Abstract
Three novel DNA-binding proteins with apparent molecular masses of 7, 10 and 30 kDa have been isolated from the hyperthermophilic methanogen Methanopyrus kandleri. The proteins were identified using a blot overlay assay that was modified to emulate the high ionic strength intracellular environment of M.kandleri proteins. A 7 kDa protein, named 7kMk, was cloned and expressed in Escherichia coli. As indicated by CD spectroscopy and computer-assisted structure prediction methods, 7kMk is a substantially α-helical protein possibly containing a short N-terminal β-strand. According to analytical gel filtration chromatography and chemical crosslinking, 7kMk exists as a stable dimer, susceptible to further oligomerization. Electron microscopy showed that 7kMk bends DNA and also leads to the formation of loop-like structures of ∼43.5 ± 3.5 nm (136 ± 11 bp for B-form DNA) circumference. A topoisomerase relaxation assay demonstrated that looped DNA is negatively supercoiled under physiologically relevant conditions (high salt and temperature). A BLAST search did not yield 7kMk homologs at the amino acid sequence level, but based on a multiple alignment with ribbon–helix–helix (RHH) transcriptional regulators, fold features and self-association properties of 7kMk we hypothesize that it could be related to RHH proteins.
INTRODUCTION
The organization and function of the genome in prokaryotic microorganisms representing the domain Archaea shows significant similarity to that in Eukarya. In many Archaea DNA is compacted into nucleosome-like particles by proteins of the HMf family, which have a structure and function very similar to that of eukaryal histones (1,2), while proteins homologous to histones have not been found in Bacteria. The transcriptional apparatus of Archaea and Eukarya are also closely related. For example, subunit complexity and amino acid sequences of individual subunits are highly conserved in archaeal and eukaryal RNA polymerases (3). Furthermore, homologs of both TATA-binding protein (TBP) and the transcription factor TFIIB, both important components of the transcription initiation complex in Eukarya, have been identified in Archaea and apparently play similar roles (4,5).
Intriguingly, despite the striking conservation of archaeal and eukaryal basal transcription, the way in which archaeal transcription is regulated at the level of single genes seems to be closely related to that in Bacteria. All archaeal regulators characterized to date bind to palindromes located close to the TATA box sequences or the transcription start sites, thus modulating transcription initiation in a way similar to bacterial repressors (6–8). Moreover, putative transcriptional regulators identified as open reading frames in archaeal genome sequences are more closely related to their bacterial than to their eukaryal counterparts (9). The majority of presumed archaeal DNA-binding regulators belong to the helix–turn–helix/winged helix–turn–helix family, although potential relatives of ribbon–helix–helix (RHH) and Zn-ribbon proteins were also identified (10). Therefore, it appears that Archaea represent a fascinating mosaic of eukaryal and bacterial elements and could provide a new view contributing to a better understanding of the separate components and their interactions.
The hyperthermophilic methanogen Methanopyrus kandleri, an archaeon with an optimal growth temperature of 98°C and an intracellular K3 cyclic 2,3-diphosphoglycerate concentration of 1.1 M (11), is positioned extremely deep within the 16S rRNA-based phylogenetic tree (12). Methanopyrus kandleri is the only archaeon that contains topoisomerase V, a type IB topoisomerase previously found only in eukaryotes and poxviruses (13,14). The microorganism contains histone MkaH with two characteristic histone fold domains (15), whose structure is homologous to the histone heterodimer consisting of eukaryotic histones H3 and H4 (16). These properties suggest that M.kandleri may be related to the last common ancestor of archaea and eukaryotes and that the study of DNA-binding proteins regulating chromatin structure and function in this microorganism might contribute to an understanding of evolutionary relationships.
We have identified new proteins in M.kandleri with strong affinity for double-stranded DNA using a blot overlay assay (17) modified to simulate the high ionic concentration in this organism. One of them, designated 7kMk, was produced in Escherichia coli and characterized. Electron microscopy (EM) imaging of the 7kMk–DNA complexes showed DNA compaction by virtue of formation of loop-like structures superficially similar to those formed by archaeal histones. The data from a relaxation assay with topoisomerase V indicated that 7kMk induces DNA negative constrained supercoiling. Because of the apparent conservation of the DNA helical periodicity upon 7kMk binding ascertained by CD spectroscopy, we hypothesize that the change in DNA topology is due to a left-handed orientation of the DNA loop.
MATERIALS AND METHODS
Cells
Methanopyrus kandleri strain AV-19 (DSM6324) has been described (11). Escherichia coli strains DH5α and BL21(DE3)/pLysS were obtained from Life Technologies (USA) and Novagen (USA), respectively. Escherichia coli XL1-Blue MRA cells carrying bacteriophage P2 (Stratagene) were used for propagation of λ phage.
Materials
Complete protease inhibitor cocktail was purchased from Roche Molecular Systems (Switzerland). Glycerol, glutamic acid (monopotassium salt) and β-mercaptoethanol were obtained from Fisher (Germany), ethidium bromide and chloroquine were from Sigma (USA) and Coomassie Brilliant Blue R-250 was from Merck (Germany). ThermoFidelase I was from Fidelity Systems (USA), while pUC19 DNA, restriction endonucleases and the Klenow fragment of E.coli DNA polymerase I were purchased from New England Biolabs (USA). Reagents for SDS–PAGE were from DIAM (Russia) and gel filtration and SDS–PAGE molecular weight standards were from Amersham Pharmacia Biotech (Sweden) and Sigma (USA), respectively.
Protein–DNA binding assay
The procedure for isolation of proteins from M.kandleri has been described elsewhere (18). Proteins from heparin chromatography fractions were separated by tricine-SDS–PAGE (19) using two gels in parallel. Proteins on one gel were transferred onto 50 cm2 nitrocellulose membrane (Protran BA85; Schleicher & Schuell, Germany) by electroblotting (20), for which transfer buffer TB (0.25 M Trizma base, 0.192 M glycine) and a semi-dry apparatus with graphite electrodes (DIAM, Russia) were used. The membrane was rinsed in the transfer buffer and incubated in two changes of renaturation buffer RB [1 M potassium glutamate (K-Glu), 0.05% Tween-20 and 20 mM Tris–HCl pH 8.0] at 37°C for 1 h. Next, the membrane was blocked with 5% BSA in buffer RB for 1 h, washed with two changes of buffer RB for 5 min and transferred to a tray that contained 25 ml of binding buffer (buffer RB supplemented with 20 ng/ml labeled DNA probe and 2 µg/ml heparin). 32P-labeled pUC19 DNA, used as a binding template, was obtained by linearization of pUC19 with EcoRI and filling in of protruding ends with the Klenow fragment of E.coli DNA polymerase I in a mixture of dGTP, dTTP, dCTP and [α-32P]dATP. After incubation with slow agitation at 37°C for 1 h, the membrane was washed with three changes of buffer RB and autoradiographed. For identification of protein bands that exhibited DNA-binding activity, the same membrane was stained with Amido Black and radioactive zones were compared to proteins in a matching gel stained with Coomassie Brilliant Blue R-250.
N-terminal protein sequencing
Proteins were separated by tricine-SDS–PAGE, using a 40-cm-long gel for better resolution of bands. Fragments of the gel containing proteins of interest were electroblotted onto ProBlott PVDF membranes (Applied Biosystems, USA) using 10 mM CAPS pH 11, 10% methanol as transfer buffer. The membranes were stained with Coomassie Brilliant Blue R-250, the protein bands of interest were excised and their N-termini were sequenced on an Applied Biosystems 477A protein sequencer.
Cloning of 7kMk protein
Based on the results of protein microsequencing, we synthesized the degenerate oligonucleotide atg gc(a/t/g/c) ag(a/g) aa(a/g) gc(a/t/g/c) ag(a/g) gt(a/t/c/g) ac(a/t/c/g) (c/t)t. This oligonucleotide was used as a primer to obtain a partial sequence of the 7kMk gene directly from M.kandleri genomic DNA using BigDye Terminator mix (Applied Biosystems, USA) and a protocol with ThermoFidelase I developed by Fidelity Systems (21). In addition, the same oligonucleotide labeled with [γ-32P]ATP and T4 polynucleotide kinase was used to screen a M.kandleri genomic λ library as described earlier (15). DNA isolated from selected plaques using a Midi λ DNA isolation kit (Qiagen, Germany) was digested with various restriction enzymes and analyzed by Southern hybridization. A 2000 bp SalI fragment found in most of the DNA samples was subcloned into pUC19 (resulting in plasmid p7k) and sequenced. The result coincided with that obtained by direct genomic sequencing. Translation of the open reading frame corresponding to the protein of interest resulted in an N-terminal amino acid sequence identical to that obtained by amino acid sequencing.
Recombinant 7kMk
The 7kMk gene was amplified by PCR using the p7k plasmid as template and inserted between NcoI and NdeI sites of the expression vector pET14b, yielding the 7kMk-expressing plasmid pE7k. Purification of recombinant 7kMk was as follows. Escherichia coli BL21DE3/pLysS cells carrying the pE7k plasmid were grown at 37°C to A600 = 0.5 and 7kMk expression was induced by adding IPTG to a final concentration of 0.4 mM, followed by further growth at 37°C for 3 h. Cells were harvested and stored at –20°C. The pellet was thawed at 37°C for 5 min, resuspended in isolation buffer IB (20 mM Tris–HCl, pH 8.0, 10 mM β-mercaptoethanol, 5% glycerol) containing 1 M NaCl and Complete protease inhibitor cocktail and then incubated for a further 5 min at 37°C, permitting lysis to occur. Following centrifugation (20 000 g, 10 min, 4°C), the cleared supernatant was incubated at 80°C for 10 min and then centrifuged again (20 000 g, 10 min, 4°C) to remove denatured proteins. DNase I (20 µg/ml) and MgCl2 (5 mM) were added to the solution and the mixture was incubated for 2 h at 37°C. DNase I was inactivated by thermal denaturation (80°C, 10 min) followed by centrifugation (20 000 g, 10 min, 4°C). The crude extract was diluted 2-fold with buffer IB and applied to a 5 ml HiTrap heparin column (Amersham Pharmacia Biotech, Sweden). 7kMk protein was eluted with a NaCl gradient (0.5–1.5 M). 7kMk-containing fractions were collected at 1–1.2 M NaCl and concentrated using Centricon-3 cartridges (Amicon, USA). The resulting concentrate was passed through a Superdex 75 gel filtration column (Amersham Pharmacia Biotech, Sweden), equilibrated with a buffer containing 1 M NaCl and 20 mM Tris–HCl, pH 8.0. The protein was concentrated further using Centricon-3 cartridges to a final concentration of 5 mg/ml.
Analytical gel filtration chromatography
Separations were performed on an analytical liquid chromatography SMART system (Amersham Pharmacia Biotech, Sweden). 7kMk was diluted to final concentrations ranging from 5.4 to 540 µM with buffer NB (10 mM Na-HEPES, pH 8.0, 1 M NaCl) and incubated at 23°C for 24 h. Then 20 µl aliquots were withdrawn and applied to a Superdex 75 gel filtration column equilibrated with NB buffer and run at a flow rate of 50 µl/min at 23°C. Alternatively, samples were subjected to chromatography immediately after dilution. The column was calibrated using RNase A, chymotrypsinogen A, ovalbumin and BSA as markers. The void volume was determined with Dextran Blue 2000.
Crosslinking assay
7kMk aliquots of 20, 100 and 400 µl in NB buffer with protein concentrations of 0.87 mg/ml (100 µM), 0.174 mg/ml (20 µM) and 0.044 mg/ml (5 µM), respectively, were incubated for 1 h at 23°C. Following incubation, the samples were mixed with glutaraldehyde (at final concentrations of 0.03, 0.1 and 0.3%). After 30 min the reactions were quenched with 100 mM Tris–HCl, pH 8.0. The samples were concentrated using Microcon-3 cartridges (Amicon) and analyzed by SDS–PAGE.
UV absorbance, fluorescence and circular dichroism (CD) spectroscopy
UV spectra were recorded on a UVIKON spectrophotometer (Kontron Instruments, Germany) with a 1 cm quartz cuvette. The 7kMk concentration was determined by absorption at 280 nm, using an extinction coefficient ɛ280 = 8480/M/cm as calculated from the amino acid composition (22). Fluorescence emission spectra were acquired on a PTI spectrofluorimeter (Photon Technology International, USA).
CD measurements were performed using a Jasco J715 spectropolarimeter. CD spectra of 7kMk (2 s response time, 50 nm/min scan rate, 5 scan average) at concentrations of 3.12 and 0.78 mg/ml in a buffer containing 1 M NaCl, 10 mM NaH2PO4/Na2HPO4, pH 7.2, were obtained using a 0.1 mm path length cuvette. Experimental data were converted to units of differential molar circular dichroic absorption. The spectra were deconvolved using the Jasco secondary structure estimation program, the CDNN program (Gerald Böhm, Martin-Luther-Universität, Halle-Wittenberg, Germany) and the CDPro software package (23,24).
Electrophoretic mobility shift assay (EMSA) of 7kMk complexes with DNA
pUC19 DNA (150 ng) linearized with BamHI was incubated with 7kMk at various concentrations in buffer GB (1 M K-Glu, 10 mM Na-HEPES, pH 8.0) or NB at 70°C for 30 min. The resulting complexes were analyzed by gel electrophoresis in 1.5% agarose at 1.5 V/cm for 16 h in TBE buffer (0.09 M Tris–borate, 0.002 M EDTA) supplemented with 100 mM sodium glutamate (Na-Glu). Bands containing DNA were monitored by staining gels in a 1 µM ethidium bromide solution.
Electron microscopy
Complexes were obtained after incubation of BamHI-linearized pUC19 DNA (150 ng) with 7kMk at various concentrations in GB buffer at 70°C for 30 min. EM samples were prepared according to Dubochet et al. (25). An aliquot (1 µl) was taken from the 10 µl reaction mixture and diluted 100-fold in incubation buffer at room temperature. Aliquots of 5–10 µl of the diluted mixture were placed on a parafilm sheet and covered with an EM grid coated with a 3–4 nm thick carbon film (the carbon films were activated by glow discharge in the presence of pentylamine vapour at a residual pressure of 150 µtorr for 30 s with a discharge current of 2–3 mA). After 2–5 min for adsorption, the samples were stained with 2% aqueous uranyl acetate for 10–15 s and blotted with filter paper. Then the samples were examined in a CM12 electron microscope (Philips, The Netherlands) in tilted dark field mode. The negatives were scanned with an Agfa Studio Scan IIsi scanner at 600 d.p.i. For printing, images were flattened using a high pass filter with a radius of 250 pixels and adjusted for contrast/brightness using Adobe Photoshop.
Topological assay
Relaxed covalently closed pUC19 was obtained by incubation of its natural negatively supercoiled form with a highly distributive mutant of topoisomerase V (Topo-75) (G.I.Belova and A.I.Slesarev, unpublished results) in 30 mM NaCl, 20 mM Tris–HCl, pH 8.0 at 25°C, at 70°C for 30 min. Following reaction, the DNA was extracted with phenol/chloroform and precipitated with ethanol. The pellet was dissolved in TE buffer (10 mM Tris–HCl, pH 7.4, 1 mM EDTA, pH 8.0) and purified on MicroSpin G-25 gel filtration columns (Pharmacia, USA).
Various amounts of 7kMk were incubated with 900 ng relaxed DNA in GB buffer (final volume 30 µl) at 80°C for 15 min. Then 20 U wild-type topoisomerase V was added to the mixture and incubation was continued for 15 or 60 min at 80°C. The reactions were terminated by incubation with 1% SDS at 90°C for 1 min, followed by treatment with proteinase K (125 µg/ml) at 40°C for 30 min. The topoisomerase reaction products were analyzed by 1.5% agarose gel electrophoresis in TBE buffer or TBE buffer containing 1 µg/ml chloroquine at 1.5 V/cm for 16 h.
RESULTS
Identification, cloning and expression of 7kMk protein
In the course of purification of DNA topoisomerase V from M.kandleri (18), a substantial change in electrophoretic mobility of supercoiled and open circular DNA was observed after incubation with early topoisomerase V fractions. The aberrant migration of DNA was tentatively ascribed to its binding with histone-like proteins. One of the proteins in the DNA-binding fractions, an archaeal histone named MkaH with an unusual two histone fold domain structure, has been previously described (15,16).
We continued the analysis of protein fractions that eluted from a heparin column with a gradient of NaCl in the range 0.5–1.5 M using the blot overlay assay (17) modified to simulate the high salt concentration in M.kandleri cells. To this end, protein fractions were subjected to tricine-SDS–PAGE and the separated proteins were then electroblotted onto nitrocellulose membrane in a buffer containing no SDS and renatured in a high salt buffer simulating the intracellular conditions of M.kandleri. Binding of a 32P-labeled double-stranded DNA probe to the membrane-bound proteins was carried out in a high salt buffer containing a 100-fold excess (w/w) of heparin over labeled DNA. Incubation conditions were adjusted using the MkaH histone as a control protein in the blot overlay assay. In a low salt buffer (140 mM NaCl, 10 mM Tris–HCl, pH 8.0) the addition of a 50-fold excess of heparin completely inhibited binding of DNA to MkaH. In contrast, in a buffer containing 1 M K-Glu, a 5000-fold excess of heparin over DNA did not change DNA binding significantly. The presence of K-Glu was speculated to facilitate natural protein–DNA contacts exploited by organisms at high intracellular salt concentrations, including hydrophobic interactions and cation bridges (26,27), thus increasing the specificity of the assay.
With this method, three novel DNA-binding proteins with apparent molecular masses of 7, 10 and 30 kDa were identified based on their mobility in SDS–PAGE. The N-terminal amino acid sequence of the 7 kDa protein was determined to be ARKAXVTLTL. Using degenerate oligonucleotides constructed on the basis of the N-terminal amino acid sequence of the protein (designated 7kMk) as primers for direct genomic sequencing, the entire open reading frame was determined. A search for homologs in a non-redundant database using the BLAST algorithm (National Center for Biotechnology Information, USA) did not reveal any related proteins. To verify that the nucleotide sequence obtained by direct sequencing did indeed encode the protein in question, a gene encoding 7kMk protein was cloned from the genomic DNA λ library of M.kandleri (15), using the degenerate oligonucleotides as probes (see Materials and Methods). The cloned nucleotide sequence coincided with that obtained by direct genomic analysis. Furthermore, its translation yielded N-terminal amino acid residues corresponding to the peptide sequence obtained by Edman degradation. This new gene encodes a polypeptide with a predicted molecular mass of 8699 Da for the protein lacking the first methionine.
The coding fragment of the 7kMk gene was cloned into the pET14b vector and expressed in E.coli BL21(DE3)/pLysS cells. A simple procedure for the purification of recombinant 7kMk was developed, which made use of its thermostability and included elimination of the major part of the E.coli proteins by heat precipitation. The final yield was >30 mg 7kMk/l culture. Recombinant protein had an electrophoretic mobility identical to the original protein isolated from archaebacteria, corresponding to an apparent molecular weight of ∼7 kDa (see Fig. 3B, lane 1). The difference between the apparent molecular weight and the actual size (8699 Da) of the 7kMk protein is not unusual and a similar effect has been reported for other small proteins (28). N-terminal amino acid sequencing revealed that the first methionine of recombinant 7kMk was processed, as was the case for the protein isolated from M.kandleri.
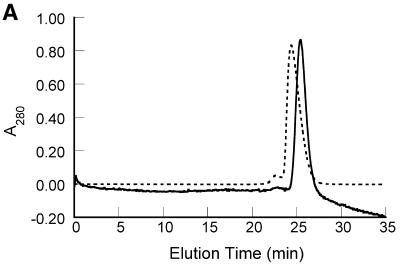
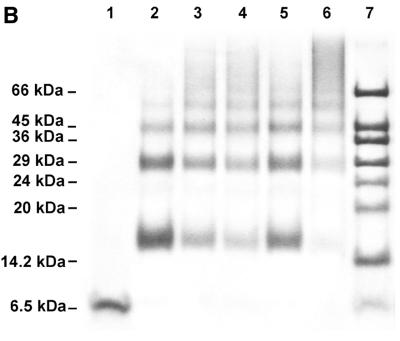
Figure 3.
Self-association behavior of 7kMk protein. (A) Elution profiles of gel filtration chromatography. 7kMk was loaded onto a Superdex 75 column at concentrations of 5.4 µM (continuous line) and 540 µM (broken line). (B) Chemical crosslinking of 7kMk protein. 7kMk samples diluted with NB buffer to final concentrations of 5 µM (lane 2), 20 µM (lanes 3–5) and 100 µM (lane 6) were digested with glutaraldehyde at final concentrations of 0.03% (lane 5), 0.1% (lanes 2, 4 and 6) and 0.3% (lane 3). Lane 1, 7kMk without fixation.
Structure and self-association properties of 7kMk
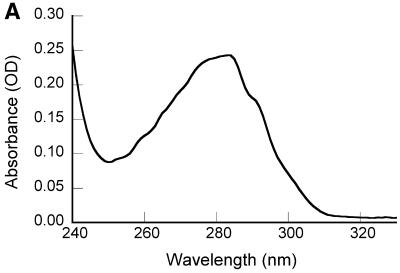
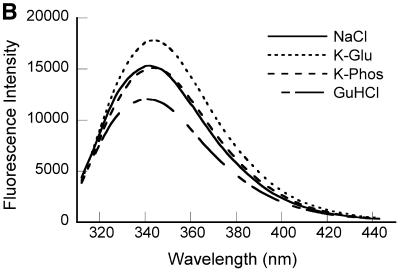
The UV absorbance spectrum of 7kMk protein (Fig. 1A) reflected the single Trp and two Tyr residues in the protein. Calculation of the extinction coefficient ɛ280 from the amino acid composition (22) resulted in a value of 8480/M/cm, which was used to determine the 7kMk concentration. Fluorescence spectroscopy of 7kMk solutions in the presence of 1 M NaCl, 1 M K-Glu or 1 M K2HPO4/KH2PO4 indicated that the sole Trp69 residue was partially solvent exposed. We came to this conclusion because the emission maximum of 7kMk was shifted from 320–330 nm, characteristic of the emission of Trp in a hydrophobic environment, to 340–350 nm, typical of this amino acid in aqueous solutions, and was not affected significantly by 6 M guanidine HCl (Fig. 1B). The shape of the fluorescence emission spectrum did not vary in the presence of the highly lyotropic anion PO43–, which simulates K3 cyclic 2,3-diphosphoglycerate and stabilizes hydrophobic interactions (29,30), suggesting that Trp69 is likely to be exposed in its physiological environment.
Figure 1.
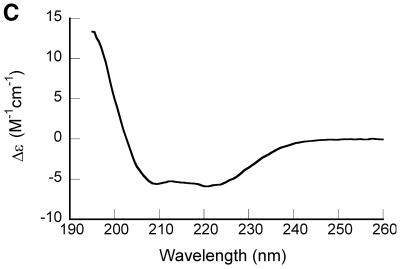
Spectroscopic characterization of purified recombinant 7kMk protein. (A) UV absorption spectrum of 7kMk protein (0.24 mg/ml) in NB buffer. (B) Fluorescence emission spectra of 7kMk (0.14 mg/ml) acquired at an excitation wavelength of 295 nm in buffers NB (‘NaCl’), 1 M K2HPO4/KH2PO4 pH 7.2 (‘K-Phos’), GB (‘K-Glu’) and NB supplemented with 6 M guanidine HCl (‘GuHCl’). (C) CD spectrum of 0.78 mg/ml 7kMk in a buffer containing 1 M NaCl, 10 mM NaH2PO4/Na2HPO4, pH 7.2, obtained with a 0.1 mm path length cuvette.
The overall shape of the CD spectrum of 7kMk (Fig. 1C) with minima of molar ellipticity at 208 and 222 nm indicated significant α-helical content in the secondary structure of the protein. The quantitative estimations of secondary structure fractions obtained with various deconvolution programs are given in Table 1. All four methods predicted similar values for α-helicity, 65–66%. We were not able to obtain high quality CD spectra at wavelengths <195 nm due to the presence of high salt concentrations in the buffer (necessary to suppress 7kMk aggregation), therefore the predicted content of β-strands should be interpreted with care. However, the programs CONTIN/LL and SELCON3, which use an expanded set of reference spectra, are supposed to provide reliable predictions for β-sheets based on a truncated wavelength range (31). Their estimates for the sum of β-strand and β-turn fractions are in the range 12–14%, suggesting that 7kMk contains a β-strand in its structure.
Table 1. Comparison of 7kMk secondary structure fractions determined by various deconvolution programs.
| α | β | β-T | U | αR | αD | Nα | |
|---|---|---|---|---|---|---|---|
| Jasco software | 0.660 | 0.247 | 0.006 | 0.080 | – | – | – |
| CDNN | 0.660 | 0.051 | 0.117 | 0.174 | – | – | – |
| CONTIN/LL | 0.652 | 0.045 | 0.092 | 0.212 | 0.441 | 0.211 | 4.05 |
| SELCON3 | 0.659 | 0.040 | 0.086 | 0.207 | 0.448 | 0.211 | 4.18 |
α, α-helix; β, β-strand; β-T, β-turn; αR, α-helix in regular conformation, αD, α-helix in distorted conformation; Nα, number of α-helical segments.
As shown in Table 1, the fractions of regular and distorted α-helical conformations predicted by the CONTIN/LL (24,32) and SELCON3 (23,24) algorithms were almost identical. The number of amino acids in distorted conformations was found to be approximately four per α-helical segment (33); based on our CD data we conclude that 7kMk contains four α-helices.
Computer-assisted prediction of the secondary structure, based on the amino acid sequence, also assigned a significant α-helical content in 7kMk, placing a short β-strand close to the N-terminus of the protein. An N-terminal β-strand followed by α-helices represents a fold reminiscent of that of the proteins of the RHH family of bacterial transcription factors (also known as the MetJ/Arc family). As shown in Figure 2, multiple alignment of the first half of the 7kMk sequence with several RHH regulators revealed conservation of important characteristic residues in the protein, in particular those that form a β-ribbon, the turn connecting α-helices and hydrophobic residues of α-helices participating in dimerization (10). In spite of their conserved tertiary structure, RHH proteins are very divergent in their sequences and it is difficult to identify a novel member of the family through a simple sequence comparison approach (10), explaining why the 7kMk homology was not evident from the BLAST search.
Figure 2.
Multiple alignment of 7kMk, RHH proteins with solved crystal structures (names in bold) and several putative archaeal RHH proteins. Hydrophobic (LIYFWVMA), polar (STNREQHD) and small (GASNSTCP) residues conserved in >80% of the sequences are colored red, green and blue, respectively. The conserved glycine residue of the turn connecting α-helices is colored violet and highlighted in yellow. The names of proteins, origins, accession codes and numbering of residues are indicated. Elements of secondary structure are given below the alignment. A, α-helix; B, β-strand; U, unordered.
In the course of 7kMk purification we found that the protein exhibited unusually high mobility in the preparative gel filtration column, suggesting its possible oligomerization. In order to understand the process of 7kMk self-association better, we used analytical gel filtration chromatography at various protein concentrations (Fig. 3A). The elution profile at low 7kMk concentration (5.4 µM) showed a major peak that corresponded to a protein with an apparent molecular weight of ∼25 kDa (elution time ∼25.5 min). Raising the protein concentration by two orders of magnitude led to an increase in the apparent molecular weight of the protein, which eluted as a major fraction of ∼32 kDa (elution time ∼24.3 min). The variable position of the major peak suggested the existence of a rapid equilibrium between species with different states of oligomerization. In this case, species with different molecular weights may not form individual fractions but migrate as a single pool, where the elution time of the pool changes with the shift in equilibrium induced by the change in protein concentration (34). Thus, the apparent molecular weight represents not a single species, but a mass averaged function of self-association products. In our case, we interpret the apparent molecular weights of 25 and 32 kDa not as two different species, but as populations with different distributions of oligomers.
The self-association of 7kMk was further characterized by means of chemical fixation. The results of chemical crosslinking with the homobifunctional amino-reactive reagent glutaraldehyde are shown in Figure 3B. Chemical fixation with 0.1% glutaraldehyde at a 7kMk concentration of 5 µM resulted in products with an apparent molecular weight of ∼15, ∼30 and ∼45 kDa (Fig. 3B, lane 2), corresponding to the dimer, tetramer and hexamer of the protein, respectively. At protein concentrations of 20 and 100 µM (lanes 3 and 6) glutaraldehyde treatment led to the appearance of high molecular weight oligomers. Fixation resulted only in dimers, tetramers, hexamers, etc. and not in trimers or pentamers. This means that the monomer is not highly populated or that it does not participate in the formation of oligomers larger than dimer. Combining the gel filtration and crosslinking data, we hypothesize that there is little or no monomer in the mixture and that 7kMk exists as a tightly bound dimer, which is apparently prone to further aggregation.
7kMk DNA-binding properties: loops and bent regions
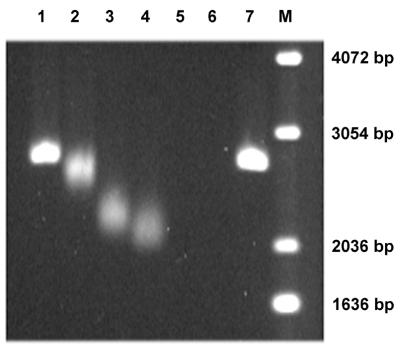
The DNA-binding properties of 7kMk were characterized by EMSA. Complexes of 7kMk with linear pUC19/BamHI DNA were obtained in a high salt buffer (1 M K-Glu, 10 mM Na-HEPES, pH 8.0) at 70°C. The problem of 7kMk aggregation at low salt concentrations, typical of M.kandleri proteins (N.Pavlov and A.Slesarev, unpublished results), was partially solved by performing agarose gel electrophoresis in a buffer supplemented with 100 mM K-Glu. The complexes were detected in the form of bands migrating faster than free DNA (Fig. 4). The mobility of the complexes increased gradually with increasing 7kMk:DNA weight ratio (Rw) in the range 1–2. At Rw values >2, complexes disappeared from the gel, which was probably due to aggregation of the protein and DNA occurring after transfer of the sample to the low salt conditions required for electrophoresis.
Figure 4.
EMSA of 7kMk–DNA complexes. BamHI linearized pUC19 DNA (150 ng) was incubated with various concentrations of 7kMk in GB buffer at 70°C for 30 min. The Rw (protein:DNA weight ratio) values of the samples loaded in lanes 1–7 were 0.5, 1, 1.5, 2, 3, 10 and 0, respectively. M, 1 kb DNA ladder (Gibco BRL). Electrophoresis was performed in 1.5% agarose at 1.5 V/cm for 16 h in TBE buffer containing 100 mM Na-Glu.
The effect of increasing DNA migration promoted by 7kMk is reminiscent of the analogous property of MkaH and other archaeal histones. These were demonstrated to compact DNA through the formation of regular loop-like structures with a perimeter of 60–75 bp (1). Therefore, it was tempting to suggest a similar mode of DNA compaction by 7kMk.
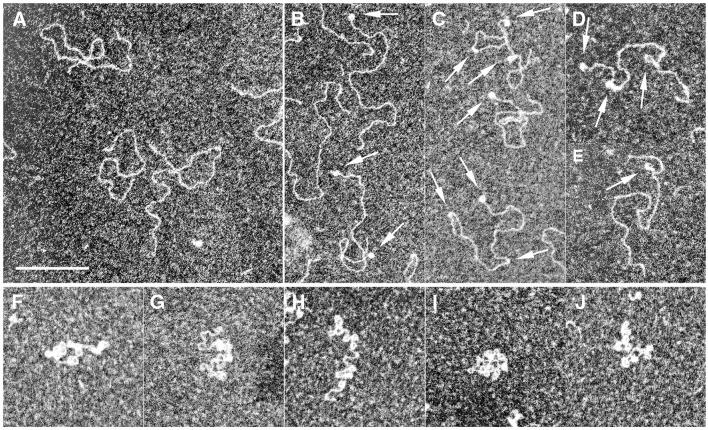
An EM study of the complexes formed with linear pUC19 DNA revealed that 7kMk did constrain DNA into regular loop-like structures (Fig. 5). At moderate Rw values (1.5), binding of 7kMk led to the formation of loops and short bent regions, with approximately one bent region or loop per single DNA molecule (Fig. 5B–E). Within bent regions, DNA was apparently thickened, most probably due to the presence of bound proteins. DNA bending might occur due to formation of an intermediate complex preceding DNA looping; however, partial disruption of the complexes in the course of EM sample preparation was also possible. At high Rw values (10), the majority of complexes appeared as DNA loops, with 10–15 loops per DNA molecule (Fig. 5F–J). The apparent circumference of a single loop was ∼43.5 ± 3.5 nm, corresponding to 136 ± 11 bp (assuming B-form DNA).
Figure 5.
EM of the complexes formed of 7kMk with pUC19/BamHI DNA. Complexes were obtained after incubation in the presence of 1 M K-Glu at 70°C at a Rw of 0 (A), 1.5 (B–E) and 10 (F–J). Arrows indicate DNA loops and bends. The scale bar represents 200 nm.
At moderate Rw, a significant fraction of loops and bends was formed in close proximity to DNA termini; in some instances both DNA termini were occupied, suggesting that there was no sequence specificity for complex formation (Fig. 5B–D). Bends and loops were also found along the entire length of the DNA molecule, most likely without any preference for a certain nucleotide sequence.
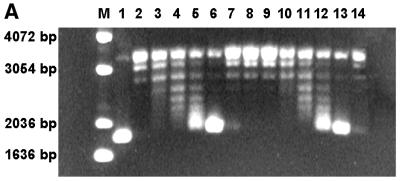
The overall topology of the complexes was examined with a topoisomerase relaxation assay using topoisomerase V (18). The advantage of this topoisomerase in comparison to other relaxation enzymes is that it relaxes both positively and negatively supercoiled DNA at K-Glu concentrations in the range 0.05–3 M and at temperatures up to 100°C. Complexes of 7kMk with relaxed covalently closed pUC19 DNA were obtained at various 7kMk:DNA ratios. Analysis of the DNA topoisomers (Fig. 6A) clearly indicated that 7kMk introduced supercoils into the DNA molecule. Formation of supercoils was observed at Rw values ranging from 0.8 (lane 4) to 3.3 (lane 6), reaching a very high absolute value of linking number difference (ΔLk) at a Rw of 3.3. At higher Rw, DNA was found to be topologically neutral, presumably due to its low accessibility to the topoisomerase (lanes 7–9), in accordance with previously reported data (35). Prolonged topoisomerase digestion of 7kMk–DNA complexes at Rw = 3.3 did not change the distribution of topoisomers significantly, indicating that the relaxation reached an equilibrium (lanes 11–14).
Figure 6.
DNA topology assay of 7kMk protein. Relaxed pUC19 (300 ng) was incubated with various concentrations of 7kMk in GB buffer at 70°C for 30 min. 7kMk was added at a Rw of 0 (lane 10), 0.4 (lane 3), 0.8 (lanes 4 and 11), 1.6 (lanes 5 and 12), 3.3 (lanes 6 and 13) 5 (lane 7), 6.7 (lanes 8 and 14) and 12 (lane 9). The resulting complexes were digested with topoisomerase V (100 ng) at the same temperature for 15 min (lanes 3–10) or 1 h (lanes 11–14). Lanes M, 1 and 2 were a 1 kb DNA ladder (Gibco BRL), negatively supercoiled pUC19 DNA isolated from E.coli and relaxed pUC19 DNA, respectively. The reaction products were analyzed by 1.5% agarose gel electrophoresis in TBE buffer (A) or TBE buffer containing 1 µg/ml chloroquine (B) at 1.5 V/cm for 16 h, stained with ethidium bromide and digitized.
The sign of supercoiling produced by 7kMk was determined by electrophoresis in the presence of 1 µg/ml chloroquine, which is known to unwind DNA and thus introduce positive supercoiling (Fig. 6B). In the presence of chloroquine the linking number difference of DNA relaxed in the absence of 7kMk (ΔLkc0) is ∼+6 (Fig. 6B, lane 10). Comparison of lanes 4–6 in Figure 7A and B shows that ΔLkc of the DNA sample in lane 6 of Figure 6B is still negative and close to ∼–8. Therefore, the ΔLk value of DNA in lane 6 equals ∼ –14 (ΔLkc – ΔLkc0).
Figure 7.
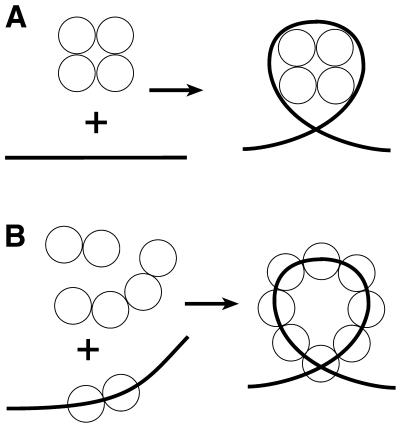
Two models of 7kMk–DNA complex formation. (A) Nucleosome-like wrapping of DNA by the 7kMk oligomer. (B) Cooperative binding of 7kMk to the DNA molecule resulting in DNA bending and looping.
We suppose that the large number of supercoils was due to formation of the DNA loops observed by EM. The negative sign of ΔLk indicates that in the loops the DNA helix formed a left-handed turn. The difference in the number of supercoils induced in relaxed DNA at Rw = 3.3 (∼14) and the number of loops formed in linear DNA at the same Rw visualized by EM (2–3, not shown) can be accounted for by the difference in DNA configuration (closed circular versus linear), inequality in salt/temperature conditions and a shift in the binding equilibrium in the EM sample due to the 100-fold dilution prior to its adsorption to the carbon film. Nevertheless, we believe that the loop is the dominant form of the 7kMk–DNA complex at high temperature and salt concentration. Alterations in DNA helical pitch upon 7kMk binding could also potentially change the DNA topology and induce the constrained supercoiling observed in the assay. However, CD spectroscopy did not reveal any changes in DNA ellipticity at 275 nm in complexes with 7kMk (70°C, Rw = 3.3), indicating that unwinding of the DNA was unlikely.
DISCUSSION
We have identified and characterized a novel DNA-binding protein, 7kMk, from the hyperthermophilic methanogen M.kandleri. The protein forms a stable dimer, which is probably capable of oligomerizing further, thus forming higher order aggregates with a rapid equilibrium between the species. The degree of oligomerization is dependent on protein concentration and possibly on the presence of DNA.
There are two different conceivable models for formation of the 7kMk–DNA complex. According to the first model, DNA wrapping occurs through its interaction with a preformed compact multimeric protein core (Fig. 7A), in a way similar to that of the tetrasome, the complex of DNA with the (H3–H4)2 tetramer of eukaryal histones (36) or archaeal nucleosomes. Alternatively, 7kMk binds to DNA in a cooperative manner, inducing its gradual bending and finally formation of the left-handed superhelical DNA loop (Fig. 7B). An in-phase DNA bending mechanism seems to be plausible, due to the presence of intermediates imaged by EM as DNA bends and the variety of protein oligomerization states.
The predicted secondary structure of the protein and features of the amino acid sequence permit us to suggest that 7kMk is closely related to the RHH family of transcription factors, although a final classification will require high resolution structure determination. Crystal structures of several RHH regulators, both in the unliganded state and in a complex with operator DNA, have been determined (37–41). The proteins from this family contain an N-terminal β-strand followed by two α-helices. Symmetrical dimers formed via the interaction of β-strands (constituting a DNA-recognizing antiparallel β-sheet) and of both α-helices are prone to further oligomerization. For example, the Arc transcriptional repressor binds to its cognate site as two individual dimers, but in a strongly cooperative manner, whereas Mnt repressor exists as a stable tetramer in solution. Interestingly, the plasmid-encoded transcriptional repressor CopG, which has a large inter-dimer interface, forms, in the crystal, a right-handed helical superstructure containing 12 protein molecules per turn, with β-ribbons pointing outwards (41). It is worth noting that CopG has been suggested to form multiprotein complexes with DNA, nucleating from the recognition sequence and extending along the DNA in both directions (41). At a high protein:DNA ratio, CopG protected up to 100 bp around the recognition site against the action of DNase I and hydroxyl radicals, contacting one face of the double helix. In this context, the apparent existence of proteins that facilitate a repressor activity of RHH regulators and mirror their abundance in various prokaryotic genomes (10,42) is potentially interesting. One explanation of this co-repression could be a stabilization of multiprotein structures formed by RHH proteins.
The 7kMk–DNA complex adds to the list of nucleoprotein structures in which bound DNA has a loop-like configuration and which are thought to be involved in regulation of transcription. The best known example is the eukaryal nucleosome, a particle containing 146 bp of DNA wrapped into a left-handed superhelix around a histone octamer (43). The nucleosome is a primary DNA packaging unit in the eukaryal cell; acetylation of histones and nucleosome remodeling play an important role in the control of gene expression (44).
Another protein with the ability to compact DNA into nucleosome-like particles is the bacterial nucleoid-associated protein HU (45). The HU dimer interacts in a non-sequence-specific manner with the minor groove of DNA, inducing its bending and supercoiling (46–48). It has been proposed that the circular complex formed by HU and DNA in vitro consists of 8–10 HU dimers and involves 80–100 bp per turn (46,49). The ability of HU to distort DNA structure is suggested to be important for activity regulation of a number of promoters (50).
Remarkably, homologs of the E.coli global transcriptional regulator Lrp also induce nucleosome-like DNA compaction. It has been shown that Lrp-like proteins bind one face of the DNA helix of very long (100–200 bp) regions upstream of promoters (51–53). For instance, the Lrp-type protein from Agrobacterium tumefaciens compacts bound DNA by the equivalent of ∼115 bp, as demonstrated by atomic force microscopy (54). It is conceivable that the mechanisms of DNA compaction by the 7kMk and Lrp proteins could be related.
The role of 7kMk remains to be elucidated. For this protein we cannot exclude the existence of specific target DNA sequences. Nevertheless, the affinity of 7kMk for random DNA is unusually high, approaching that of MkaH, a true histone from the same organism. Interestingly, homologs of sequence-specific transcription regulators exhibiting a sequence-independent mode of DNA binding have been identified in both Bacteria and Archaea and it has been suggested that they play a role in nucleoid organization/interactions (55,56). Accurate determination of the intracellular 7kMk concentration and detailed characterization of its DNA binding activity will likely permit more straightforward conclusions about its function to be made and are the subjects of further investigations.
Acknowledgments
ACKNOWLEDGEMENTS
The authors gratefully thank Dr Thomas Jovin and Dr Alexei Bogdanov for support of this work, Dr Galina Belova for her generous gift of wild-type topoisomerase V and its T75 mutant preparations, Olga Ustinova for help with CD spectroscopic characterization of 7kMk, and Uwe Plessmann for N-terminal sequencing of the recombinant 7kMk. This work was partially supported by grants from the Howard Hughes Medical Institute and the Russian Foundation for Basic Research to A.I.S and N.A.P and short-term FEBS and Max Planck Society fellowships to N.A.P.
DDBJ/EMBL/GenBank accession no. AY037781
REFERENCES
- 1.Sandman K., Krzycki,J.A., Dobrinski,B., Lurz,R. and Reeve,J.N. (1990) HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl Acad. Sci. USA, 87, 5788–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandman K. and Reeve,J.N. (2000) Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol., 173, 165–169. [DOI] [PubMed] [Google Scholar]
- 3.Langer D., Hain,J., Thuriaux,P. and Zillig,W. (1995) Transcription in archaea: similarity to that in eucarya. Proc. Natl Acad. Sci. USA, 92, 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowlands T., Baumann,P. and Jackson,S.P. (1994) The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science, 264, 1326–1329. [DOI] [PubMed] [Google Scholar]
- 5.Ouzounis C. and Sander,C. (1992) TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell, 71, 189–190. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Kupiec R., Blank,C. and Leigh,J.A. (1997) Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc. Natl Acad. Sci. USA, 94, 1316–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell S.D. and Jackson,S.P. (2000) Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem., 275, 31624–31629. [DOI] [PubMed] [Google Scholar]
- 8.Bell S.D., Cairns,S.S., Robson,R.L. and Jackson,S.P. (1999) Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell, 4, 971–982. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E.V., Mushegian,A.R., Galperin,M.Y. and Walker,D.R. (1997) Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol., 25, 619–637. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L. and Koonin,E.V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurr M., Huber,R., König,H., Jannasch,H.W., Fricke,H., Trincone,A., Kristjansson,J.K. and Stetter,K.O. (1991) Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch. Microbiol., 156, 239–247. [Google Scholar]
- 12.Burggraf S., Stetter,K.O., Rouviere,P. and Woese,C.R. (1991) Methanopyrus kandleri—an archael methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol., 14, 346–351. [DOI] [PubMed] [Google Scholar]
- 13.Slesarev A.I., Stetter,K.O., Lake,J.A., Gellert,M., Krah,R. and Kozyavkin,S.A. (1993) DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature, 364, 735–737. [DOI] [PubMed] [Google Scholar]
- 14.Belova G.I., Prasad,R., Kozyavkin,S.A., Lake,J.A., Wilson,S.H. and Slesarev,A.I. (2001) A type IB topoisomerase with DNA repair activities. Proc. Natl Acad. Sci. USA, 98, 6015–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slesarev A.I., Belova,G.I., Kozyavkin,S.A. and Lake,J.A. (1998) Evidence for an early prokaryotic origin of histones H2A and H4 prior to the emergence of eukaryotes. Nucleic Acids Res., 26, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahrner R.L., Cascio,D., Lake,J.A. and Slesarev,A. (2001) An ancestral nuclear protein assembly: crystal structure of the Methanopyrus kandleri histone. Protein Sci., 10, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen B., Steinberg,J., Laemmli,U.K. and Weintraub,H. (1980) The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res., 8, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slesarev A.I., Lake,J.A., Stetter,K.O., Gellert,M. and Kozyavkin,S.A. (1994) Purification and characterization of DNA topoisomerase V. An enzyme from the hyperthermophilic prokaryote Methanopyrus kandleri that resembles eukaryotic topoisomerase I. J. Biol. Chem., 269, 3295–3303. [PubMed] [Google Scholar]
- 19.Schagger H. and von Jagow,G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- 20.Anderson N.L., Nance,S.L., Pearson,T.W. and Anderson,N.G. (1982) Specific antiserum staining of two-dimensional electrophoretic patterns of human-plasma proteins immobilized on nitrocellulose. Electrophoresis, 3, 135–142. [Google Scholar]
- 21.Slesarev A.I., Belova,G.I., Lake,J.A. and Kozyavkin,S.A. (2001) Topoisomerase V from Methanopyrus kandleri. Methods Enzymol., 334, 179–192. [DOI] [PubMed] [Google Scholar]
- 22.Pace C.N., Vajdos,F., Fee,L., Grimsley,G. and Gray,T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci., 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sreerama N. and Woody,R.W. (1993) A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem., 209, 32–44. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama N. and Woody,R.W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal. Biochem., 287, 252–260. [DOI] [PubMed] [Google Scholar]
- 25.Dubochet J., Ducommun,M., Zollinger,M. and Kellenberger,E. (1971) A new preparation method for dark-field electron microscopy of biomacromolecules. J. Ultrastruct. Res., 35, 147–167. [DOI] [PubMed] [Google Scholar]
- 26.O’Brien R., DeDecker,B., Fleming,K.G., Sigler,P.B. and Ladbury,J.E. (1998) The effects of salt on the TATA binding protein-DNA interaction from a hyperthermophilic archaeon. J. Mol. Biol., 279, 117–125. [DOI] [PubMed] [Google Scholar]
- 27.Bergqvist S., O’Brien,R. and Ladbury,J.E. (2001) Site-specific cation binding mediates TATA binding protein-DNA interaction from a hyperthermophilic archaeon. Biochemistry, 40, 2419–2425. [DOI] [PubMed] [Google Scholar]
- 28.Ronimus R.S. and Musgrave,D.R. (1996) Purification and characterization of a histone-like protein from the Archaeal isolate AN1, a member of the Thermococcales. Mol. Microbiol., 20, 77–86. [DOI] [PubMed] [Google Scholar]
- 29.Shima S., Tziatzios,C., Schubert,D., Fukada,H., Takahashi,K., Ermler,U. and Thauer,R.K. (1998) Lyotropic-salt-induced changes in monomer/dimer/tetramer association equilibrium of formyltransferase from the hyperthermophilic Methanopyrus kandleri in relation to the activity and thermostability of the enzyme. Eur. J. Biochem., 258, 85–92. [DOI] [PubMed] [Google Scholar]
- 30.Lanyi J.K. (1974) Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev., 38, 272–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenfield N.J. (1996) Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal. Biochem., 235, 1–10. [DOI] [PubMed] [Google Scholar]
- 32.Provencher S.W. and Glockner,J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry, 20, 33–37. [DOI] [PubMed] [Google Scholar]
- 33.Sreerama N., Venyaminov,S.Y. and Woody,R.W. (1999) Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci., 8, 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildebrand E.L. and Grossman,L. (1999) Oligomerization of the UvrB nucleotide excision repair protein of Escherichia coli. J. Biol. Chem., 274, 27885–27890. [DOI] [PubMed] [Google Scholar]
- 35.Musgrave D., Forterre,P. and Slesarev,A. (2000) Negative constrained DNA supercoiling in archaeal nucleosomes. Mol. Microbiol., 35, 341–349. [DOI] [PubMed] [Google Scholar]
- 36.Alilat M., Sivolob,A., Revet,B. and Prunell,A. (1999) Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle. J. Mol. Biol., 291, 815–841. [DOI] [PubMed] [Google Scholar]
- 37.Rafferty J.B., Somers,W.S., Saint-Girons,I. and Phillips,S.E. (1989) Three-dimensional crystal structures of Escherichia coli met repressor with and without corepressor. Nature, 341, 705–710. [DOI] [PubMed] [Google Scholar]
- 38.Somers W.S. and Phillips,S.E. (1992) Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by beta-strands. Nature, 359, 387–393. [DOI] [PubMed] [Google Scholar]
- 39.Burgering M.J., Boelens,R., Gilbert,D.E., Breg,J.N., Knight,K.L., Sauer,R.T. and Kaptein,R. (1994) Solution structure of dimeric Mnt repressor (1-76). Biochemistry, 33, 15036–15045. [DOI] [PubMed] [Google Scholar]
- 40.Raumann B.E., Rould,M.A., Pabo,C.O. and Sauer,R.T. (1994) DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature, 367, 754–757. [DOI] [PubMed] [Google Scholar]
- 41.Gomis-Ruth F.X., Sola,M., Acebo,P., Parraga,A., Guasch,A., Eritja,R., Gonzalez,A., Espinosa,M., del Solar,G. and Coll,M. (1998) The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J., 17, 7404–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabuchi A., Min,Y.N., Womble,D.D. and Rownd,R.H. (1992) Autoregulation of the stability operon of IncFII plasmid NR1. J. Bacteriol., 174, 7629–7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 44.Gregory P.D. and Horz,W. (1998) Life with nucleosomes: chromatin remodelling in gene regulation. Curr. Opin. Cell Biol., 10, 339–345. [DOI] [PubMed] [Google Scholar]
- 45.Azam T.A. and Ishihama,A. (1999) Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem., 274, 33105–33113. [DOI] [PubMed] [Google Scholar]
- 46.White S.W., Wilson,K.S., Appelt,K. and Tanaka,I. (1999) The high-resolution structure of DNA-binding protein HU from Bacillus stearothermophilus. Acta Crystallogr. D, 55, 801–809. [DOI] [PubMed] [Google Scholar]
- 47.Craig N.L. and Nash,H.A. (1984) E. coli integration host factor binds to specific sites in DNA. Cell, 39, 707–716. [DOI] [PubMed] [Google Scholar]
- 48.Drlica K. and Rouviere-Yaniv,J. (1987) Histonelike proteins of bacteria. Microbiol. Rev., 51, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouviere-Yaniv J., Yaniv,M. and Germond,J.E. (1979) E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell, 17, 265–274. [DOI] [PubMed] [Google Scholar]
- 50.Kar S. and Adhya,S. (2001) Recruitment of HU by piggyback: a special role of GalR in repressosome assembly. Genes Dev., 15, 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin R., Ernsting,B., Hirshfield,I.N., Matthews,R.G., Neidhardt,F.C., Clark,R.L. and Newman,E.B. (1992) The lrp gene product regulates expression of lysU in Escherichia coli K-12. J. Bacteriol., 174, 2779–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiese D.E., Ernsting,B.R., Blumenthal,R.M. and Matthews,R.G. (1997) A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J. Mol. Biol., 270, 152–168. [DOI] [PubMed] [Google Scholar]
- 53.Madhusudhan K.T., Huang,N. and Sokatch,J.R. (1995) Characterization of BkdR-DNA binding in the expression of the bkd operon of Pseudomonas putida. J. Bacteriol., 177, 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jafri S., Evoy,S., Cho,K., Craighead,H.G. and Winans,S.C. (1999) An Lrp-type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J. Mol. Biol., 288, 811–824. [DOI] [PubMed] [Google Scholar]
- 55.Tapias A., Lopez,G. and Ayora,S. (2000) Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res., 28, 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Napoli A., Kvaratskelia,M., White,M.F., Rossi,M. and Ciaramella,M. (2001) A novel member of the bacterial-archaeal regulator family is a nonspecific DNA-binding protein and induces positive supercoiling. J. Biol. Chem., 276, 10745–10752. [DOI] [PubMed] [Google Scholar]