Abstract
Rationale and Objectives:
Irreversible electroporation (IRE) is a promising non-thermal ablation technique for the treatment of patients with hepatocellular carcinoma. Early differentiation of the IRE zone from surrounding reversibly electroporated (RE) penumbra is vital for the evaluation of treatment response. In this study, an advanced statistical learning framework was developed by evaluating standard MRI data to differentiate IRE ablation zones, to correlate with histological tumor biomarkers.
Materials and Methods:
Fourteen rabbits with VX2 liver tumors were scanned following IRE ablation and forty-six features were extracted from T1w and T2w MRI. Following identification of key imaging variables through two-step feature analysis, multivariable classification and regression models were generated for differentiation of IRE ablation zones, and correlation with histological markers reflecting viable tumor cells, microvessel density, and apoptosis rate. The performance of the multivariable models was assessed by measuring accuracy, receiver operating characteristics curve analysis, and Spearman correlation coefficients.
Results:
The classifiers integrating four radiomics features of T1w, T2w, and T1w+T2w MRI data distinguished IRE from RE zones with an accuracy of 97%, 80%, and 97%, respectively. Also, pixelwise classification models of T1w, T2w, and T1w+T2w MRI labeled each voxel with an accuracy of 82.8%, 66.5%, and 82.9%, respectively. Regression models obtained a strong correlation with behavior of viable tumor cells (0.62≤r2≤0.85, p<0.01), apoptosis (0.40≤r2≤0.82, p<0.01), and microvessel density (0.48≤r2≤0.58, p<0.01).
Conclusion:
MRI radiomics features provide descriptive power for early differentiation of IRE and RE zones while observing strong correlations among multivariable MRI regression models and histological tumor biomarkers.
Keywords: Hepatocellular carcinoma, Irreversible electroporation, Magnetic resonance imaging, Random Forest, Texture analysis
Introduction
Hepatocellular carcinoma (HCC) is one of the deadliest cancers affecting 10.4 per 100,000 men and 2.9 per 100,000 women in the United States (1). Potentially curative treatments including surgical resection and partial hepatectomy are suitable for only 30% of the patients at the time of diagnosis due to advanced disease stages (2). Atezolizumab (immune checkpoint inhibitor, ICI) in combination with bevacizumab (anti-vascular endothelial growth factor, VGEF) has recently received FDA approval as the first line of therapy for the treatment of patients with unresectable or metastatic HCC, after showing improved overall survival compared to sorafenib (tyrosine kinase inhibitor, TKI) (3). Sorafenib was approved by FDA more than a decade ago for the treatment of advanced HCC, but genetic heterogeneity (4), adverse events (5), and drug resistance (6) have been its limitations. Lenvatinib, an FDA-approved TKI, has demonstrated an improved overall survival ratio compared to sorafenib but a non-superior overall survival benefit (7).
Local ablation techniques have been developed during the last three decades for patients who are not suitable to receive surgical resection (8, 9). Ablation techniques have gained importance for the treatment of very early to early-stage HCC patients (10). However, the efficacy of thermal ablation methods is restricted by heat-sink effect and tumor location (11). Irreversible electroporation (IRE) is a novel and non-thermal ablation method with a proven safety profile (12) that destructs malignant tissues by delivering high-frequency electrical current in short pulses without damaging surrounding tissues (13). Following IRE procedure, cellular level structure of the tissues is either permanently (irreversibly electroporated, IRE zone) or temporarily (reversibly electroporated, RE zone) destructed according to the selected application parameters (electrode distance, power of electric pulses, and duration). Despite biological changes caused by IRE, region boundaries are difficult to determine immediately post IRE using standard MRI imaging modalities (14). A recent study has demonstrated that advanced MRI modalities can be performed for immediate assessment of ablation zones using advanced MRI techniques (15); however, these procedures rely on complicated MRI sequences with a lack of reproducibility and are restricted to certain centers due to complex non-standard analysis approaches (16).
Recent technological developments have enabled quantitative radiomics analysis of clinical imaging data for diagnosis and prognosis of various diseases, prediction of genomic architectures, and personalized disease staging (17-20). Advanced statistical learning methods equipped with descriptive image features have been utilized to develop expert models for improving our understanding of disease characteristics and predicting therapeutic outcomes (21). Despite the benefits emphasized by previous studies, texture-based quantitative analysis of MRI data has not been performed to differentiate IRE from RE zones for early evaluation of the IRE ablation in our knowledge. In this study, we investigated potential benefits of quantitative radiomics features computed from structural MRI data to generate multivariable models for early assessment of IRE ablation through differentiation of ablation zones. Also, we developed multiparametric regression models by utilizing noninvasive MRI features to associate characteristics of the biological changes captured with histological tumor markers.
Materials and Methods
Our pre-clinical experiment was approved and performed according to Institutional Animal Care and Use Committee regulations, and all the animals were treated under humane conditions with regular monitoring at the animal facility.
Animal Model and Treatment
Of 16 New Zealand white rabbits, two were used as a donor for VX2 incubation, and others were used for performing the study. Under basic anesthesia that was initiated via injection of ketamine (100 mg/kg) and xylazine (5 mg/kg), and continued via isoflurane (2-3% in oxygen, 3 L/min), VX2 tumor cells were injected into the hindlimbs of donor rabbits. The rabbits were monitored using MRI and tumors were allowed to grow until reaching a size of 3 cm in the longest diameter. Afterward, tumor tissues were dissected and tumor fragments with a size of about 1 mm3 were implanted in the left lobes of the remaining rabbits using a biopsy needle under percutaneous ultrasound guidance. The tumors were allowed to grow for 7-10 days as reaching a size of at least 10 mm under the guidance of MRI.
During IRE ablation, rabbits were stabilized in supine orientation and HCC tumors were revealed with mini laparotomy with anesthesia performed via isoflurane (2-3% in oxygen, 3 L/min). MRI compatible 2-electrode platinum-iridium array with a thickness of 0.4 mm in diameter was placed about 5 mm inside of the tumor. IRE ablation was performed by two researchers (2+ years of experience) using ECM830 BTX Electroporator (Harvard Apparatus, Holliston, MA) with a predefined sequence (8 pulses at 2 kV of power with a duration of 100 μs and 100 ms interval) (22). Following IRE ablation, double-layer closure was performed, and MRI scanning was performed under anesthesia.
MRI Acquisition and Analysis
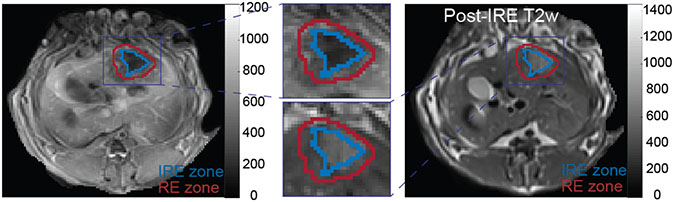
The rabbits were scanned using a 3T Siemens Magnetom Skyra MRI scanner before (only T1w and T2w MRI as baseline) and immediately after the IRE ablation procedure (T1w, T2w, and transcatheter intra-arterial perfusion (TRIP) MRI) with acquisition parameters described in Table 1. In our study, TRIP MRI was utilized to characterize perfusion properties of the tissues following IRE ablation and immediate assessment of IRE ablation zones (23). The regions of interest (ROIs: RE and IRE zones) were outlined on a single post-IRE TRIP MRI slice with maximum tumor diameter by consensus of two radiologists (6+ years of experience) under the instruction of a senior radiologist (15+ years of experience) using ITK-SNAP (v3.8). The generated mask images (IRE and RE zones) were translated to registered T1w, and T2w MRI images and corresponding images were utilized for radiomics analysis. In Figure 1, representative RE and IRE zones were shown on T1w and T2w MRI slices.
Table 1.
MRI acquisition parameters for conventional and perfusion MRI sequences
| Sequence | TR (ms) | TE (ms) | Flip Angle(°) | # of Slices | FOV (mm3) | Matrix | Thickness/Gap (mm) |
|---|---|---|---|---|---|---|---|
| T1w | 200 | 2.93 | 70 | 17 | 180×180 | 224×224 | 2/0.6 |
| T2w | 4000 | 39 | 150 | 17 | 180×180 | 192×192 | 2/0.6 |
| TRIP MRI | 2.6 | 1.02 | 8 | 20 | 128×112 | 128×112 | 2/0.6 |
Note: Transcatheter Intraarterial Perfusion Magnetic Resonance Imaging, (TRIP MRI)
Figure 1.
Representative regions corresponding to RE and IRE zones on T1w and T2w MRI slices after performing IRE ablation. After registration of MRI data, ablation regions were drawn by an experienced radiologist using TRIP MRI and translated to T1w and T2w MRI slices for further analysis.
Note (RE: Reversible electroporation; IRE: Irreversible electroporation)
Histology Analysis
Following completion of post-IRE MRI acquisition, rabbits were euthanized and liver tissue sections including irreversibly and reversibly electroporated tissue regions were collected. Afterward, tissue samples were fixed in 10% formaldehyde solution and sent to the pathology core facility for histopathological analysis. Two researchers (2+ years of experience) examined hematoxylin-eosin (H&E) staining for counting viable cells, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining for assessment of cell apoptosis, and CD31 staining for evaluation of microvessel density. The number of tumor cells, apoptosis fraction, and microvessel density were evaluated by two blinded researchers (5+ years of experience) through at least five selected regions and then averaged to define representative analysis results.
Feature Extraction and Selection
To overcome MRI intensity relativity, images were quantized using a fixed-bin-size quantization approach in which optimal bin size was empirically determined among three candidates (4, 8, and 16). A total of forty-six region-based and pixelwise features were computed from T1w and T2w MRI images through analysis of statistical characteristics of the intensity (first-order statistics, FOS, 6 features) and texture (gray-level co-occurrence matrix, GLCM, 9 features; gray-level run-length matrix, GLRLM, 13 features; gray-level size-zone matrix, GLSZM, 13 features; neighborhood gray-tone difference matrix, NGTDM, 5 features) (24-26). The complete list of the computed features is presented in Supplementary Table 1. The region-based classifiers were generated by integrating structural aspects of all the voxels in RE and IRE zones while pixelwise classification models determined the label of each voxel within RE and IRE zones with features computed over three-by-three neighborhood window. GLCM and GLRLM features were computed through four gradient directions (0°, 45°, 90°, and 135°) and combined by averaging. Afterward, features were transformed into the range of [0, 1] with min-max normalization.
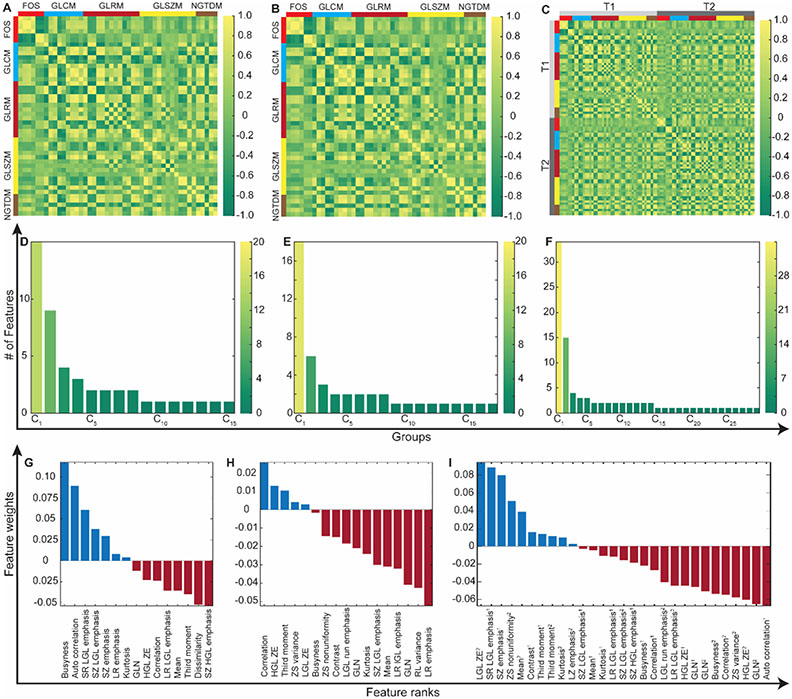
To identify key predictors, a two-step feature selection framework was followed. First, features were split into clusters by evaluating inter-relation among the features measured with Pearson correlation coefficient (Figure 2A-C), and an individual feature with the largest average correlation coefficient was selected as the representative of each cluster (Figure 2D-F). Afterward, the rankings of the candidate variables were computed with RELIEFF algorithm by evaluating the interaction between candidate MRI features and therapeutic variables (outcome and histological tumor biomarker measurements) using the ten nearest neighbors approach (Figure 2G-I) (27). Top-ranked eight variables were further analyzed to determine key predictors for treatment response and histological tumor markers while optimizing parameters of random forest (RF) classification and regression models empirically.
Figure 2.
The framework for selection of the features extracted from T1w and T2w MRI, and their concatenation. (A)-(C) visualizes pairwise correlation coefficients of the computed variables with five different feature extraction methods. (D)-(F) demonstrates the number of features in each cluster generated based on Pearson correlation coefficients. The number of clusters increases with broadening heterogeneity among image features. The importance of the features was determined via RELIEFF algorithm that evaluates the interaction between MRI features and therapeutic measurements (IRE ablation regions and histopathological tumor marker measurements (G-I).
Note: FOS, first-order statistics features; GLCM, gray-level co-occurrence matrix features; GLRM, gray-level run-length matrix features; GLSZM, gray-level size-zone matrix features; and NGTDM, neighborhood gray-tone difference matrix features. T1, T1w MRI; and T2, T2w MRI.
Statistical Analysis
The biological changes related to IRE ablation therapy were characterized through region-based and pixelwise features by generating three RF classification models: T1w, T2w, and T1w+T2w MRI (combination MRI). For individual models, candidate features were identified by performing a two-step feature-selection process. The features for combined MRI were identified by following the same two-step approach after merging the features into one set. During region-based classification experiments, three models were generated using twenty-eight images with leave-one-out cross-validation to prevent overfitting. For pixelwise classifiers, image data (2480 voxels) were separated into training and test sets with a ratio of 80%/20%, and ten-fold cross-validation was performed during training. The performance of the classification models was measured with accuracy, specificity, sensitivity, and area under the receiver operating characteristics curve (AUC).
To associate characteristics of IRE ablation regions reflected on MRI and biological changes captured with histological tumor markers, three multivariable regression models were generated using RF method with leave-one-out cross-validation. The performance of regression models was assessed with root-mean-squared-error (RMSE), Spearman correlation coefficient, and relative-error. A two-tailed Student T-test was used to evaluate statistical difference while accepting p<0.05 significant. The analysis of the data was completed using MATLAB® v9.8 (MathWorks, Natick, MA). The datasets generated during and/or analyzed during the current study will be available from the corresponding author on reasonable request.
Results
Feature Selection
The candidate features were identified by following a two-step procedure. First, T1w, T2w, and T1w+T2w MRI radiomics features were clustered based on their similarity, and representative of each cluster was determined for region-based classification model (15, 17, and 29 features) and regression models (15, 14, and 26 features). During the assessment of pixelwise features, a lower number of clusters and representative features (11, 10, and 21 features) were identified. Second, all features were sorted according to their rankings computed with RELIEFF method within their groups, and top-ranked eight features were selected for each experiment to perform classification and regression tasks.
Evaluation of the Classification Models
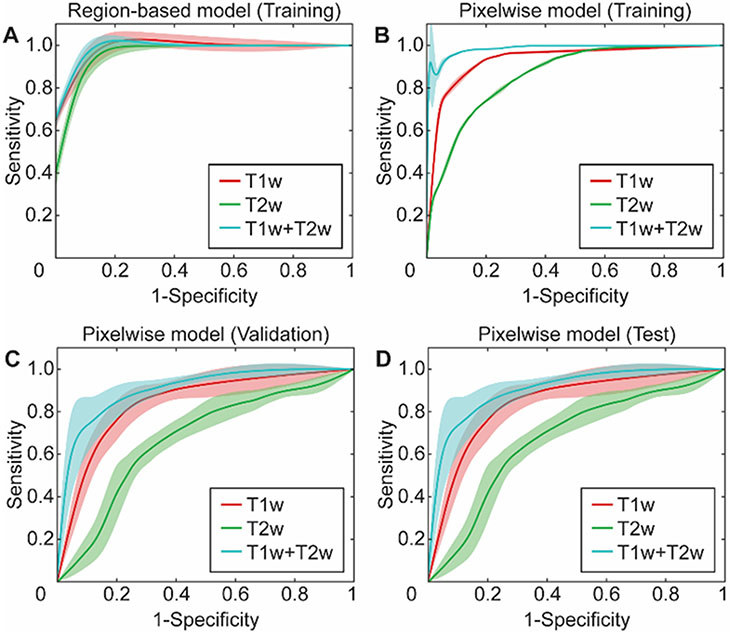
Individual RF classification models were constructed with features computed from T1w, T2w, and T1w+T2w MRI data to distinguish IRE and RE ablation zones. RF classifiers were generated with four quantitative region-based features (Table 2) that were selected through exhaustive search experiments while evaluating validation accuracy of the decision models. T1w MRI classifier integrating four features had an accuracy of 1.0 for training and 0.97 for validation experiment. Moreover, it had a specificity of (1.0, 1.0) and sensitivity of (1.0, 0.93) for training and validation experiments. On the other hand, T2w MRI classification model obtained an accuracy of (0.90, 0.80), a specificity of (0.89, 0.80), and a sensitivity of (0.90, 0.80) for training and validation sets. Moreover, T1w+T2w MRI model demonstrated slightly better performance as presenting an accuracy of (0.99, 0.97), specificity of (1.0, 0.93), and sensitivity of (0.99, 1.0) for training and validation experiments. The receiver operating characteristics (ROC) curves were shown in Figure 3A to visualize classifier behavior during training experiments.
Table 2.
The list of features used to generate classification models for differentiation of irreversible and reversible electroporation zones.
| T1w | T2w | T1w1+T2w2 | |
|---|---|---|---|
| Region-based Features | Busyness (NGTDM) | HGL ZE (GLSZM) | LGL ZE (GLSZM1) |
| Autocorrelation (GLCM) | Third moment (FOS) | SR LGL emphasis (GLRLM1) | |
| SR LGL emphasis (GLRLM) | Zone-size variance (GLSZM) | SZ emphasis (GLSZM1) | |
| GL nonuniformity (GLSZM) | Busyness (NGTDM) | Third moment (FOS1) | |
| Voxelwise Features | SD (FOS) | LGL RE (GLRLM) | LGL RE (GLRLM1) |
| LGL RE (GLRLM) | Variance (GLCM) | GL nonuniformity (GLRLM1) | |
| Mean (FOS) | Entropy (FOS) | Variance (GLCM1) | |
| LR LGL emphasis (GLRLM) | GL nonuniformity (GLSZM) | SZ LGL emphasis (GLSZM1) | |
| HGL ZE (GLSZM) | RP (GLRLM) | SD (FOS1) |
Features: Gray-level (GL), high gray-level (HGL), low gray-level (LGL), Long run (LR), run emphasis (RE), run percentage (RP), Short-run (SR), small zone (SZ), standard deviation (SD), zone emphasis (ZE)
Classes: First order statistics (FOS), gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level size-zone matrix (GLSZM), neighborhood grey tone difference matrix (NGTDM).
Note Superscript highlights image that was used to compute specific feature.
Figure 3.
The receiver operating curves (ROCs) for the predictive models were generated to discriminate RE and IRE zones by developing region-based and pixelwise classification models with random forest technique. (A) represents ROCs for the training of the region-based classifier while (B-D) represents ROCs for training, validation, and test sets for the pixelwise classification model.
The pixelwise RF classification models were generated with five features selected among eight candidate variables (Table 2) while classifier parameters were optimized by assessment of validation dataset and final model performance were measured using test data. T1w MRI classifier with five predictors obtained an accuracy of (0.88, 0.79, and 0.83) for training, validation, and test datasets. Additionally, specificity of (0.83, 0.74, and 0.79), sensitivity of (0.91, 0.83, and 0.85), and AUC of (0.93, 0.83, and 0.85) were observed for training, validation, and test sets, respectively. T2w MRI classifier demonstrated comparatively lower stability as receiving accuracy of (0.78, 0.67, and 0.67), specificity of (0.69, 0.57, and 0.59), sensitivity of (0.85, 0.74, and 0.72), and also AUC of (0.86, 0.67, and 0.66) for training, validation, and test datasets, respectively. MRI model obtained slightly better performance compared to individual models (T1w and T2w MRI). It had accuracy of (0.94, 0.83, and 0.83), specificity of (0.92, 0.79, and 0.80), sensitivity of (0.95, 0.86, and 0.85), and AUC of (0.98, 0.90, and 0.90) for training, validation, and test datasets, respectively. ROC curves for training, validation, and test datasets are shown in Figure 3B-D to demonstrate overall behavior of the classifiers.
Evaluation of Correlation between MRI features and Histological Tumor Biomarkers
The regression models were generated to describe correlation between histological tumor biomarkers and MRI radiomics features with the assessment of RMSE and Spearman correlation coefficient of the training and validation during leave-one-out cross-validation. For each histological biomarker, three RF regression models were developed following a comprehensive analysis of the features computed from T1w, T2w, and T1w+T2w MRI data while optimizing model parameters through leave-one-out cross-validation.
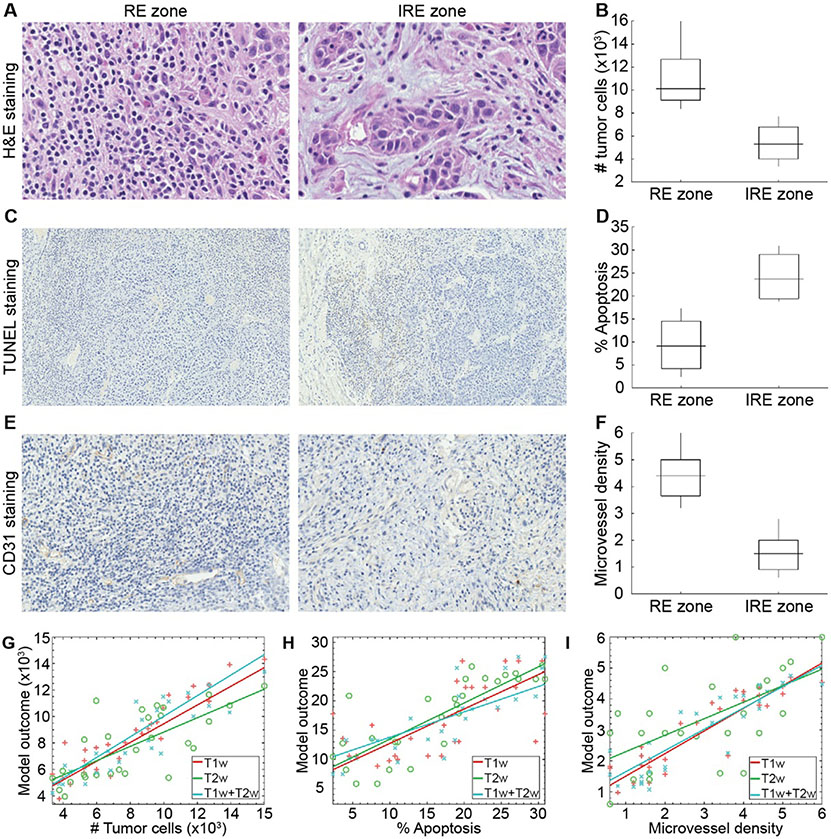
H&E-stained histological slice demonstrated significantly different (p<0.01) tumor cells in RE (10969.64±2215.64 cells) and IRE zones (5429.71±1381.72 cells) (Figure 4A-B). During multivariable analysis, three features were selected as key predictors for generating regression models to associate with tumor cell count (Table 3). T1w MRI regression model obtained an RMSE of 1657.20 that corresponds to a relative error of 0.155 and a correlation value of 0.931 (p<0.0001). T2w MRI regression model had an RMSE of 5353.67 associated with a relative error of 0.233 and a correlation value of 0.685 (p<0.0001). Furthermore, T1w+T2w MRI regression model presented an RMSE of 2318.47 that corresponded to a relative error of 0.171 and a correlation value of 0.884 (p<0.0001). The correlation of multivariable models and the tumor cell count is shown in Figure 4G.
Figure 4.
The representative histological images for visualizing IRE ablation regions (A, C, and D), statistical analysis (B, E, and F), and correlation plots (G-I). A significantly lower number of tumor cells (B), percentage of apoptosis rate (E), and higher microvessel density (F) for IRE zones compared to RE zones. The figures (G)-(I) demonstrate the behavior of the models generated with features of T1w, T2w, and T1w+T2w MRI data.
Table 3.
The list of features used to generate regression models for characterizing the behavior of histological tumor markers as reflecting biological changes in tumor tissue.
| T1w | T2w | T1w 1 + T2w 2 | |
|---|---|---|---|
| H&E | SZ LGL Emphasis (GLSZM) | LGL ZE (GLSZM) | SZ LGL Emphasis (GLSZM1) |
| SZ HGL Emphasis (GLSZM) | Kurtosis (FOS) | SZ emphasis (GLSZM1) | |
| Kurtosis (FOS) | SR LGL emphasis (GLRLM) | SZ HGL Emphasis (GLSZM1) | |
| CD31 | HGL ZE (GLSZM) | LGL ZE (GLSZM) | SZ LGL emphasis (GLSZM1) |
| SZ HGL Emphasis (GLSZM) | Kurtosis (FOS) | HGL ZE (GLSZM1) | |
| Autocorrelation (GLCM) | SR LGL emphasis (GLRLM) | Autocorrelation (GLCM1) | |
| Correlation (GLCM) | ZS nonuniformity (GLSZM) | Correlation (GLCM1) | |
| TUNEL | HGL ZE (GLSZM) | Busyness (NGTDM) | Autocorrelation (GLCM1) |
| SZ LGL Emphasis (GLSZM) | ZS variance (GLSZM) | Kurtosis (FOS2) | |
| Autocorrelation (GLCM) | SR LGL emphasis (GLRLM) | SZ LGL emphasis (GLSZM1) |
Features: Gray-level (GL), high gray-level (HGL), low gray-level (LGL), Long run (LR), run emphasis (RE), run percentage (RP), Short-run (SR), small zone (SZ), standard deviation (SD), zone emphasis (ZE), zone percentage (ZP).
Classes: First order statistics (FOS), gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level size-zone matrix (GLSZM), neighborhood grey tone difference matrix (NGTDM).
In TUNEL stained histological images, a significant difference (p<0.01) for the percentage of hepatocyte apoptosis rate was observed between ablation regions (9.28±4.98 for RE and 24.02±4.48 IRE zones) (Figure 4C-D). Throughout the multivariable analysis, a total of three quantitative features were determined as pivotal variables to generate regression models for interpreting characteristics of the apoptosis rates in ablation regions (Table 3). The regression model generated with T1w MRI features had an RMSE of 4.57×10−2 that corresponds to a relative error of 0.672 and a correlation value of 0.623 (p<0.001). T2w MRI regression model obtained an RMSE of 2.59×10−2 that was associated with a relative error of 0.529 and a correlation value of 0.828 (p=0.003). Moreover, T1w+T2w MRI regression model presented an RMSE of 2.21×10−2 that corresponded to a relative error of 0.455 and a correlation value of 0.873 (p<0.0001). The association between multivariable models and hepatocyte apoptosis rate is shown in Figure 4H.
CD31 stained histological slices presented a statistically significant microvessel density difference (p<0.001) among RE (4.36±0.77) and IRE zones (1.53±0.64) (Figure 4E-F). The multivariable regression models were generated with four MRI features for describing the behavior of microvessel density by interpreting the structures reflected in MRI (Table 3). The regression model generated with the features of T1w MRI data obtained an RMSE of 4.86×10−4 that corresponds to a relative error of 0.326 and a correlation value of 0.885 (p<0.0001). T2w MRI regression model combining texture and intensity-based features had an RMSE of 1.92×10−3 that was associated with a relative error of 0.613 and a correlation value of 0.603 (p<0.001). Moreover, T1w+T2w MRI model that integrates vital variables of T1w and T2w MRI data had an RMSE of 4.94×10−4 that corresponded to a relative error of 0.317 and a correlation value of 0.893 (p<0.001). The correlation between microvessel density and individual regression models is shown in Figure 4I.
Discussion
In this study, we investigated potential benefits of radiomics features computed from structural MRI data to develop multivariable classification models for IRE ablation assessment. The identified quantitative MRI features facilitated early discrimination of temporarily and permanently electroporated ablation zones while regression models correlated with histological tumor biomarkers for noninvasive evaluation of IRE ablation therapy. The results demonstrated that quantitative analysis of MRI texture reveals the characteristic behavior of histological tumor biomarkers and distinguishes ablation zones by interpreting early changes following IRE ablation.
IRE ablation is one of the most promising treatment modalities for cancer patients who are not eligible for potentially curative treatments e.g., surgical resection and liver transplantation (9, 28). Compared to other ablative techniques, IRE brings out several benefits including safe application near major blood vessels and minimal effects on surrounding tissues, and induction of immunological reaction against tumor cells (23). RE and IRE regions, generated following the electroporation, can be used for assessment of IRE ablation efficacy and identifying the suitability of the environment for treatments that are restricted by membrane permeability e.g., chemotherapeutic drugs and immunotherapeutic agents. Furthermore, early assessment of these permanently (IRE) and temporarily (RE) electroporated regions allows adjusting treatment framework including parameters of the ablation method. Several studies have investigated CT, US, and MRI imaging techniques for the assessment of response after IRE ablation and have concluded that MRI is the preferred modality for differentiation of the electroporation treatment zones (15, 22, 29-31). These studies demonstrated superior advantages of advanced MRI modalities over conventional MRI sequences for differentiation of treated and untreated regions with a well-demarcated boundary between them. Despite the benefits of advanced MRI modalities for reflecting IRE treatment effects, the necessity for complicated but non-standardized analysis approaches limits their usage for the early assessment of IRE ablation. Therefore, novel approaches for the analysis of conventional MRI data are required for the differentiation of temporary (RE) and permanent electroporation (IRE) regions using standard MRI data after IRE ablation.
Throughout the last decade, the potential role of texture-based computational features that capture the heterogeneity and reveal distinctive characteristics of the tissues has become more apparent (32-34). Previous studies have investigated the benefits of MRI data analysis with the aim of diagnosis and prognosis of diseases, prediction of genomic expression signatures, and stratification of tumor types (35-37). However, there is a lack of data on the role of texture analysis to characterize IRE ablation zones and show the association between MRI radiomics features and histological tumor biomarkers in this setting. In this study, we investigated the texture-based features of conventional MRI data for distinguishing IRE ablation zones. Furthermore, we evaluated the correlation of histological tumor markers and MRI radiomics features as biomarkers. Our results demonstrated that a classification model equipped with T1w MRI texture features allows accurate differentiation of IRE ablation zones immediately after the procedure. Whereas T2w MRI data is routinely used for visual assessment of the tumor characteristic for clinical practice, T2w MRI classifier obtained a lower performance compared to T1w MRI. This might be associated with underlying biological changes occurring in tumor texture captured via T1w MRI sequence. The performance of the region-based classifier is not significantly improved by combining T1w and T2w MRI features. However, combined voxelwise classifier (accuracy of 82.93% and AUC of 0.896) obtained a slight improvement compared to T1w model (accuracy of 82.80% and AUC of 0.852) and significantly for T2w model (accuracy of 66.53% and AUC of 0.657). Multivariable models were developed to analyze correlation with histopathological tumor biomarkers. The regression model examining fibrosis measurement of the tumor structure with three features of T1w MRI data (r2=0.85) had better performance compared to both T2w MRI (r2=0.62) and combined MRI models (r2=0.84). However, the performance of the multivariable models describing hepatocyte apoptosis rate (r2=0.82, p<0.01), and microvessel density (r2=0.58, p<0.01) improved with the integration of features of combined MRI data. We observed a stronger correlation between hepatocyte apoptosis rate measured with CD31 immunostaining and multivariable T1w radiomics model while characteristic of microvessel density was better captured with T2w MRI data.
Our study had limitations including the size of the subject cohort, and limited use of data-driven analysis methods. Despite the small size of our subject cohort, the sample size was comparable with previous preclinical studies (38, 39). Also, a classical statistical learning technique with less complexity was utilized to interpret IRE ablation method response with our comparatively small dataset. With the analysis of larger sample size, more complex approaches would be feasible to perform. Furthermore, conventional MRI data were included during statistical learning analysis despite available TRIP MRI data. Although advanced MRI modalities have become vital for tumor assessment (40) and were previously utilized for radiomics studies (41, 42), the lack of standardized analysis approaches restricts the reliability and reproducibility (43, 44). Therefore, clinically standard T1w and T2w MRI, utilized for diagnosis, treatment, and monitoring disease progression, were included for advanced statistical learning analyses in this study and showed very promising results. Finally, current study was performed at a single institution by using same experimental setup and devices. Further studies aiming for a comprehensive analysis of reproducibility will be beneficial.
In conclusion, our study demonstrated that characteristic tissue changes following IRE ablation can be captured with conventional MRI modalities. By developing multivariable classification models using texture-based MRI features, IRE ablation zones were successfully identified. Furthermore, imaging biomarkers reflecting underlying tissue changes show promise for noninvasive assessment of response to IRE ablation.
Supplementary Material
Grant Support:
This study was supported by the National Cancer Institute (grants R01CA196967, R01CA209886, and R01CA241532), 2019 Harold E. Eisenberg Foundation Scholar Award, and SIR Foundation Pilot Grant (PR-0000000012).
References
- 1.McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology. 2021; 73(S1):4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belghiti J, Kianmanesh R. Surgical treatment of hepatocellular carcinoma. HPB (Oxford). 2005; 7(1):42–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. New England Journal of Medicine. 2020; 382(20):1894–905. [DOI] [PubMed] [Google Scholar]
- 4.Cabral LKD, Tiribelli C, Sukowati CHC. Sorafenib Resistance in Hepatocellular Carcinoma: The Relevance of Genetic Heterogeneity. Cancers (Basel). 2020; 12(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Rajabi R, Patel S, Ketchum NS, et al. Comparative dosing and efficacy of sorafenib in hepatocellular cancer patients with varying liver dysfunction. Journal of gastrointestinal oncology. 2015; 6(3):259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang W, Chen Z, Zhang W, et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduction and Targeted Therapy. 2020; 5(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. The Lancet. 2018; 391(10126):1163–73. [DOI] [PubMed] [Google Scholar]
- 8.Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: From diagnosis to treatment. Surgical Oncology. 2016; 25(2):74–85. [DOI] [PubMed] [Google Scholar]
- 9.Eresen A, Yang J, Scotti A, Cai K, Yaghmai V, Zhang Z. Combination of natural killer cell-based immunotherapy and irreversible electroporation for the treatment of hepatocellular carcinoma. Annals of Translational Medicine. 2021; 9(13):1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. The Lancet. 2018; 391(10127):1301–14. [DOI] [PubMed] [Google Scholar]
- 11.Mauri G, Nicosia L, Varano GM, et al. Tips and tricks for a safe and effective image-guided percutaneous renal tumour ablation. Insights Imaging. 2017; 8(3):357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dollinger M, Zeman F, Niessen C, et al. Bile duct injury after irreversible electroporation of hepatic malignancies: Evaluation of MR imaging findings and laboratory values. J Vasc Interv Radiol. 2016; 27(1):96–103. [DOI] [PubMed] [Google Scholar]
- 13.Rubinsky B, Onik G, Mikus P. Irreversible electroporation: a new ablation modality--clinical implications. Technol Cancer Res Treat. 2007; 6(1):37–48. [DOI] [PubMed] [Google Scholar]
- 14.Barabasch A, Distelmaier M, Heil P, Krämer NA, Kuhl CK, Bruners P. Magnetic resonance imaging findings after percutaneous irreversible electroporation of liver metastases: A systematic longitudinal study. Investigative Radiology. 2017; 52(1):23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shangguan AJ, Zhou K, Yang J, et al. Intraprocedural Transcatheter Intraarterial Perfusion (TRIP)-MRI for Evaluation of Irreversible Electroporation Therapy Response in a Rabbit Liver Tumor Model. Clin Exp Gastroenterol. 2020; 13:543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao S-L, Metens T, Lemort M. TumourMetrics: a comprehensive clinical solution for the standardization of DCE-MRI analysis in research and routine use. Quantitative Imaging in Medicine and Surgery. 2017; 7(5):496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Long L, Wei J, et al. Radiomics for diagnosis of dual-phenotype hepatocellular carcinoma using Gd-EOB-DTPA-enhanced MRI and patient prognosis. Journal of Cancer Research and Clinical Oncology. 2019; 145(12):2995–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eresen A, Yang J, Shangguan J, et al. MRI radiomics for early prediction of response to vaccine therapy in a transgenic mouse model of pancreatic ductal adenocarcinoma. J Transl Med. 2020; 18(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li K, Xiao J, Yang J, et al. Association of radiomic imaging features and gene expression profile as prognostic factors in pancreatic ductal adenocarcinoma. Am J Transl Res. 2019; 11(7):4491–9. [PMC free article] [PubMed] [Google Scholar]
- 20.Nie P, Yang G, Guo J, et al. A CT-based radiomics nomogram for differentiation of focal nodular hyperplasia from hepatocellular carcinoma in the non-cirrhotic liver. Cancer Imaging. 2020; 20(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakabayashi T, Ouhmich F, Gonzalez-Cabrera C, et al. Radiomics in hepatocellular carcinoma: a quantitative review. Hepatol Int. 2019; 13(5):546–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, White SB, Nicolai JR, et al. Multimodality imaging to assess immediate response to irreversible electroporation in a rat liver tumor model. Radiology. 2014; 271(3):721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figini M, Zhou K, Pan L, et al. Transcatheter intra-arterial perfusion (TRIP)-MRI biomarkers help detect immediate response to irreversible electroporation of rabbit VX2 liver tumor. Magn Reson Med. 2019; 84(1):365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eresen A, Yang J, Shangguan J, Benson AB, Yaghmai V, Zhang Z. Detection of immunotherapeutic response in a transgenic mouse model of pancreatic ductal adenocarcinoma using multiparametric MRI radiomics: A preliminary investigation. Acad Radiol. 2020; 28(6):e147–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thibault G, Fertil B, Navarro C, L., et al. Texture indexes and gray level size zone matrix. Application to cell nuclei classification. 10th International Conference on Pattern Recognition and Information Processing, PRIP 2009. Minsk, Belarus 2009; 140–5. [Google Scholar]
- 26.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern B Cybern. 1989; 19(5):1264–74. [Google Scholar]
- 27.Kononenko I, Šimec E, Robnik-Šikonja M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Applied Intelligence. 1997; 7(1):39–55. [Google Scholar]
- 28.Aycock KN, Davalos RV. Irreversible Electroporation: Background, Theory, and Review of Recent Developments in Clinical Oncology. Bioelectricity. 2019; 1(4):214–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Y, Zhang Y, Nijm GM, et al. Irreversible electroporation in the liver: contrast-enhanced inversion-recovery MR imaging approaches to differentiate reversibly electroporated penumbra from irreversibly electroporated ablation zones. Radiology. 2011; 258(2):461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padia SA, Johnson GE, Yeung RS, Park JO, Hippe DS, Kogut MJ. Irreversible electroporation in patients with hepatocellular carcinoma: Immediate versus delayed findings at MR imaging. Radiology. 2015; 278(1):285–94. [DOI] [PubMed] [Google Scholar]
- 31.Felker ER, Dregely I, Chung DJ, et al. Irreversible electroporation: Defining the MRI appearance of the ablation zone with histopathologic correlation in a porcine liver model. AJR Am J Roentgenol. 2017; 208(5):1141–6. [DOI] [PubMed] [Google Scholar]
- 32.Varghese BA, Cen SY, Hwang DH, Duddalwar VA. Texture analysis of imaging: What radiologists need to know. AJR Am J Roentgenol. 2019; 212(3):520–8. [DOI] [PubMed] [Google Scholar]
- 33.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019; 380(14):1347–58. [DOI] [PubMed] [Google Scholar]
- 34.Ibrahim A, Vallières M, Woodruff H, et al. Radiomics Analysis for Clinical Decision Support in Nuclear Medicine. Semin Nucl Med. 2019; 49(5):438–49. [DOI] [PubMed] [Google Scholar]
- 35.Eresen A, Yang J, Shangguan J, et al. Prediction of therapeutic outcome and survival in a transgenic mouse model of pancreatic ductal adenocarcinoma treated with dendritic cell vaccination or CDK inhibitor using MRI texture: a feasibility study. Am J Transl Res. 2020; 12(5):2201–11. [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Min JH, Kim SK, Shin SY, Lee MW. Detection of hepatocellular carcinoma in contrast-enhanced magnetic resonance imaging using deep learning classifier: A multi-center retrospective study. Sci Rep. 2020; 10(1):9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiménez Pérez M, Grande RG. Application of artificial intelligence in the diagnosis and treatment of hepatocellular carcinoma: A review. World J Gastroenterol. 2020; 26(37):5617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang TT, Zawaski JA, Francis KN, Qutub AA, Gaber MW. Image-based classification of tumor type and growth rate using machine learning: A preclinical study. Sci Rep. 2019; 9(1):12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao M, Igata H, Takeuchi A, Sato K, Ikegaya Y. Machine learning-based prediction of adverse drug effects: An example of seizure-inducing compounds. J Pharmacol Sci. 2017; 133(2):70–8. [DOI] [PubMed] [Google Scholar]
- 40.Vohra R, Park J, Wang Y-N, et al. Evaluation of pancreatic tumor development in KPC mice using multi-parametric MRI. Cancer Imaging (BioMed). 2018; 18(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Li Z, Qu J, et al. Radiomics of multi-parametric MRI for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clinical cancer research. 2019; 25(12):3538–47. [DOI] [PubMed] [Google Scholar]
- 42.Zhou X, Yi Y, Liu Z, et al. Radiomics-based pretherapeutic prediction of non-response to neoadjuvant therapy in locally advanced rectal cancer. Annals of Surgical Oncology. 2019; 26(6):1676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padhani AR, Liu G, Koh DM, et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia (New York, NY). 2009; 11(2):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma S, Turkbey B, Muradyan N, et al. Overview of dynamic contrast-enhanced MRI in prostate cancer diagnosis and management. AJR Am J Roentgenol. 2012; 198(6):1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.