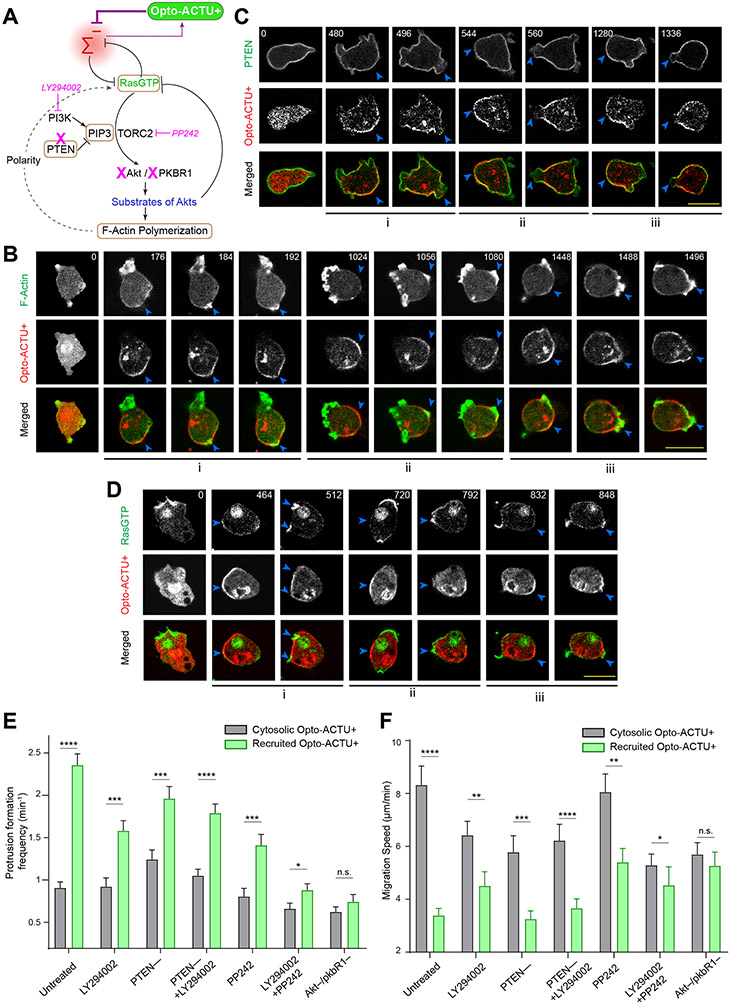
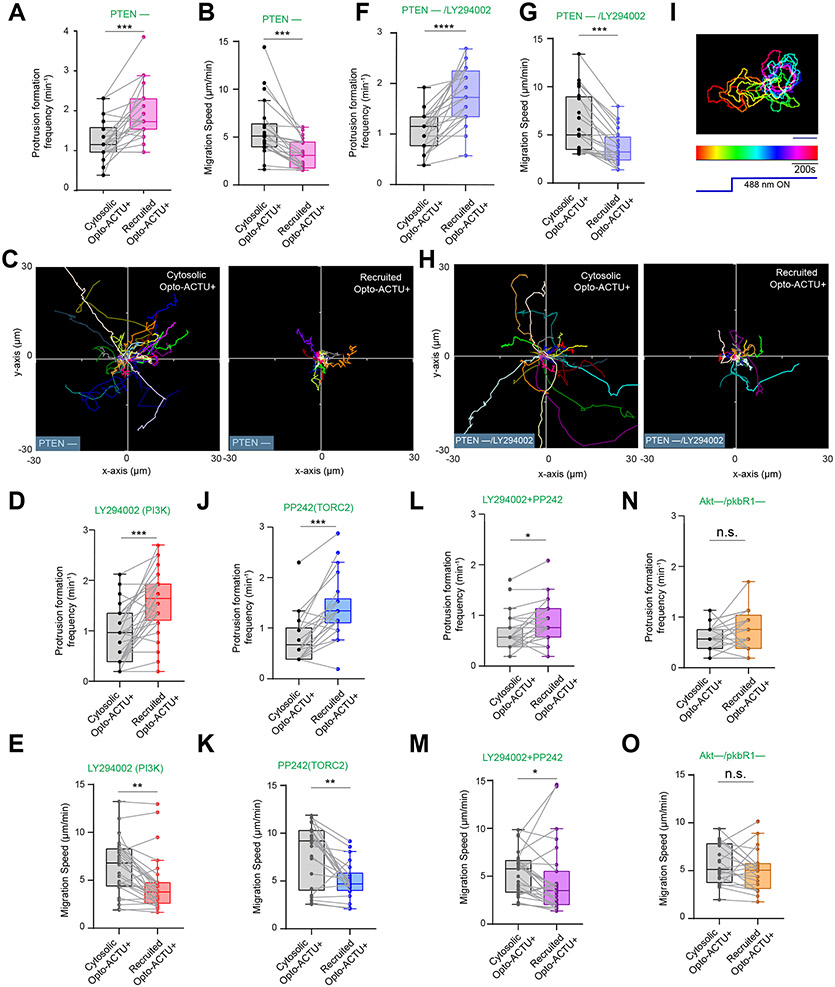
Figure 6. The phenotypic changes induced by Opto-ACTU+ recruitment is mediated by the Ras/PI3K/Akt/TORC2/F-actin network.
(A) Scheme showing nodes of signaling and cytoskeletal network that were monitored and/or perturbed, in conjunction with Opto-ACTU+ recruitment. Tan-colored rectangles: Molecules whose dynamics were recorded; Magenta blocked arrows: Pharmacological inhibitions; Magenta cross-marks: Genetic knockouts. (B) Live-cell images of Dictyostelium cells co-expressing Opto-ACTU+, cAR1-CIBN, and LimE-GFP, where recruitment was started at t=0s (first time point). The “i”, “ii”, “iii” are showing three representative actin polymerization cases. For each case, three events are shown: first, Opto-ACTU+ accumulated inside a domain of the membrane; second, F-actin polymerization was initiated there; and finally, when that domain fully turned into front state, Opto-ACTU+ moved away from that domain. Throughout this figure, Blue arrowheads: Membrane domains where Opto-ACTU+ was first accumulated and where back-states were replaced by front events; time in seconds. (C) Live-cell images of Dictyostelium cells co-expressing Opto-ACTU+ and cAR1-CIBN, along with PTEN-GFP (C) or RBD-GFP (D), where recruitment was started at t=0s (first time point). The “i”, “ii”, “iii” are showing three representative cases of PTEN dissociation from the membrane (C) or Ras activation on the membrane (D). For each case, two events are shown: first, the accumulation of recruited Opto-ACTU+ inside a domain of the membrane, and second, when that accumulation resulted in the dissociation of PTEN (as in C) and Ras activation (as in D) in that particular domain which, in turn, caused Opto-ACTU+ to move away from there. (E, F) Quantification of phenotypic changes upon Opto-ACTU+ recruitment in terms of new protrusion formation frequency (E) and migration speed (F), in presence of different pharmacological inhibitors or genetic knockouts. Untreated: nc= 28 cells; LY294002: nc= 28 cells; PP242: nc= 22 cells; PTEN –: nc= 22 cells; PTEN –+ LY294002: nc= 24 cells; LY294002+PP242: nc= 27 cells; Akt—/PKBR1—: nc= 21 cells. For each case, each of the nc cells were tracked for nf=40 frames (8 sec/frame) and time averages were taken. For pairwise comparison and more detailed data, please see Figures S9. Mean ± SEM are shown. The p-values by Mann-Whitney-Wilcoxon test.
Source numerical data are available in source data.
Next, we modified different nodes of Ras/PI3K/TORC2/Akt/F-Actin network using genetic knockouts and drug treatments to interfere with the Opto-ACTU+-mediated increased protrusion formation (Figure 6A). First, in pten– Dictyostelium cells, which have elevated PIP3 levels and increased protrusions and impaired migration80-82, recruitment of Opto-ACTU+ induced even more frequent protrusions (Figure 6E; Extended Data Figure 9A) which further impaired migration (Figure 6F; Extended Data Figures 9B,9C). Reducing PIP3 by pre-treating with LY29400261-64 only slightly inhibited the Opto-ACTU+ recruitment driven phenotypic changes in protrusion frequency (Figure 6E; Extended Data Figure 9D) and migration speed (Figure 6F; Extended Data Figure 9E) . When both PTEN and PI3K activity were impaired, recruitment of Opto-ACTU+ still increased protrusion formation (Figure 6E; Extended Data Figure 9F), slowed migration (Figure 6F; Extended Data Figures 9G,9H), and reduced polarity (Extended Data Figure 9I; Video S11). Thus, while reduction of surface charge is normally associated with PI3K activation and membrane dissociation of PTEN, in the absence of these activities, the phenotypic changes could be still induced. This observation does not completely rule out a role of PIP3 since the inhibition may not be complete and other enzymes may substitute for PTEN65, 81, 83. Standalone inhibition of TORC2 by PP2429, 84 only slightly hindered the Opto-ACTU+ recruitment driven migration phenotypic changes (Figures 6E,6F; Extended Data Figures 9J,9K). On the other hand, inhibition of both PI3K and TORC2 with cocktail of LY294002 and PP242 almost completely blocked the induced change in protrusion formation and migration speed (Figures 6E,6F; Extended Data Figures 9L,9M). Since PI3K and TORC2 act together to activate AKT and AKT-related kinase, PKBR1, 9, 62, 63, we studied the effect of optogenetic perturbation in the Akt −/PKBR1− cells. The Opto-ACTU+ recruitment induced phenotypic changes did not occur in these cells (Figure 6E,6F; Extended Data Figure 9N, 9O). Together, these findings suggest that surface charge-mediated changes in migration or protrusion formation acts via the collective action of Ras/PI3K/TORC2/AKT/F-Actin network.
Addition of actuators to excitable network simulations mimics the observed phenotypes
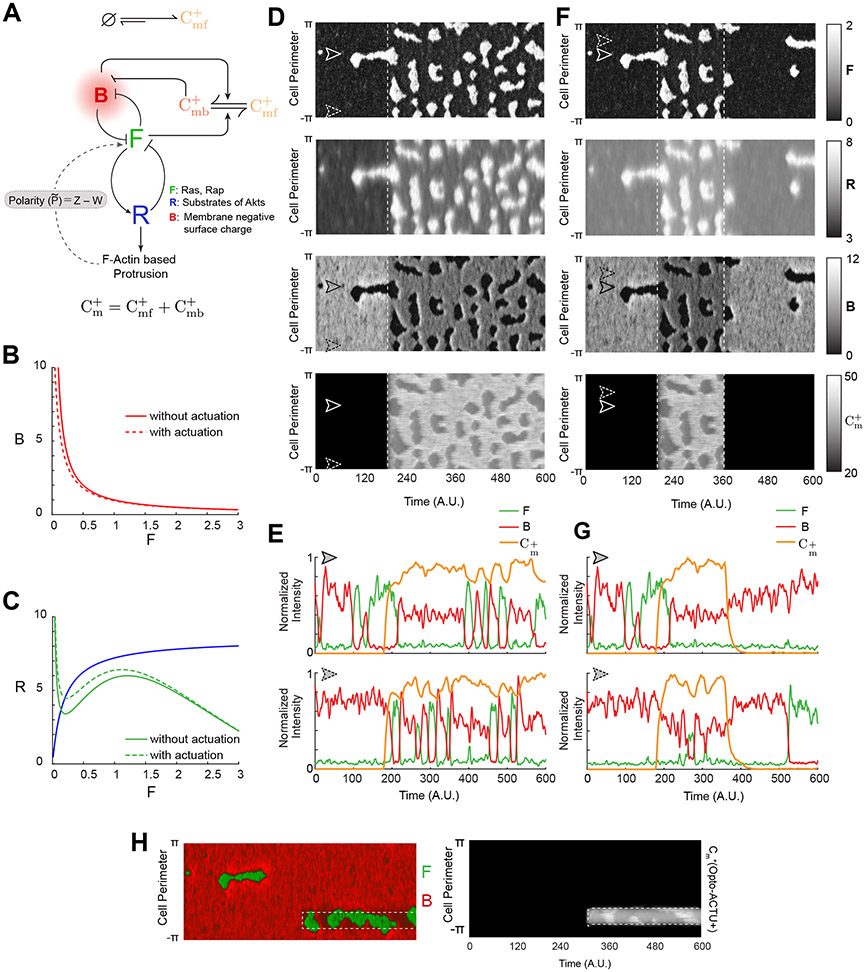
To simulate these polarity breaking and protrusion formation phenomena, we incorporated separate actuator dynamics into a model of the excitable network that includes polarity 85 (Supplementary Note 1; Figures 7A-7C). Prior to incorporating perturbations, polarity biased the excitable network, resulting in persistently localized firings which underlie protrusions (Figure 7D). Following simulated recruitment, Opto-ACTU+ lowered the threshold and caused abrupt increases in overall activity along the whole perimeter. With the development of each front-state region, Opto-ACTU+ quickly redistributed to new back-state regions where it reduced the local threshold, resulting in triggering of new protrusions (Figures 7D,7E). As in experiments, this cycle repeated. Reversibility of this actuation process was also recreated in the simulations by allowing the actuators to dissociate from the membrane (Figures 7F,7G). We also simulated the selective protrusion formation by confining the recruitment of the actuator within a back-state region and observed increased activity there which resulted a switch in polarity and usual eventual actuator rearrangement (Figure 7H). Together, these findings suggest that during polarized cell migration, higher negative surface charge at the back-state regions leads to an increased threshold which prevents protrusion formation there.
Figure 7. In silico reduction of membrane surface charge recreates the polarity breaking and demonstrates increased activity over the membrane.
(A) Schematic showing coupled system of excitable network, polarity module (involving Z and W), and Opto-ACTU+ () system. The excitable network involves membrane states F (front), B (back, defined by overall surface charge of inner membrane), and R (refractory). The polarity module comprises of local activator Z and delayed globally diffusing inhibitor W. The Opto-ACTU+ system constitutes of fast diffusing state and almost stationary membrane bound state . The total charge actuator, on the membrane is the summation of both the fast-diffusing and membrane-bound states. (B) Plot of B vs F with and without Opto-ACTU+. (C) F (green lines) and R (blue line) nullclines with and without Opto-ACTU+ (under the steady-state assumption for B). (D) The simulated kymographs of F (first), R (second), B (third) and (fourth) in response to global recruitment. The instant of recruitment is shown by the white dashed line. (E) Line scans at two locations (as denoted by dashed and solid arrows) on the simulated kymographs showing the temporal profiles of F (green), B (red) and (orange). (F, G) In silico reversible recruitment of Opto-ACTU+ demonstrating the reversibility of polarity breaking and protrusion formation events. (F) is showing the simulated kymographs and (G) is showing the line scans at two locations (as denoted by dashed and solid arrows). The color schemes of F, R, and B are same as (E). The instant of recruitment is shown by the first white dashed line and recruitment was stopped in the second dashed line. (H) Simulated kymographs of membrane states in response to the selective recruitment of Opto-ACTU+ (Cm+). Merged view of F (green) and B (red) are shown in the left panel; profile of Opto-ACTU+ () was shown in the right panel (in grayscale); White dashed box: Location of in silico selective recruitment. The same timescale is applicable in both the left and right panels. Note that, simulated selective recruitment created de novo front-state around recruitment area which, in turn, caused the recruited actuator to move away (as happened in global recruitment).
Locally elevated surface charge suppresses protrusions
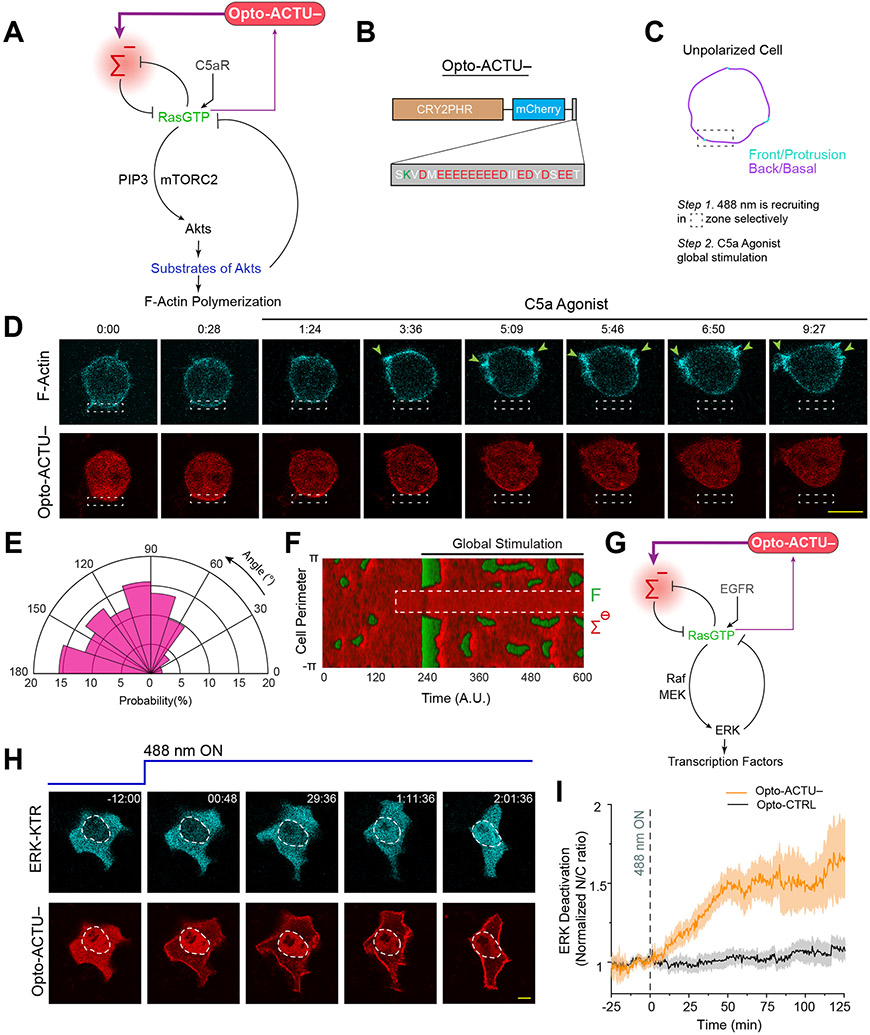
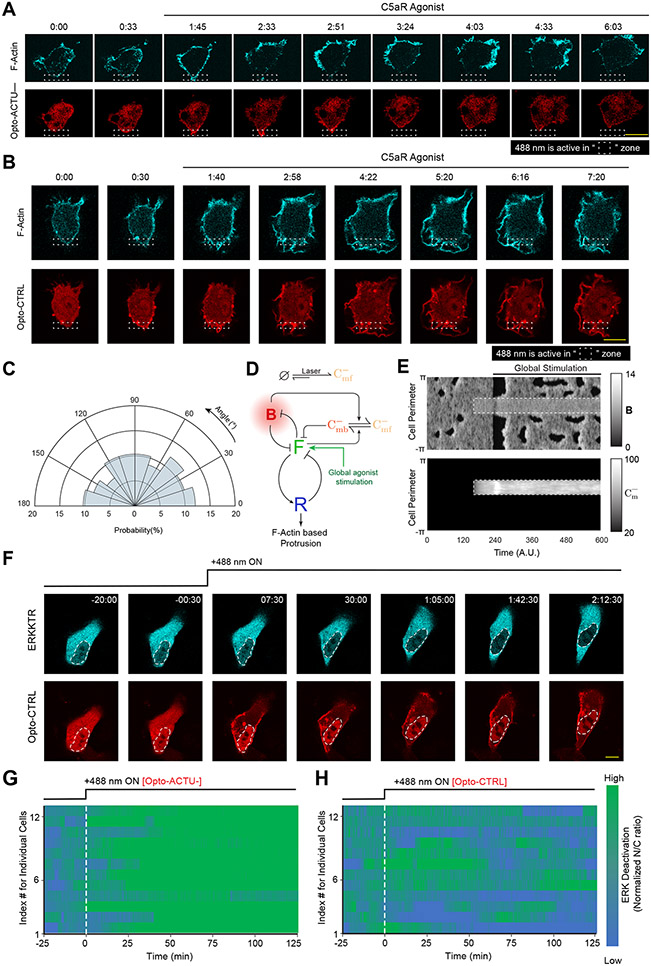
Since reduction of negative surface charge was able to activate the signaling network, we asked whether its direct elevation can deactivate the network and limit protrusions (Figure 8A). To test this, we designed a second optogenetic peptide, Opto-ACTU−, with net charge of −14 which is expected to increase the negative surface charge on the membrane upon recruitment (Figure 8B). We chose RAW 264.7 macrophages that are generally quiescent but can be globally activated by C5a receptor agonist to induce protrusions all along the membrane. We first recruited Opto-ACTU− to a confined region to increase the local negative surface charge and then globally stimulated the cell with C5a receptor agonist (Figure 8C). Protrusions formed along the membrane except in the vicinity of recruitment region (Figure 8D; Extended Data Figure 10A; Video S12). This induced a polarity in the cell and the cell slowly migrated away (Figure 8D; Extended Data Figure 10A; Video S12), resembling migration phenotype caused by direct perturbations of signaling molecules16, 86. Compared to Opto-ACTU− (Figure 8E), local recruitment of Opto-CTRL could not suppress agonist-induced protrusions (Extended Data Figure 10B; Video S12) and the protrusion formation probability remained uniform (Extended Data Figure 10C). In simulations, when we locally elevated the threshold mimicking Opto-ACTU− recruitment and superimposed a global reduction of threshold representing receptor input (Extended Data Figure 10D), activity increased everywhere except where the local suppression was enforced (Figure 8F, Extended Data Figure 10E).
Figure 8. Increase in negative surface potential in the inner membrane suppresses protrusions in macrophages and, separately, deactivates the EGF induced ERK activity in epithelial cells.
(A) Scheme for elevation of negative surface charge on membrane by the recruitment of negatively-charged optogenetic actuator, Opto-ACTU−, in context of biochemical excitable network topology with receptor input. (B) Design of Opto-ACTU− with net charge −14. Positively-charged amino acids in green, negatively-charged amino acids in red. (C-E) Experimental setup of selective Opto-ACTU−recruitment, followed by uniform C5a stimulation, in unpolarized RAW 264.7 macrophages (C); representative time-lapse images demonstrating cell migration driven by selective suppression of protrusion in the site where Opto-ACTU− was locally recruited and protrusion formation in other areas of cortex upon uniform C5a stimulation (D); polar histogram indicating higher probability of protrusion formation away from recruitment area. Time in min:sec format (D); Green arrows: F-actin-rich protrusions marked by Lifeact (D); nc=12 cells, np=51 protrusions (E). Cells were co-expressing Opto-ACTU−, CIBN-CAAX, and Lifeact-mVenus. (F) Simulated kymograph of membrane states in response to the in silico recruitment of Opto-ACTU− (Cm−), followed by global stimulation. Front or F-state is in green, back or Σ−-state is in red. (G) Scheme for elevation of negative surface charge in context of excitable network-mediated ERK regulation, along with receptor input module. (H) Representative time-lapse images of MCF10A cell displaying ERKKTR translocation from cytosol to nucleus demonstrating ERK deactivation, upon Opto-ACTU− global recruitment to membrane (cells were co-expressing CIBN-CAAX as membrane anchor); cells were pre-treated with and maintained in a saturating dose of EGF throughout the experiment; time in hr:min:sec format (H). (I) Quantification of ERK deactivation in terms of ERKKTR nucleus/cytosol ratio; n=12 cells for each case (Opto-ACTU− and Opto-CTRL), mean ± SEM; 488 nm was turned on first at t=0 min (as shown by vertical dashed line).
Source numerical data are available in source data.
Increased surface charge subverts EGF induced ERK activation
We next asked whether increasing negative membrane surface potential can override the EGF receptor mediated activation of ERK since the pathway involves Ras activation (Figure 8G). MCF10A cells were first activated with a saturating dose of EGF, which was confirmed by the predominantly cytosolic distribution of ERK-activation sensor ERK-KTR (Figure 8H first time point; Extended Data Figure 10F first time point, Video S13). When we raised the surface charge of the membrane by globally recruiting Opto-ACTU− (Figure 8G), a substantial fraction of ERK-KTR became nuclear, indicating a deactivation of ERK (Figure 8H; Video S13), whereas control Opto-CTRL recruitment caused no detectable effect (Extended Data Figure 10F; Video S13). Quantitation of multiple cells showed that, despite some fluctuations in ERK activity (Extended Data Figures 10G,10H), Opto-ACTU− recruitment provoked ~50% increase in nucleus/cytosol ratio of KTR, compared to uncharged control (Figure 8I).
DISSCUSSION
Our study suggests that negative surface charge or zeta potential on the inner leaflet of the membrane coordinates the activities of the Ras/PI3K/TORC2/F-actin networks that control cell migration and polarity. Under physiological conditions, there is a spontaneous reduction in charge during network activation at protrusions and within front-state regions on ventral waves of cells. Decreases in the levels of PI(4,5)P2, PI(3,4)P2, PS, and PA collectively contribute to this charge reduction. Altering membrane surface charge is necessary and sufficient for network activation and, not a merely a consequence of it, since recruiting a positively charged actuator to the membrane activates the network whereas recruiting a negatively charged actuator prevents the chemoattractant or growth factor stimulated activation. Our results suggest that inner membrane surface charge is a key biophysical parameter that appears within the feedback loops determining the set-point of these networks.
Our data indicate that the fluid-mosaic model of the membrane is more complex than originally envisioned. Large patches of multiple anionic lipids and peripheral membrane proteins co-segregate into defined “phase” domains which propagate as waves. For phosphoinositides and peripheral membrane proteins, wave propagation is likely mediated by sequential transient modification of the lipid headgroups and shuttling of the proteins68, 69, 87-89. For example, local transient increases in PIP3 occur where PI3K is briefly activated by Ras90, 91 and PTEN reversibly dissociates 80, 92. Detailed studies will be needed to determine which kinases and phosphatases are sequentially activated or inhibited to control PI(4,5)P2, PI(3,4)P2 or PA levels in membrane. Based on our initial data showing PS is regulated by dynamic externalization, we anticipate that flippases, floppases, or scramblases whose activation tracks closely with Ras/PI3K/TORC2/F-Actin network activation will be identified in future.
The optogenetic actuators enabled generic alteration of surface charge inside live cells with spatiotemporal control. The observed effects on cell behavior suggest that an induced biophysical change, i.e., a reduction in zeta potential, sets in motion a series of biochemical events involved in motility and polarity. To our knowledge, an effect of direct surface charge perturbation on signaling and cytoskeletal events has not been reported. Traditional approaches with isolated membranes 29, 35, 93-95 or giant unilamellar vesicles/liposomes or charged surfactant typically cannot accurately mimic the physiological environment inside cells. Other recent methods21, 22, 51, such as phosphoinositide degradation/synthesis by chemically-induced dimerization or different pharmacological perturbations (which changes calcium/ATP levels) do not provide spatial control and do not decouple the surface charge from other cell physiological changes. Since our novel actuators can work orthogonally to external cues, such as chemoattractant gradients and/or external electric fields, and can act at the inner membrane, they can potentially unravel the molecular architecture of biochemical networks in other physiological scenarios such as immune synapse formation and phagocytosis (and others that likely will be found in the future) where surface charge remodeling takes place.
Our study establishes surface potential as an organizer of signal transduction and cytoskeletal events that control cell migration and polarity. We suggest the term “action surface potentials” to describe the traveling membrane domains of transiently decreased negative surface charge. We propose that high negative surface charge corresponds to a “resting” or back-state of the membrane while the regions of decreasing surface charge are transitioning to an active/front-state which leads to protrusions. We envision that, as surface charge decreases, molecules that regulate the signaling activities respond differentially. Further studies are needed to determine which crucial network components may be directly regulated by charge. We show here that PTEN back-state localization is largely determined by its positively charged N-terminal (which is necessary for membrane binding as well81, 96) and other peripheral membrane proteins have been previously reported to associate with the membrane via charged regions20-22, 30, 32. We speculate that some of these charge-sensitive components in turn initiate downstream events which mediate further loss of multiple anionic lipids in the front regions, additionally decreasing the membrane surface charge. Such feedback loops would enable small fluctuations to expand into propagating waves and can have outsized phenotypic effects. This architecture would be analogous to the ability of transmembrane potential to regulate key ion channels which in turn regulates transmembrane potential during action potential propagation.
The “action surface potential” hypothesis can explain a series of heretofore puzzling observations: First, as we discussed, a vast number of signal transduction activities undergo a highly coordinated stereotypical transient response. This extraordinary degree of coordination within an extensive series of stepwise interactions could be facilitated if the regulation of key components depended on an organizer located on the inner surface of the membrane, such as the surface charge. Second, cells expend significant energy to sustain an asymmetric distribution of anionic lipids on inner leaflet of the membrane, which has little apparent structural advantage. Of course, PIP2 serves as a substrate for PLC and PI3K and PS as a signal for apoptosis. However, we suggest that another, perhaps larger, role of this asymmetric distribution of charged lipids is set up a basal state for triggering of action surface potentials which involve changes in multiple lipids acting in a common direction. Third, the excitable nature of the action surface potentials could underly oscillations for frequency control of gene expression and enable global control over cytoskeletal activities. In the latter case, the lateral propagation of the waves along the cortex in more or less circular patterns provide form and dimension to protrusions that enable cells to polarize and migrate, engulf nutrients, and divide. Recent reports highlight that travelling waves of cytoskeletal and signaling activities mediate a diverse range of cell physiological processes in various cells and organisms12, 67-69, 87, 97-102. Our results imply that action surface potential likely contribute in spatiotemporally orchestrating all these events.
METHODS
Cell culture
The wild-type Dictyostelium discoideum cells of axenic strain AX2 were cultured in HL-5 media at 22 °C. Hygromycin (50 μg/mL) and/or G418 (30 μg/mL) were added to the media to maintain cell lines expressing different constructs. PI3K 1−/2− and PTEN– Dictyostelium cells were cultured like AX2 whereas heat-killed Klebsiella aerogenes were added in the culture media to grow PKBA−/ PKBR1− Dictyostelium cells. Cells were usually maintained in petri dishes and were transferred to shaking culture around 2-4 days before electrofusion or differentiation experiments. PTEN– Dictyostelium cells were always grown in petri dishes. All the experiments were done within 2 months of thawing the cells from the frozen stocks.
RAW 264.7 macrophage-like cells were obtained from N. Gautam laboratory (Washington University School of Medicine in St. Louis, MO) and mammary epithelial MCF-10A cells were obtained from M. Iijima laboratory (Johns Hopkins University School of Medicine, MD). RAW 264.7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 4500mg/L glucose, L-glutamine, sodium pyruvate, and sodium bicarbonate (Sigma-Aldrich; D6429), supplemented with 10% heat-inactivated fetal bovine serum (ThermoFisher Scientific; 16140071) and 1% penicillin-streptomycin (ThermoFisher Scientific; 15140122). MCF-10A cells were cultured in DMEM/F-12 medium with GlutaMAX (ThermoFisher Scientific;10565042) supplemented with 5% heat-inactivated horse serum (ThermoFisher Scientific; 26050088), 1% penicillin-streptomycin, epidermal growth factor (EGF) (20 ng/ml) (Sigma-Aldrich; E9644), cholera toxin (100 ng/ml), hydrocortisone (0.5 mg/ml), and insulin (10 μg/ml). All cells were subcultured every 2-6 days using cell lineage-specific techniques to maintain healthy confluency. All experiments were done with low passage number cells. All mammalian cells were maintained under humidified conditions at 37 °C and 5% CO2.
DNA constructs
All sensors and actuators were usually codon-optimized if used for heterologous expression. R(+8)-Pre was obtained from the C-terminal tail of KRas4b and all the serines and threonines were mutated to alanine to prevent phosphorylation and all the lysines were mutated to arginines to avoid ubiquitination21. In R(+7)-Pre, R(+4)-Pre, and R(+2)-Pre, arginines are sequentially mutated to glutamines to reduce the positive charge of the sensor. The cAR1 is a Dictyostelium protein that works as a GPCR. Palm/Pre is the C-terminal tail of HRas. PM-LYN is the first 11 amino acids of the human Tyrosine-protein kinase LYN. In PTEN1-18-CAAX, a CAAX motif was added to the first 18 amino acids PTEN. RacGCT is the C-terminal tail of RacG which we identified when we entered the KRas4b tail as query sequence in NCBI Protein BLAST (blastp), with organism Dictyostelium specified in search set. Optogenetic actuator Opto-ACTU+, which has a net charge +16, was designed by removing CAAX tail from R(+8)-Pre (so that it becomes cytosolic) and making a dimer of it, joined by a three glycine linker. Optogenetic actuator Opto-ACTU−, which has a net charge −14, was designed using the C-terminal polyanionic tail sequence of mouse protein Rad17.
All surface charge sensors and actuators, i.e. GFP-R(+8)-Pre, GFP-R(+7)-Pre, GFP-R(+4)-Pre, GFP-R(+2)-Pre, GFP-Palm/Pre, GFP-PTEN1-18-CAAX, GFP-RacGCT, Opto-ACTU+, and Opto-ACTU− were generated by annealing the appropriate synthetic oligonucleotides, followed by restriction enzyme mediated digestion and subcloning into proper Dictyostelium or mammalian vectors. All other constructs were made by PCR amplification followed by standard restriction enzyme cloning or by site-directed mutagenesis kit (QuickChange II, Agilent Technologies, 200523). All oligonucleotides were obtained from Sigma-Aldrich. All sequences were verified by diagnostic restriction digest and by standard Sanger sequencing (JHMI Synthesis & Sequencing Facility).
The following plasmid constructs were made in this study. Selected will be deposited on Addgene or dictyBase103, 104 and rest will be available from the authors upon direct request: a) GFP-R(+8)-Pre (pDM358), b) GFP-R(+8)-Pre (pDEXG), c) GFP-R(+7)-Pre (pDEXG), d) GFP-R(+4)-Pre (pDEXG), e) GFP-R(+2)-Pre (pDEXG), f) GFP-PTEN1-18-CAAX (pDEXG), g) GFP-RacGCT (pDEXG), h) GFP-LactC2 (pTX-GFP), i) GFP-Spo20 (pDEXG), j) Opto-ACTU+ (pCV5), k) Opto-CTRL (pCV5), l) cAR1-CIBN (pDM358), m) N150PKBR1-CIBN (pDM358), n) Opto-ACTU− (pmCherryN1), o) LimEΔcoil-Halo (pCV5).
GFP-R(+8)-Pre (mammalian) was from S. Grinstein (Addgene plasmid # 17274), GFP-LactC2 (mammalian) was from S. Grinstein (Addgene plasmid # 22852), GFP-Spo20, originally made by Vitale et al. 49, was a kind gift from G. Du (McGovern Medical School, UTHealth), mPlum-LimEΔcoil was from A. Müller-Taubenberger (LMU Munich), pCRY2PHR-mCherryN1 (i.e. mammalian Opto-CTRL) was from C. Tucker (Addgene plasmid # 26866), CIBN-CAAX was from P. De Camilli and O. Idevall-Hagren (Addgene plasmid # 79574), Lifeact-mVenus was from J. Zhang (Addgene plasmid # 87613), ERKKTR-iRFP713 was from J. Toettcher (Addgene plasmid # 111510), Lyn11-FRB-CFP (Addgene plasmid # 38003) and Pseudojanin (Addgene plasmid #37999) was from Robin Irvine, and GFP-OSH2-2xPH (Addgene Plasmid #161987) as well as PH-PLCD1-GFP (Addgene Plasmid #51407) was from Tamas Balla.
Rest of the plasmids used in this study were available in Devreotes Lab.
Transfection
Dictyostelium AX2 cells were transfected by standard electroporation protocol. Briefly, for each transfection, 107 cells were pelleted, washed twice with ice-cold H-50 buffer (20 mM HEPES, 50 mM KCl, 10 mM NaCl, 1 mM MgSO4, 5 mM NaHCO3, 1 mM NaH2PO4, pH adjusted to 7.0) , and subsequently resuspended in 100 μL ice-cold H-50 buffer. Around 1-5 μg of total DNA was mixed with the cell suspension and it was transferred to an ice-cold 0.1 cm gap cuvette (Bio-Rad; 1652089) for electroporation at 850V and 25 μF twice, in a 5 second interval (Bio-Rad Gene Pulser Xcell Electroporation Systems). After a 5 min incubation on ice, electroporated cells were transferred to a 10 cm Petri dish containing HL-5 media supplemented with heat-killed Klebsiella aerogenes bacteria. Cells were selected by adding hygromycin B (50 μg/ml) and/or G418 (20-30 μg/ml) after 1-2 days, as per the antibiotic resistances of the vectors. For optogenetics and chemically-induced-dimerization experiments where three different protein co-expressions were necessary, two different pCV5 vectors and one pDM358 vector were used and cells were selected aginst both drugs.
RAW 264.7 cells were transfected by nucleofection in an Amexa Nucleofector II device, using Amexa Cell line kit V (Lonza; VACA-1003), following a pre-existing protocol105. For each transfection, 3x106 cells were harvested, resuspended in 100 μL supplemented Nucleofector Solution V. Then total 4-6 μg of DNA mixtures were added and immediately transferred to a Lonza cuvette for electroporation using program setting D-032. 500 μL of pre-warmed pH adjusted culture media was added to electroporated cells in the cuvette subsequently. Cell suspension was then transferred to a 1.5 mL vial and incubated at 37 °C and 5% CO2 for 10 mins. Next, ~50-100 μL solution containing cells were transferred to a coverslip chambers and allowed to adhere for an hour. Finally, ~400 μL of pre-warmed pH adjusted culture media was added to each chamber and cells were further incubated for 4-6 hours before imaging.
MCF-10A cells were transiently transfected using Lipofectamine 3000 Transfection Reagent (ThermoFisher Scientific; L3000001), as per manufacturer’s protocol. Briefly, ~0.8 μg DNA was mixed in 160 μL serum-free Opti-MEM (ThermoFisher Scientific; 31985062) media containing 3 μL Lipofectamine 3000 reagent and incubated for 5-7 mins at RT to allow formation of DNA-lipid complex. Post-incubation, DNA-lipid complex was added to 2x105 MCF-10A cells plated on a 2-well glass chamber. Cells were incubated at 37 °C/5% CO2 for 5 hours, after which the DNA-lipid complex was removed, cells were washed thoroughly, and incubated for a further 12-18 hours before imaging.
Drugs and reagents
Annexin V, Alexa Fluor 488 conjugate was obtained from ThermoFisher Scientific (Invitrogen; A13201) and was stored in 4 °C. Latrunculin A (Enzo Life Sciences; BML-T119-0100) was dissolved in DMSO to make a stock solution of 5 mM. Caffeine (Sigma-Aldrich; C0750) was dissolved in ddH2O to make a stock solution of 80 mM. Rapamycin (Sigma-Aldrich; 553210) was dissolved in DMSO to prepare a 10 mM stock solution. cAMP (Sigma-Aldrich; A6885) was dissolved in ddH2O to make a stock solution of 10 mM. Janelia Fluor 646 HaloTag (Promega Corporation; GA1120) was dissolved in DMSO to prepare 200 μM stock solution which was stored in 4 °C and was diluted 1000X during the experiment. C5a receptor agonist FKP-(D-Cha)-Cha-r (Anaspec; 65121) was dissolved in 1X PBS to make to 2.5 mM stock solution. Anti-BSA antibody was obtained from Sigma (Sigma-Aldrich; SAB4200688). Stock solution for EGF (Sigma-Aldrich, E9644) was prepared by dissolving it in 10 mM acetic acid to a final concentration of 1 mg/ml. Hydrocortisone (Sigma-Aldrich; H0888) was dissolved in 200 proof ethanol to make 1 mg/mL stock. Insulin (Sigma-Aldrich; I1882) was resuspended in sterile ddH2O (containing 1% glacial acetic acid) to make 10 mg/mL stock solution. Cholera toxin (Sigma-Aldrich; C-8052) was dissolved at in sterile ddH2O which resulted 1mg/mL stock solution and stored at 4°C. LY294002 was obtained from ThermoFisher Scientific (Invitrogen; PHZ1144) and dissolved in DMSO to make 40 mM solution. PP242 (Sigma-Aldrich; 475988) was dissolved in DMSO to prepare 20 mM stock solution. Unless otherwise mentioned, everything was stored as small aliquots in −20 °C.
Microscopy and live cell imaging
Unless otherwise mentioned, all experiments were performed inside a heated (37 °C) chamber with a 5% CO2 supply (in case of mammalian cell imaging), or on a 22 °C stage (in case of Dictyostelium imaging). All time-lapse live-cell images were acquired using one of these four microscopes: a) Zeiss LSM780-FCS Single-point, laser scanning confocal microscope, (Zeiss Axio Observer with 780-Quasar; 34-channel spectral, high-sensitivity gallium arsenide phosphide detectors), b) Zeiss LSM880-Airyscan FAST Super-Resolution Single-point confocal microscope (Zeiss AxioObserver with 880-Quasar (34-channel spectral, high-sensitivity gallium-arsenide phosphide detectors), c) Zeiss LSM800 GaAsP Single-point, laser scanning confocal microscope with wide-field camera, and d) Nikon Eclipse Ti-E dSTROM Total Internal Reflection Fluorescence (TIRF) Microscope (Photometrics Evolve EMCCD camera).
In Zeiss 780 and 800, 488 nm (argon laser) excitation was used for GFP; 561 nm (solid-state) excitation was used for RFP, mCherry, and mPlum; and 633 nm (diode laser) excitation was used for iRFP713 and Janelia Fluor 646 HaloTag. In Zeiss 880, 488 nm (argon laser) excitation was used for GFP; 514 nm (argon laser) excitation was used for YFP and mVenus; 594 nm (HeNe laser) excitation was used for mCherry; and 633 nm (diode laser) excitation was used for iRFP713. In Nikon TIRF, 488nm (argon laser) excitation was used for GFP and 561 nm (0.5W fiber laser) excitation was used for mCherry and RFP. In Zeiss 780, 800, and 880, 40X/1.30 Plan-Neofluar oil objective (with appropriate digital zoom) was used and in Nikon TIRF, 100x/1.4 Plan-Apo oil objective was used. Both Zeiss 780 and Zeiss 880 confocal microscopes were operated by ZEN Black software, Zeiss 800 confocal microscopes are operated by ZEN Blue software, whereas the Nikon TIRF was controlled by Nikon NIS-Elements. To visualize cortical/ventral waves in Dictyostelium and RAW 264.7 cells, either TIRF microscope was used or confocal microscopes were focused on the very bottom of the cell to capture the substrate-attached ventral surface of the cell.
Cell differentiation
For Dictyostelium cell development, 8x107 cells of exponential growth phase were collected from suspension culture and pelleted, as previously described106. After washing twice with DB (Development buffer; 5 mM Na2HPO4, 5 mM KH2PO4, supplemented with 2 mM MgSO4 and 0.2 mM CaCl2), cells were resuspended in 4 mL DB and shaken at 110 rpm for 1 hour. The cells were then pulsed with 50–100 nM cAMP (5s pulse every 6 min) using a time-controlled peristaltic pump for 5–6 h with continual shaking. This allowed the cells to become developed and polarized. After development, from the shaker, around 2-5x104 cells were transferred to an 8-well coverslip chamber, resuspended thoroughly in 450 μL of DB, and incubated for 20-30 min before starting the image acquisition.
Frustrated phagocytosis and osmotic shock
To visualize ventral waves in RAW 264.7 macrophages, we have slightly modified a pre-existing protocol43. Briefly, Nunc Lab-Tek 8-well coverslip chambers were prewashed with 30% nitric acid, coated with 1 mg/mL BSA for 3 hours, washed with PBS, and finally incubated with 5 μg/mL anti-BSA antibody (1:200 dilution) for 2 hours. Chambers were finally washed two times with PBS to remove excess antibodies. Before the imaging, transfected RAW 264.7 cells were starved in suspension in 1X Ringer’s buffer (150 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 20 mM HEPES and 2 g/L glucose, pH 7.4) for 30 min. Next, these cells were introduced to the opsonized chambers and allowed to spread on antibody-coated surface for 5-10 min, and then hypotonic shock was applied using 0.5X Ringer’s solution.
Electrofusion
Exponential growth phase Dictyostelium cells from suspension culture were collected, washed, and resuspended in 10 mL SB (17 mM Soerensen buffer containing 15 mM KH2PO4 and 2 mM Na2HPO4, pH 6.0) at a density of 1.5 × 107 cells/ml, as previously described40, 57. The cells were rolled gently in a conical tube for around 30-40 min to promote visible cluster formation. 800 μL of rolled cells were transferred to a 4-mm-gap Bio-Rad electroporation cuvette, using pipette tips with cut off edges. The electroporation was performed with 1kV, 3 μF once, then with 1kV, 1 μF twice more (with 3 s gap between two pulses) to facilitate membrane hemifusion. 35 μL of cells were transferred from the cuvette to a Nunc Lab-Tek 8 well chamber and incubated for 5 more min and then fresh SB buffer supplemented with 2 mM CaCl2 and 2 mM MgCl2, was gently added to these cells. Cells were incubated at 22 °C for next 1-1.5 hours for recovery before imaging.
Annexin V binding assay
Growth phase Dictyostelium cells were transferred to an 8-well Nunc Lab-Tek coverslip chamber and allowed to adhere for 10-15 min. Next the HL-5 medium was removed and 450 μL DB buffer was added to the cells. Cells were incubated at 22 °C for ~ 60-120 min. After that, the DB buffer was exchanged with DB buffer with excess calcium (2.5 mM of CaCl2 final concentration) and incubated for 30 min. The coverslip chambers were transferred to ice bath (to slow down the protrusion formation and withdrawal frequency) and rest of the procedure was done in ice. Next, 10 μL of Annexin V, Alexa Fluor 488 conjugate was added to each well and it was allowed to bind for 45-60 sec. Then the media (containing Annexin V, Alexa Fluor 488 conjugate) was aspirated and was quickly washed for two times using DB with excess calcium buffer to get rid of all unbound Annexin V. Cells were then fixed without permeabilization, using 2% paraformaldehyde and 0.25% Glutaraldehyde in HL-5 (with 2.5 mM of Ca2+) for 15 min and then washed twice and finally put under DB with excess calcium for imaging.
Gradient stimulation assay
Gradient stimulation assay by cAMP-filled needle was performed as per the established protocol106, 107. Dictyostelium cells co-expressing PHcrac-mCherry and GFP-R(+8)-Pre were differentiated (as described in the ‘Cell differentiation’ section) for 5.5-6.5 hours and were put under cAMP gradient. Cells were pre-treated with LatA to inhibit cytoskeletal activities. A 10 μM cAMP- filled micropipette (Femtotips, Eppendorf) connected to a FemtoJet Microinjector (Eppendorf) was used to provide the gradient stimulation. The microinjector was employed in a continuous injection mode with a compensation pressure of 70 hPa. Stimulation was induced by suddenly bringing the micropipette (using the micromanipulator) close to the (x,y,z) coordinate of the cell that was being imaged and putting it at one side of that cell.
Chemically induced dimerization
The plasmids and experimental details of CID system were mostly described in our previous reports9, 57. Here, to generate Figure 3A-E and Extended Data Figure 6A-C, two following combinations were used to express the systems: a) cAR1-FKBP-FKBP (pCV5) as membrane anchor, mCherry-FRB-Inp54p (pCV5) as recruitee, GFP-R(+8)-Pre (pDM 358) as readout; and b) cAR1-FKBP-FKBP (pDM 358) as membrane anchor, mCherry-FRB-Inp54p (pCV5) as recruitee, PHPLCδ-YFP (pCV5) as readout. Growth phase Dictyostelium cells were transferred to an 8-well coverslip chamber and incubated for 10-15 min so that they can adhere well. Then, the HL-5 medium was replaced with 450 μL of DB buffer. After 15-20 min of media change, image acquisition was started. After imaging a certain number of frames, rapamycin was added gently to the chamber (to final concentration 5μM) during image acquisition to felicitate recruitment of Inp54p to plasma membrane. In case of Figure 3E, first it was ensured that the Inp54p was recruited to the membrane by looking at a few confocal slices at the middle of the cell, then focused to the substrate-attached surface to visualize waves.
To perform CID-mediated recruitment in RAW 264.7 macrophages, Pseudojanin, Lyn11-FRB-CFP, and GFP-R(+8)-Pre/GFP-OSH2-2xPH/GFP-PH-PLCδ were nucleofected (as described above). After 4-6 hours of nucleofection, culture media replaced with 450 μL of new warm HBSS buffer (Hank’s Balanced Salt Solution) with 1 g/L of glucose and cells were incubated for another 0.5-1 hour before imaging. During imaging, after recording certain number of frames for ~2-5 min, rapamycin was introduced (as described above).
Optogenetic regulation of cell migration
To analyze the effect of surface charge lowering on cell polarity and migration pattern, Dictyostelium cells was selected against both Hygromycin and G418 to co-express LimEΔcoil-Halo (pCV5)/RBD-GFP(CV5)/PHcrac-YFP(CV5) and cAR1-CIBN (pDM 358), along with Opto-ACTU+ (pCV5) or Opto-CTRL (pCV5). Cells were properly developed as described in previous section. After 6-7 hours of development, around 2-5x104 cells were collected from the shaker and transferred to an 8-well coverslip chamber. Then cells were resuspended thoroughly in 450 μL of DB and incubated for around 20 min before starting the image acquisition. For experiments presented in Figures 6E, 6F and Extended Data figures S9D-M, developed appropriate Dictyostelium cells were incubated with 40 μM LY294002 or 20 μM PP242 or both for >45 min in DB buffer before image acquisition was started. For global recruitment experiments, after imaging ≥320 s, the 488 nm Laser was turned ON globally to initiate recruitment and was intermittently turned on at a lower intensity during image acquisition (usually for around 970 ms after each 8s) to keep Opto-ACTU+ or Opto-CTRL on the membrane throughout the imaging. For spatially confined optical recruitment, a region of interest was drawn and that particular area was illuminated was 488nm laser in multiple iteration.
For surface charge elevation experiments, RAW 264.7 macrophage cells co-expressing Lifeact-mVenus, CIBN-CAAX, and Opto-ACTU− or Opto-CTRL was used. After 4-6 hours of nucleofection, media was aspirated from chambers and cells were put in 450 μL of new warm HBSS buffer containing 1 g/L of glucose and cells were incubated for another 0.5-1 hour before starting image acquisition. During imaging, first Opto-ACTU− or Opto-CTRL was selectively recruited first using 488 nm laser (as described above) and subsequently C5a receptor agonist FKP-(D-Cha)-Cha-r was added (diluted in HBBS; final concentration of 10 μM) and image acquisition was continued.
Optogenetic deactivation of ERK
The KTR sensors enables to convert specific kinase activities into a nucleocytoplasmic shuttling equilibrium for convenient visualization and quantification11, 108. Basically, ERKKTR sensor becomes cytosolic to nuclear upon the deactivation of ERK. To analyze the effect of optogenetic perturbation of inner membrane surface charge, MCF10A cells were transfected with ERKKTR-iRFP713 (as the readout), CIBN-CAAX (as membrane anchor), along with Opto-ACTU− or Opto-CTRL (as the recruitee). Transfected cells were incubated overnight in complete culture medium containing 5% horse serum,10 μg/ml insulin, and 20 ng/mL EGF, which facilitated activation of ERK. Imaging was performed in the same media. After 25 mins of image acquisition, 488 nm Laser was globally turned ON to facilitate recruitment and was intermittently turned on during image acquisition to keep Opto-ACTU− or Opto-CTRL on the membrane throughout the imaging. ERKKTR localization profile was used to quantitate ERK activity over the time.
Image analysis
Image analysis was performed in MATLAB 2019a (MathWorks, Natick, MA, USA) and Fiji/ImageJ 1.52i (NIH). The results were plotted using MATLAB 2019a, OriginPro 9.0 (OriginLab, Northampton, MA, USA), or GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA).
Colocalization study:
Image analysis for the colocalization study was performed with custom codes written in MATLAB 2019a. As the preprocessing steps, the background subtraction and the gaussian/top hat filtering were applied to all the images. The cell area from the background and the bright patches of the protein of interest from the cell area were segmented using thresholding method.
To quantify the extent of colocalization between two proteins of interest, say A and B, in a cell, the following conditional probabilities were computed:
: the probability of finding ‘high localization’ of protein A in the regions of ‘high localization’ of protein B.
: the probability of finding ‘high localization’ of protein B in the regions of ‘high localization’ of protein A.
: the probability of finding ‘high localization’ of protein A in the regions of ‘low localization’ of protein B.
: the probability of finding ‘low localization’ of protein A in the regions of ‘high localization’ of protein B.
: the probability of finding ‘high localization’ of protein B in the regions of ‘low localization’ of protein A.
: the probability of finding ‘low localization’ of protein B in the regions of ‘high localization’ of protein A.
High and the low localization of a protein is decided by the threshold values assumed in the segmentation step. If both the proteins, A and B were colocalized in a cell, then the following inequalities would hold true:
| (a) |
| (b) |
| (c) |
| (d) |
The inequality sign would be preserved under the change of ordering. The sign reverses if the proteins do not colocalize. Clearly in case of the colocalization, the ratio of these conditional probabilities (from Eqn. (a-d)) will be greater than 1, whereas it will be less than 1 for the cases of complementary localization. Taking logarithm of these ratios further differentiates these two cases leading to positive and negative values, respectively. For the concise representation of the data, we computed the following average of the four ratios of conditional probabilities for each image frame as follows, and named it as Conditional Probability Index (henceforth CP index):
For the box and whisker plot representation, we averaged the CP index. over n frames analyzed for every cell image (henceforth Avg. CP index.) as follows:
where is the total number of frames analyzed for a sample.
Kymographs:
For the membrane kymographs the cells were segmented against the background following standard image processing steps with the custom code written in MATLAB 2019b. The kymographs were created from the segmented cells as previously described where consecutive lines over time were aligned by minimizing the sum of the Euclidean distances between the coordinates in two consecutive frames by a custom-written MATLAB function67. A linear colormap was used for the normalized intensities in the kymographs. For grayscale kymographs, black indicating the lowest intensity and white the highest whereas in colored kymographs, lowest intensity was indicated by blue and highest by yellow.
Line kymographs that accompanied ventral waves were generated in Fiji/ImageJ by drawing a thick line with line width 8-12 pixel and processing the entire stack in “KymographBuilder” plugin.
Line scan intensity profile:
Line scans were performed in Fiji/ImageJ (NIH) by drawing a straight-line segment (using line tool) inside the cells with line width 8-12 pixel so that we can obtain an average intensity value. The intensity values along that particular line were obtained in green as well as red channels using “Plot Profile” option. The values were saved and normalized, in OriginPro 9.0 (OriginLab, Northampton, MA, USA). Then the intensity profiles were graphed and finally smoothened using Savitzky-Golay or Adjacent-Averaging method, using proper boundary conditions. The plots were then normalized by dividing by maximum value. For a particular linescan, the green and red profiles were smoothed and processed using exact same parameters to maintain consistency.
Time-series plot of Membrane/Cytosol ratio:
To make the plots in Figure 3C and Extended data Figure 6A, cells were first segmented into membrane and cytosolic masks following standard image processing steps with the custom code written in MATLAB 2019b. The average intensities from the channels were computed for both the masks. The computed intensities were later corrected for the photobleaching effect by dividing the values by an exponential fit to the temporal profiles of the respective normalized average intensities in the cell. Finally, we computed the ratios of the corrected average intensities for different channels. For the plotting of the temporal profiles of the red channel data we normalized the pre-recruitment average profiles to unity, whereas for the green channel, the normalization was done using the respective steady-state value.
Cell tracking and migration analysis:
Using the “Threshold” option of Fiji/ImageJ, image stack was first thresholded properly so that the generated binary image covers all the pixels of the cells. Range was not reset and “Calculate threshold for each image” option was unchecked. Subsequently, using “Analyzed Particles” option, a size-based thresholding was applied (to exclude non-cell particles) and cell masks were generated. Next, “Fill holes”, “Erode”, and “Dilate” options were applied, sequentially and judiciously, to obtain the proper binarized mask for cells. To generate the migrating cell outlines in Figure 5G and Extended data Figure 9I, “Outline” command was operated on binarized cells and finally “Temporal-Color Code” was used. For other plots in Figure 5 and Extended data Figure 9, “Shape descriptors” and “Centroid” options (inside “Set Measurement”) were used with “Analyzed Particles” command to obtain the values of centroid coordinates and circularity values (circularity = area/perimeter2). For the centroid, the starting point was set to origin for each track by applying and for i=0 to (n-1). The new translated coordinates were plotted to generate the tracks. Same increment colormap (OriginPro) was used in before-recruitment and after-recruitment tracks to enable pairwise comparison in Figure 5D and Extended Data figure 9C, H. For velocity, displacement between each two frames were computed using , which was divided by time-interval to compute speed. These speeds were time-averaged to generate each datapoint in the migration speed box plots.
Global protrusion formation analysis:
The new protrusion formation frequency upon global recruitment of Opto-ACTU+ or Opto-CTRL was computed in Fiji/ImageJ. In the stack of the binarized cell masks (binarization was performed as per the method described in the ‘Cell tracking and migration analysis’ section), “Stack Difference” command of “Multi Kymograph” was applied. The stack was inverted and a size-based thresholding was performed. If required, the resulting stack was processed further using “Despeckle” command and then protrusion numbers were manually counted. The numbers were counted for the same frames for which the migration speed was quantified.
Selective protrusion formation analysis:
To perform the selective protrusion formation analysis upon confined recruitment of Opto-ACTU+, Opto-ACTU−, and Opto-CTRL, we segmented the recruitment area on the membrane with proper threshold. We then marked the area using the segmented line tool. Using a custom-written ImageJ Macro, “Fit spline” and “Straighten” commands were sequentially operated. The macro was then used to find and mark the midpoint. The centroid of the cell was found and marked using another macro. Using the angle tool, considering the centroid as vertex, the angles between protrusions and the midpoint of the recruitment areas were obtained. Values were imported to MATLAB and plotted using the “polarhistogram” command. Sturges’ formula was used to determine the minimum number of bins.
KTR translocation analysis:
The nucleus to cytosol ratio of ERKKTR was computed over the time to quantitate the extent of ERK deactivation upon Opto-ACTU− or Opto-CTRL recruitment. The cells were first segmented in Fiji/ImageJ (as described earlier in the ‘Cell tracking and migration analysis’ section). Using “Analyze Particles” option, the whole cell intensities were obtained. Nucleus intensities was obtained manually by selecting nucleus from the phase channels of the stack. The cytosolic intensities were found by subtracting nuclear intensity from the whole cell intensity. After ratios were found, a conditional formatting rule was applied in Microsoft Excel to generate heat maps in Extended Data Figure 10G, H. The mean and SEM of these ratios were calculated and plotted in Figure 8I.
Statistics and Reproducibility
The statistical analyses were performed using MATLAB 2019a and GraphPad Prism 8. Time-series data was shown as mean ± SEM or mean± SD, as indicated. All box plots were graphed following Tukey’s convention. Statistical significance and P-values were determined by paired or unpaired two-tailed non-parametric tests as specifically indicated in the figure captions. Sample sizes were chosen empirically as per the standard custom followed in the field and similar sample sizes were used for experiment and control group. For chemically induced dimerization and optogenetics experiments, for either control or experiment group, data were excluded when there is no recruitment because it essentially implies the absence of expression of the untagged membrane anchor in that particular cell. No other data were excluded from the study. All reported findings were reliably reproduced using at least three biological replicates. To make any experiment or control group cell lines, cells were collected randomly from the parental cell lines in culture. The time at which any drug treatment and/or imaging experiment was performed (with any custom cell line) was chosen randomly. The Investigators were not blinded to allocation during experiments and outcome assessment. The following convention was followed to show P-values: ns denotes P>0.05, * denotes P ≤ 0.05, ** denotes P ≤ 0.01, *** denotes P ≤ 0.001, and **** denotes P ≤ 0.0001.
Extended Data
Extended Data Figure 1: Cells generate two mutually exclusive dynamic states in the membrane during migration and ventral wave propagation.
(A) Coordinated localization dynamics of signaling (PIP3) and cytoskeletal components (F-actin) in migrating Dictyostelium cell protrusions. Top panel: Live-cell images (Arrowheads: Protrusions enriched in both F-actin and PIP3). Bottom panel: 360° membrane kymographs showing consistency of coordination. Here and in all other kymographs, numbers on the left denote time in seconds, unless otherwise mentioned. Throughout this study, PIP3 level is marked by PHcrac in Dictyostelium and by PHAkt in macrophages, whereas, newly polymerizing F-actin is marked by LimEΔcoil (‘LimE’ hereafter) in Dictyostelium and by Lifeact in macrophages. (B, C) Coordinated propagation of signaling (PIP3) and cytoskeletal (F-Actin) components in ventral-surface cortical waves of Dictyostelium (B) and RAW 264.7 macrophages (C). Top two panels show live-cell images and bottom panels show line-scan intensity profile along the white lines. Similar convention is followed throughout this article. (D, E) Activated Ras and PIP3 colocalizing in the protrusions (D), whereas PTEN selectively dissociating from it (E), in migrating Dictyostelium cells. Left: Live-cell images, Right: 360° membrane kymographs. Arrowheads: Protrusions/front-states. Throughout this study, Ras-Binding Domain of Raf1 (RBD) was used as a detector of Ras activation. (F, G) In propagating waves of Dictyostelium, activated Ras and PIP3 dynamically colocalized and defined the front-state regions (F), whereas PIP3 and PTEN exhibit consistent complementarity (G). Live-cell images, line-scan intensity profiles, and line-kymographs are shown. (H) Complementary distribution of PIP3 and PTEN is independent of cytoskeleton. Here Dictyostelium cells are imaged in presence of Latrunculin-A (F-actin polymerization inhibitor). Arrowheads denoting front-states. (I) Schematic showing the front-back complementary patterning in three different scenarios: migrating cell protrusions, cytoskeleton-independent cortical symmetry breaking, and propagating ventral waves. For first two cases, we could study a 1D profile, whereas for ventral waves, we observed a 2D profile at the substrate-attached surface. Several examples of established signaling and cytoskeletal components are listed and categorized. In all situations, when a front-state was created from the back/basal-state of the membrane, back markers moved away from that particular domain, maintaining complementarity. Scale bars are 10 μm.
Extended Data Figure 2: Developing conditional probability index as a metric to quantify the extent of co-localization and complementary localization.
(A) Schematics showing the application of the concepts of conditional probability in quantifying the degree of colocalization between two entities, R and G. The regions of the high enrichments of the species R and G are denoted as Rhigh and Ghigh whereas the depleted states are denoted as Rlow and Glow, respectively (top panels). The overlapped region (yellow in the bottom left panel) denotes Rhigh∩Ghigh. The other necessary probabilities are also shown which are required in the computation of the respective Conditional Probability Index (CP index). (B) The mathematical description of the CP index. As usual, P(Rhigh∣Ghigh) denotes Probability of selecting Rhigh, given Ghigh is already selected. Rest of the expression follow the same standard convention (please see methods for details). (C and D) Time series plots of CP indices of established back protein PTEN (C) and established front sensor RBD (D); number of cells nc =15 for RBD (C) and nc =17 for PTEN (D). Throughout this paper, to generate CP index time-plots, each cell was analyzed for nf =20 frames; data are mean ± SEM. Top panels show representative images of ventral waves in Dictyostelium cells co-expressing either PHCrac and RBD (C) or PHCrac and PTEN (D). Note that the sign of CP index of PTEN is negative and RBD is positive which demonstrate their back-state and front-state localization, respectively. The modulus value of CP indices indicates the extent of co- or complementary localization. Throughout this paper, all CP indices are calculated with respect to PIP3.
Extended Data Figure 3: PI(4,5)P2, PI(3,4)P2, PS, and PA exhibit consistent yet dynamic back-state distribution.
(A) Representative line-kymograph of ventral waves in RAW 264.7 macrophages co-expressing PI(4,5)P2 biosensor (GFP-PHPLCδ), and PIP3 biosensor (PHAkt-mCherry). Time-lapse images and line-scan intensity profiles were shown in Figure 1C. (B) Representative line-kymograph of ventral waves in Dictyostelium cells co-expressing PI(3,4)P2 biosensor (PHCynA-KikGR) and PIP3 biosensor (PHCrac-mCherry). Time-lapse images and line-scan intensity profiles were shown in Fig. 1D. (C) Representative line-kymographs of ventral wave pattern shown in Dictyostelium cells co-expressing PS biosensor (GFP-LactC2) and PIP3 biosensor, (PHCrac-mCherry). Time-lapse images and line-scan intensity profiles were shown in Fig. 1G. (D) Representative line-kymographs of ventral wave pattern in RAW 264.7 macrophage cells co-expressing PS biosensor (GFP-LactC2) and PIP3 biosensor, (PHCrac-mCherry). Time-lapse images and line-scan intensity profiles were shown in Figure 1H. (E) Representative line-kymographs of ventral wave pattern in Dictyostelium cells co-expressing PA biosensor (GFP-Spo20) and PIP3 biosensor (PHCrac-mCherry). Time-lapse images and line-scan intensity profiles were shown in Fig. 1J. (F) Time-lapse images of migrating Dictyostelium cells co-expressing GFP-Spo20 and PHcrac-mCherry. White arrows: Protrusions where PIP3 is enriched and PA is depleted. Blue arrows: Spo20 returned back to the membrane as protrusions were eventually retracted and membrane domain returned to its basal back-state. (G) Box and Whisker plot of time-averaged CP indices of four anionic phospholipids (PI(4,5)P2, PI(3,4)P2, PS, and PA), together with uniform membrane marker control cAR1, back protein PTEN, and front sensor RBD; nc=16 cells for PI(4,5)P2/PHPLCδ, nc=10 cells for PI(3,4)P2/PHCynA, nc=15 cells for PS/LactC2, nc=16 cells for PS/Spo20, nc =20 cells for cAR1, nc = 17 cells for PTEN, nc = 15 cells for RBD. As mentioned earlier, to generate each datapoint, nf =20 frames were averaged for the above-mentioned number of cells.
Extended Data Figure 4: Spatiotemporal organization of different mutated charge sensors and uniform membrane controls.
(A) Representative live cell images of Dictyostelium cells co-expressing GFP-R(+8)-Pre and PHCrac-mCherry under chemotactic gradient stimulation. Solid magenta arrowhead indicates the direction of micropipette (filled with 1 μM cAMP) for gradient stimulation. Dashed magenta arrowhead indicates the introduction of needle (t=0s) which is manifested by the transient global response in PHCrac channel. Cells were pre-treated with Latrunculin A. (B-D) Live-cell time-lapse images and line scan intensity profiles of Dictyostelium cells expressing PHcrac-mCherry, along with GFP-R(+7)-Pre (B) or GFP-R(+4)-Pre (C) or GFP-R(+2)-Pre (D), during ventral wave propagation, displaying decreasing extent of back-state preference of the surface charge sensors. The first time points were showed in Figure 2J (in grayscale colormap). (E and F) The 360° membrane kymographs of cells shown in Figure 2K, indicating R(+7)-Pre consistently moves away from PIP3-rich protrusions (E), whereas R(+2)-Pre is uniform over the cortex (F). (G) Live-cell images, line scan intensity profiles, and representative line-kymographs of ventral waves in Dictyostelium cells co-expressing PHcrac-mCherry and membrane marker cAR1-GFP, demonstrating that cAR1 does not distribute to front- or back- state regions and it is consistently uniform over the membrane. (H) Live-cell time-lapse images of migrating Dictyostelium cells co-expressing PHcrac-mCherry and cAR1-GFP showing cAR1 is symmetric over the membrane. Black arrows: PIP3- rich protrusions where cAR1 was present as well. (I) Live-cell images, linescan intensity profiles, and representative line-kymographs of ventral waves in RAW 264.7 cells co-expressing PHAKT-mCherry and membrane marker, LYN-GFP, showing consistent uniform profile of LYN over the membrane and no depletion in front-state area. (J) Live-cell time-lapse images of migrating Dictyostelium cells co-expressing PHcrac-mCherry and GFP-Palm/Pre, showing a symmetric profile of Palm/Pre over the membrane. Black arrows: Protrusions/front-states. In (G-J), the “Fire invert” LUT of Fiji/ImageJ was used so that it can clearly show any small inhomogeneity.
Extended Data Figure 5: Different polybasic sequences localize to back-state regions depending on their charge, irrespective of their exact amino acid sequences.
(A, B) Representative live-cell images, lines can intensity profiles, and representative line-kymographs of Dictyostelium cells co-expressing PHCrac-mCherry and GFP-RacGCT (A) or PTEN1-18-CAAX (B), demonstrating consistent dynamic back distribution for RacGCT and limited back distribution for PTEN1-18-CAAX in ventral waves. For exact sequence details, please see Table S1. (C and D) Representative live-cell time-lapse images showing distribution of RacGCT (C) or PTEN1-18-CAAX (D) in migrating Dictyostelium cells (co-expressing PHCrac-mCherry), demonstrating localization profiles analogous to (A, B). (E, F) Time series plots of CP index of RacGCT (E) and PTEN1-18-CAAX (F) show the extent of back localization; nc=17 for RacGCT (E), nc= 12 for PTEN1-18-CAAX (F); mean ± SEM. (G) Comparison of localization profile by box plot of time-averaged CP indices of all surface charge sensors, together with uniform membrane marker controls, back protein PTEN, and front sensor RBD; R(+8)-Pre: nc =30, R(+7)-Pre: nc =23, R(+4)-Pre: nc =20, R(+2)-Pre: nc =12, RacGCT: nc =17, PTEN1-18-CAAX :nc =12, cAR1: nc =20, Palm/Pre: nc =11, PTEN: nc =17, RBD: nc =15. Box and whiskers are graphed as per Tukey’s method. All p-values by Mann-Whitney-Wilcoxon test.
Extended Data Figure 6: Dynamics of surface charge sensor in PI(4,5)P2 and PI(3,4)P2 depleted cells.
(A) Time course of membrane/cytosol ratio of PHPLCδ and Inp54p upon rapamycin addition (indicated by black dashed vertical line), in Dictyostelium cells co-expressing cAR1-FKBP-FKBP, mCherry-FBP-Inp54p, and PHPLCδ-GFP, demonstrating PHPLCδ dissociated from membrane upon PI(4,5)P2 depletion; n=17 cells; mean ± SEM. (B and C) Cell tracks show the migration profile of Dictyostelium cells expressing chemically induced dimerization system cAR1-FKBP-FKBP and mCherry-FBP-Inp54p, along with PHPLCδ-GFP (B) or GFP-R(+8)-Pre (C), before and after rapamycin induced recruitment. Tracks demonstrating similar change in migration profile in both cases, as quantified in terms of migration speed in Figure 3D. To generate each track for nc=32 cells (in each case), cells were followed for nf=60 frames (7s/frame). (D) Representative image of Dd5p4− Dictyostelium cell (where PI(3,4)P2 level is low) co-expressing GFP-R(+8)-Pre and LimE-mRFP displaying characteristic membrane association and back localization of R(+8)-Pre; white arrows denote F-actin rich protrusions. (E) Quantification of membrane association of R(+8)-Pre in wild type and Dd5p4- cells, in terms of membrane/cytosol ratio; n=29 cells in each case; p-value by Mann-Whitney-Wilcoxon test. (F) Example of a quiet or non-protrusion forming Dictyostelium cells expressing LimE-mRFP, whose outer leaflet of membrane was allowed to transiently bind with Annexin V, Alexa Fluor 488 conjugate.
Extended Data Figure 7: Local recruitment of Opto-ACTU+ in polarized cells induce de novo generation of protrusion near recruitment area, whereas neither local nor global recruitment of uncharged control Opto-CTRL elicit any phenotypic changes.
(A) A representative example of de novo formation of protrusion from a position of choice in the back-state region of the membrane by spatially confined recruitment of Opto-ACTU+. Magenta arrows: Old protrusions, Green arrows: New protrusions. (B) A representative example of spatially confined optogenetic recruitment of Opto-CTRL demonstrating no increase in protrusion generation from the site of recruitment. In (A-B), along with the Opto-ACTU+ (A) or Opto-CTRL(B), cells were co-expressing cAR1-CIBN and LimE-Halo; the numbers on the images denote time in seconds. (C-D) Time-lapse snapshots (C) and time-stack (D) of Dictyostelium cells co-expressing Opto-CTRL and cAR1-CIBN, demonstrating the unaltered cell morphology and migration behavior in polarized Dictyostelium cells, upon optogenetic recruitment. Numbers are time in seconds (C). 488 nm switched ON globally at t=0s. Yellow arrows: Opto-CTRL is uniform over cortex and did not move away from protrusions (D). (E-H) Quantification of cell morphology and migration mode in terms of cell circularity index (E), cell tracks (F), migration speed (G), and new protrusion formation frequency (H), upon Opto-CTRL recruitment (n=25 cells). Data shown as mean ± SEM over time in (E). In (F-H), for either before or after recruitment tracks, each cell tracked for nf=40 frames (t=320 s). Tracks were reset to the same origin in (F). For pairwise comparison, tracks are color-coded in (F) and data from same cell are connected by gray line in (G) and (H). The p-values by Mann-Whitney-Wilcoxon test.
Extended Data Figure 8: Global recruitment of Opto-ACTU+ can cause spatiotemporally confined activation of Ras/PI3K/Akt/TORC2/F-actin network components.
(A) Intensity profiles of LimE-GFP and Opto-ACTU+ along the white lines (the images are same as shown in Figure 6B) demonstrate that F-actin polymerizes in the domains of membrane where Opto-ACTU+ accumulates and when that leads to a protrusion, Opto-ACTU+ moves away with a short time delay. (B) 360° membrane kymograph of cell shown in Figure 6B. (C) 360° membrane kymograph of cell shown in Figure 6C. (D) Time-lapse live cell images of Dictyostelium cells co-expressing Opto-ACTU+, cAR1-CIBN, and PHCrac-YFP where recruitment was started at t=0s. Numbers show time in seconds. The “i” and “ii” are showing two different PIP3 production events which eventually lead protrusion formation. For each event, blue arrowheads are showing the areas where Opto-ACTU+ was first accumulated which in turn became the areas of PIP3 production and eventually, after protrusion formation, Opto-ACTU+ moved away to a newer back-state area. (E) 360° membrane kymograph of cell shown in (D). (F) 360° membrane kymograph of cell shown in Figure 6D.
Extended Data Figure 9: The effect of specific genetic and pharmacological perturbations upon the phenotypic changes induced by Opto-ACTU+ recruitment.
(A-C) Quantification of migration profile changes in terms of new protrusion formation frequency (A), speed (B), and cell tracks (C), upon Opto-ACTU+ recruitment, in PTEN– Dictyostelium cells; nc= 22 cells. (D, E) Quantification of migration profile changes in terms of new protrusion formation frequency (D) and speed (E), upon Opto-ACTU+ recruitment in Dictyostelium cells, pre-treated with PI3K inhibitor LY294002; nc= 28 cells. (F-H) Quantification of migration profile changes in terms of new protrusion formation frequency (F), speed (F), and cell tracks (H), upon Opto-ACTU+ recruitment in PTEN– Dictyostelium cells, pre-treated with LY294002; nc= 24 cells. (I) Temporally color-coded cell outlines of a representative migrating PTEN– Dictyostelium cells, pre-treated with PI3K inhibitor LY294002, showing cell morphology and migratory profile before and after 488nm was turned on to recruit Opto-ACTU+ (corresponding to Video S11). (J-O) Quantification of migration profile changes in terms of new protrusion formation frequency (J, L, N) and migration speed (K, M, O) upon Opto-ACTU+ recruitment under different genetic and pharmacological inhibitions. In (J-K) cells were pre-treated with PP242 to inhibit TORC2 (nc= 22 cells); in (L-M) cells were pre-treated with both LY294002 and PP242 to simultaneously block PI3K and PP242 (nc= 27 cells); in (N-O), Akt—/PKBR1— double knockout cell line was used (nc= 21 cells). For each case, each of the nc cells were tracked for nf=40 frames (8 sec/frame was imaging frequency) and time averages were taken. Tracks were reset to the same origin in (C) and (H). For pairwise comparison, tracks are color-coded in (C) and (H). In all box plots here, for pairwise comparison, data from same cell are connected by gray lines. The p-values by Mann-Whitney-Wilcoxon test.
Extended Data Figure 10: Selective recruitment of uncharged control Opto-CTRL cannot suppress protrusion in RAW 264.7 macrophages or global recruitment of Opto-CTRL cannot deactivate ERK in MCF10A cells, unlike Opto-ACTU−.
(A) Representative live-cell time-lapse images of RAW 264.7 cells undergoing light-triggered spatially confined recruitment of Opto-ACTU−, followed by global stimulation with C5a receptor agonist, demonstrating selective protrusion suppression in the site where Opto-ACTU− was locally recruited and robust protrusion formation in other areas of cortex. Time in min:sec format. Cells were co-expressing Opto-ACTU−, CIBN-CAAX, and Lifeact-mVenus. (B) Representative live-cell time-lapse images of RAW 264.7 cells undergoing light-triggered spatially confined recruitment of Opto-CTRL, followed by global stimulation by C5a receptor agonist. Time in min:sec format. Cells were co-expressing Opto-CTRL, CIBN-CAAX, and Lifeact-mVenus. (C) Polar histogram indicating probability of protrusion formation is essentially uniform over the cortex upon global stimulation with C5a receptor agonist, in spatially confined Opto-CTRL recruited cells; nc=12 cells, np=59 protrusions. (D) Schematic showing coupled system of excitable network, polarity module, and Opto-ACTU− system input along with global agonist stimulation. (E) The simulated kymographs of B (top) and (bottom) in response to local recruitment of Opto-ACTU−. The location of recruitment is denoted by the white dashed box. The solid black line denotes the span of global agonist stimulation. (F) Representative live-cell time-lapse images of a MCF10A cell displaying ERKKTR maintaining its cytosolic distribution upon Opto-CTRL recruitment, demonstrating no substantial ERK deactivation; cells were pre-treated with and maintained in a saturated dose of EGF throughout the experiment. Time in hr:min:sec format; 488 nm laser was first turned ON at t=0 min. (G, H) Individual cell level changes in the nuclear/cytosolic ratio of ERKKTR over time, upon recruitment of Opto-ACTU− (G) or Opto-CTRL (H). Population average is in Figure 8I. The color scale shown in right is applicable to both panels.
Supplementary Material
Table S1: Different charge sensors and membrane marker controls. For each one, the net charge, sequence length, and exact sequence or reference is listed. Green color denotes positively charged amino acids and red color denotes negatively charged amino acids.
Table S2: Parameter values for the computational model.
Video S1. Dynamic localization of PI(4,5)P2 to the back-state of the membrane in both Dictyostelium and RAW 264.7 macrophages. The spatiotemporal complementary patterns were observed during protrusion formation in Dictyostelium (A), ventral wave propagation in Dictyostelium (B), and ventral wave propagation in RAW 264.7 macrophages (C). Left panels show PHPLCδ-GFP, middle panels show PHCrac-mCherry (in A and B) or PHAkt-mCherry (in C), and the right panels show the merged view. Top left corners show time in mm:ss format.
Video S2. During ventral wave propagation in Dictyostelium, PI(3,4)P2 dynamically localizes to the back-state of the membrane. Left panels show PHCynA-KikGR, middle panels show PHcrac-mCherry, and the right panels show the merged view. Top left corners show time in mm:ss format.
Video S3. Dynamic preferential back-state distribution of phosphatidylserine in membrane during ventral wave propagation in Dictyostelium and RAW 264.7 macrophages. (A) Ventral wave patterns in Dictyostelium. Left panel shows GFP-LactC2, middle panel shows PHCrac-mCherry, and the right panel shows the merged view. (B) Ventral wave propagation in RAW 264.7 macrophages. Left panels show overall pattern in the cell; right panels show the pattern in the zoomed-in area corresponding to the white rectangular boxes in the left channel. In either left or right panels, top channels show GFP-LactC2 and bottom channels show PHAkt-mCherry. Scale bar is 10 μm. In both (A) and (B), Top left corners show time in mm:ss format.
Video S4. Dynamic preferential distribution of generic negative surface charge sensor to the back-state of the membrane in Dictyostelium. The spatiotemporal patterns were observed during ventral wave propagation (A) and protrusion formation (B). Left panels show GFP-R(+8)-Pre, middle panels show PHcrac-mCherry, and the right panels show the merged view. Top left corners show time in mm:ss format.
Video S5. Dynamic preferential distribution of generic negative surface charge sensor to the back-state of the membrane in ventral waves of RAW 264.7 macrophages. Left panel shows GFP-R(+8)-Pre, middle panel shows PHAkt-mCherry, and the right panel shows the merged view. Top left corners showing time in mm:ss format.
Video S6. Dissociation of PI4P sensor PHOSH2X2 and PI(4,5)P2 sensor PHPLCδ upon recruitment of pseudojanin by chemically induced dimerization system in RAW 264.7 macrophages. (A and B) Left panels: GFP-PHOSH2X2, Right Panels: Pseudojanin. (C and D) Left panels: GFP-PHPLCδ, Right Panels: Pseudojanin. All RAW 264.7 cells are expressing Lyn-FRB as well. In all cases, rapamycin addition time was indicated by the appearance of white-colored text “+Rapamycin” in the top middle. Top left corners showing time in mm:ss format.
Video S7. Profile of membrane surface charge sensor R(+8)-Pre, upon recruitment of pseudojanin by chemically induced dimerization system in RAW 264.7 macrophages. (A and B). Two examples of Pseudojanin recruitment in RAW 264.7 cells co-expressing GFP-R(+8)-Pre and Lyn-FRB. Left Panels: GFP-R(+8)-Pre; Right panels: Pseudojanin. Rapamycin addition time was indicated by the appearance of white-colored text “+Rapamycin” in the top middle. Top left corners showing time in mm:ss format.
Video S8. Optically confined recruitment of Opto-ACTU+ can trigger protrusions de novo. In (A-D), 488 nm laser was selectively irradiated inside the white rectangular boxes; top left corners showing time in seconds; untagged cAR1-CIBN was expressed. (A and B) Two examples of Opto-ACTU+ recruitment into the back-state regions of the membrane showing the generation of new protrusions in the vicinity of the recruitment. In (A) stage was moved to start recruitment at time 0s. Left panels: LimEΔcoil (newly-polymerized F-Actin sensor), right panels: Opto-ACTU+. (C and D) Two examples of Opto-CTRL recruitment in the back-states of the membrane, showing no generation of new protrusions from the recruitment areas. Left panels: LimEΔcoil (newly-polymerized F-Actin sensor), right panels: Opto-CTRL.
Video S9. Global recruitment of Opto-ACTU+ to the membrane in polarized Dictyostelium cell elicits random protrusions and abrogates pre-existing polarity and persistent migration. (A) Global optical recruitment of Opto-ACTU+. Left panel: Opto-ACTU+, right panel: DIC. (B) Global optical recruitment of Opto-CTRL. Left panel: Opto-CTRL, right panel: DIC. For both (A) and (B): Untagged cAR1-CIBN was expressed. Top left corners show time in seconds. The time before recruitment shown with a negative sign. The 488 nm laser was turned ON at 0s to initiate the recruitment, as depicted by the appearance of green-colored text “488 nm ON” in the bottom middle.
Video S10. Cell morphology and migration mode changes upon global recruitment of Opto-ACTU+ are reversible. (A) and (B) demonstrate two examples where Opto-ACTU+ was first recruited to the membrane by turning on 488 nm laser (recruitment initiated when white-colored text “488 nm OFF” switched to green-colored text “488 nm ON” in the videos) which resulted increased protrusion formation, loss of polarity, and impaired migration; then to reverse the process, the laser was turned off (when the green-colored text “488 nm ON” switched to white colored text “488 nm OFF”), Opto-ACTU+ returned to cytosol, and the cell eventually regained its polarized morphology and migration behavior. Left panels: Opto-ACTU+, right panels: DIC; untagged cAR1-CIBN was expressed. Top left corners show time in mm:ss format in (A) and in hh:mm:ss in (B).
Video S11. Cell migration mode and morphology changes in a PTEN– Dictyostelium cell, pretreated with PI3K inhibitor LY294002, upon global recruitment of Opto-ACTU+. Left panels show Opto-ACTU+ and right panels show DIC channel. Untagged cAR1-CIBN was expressed. Top left corners show time in mm:ss format. The 488 nm laser was turned ON to initiate the recruitment, as depicted by the appearance of white text “488 nm ON” in the bottom middle.
Video S12. Optically confined recruitment of Opto-ACTU− can locally suppress protrusions in RAW 264.7 macrophages. In both (A)-(C), 488 nm laser was selectively irradiated inside the white rectangular boxes; untagged CIBN-CAAX was expressed; top left corners show time in mm:ss format. Either Opto-ACTU− (A and B) or Opto-CTRL (C) was first locally recruited and then cells were globally stimulated with C5a receptor agonist. The appearance of white-colored text “+C5aR Agonist” in the videos denoting the addition of the agonist. Left panels: Lifeact-mVenus; Right panels: Opto-ACTU− (A and B) or Opto-CTRL (C).
Video S13. Global recruitment of Opto-ACTU− to the membrane in MCF 10A epithelial cells can deactivate the EGF induced ERK activation. (A) Translocation of ERK-KTR from cytosol to nucleus upon Opto-ACTU− recruitment. Left panel: ERK-KTR-iRFP713, right panel: Opto-ACTU−. (B) ERKKTR maintained its cytosolic distribution upon Opto-CTRL recruitment. Left panel: ERK-KTR-iRFP713, right panel: Opto-CTRL. In both (A and B), top left corners showing time in hh:mm:ss format. 488 nm laser was globally turned ON at t=0 (as shown by the appearance of green-colored text “488 nm ON”). Cells were pre-treated with a saturated dose of EGF and they were maintained in that condition throughout the experiment.
Acknowledgments:
We thank S. Grinstein (Hospital for Sick Children/University of Toronto) for stimulating discussions. We are grateful to M. Kozlov (Tel Aviv University) for helping us with resident charge calculation. We thank D. Andrew, C.-H. Huang, all of the members of the Devreotes and Iglesias laboratories as well as the members of the D. Robinson and M. Iijima laboratories (School of Medicine, Johns Hopkins University) for their valuable suggestions. We are grateful to Y. Long and Y. Deng (Devreotes lab) for helping with some experiments. We would like to thank N. Gautam (Washington University School of Medicine in St. Louis), and R. R. Kay (MRC LMB) for providing cells. We thank G. Du (McGovern Medical School, UTHealth) and A. Müller-Taubenberger (LMU Munich) for sharing plasmids. We thank Addgene and dictyBase for providing the plasmids and resources. This work was supported by NIH grant R35 GM118177 (to P.N.D.), DARPA HR0011-16-C-0139 (to P.A.I. and P.N.D.), AFOSR MURI FA95501610052 (to P.N.D.), as well as NIH grant S10 OD016374 (to S. Kuo of the JHU Microscope Facility).
Footnotes
Competing interests: The authors declare no competing interests.
Data availability:
Source data have been provided in Source Data. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Code availability:
Computational simulation codes are available on GitHub: https://github.com/tatsatb/Simulation-of-Membrane-Surface-Charge-Perturbation. Any additional information will be available from the corresponding author upon reasonable request.
References
- 1.Saha S, Nagy TL & Weiner OD Joining forces: crosstalk between biochemical signalling and physical forces orchestrates cellular polarity and dynamics. Philos. Trans. R. Soc. Lond. B. Biol. Sci 373, 10.1098/rstb.2017.0145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devreotes PN et al. Excitable Signal Transduction Networks in Directed Cell Migration. Annu. Rev. Cell Dev. Biol 33, 103–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoxhaj G & Manning BD The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridley AJ et al. Cell migration: integrating signals from front to back. Science 302, 1704–1709 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Pal DS, Li X, Banerjee T, Miao Y & Devreotes PN The excitable signal transduction networks: movers and shapers of eukaryotic cell migration. Int. J. Dev. Biol 63, 407–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaney KF, Huang CH & Devreotes PN Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys 39, 265–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shellard A, Szabo A, Trepat X & Mayor R Supracellular contraction at the rear of neural crest cell groups drives collective chemotaxis. Science 362, 339–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao Y et al. Altering the threshold of an excitable signal transduction network changes cell migratory modes. Nat. Cell Biol 19, 329–340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houk AR et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell 148, 175–188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JM et al. Integrating chemical and mechanical signals through dynamic coupling between cellular protrusions and pulsed ERK activation. Nat. Commun 9, 4673–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan H et al. An Excitable Ras/PI3K/ERK Signaling Network Controls Migration and Oncogenic Transformation in Epithelial Cells. Dev. Cell 54, 608–623.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai W et al. Tissue topography steers migrating Drosophila border cells. Science 370, 987–990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graziano BR et al. Cell confinement reveals a branched-actin independent circuit for neutrophil polarity. PLoS Biol. 17, e3000457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang I et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503, 281–284 (2013). [DOI] [PubMed] [Google Scholar]
- 16.O'Neill PR et al. Membrane Flow Drives an Adhesion-Independent Amoeboid Cell Migration Mode. Dev. Cell 46, 9–22.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampert TJ et al. Shear force-based genetic screen reveals negative regulators of cell adhesion and protrusive activity. Proc. Natl. Acad. Sci. U. S. A 114, E7727–E7736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fort L et al. Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat. Cell Biol 20, 1159–1171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kholodenko BN, Hancock JF & Kolch W Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol 11, 414–426 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaughlin S & Murray D Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Yeung T et al. Receptor activation alters inner surface potential during phagocytosis. Science 313, 347–351 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Ma Y, Poole K, Goyette J & Gaus K Introducing Membrane Charge and Membrane Potential to T Cell Signaling. Front. Immunol 8, 1513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldenberg NM & Steinberg BE Surface charge: a key determinant of protein localization and function. Cancer Res. 70, 1277–1280 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Rickert P, Weiner OD, Wang F, Bourne HR & Servant G Leukocytes navigate by compass: roles of PI3Kgamma and its lipid products. Trends Cell Biol. 10, 466–473 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parent CA & Devreotes PN A cell's sense of direction. Science 284, 765–770 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Senju Y & Lappalainen P Regulation of actin dynamics by PI(4,5)P2 in cell migration and endocytosis. Curr. Opin. Cell Biol 56, 7–13 (2019). [DOI] [PubMed] [Google Scholar]
- 27.McLaughlin S The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem 18, 113–136 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg S et al. Mapping the electrostatic profile of cellular membranes. Mol. Biol. Cell, mbcE19080436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray D et al. Electrostatic properties of membranes containing acidic lipids and adsorbed basic peptides: theory and experiment. Biophys. J 77, 3176–3188 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaughlin S The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem 18, 113–136 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Aguilella VM, Verdia-Baguena C & Alcaraz A Lipid charge regulation of non-specific biological ion channels. Phys. Chem. Chem. Phys 16, 3881–3893 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Heo WD et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314, 1458–1461 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander RT et al. Membrane surface charge dictates the structure and function of the epithelial Na+/H+ exchanger. EMBO J. 30, 679–691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green WN & Andersen OS Surface charges and ion channel function. Annu. Rev. Physiol 53, 341–359 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Ma Y et al. A FRET sensor enables quantitative measurements of membrane charges in live cells. Nat. Biotechnol 35, 363–370 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Shi X et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature 493, 111–115 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Maxson ME & Grinstein S The Role of Membrane Surface Charge in Phagocytosis. Adv. Exp. Med. Biol 1246, 43–54 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Li X et al. Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration. Proc. Natl. Acad. Sci. U. S. A 115, E9125–E9134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagorda A & Parent CA Eukaryotic chemotaxis at a glance. J. Cell. Sci 121, 2621–2624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerhardt M et al. Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state. J. Cell. Sci 127, 4507–4517 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Matsuoka S & Ueda M Mutual inhibition between PTEN and PIP3 generates bistability for polarity in motile cells. Nat. Commun 9, 4481–0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arai Y et al. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc. Natl. Acad. Sci. U. S. A 107, 12399–12404 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masters TA, Sheetz MP & Gauthier NC F-actin waves, actin cortex disassembly and focal exocytosis driven by actin-phosphoinositide positive feedback. Cytoskeleton 73, 180–196 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Gerisch G, Schroth-Diez B, Muller-Taubenberger A & Ecke M PIP3 waves and PTEN dynamics in the emergence of cell polarity. Biophys. J 103, 1170–1178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaney KF, Borleis J, Iglesias PA & Devreotes PN Novel protein Callipygian defines the back of migrating cells. Proc. Natl. Acad. Sci. U. S. A 112, 3845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeung T et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Yeung T et al. Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J. Cell Biol 185, 917–928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeniou-Meyer M et al. Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J. Biol. Chem 282, 21746–21757 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Kassas N et al. Genetically encoded probes for phosphatidic acid. Methods Cell Biol. 108, 445–459 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Dickson EJ & Hille B Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J 476, 1–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fairn GD et al. An electrostatic switch displaces phosphatidylinositol phosphate kinases from the membrane during phagocytosis. J. Cell Biol 187, 701–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolfsson HI et al. Lipid organization of the plasma membrane. J. Am. Chem. Soc 136, 14554–14559 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Leventis R & Silvius JR Lipid-binding characteristics of the polybasic carboxy-terminal sequence of K-ras4B. Biochemistry 37, 7640–7648 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Fukushima S, Matsuoka S & Ueda M Excitable dynamics of Ras triggers spontaneous symmetry breaking of PIP3 signaling in motile cells. J. Cell. Sci 132, jcs224121. doi: 10.1242/jcs.224121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin T, Zhang N, Long Y, Parent CA & Devreotes PN Localization of the G protein betagamma complex in living cells during chemotaxis. Science 287, 1034–1036 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Janetopoulos C, Ma L, Devreotes PN & Iglesias PA Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A 101, 8951–8956 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miao Y et al. Wave patterns organize cellular protrusions and control cortical dynamics. Mol. Syst. Biol 15, e8585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hammond GR et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin S et al. Immunologic stimulation of mast cells leads to the reversible exposure of phosphatidylserine in the absence of apoptosis. Int. Arch. Allergy Immunol 123, 249–258 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Balasubramanian K, Mirnikjoo B & Schroit AJ Regulated externalization of phosphatidylserine at the cell surface: implications for apoptosis. J. Biol. Chem 282, 18357–18364 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Chung CY, Potikyan G & Firtel RA Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7, 937–947 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Meili R, Ellsworth C & Firtel RA A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr. Biol 10, 708–717 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Kamimura Y et al. PIP3-independent activation of TorC2 and PKB at the cell's leading edge mediates chemotaxis. Curr. Biol 18, 1034–1043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Haastert PJ, Keizer-Gunnink I & Kortholt A Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J. Cell Biol 177, 809–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoeller O et al. Two distinct functions for PI3-kinases in macropinocytosis. J. Cell. Sci 126, 4296–4307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhattacharya S et al. Traveling and standing waves mediate pattern formation in cellular protrusions. Sci. Adv 6, eaay7682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang CH, Tang M, Shi C, Iglesias PA & Devreotes PN An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat. Cell Biol 15, 1307–1316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiner OD, Marganski WA, Wu LF, Altschuler SJ & Kirschner MW An actin-based wave generator organizes cell motility. PLoS Biol. 5, e221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bement WM et al. Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat. Cell Biol 17, 1471–1483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong Y, Huang CH, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci U S A. 2010; 107:17079–17086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Biswas D, Devreotes PN & Iglesias PA Three-dimensional stochastic simulation of chemoattractant-mediated excitability in cells. PLoS Comput. Biol 17, e1008803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wittmann T, Dema A & van Haren J Lights, cytoskeleton, action: Optogenetic control of cell dynamics. Curr. Opin. Cell Biol 66, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tischer D & Weiner OD Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol 15, 551–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karunarathne WK, O'Neill PR & Gautam N Subcellular optogenetics - controlling signaling and single-cell behavior. J. Cell. Sci 128, 15–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeRose R, Miyamoto T & Inoue T Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 465, 409–417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue T, Heo WD, Grimley JS, Wandless TJ & Meyer T An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods 2, 415–418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu YI et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karunarathne WK, Giri L, Patel AK, Venkatesh KV & Gautam N Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc. Natl. Acad. Sci. U. S. A 110, 1575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Neill PR, Kalyanaraman V & Gautam N Subcellular optogenetic activation of Cdc42 controls local and distal signaling to drive immune cell migration. Mol. Biol. Cell 27, 1442–1450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iijima M & Devreotes P Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599–610 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Iijima M, Huang YE, Luo HR, Vazquez F & Devreotes PN Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J. Biol. Chem 279, 16606–16613 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Wessels D, Lusche DF, Kuhl S, Heid P & Soll DR PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J. Cell. Sci 120, 2517–2531 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Takeda K, Sasaki AT, Ha H, Seung HA & Firtel RA Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J. Biol. Chem 282, 11874–11884 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Cai H et al. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J. Cell Biol 190, 233–245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi C, Huang CH, Devreotes PN & Iglesias PA Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput. Biol 9, e1003122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O'Neill PR & Gautam N Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol. Biol. Cell 25, 2305–2314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu M & Liu J Mechanobiology in cortical waves and oscillations. Curr. Opin. Cell Biol 68, 45–54 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Wu Z, Su M, Tong C, Wu M & Liu J Membrane shape-mediated wave propagation of cortical protein dynamics. Nat. Commun 9, 136–1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gerhardt M et al. Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state. J. Cell. Sci 127, 4507–4517 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Rodriguez-Viciana P et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370, 527–532 (1994). [DOI] [PubMed] [Google Scholar]
- 91.Pacold ME et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Myers MP et al. The Lipid Phosphatase Activity of PTEN is Critical for Its Tumor Supressor Function. Proceedings of the National Academy of Sciences - PNAS 95, 13513–13518 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van den Bogaart G et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dathe M et al. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry 35, 12612–12622 (1996). [DOI] [PubMed] [Google Scholar]
- 95.Parthasarathy R, Cripe PA & Groves JT Electrostatically driven spatial patterns in lipid membrane composition. Phys. Rev. Lett 95, 048101 (2005). [DOI] [PubMed] [Google Scholar]
- 96.Vazquez F et al. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. U. S. A 103, 3633–3638 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inagaki N & Katsuno H Actin Waves: Origin of Cell Polarization and Migration? Trends Cell Biol. 27, 515–526 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Cheng X & Ferrell JE Apoptosis propagates through the cytoplasm as trigger waves. Science 361, 607–612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang HW, Collins SR & Meyer T Locally excitable Cdc42 signals steer cells during chemotaxis. Nat. Cell Biol 18, 191–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Haastert PJ, Keizer-Gunnink I & Kortholt A Coupled excitable Ras and F-actin activation mediates spontaneous pseudopod formation and directed cell movement. Mol. Biol. Cell 28, 922–934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lange M, Prassler J, Ecke M, Muller-Taubenberger A & Gerisch G Local Ras activation, PTEN pattern, and global actin flow in the chemotactic responses of oversized cells. J. Cell. Sci 129, 3462–3472 (2016). [DOI] [PubMed] [Google Scholar]
- 102.Xiong D et al. Frequency and amplitude control of cortical oscillations by phosphoinositide waves. Nat. Chem. Biol 12, 159–166 (2016). [DOI] [PubMed] [Google Scholar]
Methods-only references:
- 103.Kreppel L et al. dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res. 32, 332 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fey P, Dodson RJ, Basu S & Chisholm RL One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Center in 2012. Methods Mol. Biol 983, 59–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meshik X, O'Neill PR & Gautam N Optogenetic Control of Cell Migration. Methods Mol. Biol 1749, 313–324 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Kamimura Y, Tang M & Devreotes P Assays for chemotaxis and chemoattractant-stimulated TorC2 activation and PKB substrate phosphorylation in Dictyostelium. Methods Mol. Biol 571, 255–270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cai H, Huang CH, Devreotes PN & Iijima M Analysis of chemotaxis in Dictyostelium. Methods Mol. Biol 757, 451–468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de la Cova C, Townley R, Regot S & Greenwald I A Real-Time Biosensor for ERK Activity Reveals Signaling Dynamics during C. elegans Cell Fate Specification. Dev. Cell 42, 542–553.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Different charge sensors and membrane marker controls. For each one, the net charge, sequence length, and exact sequence or reference is listed. Green color denotes positively charged amino acids and red color denotes negatively charged amino acids.
Table S2: Parameter values for the computational model.
Video S1. Dynamic localization of PI(4,5)P2 to the back-state of the membrane in both Dictyostelium and RAW 264.7 macrophages. The spatiotemporal complementary patterns were observed during protrusion formation in Dictyostelium (A), ventral wave propagation in Dictyostelium (B), and ventral wave propagation in RAW 264.7 macrophages (C). Left panels show PHPLCδ-GFP, middle panels show PHCrac-mCherry (in A and B) or PHAkt-mCherry (in C), and the right panels show the merged view. Top left corners show time in mm:ss format.
Video S2. During ventral wave propagation in Dictyostelium, PI(3,4)P2 dynamically localizes to the back-state of the membrane. Left panels show PHCynA-KikGR, middle panels show PHcrac-mCherry, and the right panels show the merged view. Top left corners show time in mm:ss format.
Video S3. Dynamic preferential back-state distribution of phosphatidylserine in membrane during ventral wave propagation in Dictyostelium and RAW 264.7 macrophages. (A) Ventral wave patterns in Dictyostelium. Left panel shows GFP-LactC2, middle panel shows PHCrac-mCherry, and the right panel shows the merged view. (B) Ventral wave propagation in RAW 264.7 macrophages. Left panels show overall pattern in the cell; right panels show the pattern in the zoomed-in area corresponding to the white rectangular boxes in the left channel. In either left or right panels, top channels show GFP-LactC2 and bottom channels show PHAkt-mCherry. Scale bar is 10 μm. In both (A) and (B), Top left corners show time in mm:ss format.
Video S4. Dynamic preferential distribution of generic negative surface charge sensor to the back-state of the membrane in Dictyostelium. The spatiotemporal patterns were observed during ventral wave propagation (A) and protrusion formation (B). Left panels show GFP-R(+8)-Pre, middle panels show PHcrac-mCherry, and the right panels show the merged view. Top left corners show time in mm:ss format.
Video S5. Dynamic preferential distribution of generic negative surface charge sensor to the back-state of the membrane in ventral waves of RAW 264.7 macrophages. Left panel shows GFP-R(+8)-Pre, middle panel shows PHAkt-mCherry, and the right panel shows the merged view. Top left corners showing time in mm:ss format.
Video S6. Dissociation of PI4P sensor PHOSH2X2 and PI(4,5)P2 sensor PHPLCδ upon recruitment of pseudojanin by chemically induced dimerization system in RAW 264.7 macrophages. (A and B) Left panels: GFP-PHOSH2X2, Right Panels: Pseudojanin. (C and D) Left panels: GFP-PHPLCδ, Right Panels: Pseudojanin. All RAW 264.7 cells are expressing Lyn-FRB as well. In all cases, rapamycin addition time was indicated by the appearance of white-colored text “+Rapamycin” in the top middle. Top left corners showing time in mm:ss format.
Video S7. Profile of membrane surface charge sensor R(+8)-Pre, upon recruitment of pseudojanin by chemically induced dimerization system in RAW 264.7 macrophages. (A and B). Two examples of Pseudojanin recruitment in RAW 264.7 cells co-expressing GFP-R(+8)-Pre and Lyn-FRB. Left Panels: GFP-R(+8)-Pre; Right panels: Pseudojanin. Rapamycin addition time was indicated by the appearance of white-colored text “+Rapamycin” in the top middle. Top left corners showing time in mm:ss format.
Video S8. Optically confined recruitment of Opto-ACTU+ can trigger protrusions de novo. In (A-D), 488 nm laser was selectively irradiated inside the white rectangular boxes; top left corners showing time in seconds; untagged cAR1-CIBN was expressed. (A and B) Two examples of Opto-ACTU+ recruitment into the back-state regions of the membrane showing the generation of new protrusions in the vicinity of the recruitment. In (A) stage was moved to start recruitment at time 0s. Left panels: LimEΔcoil (newly-polymerized F-Actin sensor), right panels: Opto-ACTU+. (C and D) Two examples of Opto-CTRL recruitment in the back-states of the membrane, showing no generation of new protrusions from the recruitment areas. Left panels: LimEΔcoil (newly-polymerized F-Actin sensor), right panels: Opto-CTRL.
Video S9. Global recruitment of Opto-ACTU+ to the membrane in polarized Dictyostelium cell elicits random protrusions and abrogates pre-existing polarity and persistent migration. (A) Global optical recruitment of Opto-ACTU+. Left panel: Opto-ACTU+, right panel: DIC. (B) Global optical recruitment of Opto-CTRL. Left panel: Opto-CTRL, right panel: DIC. For both (A) and (B): Untagged cAR1-CIBN was expressed. Top left corners show time in seconds. The time before recruitment shown with a negative sign. The 488 nm laser was turned ON at 0s to initiate the recruitment, as depicted by the appearance of green-colored text “488 nm ON” in the bottom middle.
Video S10. Cell morphology and migration mode changes upon global recruitment of Opto-ACTU+ are reversible. (A) and (B) demonstrate two examples where Opto-ACTU+ was first recruited to the membrane by turning on 488 nm laser (recruitment initiated when white-colored text “488 nm OFF” switched to green-colored text “488 nm ON” in the videos) which resulted increased protrusion formation, loss of polarity, and impaired migration; then to reverse the process, the laser was turned off (when the green-colored text “488 nm ON” switched to white colored text “488 nm OFF”), Opto-ACTU+ returned to cytosol, and the cell eventually regained its polarized morphology and migration behavior. Left panels: Opto-ACTU+, right panels: DIC; untagged cAR1-CIBN was expressed. Top left corners show time in mm:ss format in (A) and in hh:mm:ss in (B).
Video S11. Cell migration mode and morphology changes in a PTEN– Dictyostelium cell, pretreated with PI3K inhibitor LY294002, upon global recruitment of Opto-ACTU+. Left panels show Opto-ACTU+ and right panels show DIC channel. Untagged cAR1-CIBN was expressed. Top left corners show time in mm:ss format. The 488 nm laser was turned ON to initiate the recruitment, as depicted by the appearance of white text “488 nm ON” in the bottom middle.
Video S12. Optically confined recruitment of Opto-ACTU− can locally suppress protrusions in RAW 264.7 macrophages. In both (A)-(C), 488 nm laser was selectively irradiated inside the white rectangular boxes; untagged CIBN-CAAX was expressed; top left corners show time in mm:ss format. Either Opto-ACTU− (A and B) or Opto-CTRL (C) was first locally recruited and then cells were globally stimulated with C5a receptor agonist. The appearance of white-colored text “+C5aR Agonist” in the videos denoting the addition of the agonist. Left panels: Lifeact-mVenus; Right panels: Opto-ACTU− (A and B) or Opto-CTRL (C).
Video S13. Global recruitment of Opto-ACTU− to the membrane in MCF 10A epithelial cells can deactivate the EGF induced ERK activation. (A) Translocation of ERK-KTR from cytosol to nucleus upon Opto-ACTU− recruitment. Left panel: ERK-KTR-iRFP713, right panel: Opto-ACTU−. (B) ERKKTR maintained its cytosolic distribution upon Opto-CTRL recruitment. Left panel: ERK-KTR-iRFP713, right panel: Opto-CTRL. In both (A and B), top left corners showing time in hh:mm:ss format. 488 nm laser was globally turned ON at t=0 (as shown by the appearance of green-colored text “488 nm ON”). Cells were pre-treated with a saturated dose of EGF and they were maintained in that condition throughout the experiment.
Data Availability Statement
Source data have been provided in Source Data. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Computational simulation codes are available on GitHub: https://github.com/tatsatb/Simulation-of-Membrane-Surface-Charge-Perturbation. Any additional information will be available from the corresponding author upon reasonable request.