Abstract
The knowledge on the genetic component of non-alcoholic fatty liver disease (NAFLD) has grown exponentially over the last 10 to 15 years. This review summarizes the current evidence and the latest developments in the genetics of NAFLD and non-alcoholic steatohepatitis (NASH) from the immune system’s perspective. Activation of innate and or adaptive immune response is an essential driver of NAFLD disease severity and progression. Lipid and immune pathways are crucial in the pathophysiology of NAFLD and NASH. Here, we highlight novel applications of genomic techniques, including single-cell sequencing and the genetics of gene expression, to elucidate the potential involvement of NAFLD/NASH-risk alleles in modulating immune system cells. Together, our focus is to provide an overview of the potential involvement of the NAFLD/NASH-related risk variants in mediating the immune-driven liver disease severity and diverse systemic pleiotropic effects.
Keywords: Nonalcoholic steatohepatitis, Genetics, PNPLA3, HSD17B13, Immune system
INTRODUCTION
The global trends in the prevalence and incidence of non-alcoholic fatty liver disease (NAFLD) represent a significant public health challenge. The disease prevalence has reached alarming figures not only in adults but also in the children’s population [1,2]. Knowledge regarding the genetic component of NAFLD has grown exponentially over the last 10–15 years [3-7]. With this knowledge, it has become possible to translate information of risk alleles and its effects on the disease biology into clinical application [6,8]. Most importantly, knowledge on the genetic component of NAFLD may be leveraged to identify individuals at risk and/or to estimate the risk of severe histological outcomes, including non-alcoholic steatohepatitis (NASH)-fibrosis, cirrhosis, and hepatocellular carcinoma [6,8].
While NAFLD is a disorder characterized by excess accumulation of fat in hepatocytes, in up to 40% of individuals with NAFLD, there are additional findings of portal and lobular inflammation and hepatocyte injury which characterize the severe histological forms of the disease [3]. Therefore, activation of the immune system is a key feature of the disease severity and progression [3].
Furthermore, progressive clinical forms of NAFLD, including NASH-fibrosis, NASH-cirrhosis, and eventually hepatocellular carcinoma, are the main drivers of liver disease-associated mortality worldwide [1,2].
Although remarkable progress has been made in understanding the disease biology, it remains unclear how to link NAFLD/NASH-associated variants with immune-specific cells mechanistically and how to explain the role of genetics in immune-driven disease progression.
In this review, we summarize the current evidence and the latest developments in the field of genetics of NAFLD and NASH—the disease’ severe histological form—from the perspective of the role of risk alleles in modulating gene expression of cells of the immune system. Our focus is to provide an overview of the potential involvement of the NAFLD/NASH-related risk variants in mediating the immune-driven disease severity.
A SHORT OVERVIEW OF VARIANTS INFLUENCING THE RISK AND PROTECTION AGAINST NAFLD AND THE HISTOLOGICAL DISEASE SEVERITY
Genetic discoveries in the field of NAFLD have mainly been motorized by the use of genome-wide (GWAS) [9,10], exome-wide (EWAS) [11], and more recently, phenome-wide (PHEWAS) association studies using electronic health records [12], as well as high-throughput sequencing technologies, which allowrefining and mapping of the discovered variants [13].
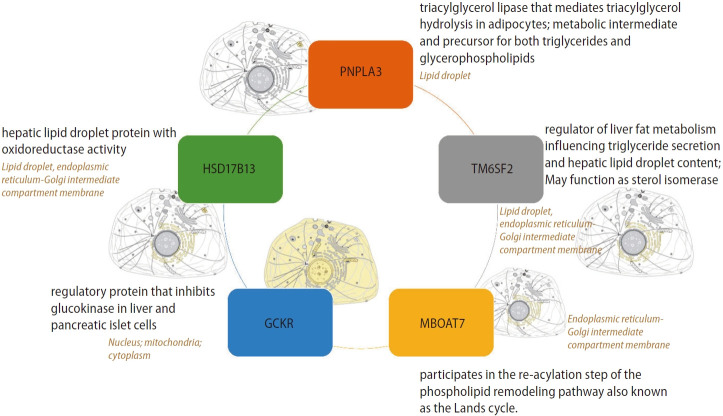
Most relevant and replicated targets associated with the genetic component of NAFLD are illustrated in Figure 1, which depicts the primary protein function and subcellular localization. Notably, major candidate gene variants function in metabolic pathways.
Figure 1.
Most relevant and replicated targets associated with the genetic component of NAFLD. Figure depicts primary protein function and subcellular localization. Information was retrieved from UniProt, a comprehensive and freely accessible resource of protein sequence and functional information available at https://www.uniprot.org. NAFLD, non-alcoholic fatty liver disease; PNPLA3, patatin-like phospholipase domain containing 3; HSD17B13, hydroxysteroid 17-beta dehydrogenase 13; MBOAT7, membrane bound O-acyltransferase domain containing 7; GCKR, glucokinase regulator; TM6SF2, transmembrane 6 superfamily member 2.
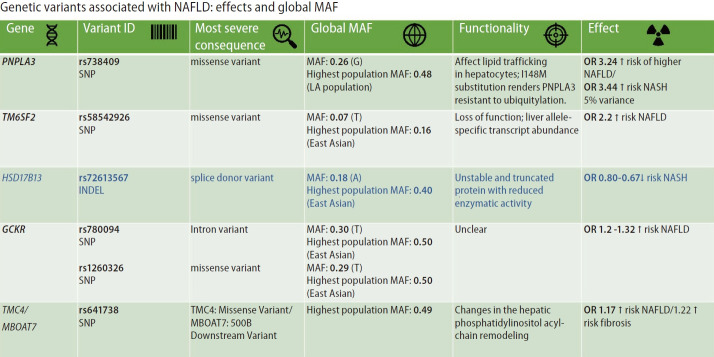
Figure 2 summarizes the most replicated variants associated with NAFLD and NASH, including the global minor allele frequency, the variant’s most severe consequence, the variant functionality, and the variant effect on the disease traits. It is interesting to point out that most of the variants associated with NAFLD and NASH are mapped to coding regions of the genome facilitating the variants’ functional assessment.
Figure 2.
Summary of variants influencing the risk and protection against NAFLD and the histological disease severity. The figure depicts the most replicated variants associated with NAFLD and NASH, including the global minor allele frequency, the most severe consequence of the variant, and the linked variant functionality. In addition, the figure highlights the main effect(s) on the risk and/or protection against NAFLD and NASH. Information was retrieved from Ensembl (available at https://www.ensembl.org/). NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; PNPLA3, patatin-like phospholipase domain containing 3; HSD17B13, hydroxysteroid 17-beta dehydrogenase 13; MBOAT7, membrane bound O-acyltransferase domain containing 7; GCKR, glucokinase regulator; TM6SF2, transmembrane 6 superfamily member 2; TMC4, transmembrane channel-like 4; OR, odds ratio; MAF, minor allele frequency; SNP, single nucleotide polymorphism; LA, Latino population.
The variants and single nucleotide polymorphisms (SNPs) identified in GWAS, EWAS, and PHEWAS, that were further replicated in extensive studies across the world as being associated with the NAFLD phenotype and the disease severity (NASH and NASH fibrosis), explain only approximately 30–50% of the estimated heritability of the disease. The effect of each SNP on NAFLD and disease-associated traits is relatively modest (Fig. 2).
However, the effect of rs738409 C/G variant located in PNPLA3 (patatin-like phospholipase domain containing 3) on the risk of NAFLD and the disease progression is probably the strongest effect for a common variant modifying the genetic susceptibility of NAFLD and NASH (explaining ~5.3% of the total variance) [14]. The evidence indicates that homozygous carriers of the G-risk allele of rs738409 present 3.24-fold greater risk of higher liver necroinflammatory scores and 3.2-fold greater risk of developing fibrosis when compared with homozygous CC [14,15].
The rs58542926 C/T variant located in TM6SF2 (Transmembrane 6 Superfamily Member 2) that was initially associated with liver fat accumulation and aminotransferase levels in a large GWAS study [11] and further replicated in subsequent candidate gene association studies [16,17] encodes for a protein involved in lipid metabolism. The rs58542926 is an important modifier of blood lipid traits in different populations. As a challenge in personalized medicine, the C-allele, which has an overall frequency as high as 93%, is associated with higher blood lipids, whereas the T allele confers a moderate risk for NAFLD (carriers of the risk allele present approximately ∼2.2% higher lipid fat content) but lower blood lipids [18].
Likewise, the rs72613567 insertion/deletion variant in HSD17B13 (hydroxysteroid 17-beta-dehydrogenase 13), the functional consequence of which is a splice donor variant of the HSD17B13 [12], presents protective effect against NAFLD and severe histologic outcomes [12,19,20].
The modest effects on NAFLD risk of the rs780094 in GCKR (glucokinase regulator)—odds ratio(OR) ~1.2 [21] and rs641738 located in TMC4 (transmembrane channel-like 4) exon 1 (p.Gly17Glu) and 500 bases downstream of the MBOAT7 (TMC4/MBOAT7)—~OR 1.17 [22], are also highlighted in Figure 2.
In addition, the genetic architecture of NAFLD and NASH involves rare variants in other loci, for example, the recently discovered p.P426L loss-of-function variant (rs143545741 C>T) located in autophagy-related 7 (ATG7) [23]. Furthermore, a rare nonsense mutation (rs149847328, p.Arg227Ter) in the glucokinase regulator (GCKR) has also been recently reported in an adult patient with NAFLD, morbid obesity, and type 2 diabetes. The p.Arg227Ter was associated with a rapidly progressive histological form of the disease [24].
Besides, the genetic component of NAFLD and NASH involves mutations in genes of the oxidative phosphorylation (OXPHOS) chain of the mitochondrial DNA (mtDNA) [25,26], and variants in long noncoding RNAs (lncRNAs), which have a remarkable role in transcriptional and epigenetic regulation [27,28]. Moreover, we reported that deregulated expression of a particular lncRNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), stratifies patients into the histologic phenotypes associated with NAFLD severity [28]. MALAT1 up-regulation seems to be a common molecular mechanism in immune-mediated chronic inflammatory liver damage, which suggests that convergent pathophenotypes (inflammation and fibrosis) share similar molecular mediators leading to cancer [28].
NOVEL ASPECTS OF GENETICS IN NAFLD: GENE VARIANTS AND INTERACTION EFFECTS
The nonsynonymous rs738409 variant in PNPLA3 is regarded as the major genetic component of NAFLD and NASH [9,14,15]. The risk effect of this variant on developing fatty liver is the strongest ever reported for a common variant modifying the genetic susceptibility of NAFLD (5% of the total variance) [14,15]. A recent two-stage (discovery and replication) GWAS that included NAFLD patients characterized by liver biopsy confirmed the rs738409 variant in PNPLA3 as a risk factor for the full histological spectrum in patients of European ancestry [29]. Likewise, this large GWAS confirmed important contributions from variants in TM6SF2 (rs58542926) and HSD17B13 (rs72613567), but not MBOAT7 (rs641738), in the disease biology [29].
Like many other complex diseases, NAFLD results from the interaction between genes and environmental factors [5-7]. Hence, in addition to individual genetic susceptibility, other important factors contribute to the phenotypic expression of NAFLD and NASH, including dietary patterns and food.
There have been attractive studies which focused on gene-diet interaction effect/s, for example, a recent study assessing a gene-diet interaction among rs738409, nutrient intake, and liver histology severity [30]. Vilar-Gomez et al. [30] showed that PNPLA3 rs738409 G-allele might modulate the effect of specific dietary nutrients on the risk of fibrosis in patients with NAFLD.
Other studies have explored gene-gene interaction effects, which are also known as epistasis. For example, Vilar-Gomez et al. [31] found that the protection conferred by HSD17B13 rs72613567 A-allele on severe histological outcomes may be limited to selected subgroups of individuals. Specifically, the protective effects of rs72613567 A-allele on the risk of inflammation and fibrosis seem to be notably stronger in women, persons aged 45 or older, individuals with diabetes, or those with body mass index ≥35, even after adjusting for the other relevant confounders [31].
Other human studies have explored the direct effect of the PNPLA3 rs738409 on developing liver fibrosis in relation to liver histologic traits. Specifically, Vilar-Gomez et al. [32] recently reported that a large proportion of the indirect effect of rs738409 on fibrosis severity is mediated through portal inflammation.
Finally, recent studies have highlighted the influence of genetic variants, including variants influencing the risk and protection against NAFLD-histological severity (PNPLA3-rs738409, TM6SF2-rs58542926, MBOAT7-rs641738, and HSD17B13-rs72613567) and a variant influencing macronutrient intake (FGF21-rs838133), on the liver microbial DNA composition [33]. For example, Pirola et al. [33] found that members of the Gammaproteobacteria class were significantly enriched in carriers of the rs738409 and rs58542926 risk-alleles, including Enterobacter and Pseudoalteromonas genera, respectively.
GWAS ON NAFLD AND VARIANTS IN IMMUNE-RELATED LOCI
The analysis of the GWAS catalog using the EMBL-EBI dataset (EMBL’s European Bioinformatics Institute) has shown interesting associations between variants in immune-related loci and NAFLD (Table 1). The human major histocompatibility complex on chromosome 6p21 has been associated with susceptibility to many liver diseases. GWAS confirmed the potential association of NAFLD with many variants in HLA genes and interleukin 36 alpha (IL36A) and beta (IL36B) (Table 1).
Table 1.
GWAS catalog and associations between variants in immune-related loci and NAFLD susceptibility
| Mapped loci | Variant ID | P-value | Study accession | Chromosome location |
|---|---|---|---|---|
| IL36A, IL36B | rs28946269 | 9×10-6 | GCST008468 | 2:113011237 |
| HLA-DRB5, HLA-DRB9 | rs7748270 | 4×10-12 | GCST90094908 | 6:32480822 |
| HLA-DRB1, HLA-DQA1 | rs5021727 | 3×10-9 | GCST90094908 | 6:32610856 |
| HLA-DQA1, HLA-DRB1 | rs9271325 | 2×10-8 | GCST90094908 | 6:32614736 |
| HLA-DQA1, HLA-DRB1 | rs9271406 | 2×10-8 | GCST90094908 | 6:32619811 |
| HLA-DQA1 | rs2213287 | 5×10-9 | GCST90094908 | 6:32637731 |
| HLA-DQA1 | rs9272699 | 2×10-8 | GCST90094908 | 6:32641452 |
GWAS, genome-wide association study; NAFLD, non-alcoholic fatty liver disease.
Source: https://www.ebi.ac.uk/ EMBL’s European Bioinformatics Institute.
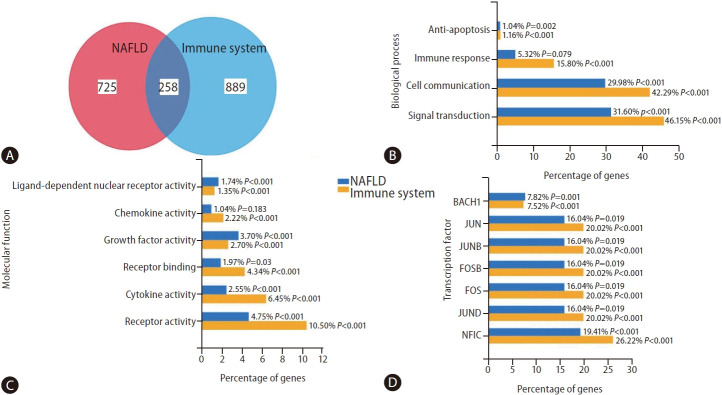
To obtain a more comprehensive view of the overlap between NAFLD and immune system-associated genes, we searched the literature with the query “NAFLD” and “immune system” using the web-based platform Genie (available at cbdm.mdc-berlin.de/tools/genie/) [34]. Using a cutoff of 0.01 for abstracts and a false discovery rate <0.01 for genes, we retrieved 941/983 and 975/1,524 abstracts/genes, corresponding to NAFLD and the immune system, respectively. Two hundred fifty-eight genes were associated with both NAFLD and the immune system (Fig. 3A). As shown in Supplementary Figure 1, some of the 258 overlapping genes are expressed preferentially in cells of the immune system, for example, MPO (myeloperoxidase), a major component of neutrophil azurophilic granules. In contrast, certain genes, such as C3 (complement C3), SERPINA1 (serpin family A member 1, a serine protease inhibitor), or KART18/19 (keratin 18 and 19, intermediate filament chain keratins), are expressed in different adult tissues, including liver, heart, ovary, lung, or colon (Supplementary Fig. 1). Only a few are expressed in any cells, for example, KRT8, HSPD1 (heat shock protein family D member 1) or HSPA5 (heat shock protein family D member 5, encoding a mitochondrial protein which may function as a signaling molecule in the innate immune system).
Figure 3.
Overlapping between gene variants associated with NAFLD and the immune system. This figure provides a functional enrichment analysis of the genes and proteins associated with NAFLD and the immune system, the information of which has been extracted from heterogeneous genomic and proteomic resources using the FunRich program. [55,56] The literature was searched with the query “NAFLD” and “immune system” by using the platform Genie (available at https://edoc.mdc-berlin.de/11677/) [34] with a cutoff of 0.01 for abstracts and an FDR <0.01 for genes. Pathways were ranked according to the P-value, whereby P<0.05 was considered statistically significant. % indicates the percentage of altered genes in the whole pathway. (A) Venn diagram of the corresponding gene lists with overlapping genes obtained by the FunRich program, a user-friendly tool, which performs functional enrichment analysis on the generated datasets (available at http://www.funrich.org/). [55] (B) Enrichment of genes of both lists in Biological Processes. (C) Enrichment of genes of both lists in Molecular Function. (D) Associated transcription factors. NAFLD, non-alcoholic fatty liver disease; FDR, false discovery rate.
Both gene groups were significantly enriched in anti-apoptotic, cell communication, and signal transduction biological processes (Fig. 3B). As expected, the molecular function characterizing NAFLD-the immune system-shared genes are significantly similar (i.e., ligand-dependent nuclear receptor, chemokine, growth factor, cytokine, and receptor activities) (Fig. 3C). Finally, Figure 3D shows shared genes-associated transcription factors (TF). As novel findings, we found BACH1, which encodes a TF that belongs to the Cap’n’collar (CNC) type of basic region leucine zipper factor family (CNC-bZip) associated with cancer metastasis [35]. On the other hand, we also found NFIC, whose encoded protein belongs to the CTF/NF-I family. These are dimeric DNA-binding proteins that function as cellular TFs and as replication factors for adenoviruses, which also play a role in cancer cell proliferation and metastasis through an epithelial-to-mesenchymal transition process [36].
Finally, results from a recent study using multicellular liver culture that recapitulates many key features of NAFLD suggested a potential causal link between elevated interleukin 6 (IL6)/STAT3 activity and rs738409-mediated susceptibility to NAFLD [37]. Park et al. [37] showed that dampening IL6-STAT3 activity alleviated the rs738409-G risk allele-mediated risk of NAFLD. This effect was attributed to the elevated IL6-STAT3 activity in liver cultures carrying the rs738409 G-risk allele that increased NF-kB activity [37]. This finding has clinical implications. For instance, a network-based druggability assessment for STAT3, which examines the structure or the protein-protein interaction around the target, suggests that STAT3 is a good drug target presenting a ligand-based druggability score of 97% [6]. In addition, this finding is particularly relevant in light of the association between NAFLD-predisposing risk factors, including obesity and insulin-resistance, and STAT3 gene variants [38].
Interestingly, from the above-described approach of clustering NAFLD and the immune system-associated genes, we retrieved a long list of potential drugs to target the disease (data not shown). Among the obvious repurposed drug candidates, such as non-steroid anti-inflammatory drugs, statins, antidiabetic drugs, etc., auranofin emerged. Hwangbo et al. [39] reported that auranofin ameliorates the characteristics of NAFLD through the inhibition of NLRP3 inflammasome, and Lee et al. [40] recently found that auranofin attenuates hepatic steatosis and fibrosis in NAFLD via NRF2 and NF-kappaB signaling pathways.
RISK ALLELES IN COMMON VARIANTS ASSOCIATED WITH NAFLD/ NASH AND GENE REGULATION OF IMMUNE SYSTEM: eQTLs
Activation of the immune system, including innate and or adaptive immune response, is an essential driver of the disease severity and progression [3]. While various immune-responsive cells are involved in the pathogenesis of NASH, including T cells and natural killer T cells, the classical effectors of NASH-linked inflammation are Kupffer cells and recruited macrophages [3]. In addition, the infiltrated immune cells play several roles in the liver of NASH patients, including the release of cytokines, chemokines, and eicosanoids, among other inflammatory factors [3,41].
Analysis of genetic pathways in NASH has shown that the immune system is significantly enriched with the sub-pathway “innate immune system” and “cytokine signaling in the immune system" [7]. However, much remains to be understood in how risk alleles modify the immune system.
The genomic tools, including GWAS complemented by expression quantitative trait locus (eQTL) analyses, are powerful instruments for understanding how disease-linked variants regulate the expression of quantitative molecular phenotypes across diverse tissues.
GWAS of complex diseases, including NAFLD and NASH, showed that some gene variants are implicated in the susceptibility of multiple traits—a phenomenon known as pleiotropy [42]. This feature involves not only the rs738409 variant in PNPLA3 but also variants in TM6SF2, HSD17B13, and MBOAT7 that are associated with diverse laboratory measurements related to hematological traits [42].
In addition, the rs738409 has been shown to be associated with the soluble intercellular adhesion molecule 1 (sICAM-1) concentration in a large GWAS involving 22,435 healthy women from the Women’s Genome Health Study [43]. ICAM-1 is an endothelium and cells of the immune system-derived inflammatory marker. This finding is particularly relevant, as previous studies demonstrated that NAFLD is associated with elevated circulating levels of sICAM-1 and abnormal liver expression of ICAM-1 [44]. Furthermore, it was found that liver ICAM-1 expression levels are significantly correlated with liver lobular inflammatory infiltrate and the severity of necroinflammatory activity [44].
Another important aspect is the exploration of the influence of genetic variation on gene expression across tissues and cell types. For example, Table 2 shows the associations of major variants in NAFLD-NASH genes with gene expression levels in non-liver tissues, of which information has been extracted from PhenoScanner, a curated database holding publicly available results from large-scale genome-wide association studies [45,46].
Table 2.
Variants in NAFLD/NASH-associated genes and regulation of gene expression in immune related loci
| Tissue | Gene expression | Sample size | P-value |
|---|---|---|---|
| SNP ID: rs738409 hg19_coordinates: chr22:44324727 (PNPLA3) | |||
| Adipose subcutaneous | SAMM50 | 385 | 9.60E-07 |
| Whole blood | SAMM50 | 5,257 | 5.67E-106 |
| Whole blood | SAMM50 | 31,300 | 3.15E-18 |
| Whole blood | FAM89B | 31,300 | 5.49E-06 |
| SNP ID: rs58542926 hg19_coordinates: chr19:19379549 (TM6SF2) | |||
| Whole blood | CXCL9 | 5,257 | 3.13E-06 |
| Whole blood | CXCL16 | 28,533 | 5.71E-06 |
| SNP ID: rs641738 hg19_coordinates: chr19:54676763 (TMC4/MBOAT7) | |||
| Whole blood | LILRP1 | 5,417 | 8.87E-06 |
Variant: rs738409, gene: PNPLA3. Variant: rs58542926, gene: TM6SF2. Variant: rs72613567, gene: HSD17B13. Variant: rs641738, gene: MBOAT4/TMC4.
SNP, single nucleotide polymorphism; PNPLA3, patatin-like phospholipase domain containing 3; TM6SF2, transmembrane 6 superfamily member 2; MBOAT7, membrane bound O-acyltransferase domain containing 7; TMC4, transmembrane channel-like 4; NAFLD, nonalcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
Information was retrieved from http://www.phenoscanner.medschl.cam.ac.uk/, a curated database holding publicly available results from large-scale genome-wide association studies. [45,46]
The rs738409 is associated with adipose tissue and blood expression levels of SAMM50, which plays a crucial role in the maintenance of the structure of mitochondrial cristae, the proper assembly of the mitochondrial respiratory chain complexes, and/or the maintenance of mtDNA [47]. In addition, the rs738409 is associated with whole blood expression levels of FAM89B (Family With Sequence Similarity 89 Member B), which negatively regulates TGFb-induced signaling—a key factor involved in the regulation of immune response [48].
The rs58542926 in TM6SF2 is associated with blood expression levels of CXCL9 (C-X-C Motif Chemokine Ligand 9)—a member of the chemokine superfamily that encodes secreted proteins involved in immunoregulatory and inflammatory processes, and expression levels of CXCL16 (C-X-C Motif Chemokine Ligand 16), which is involved in several processes, including positive regulation of cell growth, response to interferon-gamma, and response to tumor necrosis factor.
The rs641738 in MBOAT7 is associated with the whole blood expression levels of LILRP1 (leukocyte immunoglobulin-like receptor pseudogene)—also known as leukocyte-expressed receptors of the immunoglobulin superfamily.
RISK ALLELES IN COMMON VARIANTS ASSOCIATED WITH NAFLD/NASH AND ITS RELATIONSHIP WITH IMMUNE SYSTEM CELLS TYPES
In the last few years, novel molecular approaches have allowed the differentiation between eQTLs in “bulk” samples of different tissues and “single cell” eQTLs. The difference is that eQTLs from bulk samples represent the average gene expression across all cells in a given tissue. Conversely, eQTLs using single-cell sequencing technology (scRNA-seq) allow the cell-specific gene expression signature (cell type-specific eQTLs).
Although technological advances illuminate the pathophysiology of NAFLD, how the major genetic variants associated with the risk (rs738409) and protection (rs72613567) against NAFLD and NASH affect the gene expression of specific immune cells remains largely unknown. To gain further insight into this aspect, we explored the DICE database (database of immune cell expression, eQTLs, and epigenomics), which helped to reveal the effects of disease risk-associated genetic polymorphisms on specific immune cell types (https://dice-database.org) [49].
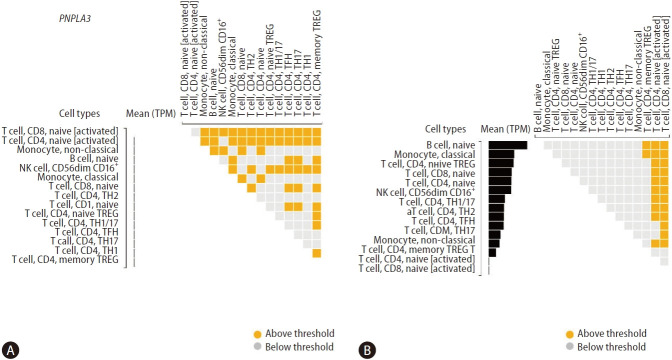
Figure 4 shows differential gene expressions of PNPLA3 and HSD17B13 across specific immune cell types. We found very modest levels of PNPLA3 expression in T cell, CD8, naïve [activated], and T cell, CD4, naive [activated] (Fig. 4A). In addition, we explored the genetic variants directly associated with PNPLA3 gene expression level (SNP located within +/– 1 Mb of the TSS) or eQTLs, and found three single nucleotide polymorphisms in chromosome 22 influencing T cell, CD4, memory TREG, including rs5766088, rs9626589, and rs9626589.
Figure 4.
Gene expression levels of PNPLA3 and HSD17B13 in immune cells. Differential gene expressions of PNPLA3 (A) and HSD17B13 (B) across cell types as calculated by the DESeq package (version 1.6.3). Cells are sorted based on the median gene expression from the highest to the lowest. Squares in the upper diagonal matrix indicate results from pair-wise comparisons of two cell types on the x-axis and y-axis. The figure shows 2 log2 fold-change. Findings were retrieved from the DICE (Database of Immune Cell Expression, Expression quantitative trait loci [eQTLs] and Epigenomics) project (available at https://dice-database.org). PNPLA3, patatin-like phospholipase domain containing 3; HSD17B13, hydroxysteroid 17-beta dehydrogenase 13.
Conversely, we found significant levels of HSD17B13 expression across a variety of immune cells, including B cell, naïve monocyte, classical T cell, CD4, naive TREGT cell, CD8, naïve T cell, CD4, naïve natural killer (NKO cell, CD56dim CD16+T cell), CD4, TH1/17T cell, CD4, TH1T cell, CD4, TH2T cell, CD4, TFHT cell, CD4, TH17 monocyte, non-classical, and T cell, CD4, memory TREG (Fig. 4B). More importantly, in addition to these cells being relevant effectors of cytotoxicity, these findings were also aligned with our previous results on the effect/s of the splice variant rs72613567 in HSD17B13 on the liver transcriptome. Specifically, we found that the most significant changes in the liver gene expression are enriched by biological pathways related to the immune system, including antigen presentation and interferon-related processes, cytokine signaling, and signal transduction [19].
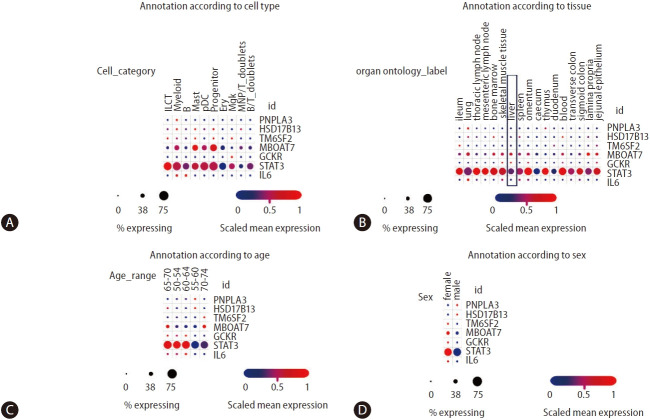
More recent studies on multi-tissue single-cell transcriptomics have allowed a broader understanding of the genetic architecture of complex diseases concerning the cross-talk between genetic variants and immune cells [50]. Domínguez Conde et al. [50] profiled immune cell populations isolated from a wide range of donor-matched tissues, generating nearly 360,000 single cell transcriptomes. Using data from the study by Domínguez Conde et al. [50], which can be freely retrieved from the Single Cell Portal, we explored the distribution of PNPLA3, HSD17B13, STAT3, and IL6 expressions across tissues and immune cell types using (Fig. 5). On the one hand, we observed that the expression levels of PNPLA3 are generally very modest across cell types compared to HSD17B13 levels, which present higher levels of expression in innate lymphoid cells, myeloid, mast, and progenitor cells, as well as megakaryocytes (Fig. 5A)—despite the relatively low percentage of gene-expressing cells. On the other hand, MBOAT7 presents a relatively high level of expressions in myeloid, mast, and progenitor cells, with more than 50% of cells expressing the gene (Fig. 5A). As expected, STAT3 presents not only very high levels of expression across diverse immune cells in all conditions, but also significant levels of expression in the liver tissue (Fig. 5B).
Figure 5.
Analysis of NAFLD/NASH-risk alleles and cross-tissue immune cell expression. Information was retrieved from Single Cell Portal (available at https://singlecell.broadinstitute.org/single_cell/study/SCP1845/). [50] Panels depict annotation of cell population type (A), organ/tissue distribution (B), age (C), and sex of donors (D). Scaling is relative to each gene’s expression across all cells in a given annotation selection (i.e., cells associated with each column label in the dot plot). Gene targets were arbitrarily selected, including major NAFLD/NASH-related loci and two immune-related genes (IL6 and STAT3). ILCT: innate lymphoid cells, pDCs: plasmacytoid dendritic cells, which are a unique subset of dendritic cells specialized in secreting high levels of type I interferons, myeloid: myeloid cells are granulocytic and phagocytic leukocytes that traverse blood and solid tissues, B: B lymphocytes, also called B cells, mast: mast cells are immune cells of the myeloid lineage, progenitor: the common B- and T-cell progenitor can be found in the bone marrow, ery: erythroid cells, mgk: megakaryocytes/platelets, MNP/RT doublets cells: mononuclear phagocytes, B/T doublets: B and T cells stuck together as a “doublet.” NAFLD, non-alcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; IL6, interleukin 6; PNPLA3, patatin-like phospholipase domain containing 3; HSD17B13, hydroxysteroid 17-beta dehydrogenase 13; MBOAT7, membrane bound O-acyltransferase domain containing 7; GCKR, glucokinase regulator.
Remarkable differences in gene expressions across different age groups are also present in Figure 5C, the biological meaning of which remains unknown. However, these differences might explain differences in disease outcomes and sexual dimorphism (Fig. 5D).
CONCLUSION
Recent findings based on GWAS, single cells transcriptomics, and analysis of eQTLs may prime future studies that can help to understand the functional basis of shared loci between NAFLD and NASH and immune-mediated mechanisms of the disease severity.
Likewise, while translating GWAS, EWAS, and PHEWAS signals into clinical applications has been slow, genetic knowledge in NAFLD and NASH may significantly improve disease management and monitoring. The accumulated genetic knowledge is now being used to predict disease outcomes and personalized medicine in the field of NAFLD [8,51,52], and to repurpose drugs and/or select potential actionable targets to treat the disease [4,53,54].
Acknowledgments
This study was partially supported by grants PICT 2018-889, PICT 2019-0528, PICT 2018-00620, PICT 2020-799 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT).
Abbreviations
- GCKR
glucokinase regulator
- GWAS
genome-wide association study
- HSD17B13
hydroxysteroid 17-beta dehydrogenase 13
- MBOAT7
membrane bound O-acyltransferase domain containing 7
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- PNPLA3
patatin-like phospholipase domain containing 3
- SNP
single nucleotide polymorphism
- TM6SF2
transmembrane 6 superfamily member 2
Footnotes
Authors’ contribution
C.J.P concept of the work, manuscript writing and approval. S.S. concept of the work, manuscript writing and approval.
Conflicts of Interest
The authors have no conflicts to disclose.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Clinical and Molecular Hepatology website (http://www.e-cmh.org).
Tissue-specific expression patterns of the 258 overlapping genes associated with both NAFLD and the immune system from Figure 3, as described in the main text. The heat plot was also obtained by the FunRich program described in the legend of Figure 3. [55] NAFLD, non-alcoholic fatty liver disease.
REFERENCES
- 1.Paik JM, Golabi P, Younossi Y, Mishra A, Younossi ZM. Changes in the global burden of chronic liver diseases from 2012 to 2017: the growing impact of NAFLD. Hepatology. 2020;72:1605–1616. doi: 10.1002/hep.31173. [DOI] [PubMed] [Google Scholar]
- 2.Yip TC, Vilar-Gomez E, Petta S, Yilmaz Y, Wong GL, Adams LA, et al. Geographical similarity and differences in the burden and genetic predisposition of NAFLD. Hepatology. 2022 Sep 5; doi: 10.1002/hep.32774. doi: [DOI] [PubMed] [Google Scholar]
- 3.Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 4.Pirola CJ, Sookoian S. The lipidome in nonalcoholic fatty liver disease: actionable targets. J Lipid Res. 2021;62:100073. doi: 10.1016/j.jlr.2021.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sookoian S, Pirola CJ. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol. 2017;23:1–12. doi: 10.3350/cmh.2016.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sookoian S, Pirola CJ. Genetics of nonalcoholic fatty liver disease: from pathogenesis to therapeutics. Semin Liver Dis. 2019;39:124–140. doi: 10.1055/s-0039-1679920. [DOI] [PubMed] [Google Scholar]
- 7.Sookoian S, Pirola CJ, Valenti L, Davidson NO. Genetic pathways in nonalcoholic fatty liver disease: insights from systems biology. Hepatology. 2020;72:330–346. doi: 10.1002/hep.31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirola CJ, Sookoian S. Personalized medicine in nonalcoholic fatty liver disease. Clin Mol Hepatol. 2022;28:935–938. doi: 10.3350/cmh.2022.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salatino A, Sookoian S, Pirola CJ. Computational pipeline for next-generation sequencing (NGS) studies in genetics of NASH. Methods Mol Biol. 2022;2455:203–222. doi: 10.1007/978-1-0716-2128-8_16. [DOI] [PubMed] [Google Scholar]
- 14.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 15.Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 17.Sookoian S, Castaño GO, Scian R, Mallardi P, Fernández Gianotti T, Burgueño AL, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61:515–525. doi: 10.1002/hep.27556. [DOI] [PubMed] [Google Scholar]
- 18.Pirola CJ, Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: a meta-analysis. Hepatology. 2015;62:1742–1756. doi: 10.1002/hep.28142. [DOI] [PubMed] [Google Scholar]
- 19.Pirola CJ, Garaycoechea M, Flichman D, Arrese M, San Martino J, Gazzi C, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60:176–185. doi: 10.1194/jlr.P089953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Belyaeva OV, Brown PM, Fujita K, Valles K, Karki S, et al. 17-Beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69:1504–1519. doi: 10.1002/hep.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zain SM, Mohamed Z, Mohamed R. Common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: a meta-analysis. J Gastroenterol Hepatol. 2015;30:21–27. doi: 10.1111/jgh.12714. [DOI] [PubMed] [Google Scholar]
- 22.Teo K, Abeysekera KWM, Adams L, Aigner E, Anstee QM, Banales JM, et al. rs641738C>T near MBOAT7 is associated with liver fat, ALT and fibrosis in NAFLD: a meta-analysis. J Hepatol. 2021;74:20–30. doi: 10.1016/j.jhep.2020.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baselli GA, Jamialahmadi O, Pelusi S, Ciociola E, Malvestiti F, Saracino M, et al. Rare ATG7 genetic variants predispose patients to severe fatty liver disease. J Hepatol. 2022;77:596–606. doi: 10.1016/j.jhep.2022.03.031. [DOI] [PubMed] [Google Scholar]
- 24.Pirola CJ, Flichman D, Dopazo H, Fernández Gianotti T, San Martino J, Rohr C, et al. A rare nonsense mutation in the glucokinase regulator gene is associated with a rapidly progressive clinical form of nonalcoholic steatohepatitis. Hepatol Commun. 2018;2:1030–1036. doi: 10.1002/hep4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirola CJ, Garaycoechea M, Flichman D, Castaño GO, Sookoian S. Liver mitochondrial DNA damage and genetic variability of Cytochrome b - a key component of the respirasome - drive the severity of fatty liver disease. J Intern Med. 2021;289:84–96. doi: 10.1111/joim.13147. [DOI] [PubMed] [Google Scholar]
- 26.Sookoian S, Flichman D, Scian R, Rohr C, Dopazo H, Gianotti TF, et al. Mitochondrial genome architecture in non-alcoholic fatty liver disease. J Pathol. 2016;240:437–449. doi: 10.1002/path.4803. [DOI] [PubMed] [Google Scholar]
- 27.Sookoian S, Rohr C, Salatino A, Dopazo H, Fernandez Gianotti T, Castaño GO, et al. Genetic variation in long noncoding RNAs and the risk of nonalcoholic fatty liver disease. Oncotarget. 2017;8:22917–22926. doi: 10.18632/oncotarget.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sookoian S, Flichman D, Garaycoechea ME, San Martino J, Castaño GO, Pirola CJ. Metastasis-associated lung adenocarcinoma transcript 1 as a common molecular driver in the pathogenesis of nonalcoholic steatohepatitis and chronic immune-mediated liver damage. Hepatol Commun. 2018;2:654–665. doi: 10.1002/hep4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol 2020;73:505-515. Erratum in: J Hepatol. 2021;74:1274–1275. doi: 10.1016/j.jhep.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Vilar-Gomez E, Pirola CJ, Sookoian S, Wilson LA, Belt P, Liang T, et al. Impact of the association between PNPLA3 genetic variation and dietary intake on the risk of significant fibrosis in patients with NAFLD. Am J Gastroenterol. 2021;116:994–1006. doi: 10.14309/ajg.0000000000001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilar-Gomez E, Pirola CJ, Sookoian S, Wilson LA, Liang T, Chalasani N. The protection conferred by HSD17B13 rs72613567 polymorphism on risk of steatohepatitis and fibrosis may be limited to selected subgroups of patients with NAFLD. Clin Transl Gastroenterol. 2021;12:e00400. doi: 10.14309/ctg.0000000000000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilar-Gomez E, Pirola CJ, Sookoian S, Wilson LA, Liang T, Chalasani N. PNPLA3 rs738409 and risk of fibrosis in NAFLD: exploring mediation pathways through intermediate histological features. Hepatology. 2022;76:1482–1494. doi: 10.1002/hep.32491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirola CJ, Salatino A, Quintanilla MF, Castaño GO, Garaycoechea M, Sookoian S. The influence of host genetics on liver microbiome composition in patients with NAFLD. EBioMedicine. 2022;76:103858. doi: 10.1016/j.ebiom.2022.103858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontaine JF, Priller F, Barbosa-Silva A, Andrade-Navarro MA. Génie: literature-based gene prioritization at multi genomic scale. Nucleic Acids Res. 2011;39(Web Server issue):W455–W461. doi: 10.1093/nar/gkr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiel C, Le Gal K, Ibrahim MX, Jahangir CA, Kashif M, Yao H, et al. BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell. 2019;178:330–345. doi: 10.1016/j.cell.2019.06.005. e22. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Shi X, Wu S. miR-550a-3/NFIC plays a driving role in esophageal squamous cell cancer cells proliferation and metastasis partly through EMT process. Mol Cell Biochem. 2020;472:115–123. doi: 10.1007/s11010-020-03790-y. [DOI] [PubMed] [Google Scholar]
- 37.Park J, Zhao Y, Zhang F, Zhang S, Kwong AC, Zhang Y, et al. IL-6/STAT3 axis dictates the PNPLA3-mediated susceptibility to nonalcoholic fatty liver disease. J Hepatol. 2023;78:45–56. doi: 10.1016/j.jhep.2022.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gianotti TF, Sookoian S, Gemma C, Burgueño AL, González CD, Pirola CJ. Study of genetic variation in the STAT3 on obesity and insulin resistance in male adults. Obesity (Silver Spring) 2008;16:1702–1707. doi: 10.1038/oby.2008.250. [DOI] [PubMed] [Google Scholar]
- 39.Hwangbo H, Kim MY, Ji SY, Kim SY, Lee H, Kim GY, et al. Auranofin attenuates non-alcoholic fatty liver disease by suppressing lipid accumulation and NLRP3 inflammasome-mediated hepatic inflammation in vivo and in vitro. Antioxidants (Basel) 2020;9:1040. doi: 10.3390/antiox9111040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SM, Koh DH, Jun DW, Roh YJ, Kang HT, Oh JH, et al. Auranofin attenuates hepatic steatosis and fibrosis in nonalcoholic fatty liver disease via NRF2 and NF-κB signaling pathways. Clin Mol Hepatol. 2022;28:827–840. doi: 10.3350/cmh.2022.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huby T, Gautier EL. Immune cell-mediated features of nonalcoholic steatohepatitis. Nat Rev Immunol. 2022;22:429–443. doi: 10.1038/s41577-021-00639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirola CJ, Salatino A, Sookoian S. Pleiotropy within gene variants associated with nonalcoholic fatty liver disease and traits of the hematopoietic system. World J Gastroenterol. 2021;27:305–320. doi: 10.3748/wjg.v27.i4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paré G, Ridker PM, Rose L, Barbalic M, Dupuis J, Dehghan A, et al. Genome-wide association analysis of soluble ICAM-1 concentration reveals novel associations at the NFKBIK, PNPLA3, RELA, and SH2B3 loci. PLoS Genet. 2011;7:e1001374. doi: 10.1371/journal.pgen.1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sookoian S, Castaño GO, Burgueño AL, Rosselli MS, Gianotti TF, Mallardi P, et al. Circulating levels and hepatic expression of molecular mediators of atherosclerosis in nonalcoholic fatty liver disease. Atherosclerosis. 2010;209:585–591. doi: 10.1016/j.atherosclerosis.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ott C, Ross K, Straub S, Thiede B, Götz M, Goosmann C, et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1715. doi: 10.1016/j.cell.2018.10.022. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Domínguez Conde C, Xu C, Jarvis LB, Rainbow DB, Wells SB, Gomes T, et al. Cross-tissue immune cell analysis reveals tissuespecific features in humans. Science. 2022;376:eabl5197. doi: 10.1126/science.abl5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jun DW. An analysis of polygenic risk scores for non-alcoholic fatty liver disease. Clin Mol Hepatol. 2021;27:446–447. doi: 10.3350/cmh.2021.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sookoian S, Pirola CJ. Precision medicine in nonalcoholic fatty liver disease: new therapeutic insights from genetics and systems biology. Clin Mol Hepatol. 2020;26:461–475. doi: 10.3350/cmh.2020.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HY. Recent advances in nonalcoholic fatty liver disease metabolomics. Clin Mol Hepatol. 2021;27:553–559. doi: 10.3350/cmh.2021.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sookoian S, Pirola CJ. Repurposing drugs to target nonalcoholic steatohepatitis. World J Gastroenterol. 2019;25:1783–1796. doi: 10.3748/wjg.v25.i15.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonseka P, Pathan M, Chitti SV, Kang T, Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. 2021;433:166747. doi: 10.1016/j.jmb.2020.166747. [DOI] [PubMed] [Google Scholar]
- 56.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tissue-specific expression patterns of the 258 overlapping genes associated with both NAFLD and the immune system from Figure 3, as described in the main text. The heat plot was also obtained by the FunRich program described in the legend of Figure 3. [55] NAFLD, non-alcoholic fatty liver disease.