Abstract
The exosome, an evolutionarily conserved complex of multiple 3′→5′ exoribonucleases, is responsible for a variety of RNA processing and degradation events in eukaryotes. In this report Arabidopsis thaliana AtRrp4p is shown to be an active 3′→5′ exonuclease that requires a free 3′-hydroxyl and degrades RNA hydrolytically and distributively, releasing nucleoside 5′-monophosphate products. AtRrp4p behaves as an ∼500 kDa species during sedimentation through a 10–30% glycerol gradient, co-migrating with AtRrp41p, another exosome subunit, and it interacts in vitro with AtRrp41p, suggesting that it is also present in the plant cell as a subunit of the exosome. We found that, in addition to a previously reported S1-type RNA-binding domain, members of the Rrp4p family of proteins contain a KH-type RNA-binding domain in the C-terminal half and show that either domain alone can bind RNA. However, only the full-length protein is capable of degrading RNA and interacting with AtRrp41p.
INTRODUCTION
The exosome is a complex of at least 10 3′→5′ exoribonucleases (and possibly other proteins) that is responsible for a variety of RNA processing events, as well as for complete degradation of certain RNA species (1–3). Two compositionally and functionally distinct exosome forms, nuclear and cytoplasmic, have been described in yeast and human cells (1,4,5) and an exosome-related complex has been described in the nuclei and cytoplasm of Trypanosoma brucei (6). It has been proposed that the origin of the exosome complex predates the divergence of eukaryotes and Archaea (7). Autoantibodies against the exosome subunits are found in patients suffering from poliomyositis–scleroderma overlap syndrome, a severe autoimmune disease. Every exosome subunit is essential in yeast except for Rrp6p, loss of which is conditionally lethal. The phenotypes of partial loss of function mutations in different subunits are often similar, suggesting that an intact complex is required for exosome-mediated RNA processing and degradative reactions, as well as for cell viability. The full substrate range of the exosome, its structural organization and mechanisms of regulation and even why exactly it is essential for cell viability remain to be established.
Strikingly, almost all of the core exosome subunits are potentially catalytic entities. In the yeast exosome, characterized in most detail, 6 of the 10 ‘core’ subunits are RNase PH-like phosphorolytic exonucleases (Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p and Mtr3p), while Rrp4p and possibly Rrp40p belong to a novel type of hydrolytic exonucleases. Rrp44p is a RNase II-like hydrolytic enzyme, and only Csl4p has no obvious catalytic function (but has a S1-type RNA-binding domain; 1,3). Rrp6p, a protein that is found exclusively in the nuclear form of the exosome, is related to Escherichia coli RNase D (4,8,9). Curiously, purified yeast exosomes have rather low exonucleolytic activity, perhaps hinting that additional cofactors might be needed to activate the catalytic sites to act on particular substrate RNAs (1,3). The Ski2/3/8p complex (10), which is involved in cytoplasmic (i.e. mRNA 3′→5′ decay), but not nuclear (e.g. processing of 7S rRNA precursor into mature 5.8S rRNA), reactions of the exosome (11), appears to be one such cofactor, while Ski7p may act as a bridge between the exosome core and the Ski2/3/8p complex (12).
We are pursuing an analysis of exosome structure and function in a model plant species Arabidopsis thaliana. We have previously demonstrated that the plant homolog of the Rrp41p subunit, AtRrp41p, rescues the lethal phenotype of a yeast rrp41 null mutant, corrects the defects of a conditional rrp41 (ski6-100) allele in mRNA 3′→5′ degradation and 5.8S rRNA processing, interacts in vitro with two of the yeast exosomal proteins and resides in an ∼500 kDa complex, which is most likely the plant exosome (13). In this manuscript characterization of another putative Arabidopsis exosome subunit, AtRrp4p, is presented.
MATERIALS AND METHODS
Materials
Arabidopsis thaliana (var. Columbia) was from Lehle Seeds. Cauliflower heads were purchased at local supermarkets. Antiserum against full-length soluble GST–AtRrp4p fusion protein was raised at Covance Co., purified on protein A–agarose and subsequently affinity purified using GST–AtRrp4p immobilized on nitrocellulose, as described (14).
Computational analysis
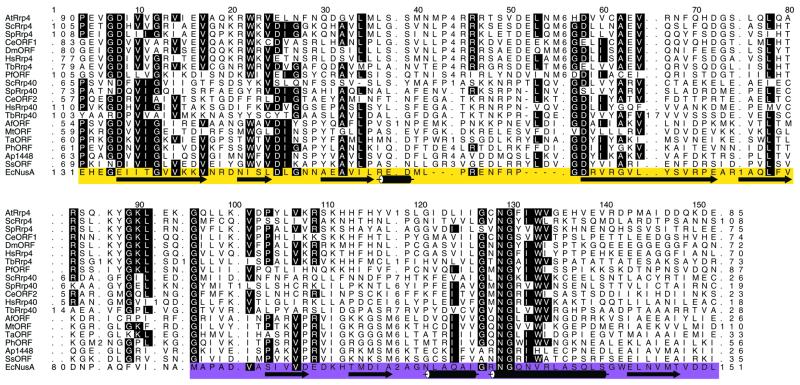
We identified sequence homologs of AtRrp4p in other species using PSI-BLAST as implemented at the NCBI World Wide Web page interface to protein–protein BLAST. Using the default parameter settings, the program was run until convergence such that members of the Rrp4/Rrp40 sequence family had E values <0.001. A hidden Markov model (HMM) for this family was trained (http://www.cse.ucsc.edu/research/compbio/sam.html) using PSI-BLAST pairwise alignment as the starting point. The HMM-generated multiple sequence alignment shows that the Rrp4/Rrp40 family is similar over ∼180 amino acids. The regions of highest similarity (shown in Fig. 1) encompass an S1 domain (yellow) separated from a KH domain (magenta) by a short linker which differs in the Rrp4 and Rrp40 sequence families.
Figure 1.
An HMM-generated multiple sequence alignment of the Rrp4/Rrp40 sequence family. Highly conserved alignment positions are highlighted. Numbers within the alignment denote the number of residues not shown explicitly and which align to the insert state of the HMM. The sequences shown are as follows. AtRRP4, A.thaliana Rrp4p or gene F15K9.4 (locus name AAC72108); ScRRP4, S.cerevisiae Rrp4p (RRP4_YEAST); SpRRP4, S.pombe Rrp4p (RRP4_SCHPO); CeORF1, C.elegans open reading frame (ORF) Y73B6BL.3 (C084197_22); DmORF, Drosophila melanogaster ORF CG3931 (AAF47028); HsRRP4, Homo sapiens Rrp4p (RRP4_HUMAN); TbRRP4, T.brucei Rrp4p (CAC39255); PfORF, P.falciparum ORF MAL4P2.38 (CAB62879); ScRRP40, S.cerevisiae Rrp40p (RR40_YEAST); SpRRP40, S.pombe Rrp40p (CAB16582); CeORF2, C.elegans ORF F59C6.4 (T22989); HsRRP40, Homo sapiens Rrp40p (RR40_HUMAN); TbRRP40, T.brucei Rrp40p (CAC39257); AfORF, Archaeoglobus fulgidus ORF (AF0492); MtORF, Methanobacterium thermoautotrophicum ORF MTH684 (B69191); TaORF, Thermoplasma acidophilum ORF Ta1292 (CAC12414); PhORF, Pyrococcus horikoshii ORF PH1551 (G71032); ApORF, Aeropyrum pernix ORF APE1448 (H72623); SsORF, Sulfolobus solfataricus ORF c20_026 (CAB57567); EcNusA, E.coli NusA (NUSA_ECOLI). Secondary structure elements of the S1-type RNA-binding domain and of the βααβ core of the second KH RNA-binding domain, from the crystal structure of T.maritima NusA protein (19), which is highly homologous to E.coli NusA, are shown below the alignment in yellow and magenta shading, respectively. The Rrp4/Rrp40 sequence family contains members from all three kingdoms: Eukarya (sequence names beginning with At, Sc, Sp, Ce, Dm, Hs, Tb and Pf), Archaea (Af, Mt, Ta, Ph and Ap) and Bacteria (Ec).
Expression survey
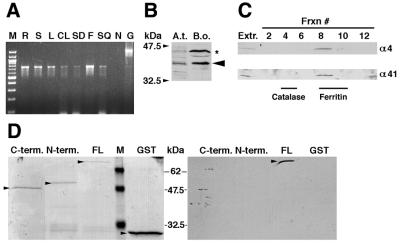
Total RNA was extracted from the Arabidopsis tissues indicated in Figure 3 as described (15) and reverse transcribed using an oligo(dT) primer and SuperScript II (Life Technologies). Reactions were spiked with a small amount of [α-32P]dATP in order to confirm that cDNA synthesis efficiency was similar between the tissues. Equal amounts of the resulting cDNAs were PCR amplified using primers oDB503 (5′-ACATGGAACTTCCGAGGTCGATGG-3′) and oDB504 (5′-TCTGCATATGGTTTGTCGAGTCTC-3′) for 30 cycles and resolved by agarose gel electrophoresis.
Figure 3.
Arabidopsis AtRrp4p is expressed widely, is present in a large complex and interacts with AtRrp41p. (A) Results of the RT–PCR expression survey. Tissues tested are as follows: roots (R), stems (S), rosette leaves (L), cauline leaves (CL), total seedlings (SD), flowers (F) and siliques (SQ). M, molecular weight marker (NEB 100 bp ladder); N, negative control (no template); G, genomic DNA. (B) The antibody raised against GST–AtRrp4p reacts with the same size polypeptide in Arabidopsis and cauliflower extracts. The band marked with an asterisk is a cross-reacting cauliflower protein. (C) Western blot analysis of the total cauliflower extract resolved by sedimentation through a 10–30% glycerol gradient, with antibodies against AtRrp4p (top) and AtRrp41p (bottom). Fraction numbers tested (top to bottom) are indicated above the lanes. Fraction 8 corresponds to an ∼500 kDa complex, based on the behavior of BSA (67 kDa, migrates close to the top of the gradient), catalase (240 kDa) and ferritin (480 kDa) markers fractionated in a parallel gradient. Lanes marked Extr. were loaded with total cauliflower extract. (D) Analysis of AtRrp4p–AtRrp41p interactions by GST pulldown assay. GST fusions to the AtRrp4p C-terminal domain, N-terminal domain, full-length AtRrp4p or GST alone (10 µg each polypeptide, indicated above the respective lanes) were bound to glutathione–agarose beads. To control for equal loading of the resin, 3% of the bound material was resolved by SDS–PAGE and stained with Coomassie brilliant blue (left, respective species indicated by arrowheads), while the rest was challenged with MBP–AtRrp41p fusion protein (5 µg). The bound MBP–AtRrp41p was detected by western blotting using antibody against the C-terminus of AtRrp41p (right). While the MBP–AtRrp41p fusion almost co-migrates with GST–AtRrp4p on SDS–PAGE, the observed signal (arrowhead) is due to MBP–AtRrp41p, since the antibody against AtRrp41p used here does not cross-react with GST–AtRrp4p (data not shown).
Exonuclease substrates and assays
Oligo(rA)23 was labeled at the 5′-end by polynucleotide kinase and gel purified. A radiolabeled in vitro transcript was made from the polylinker of plasmid pSP65(A38) (16) linearized with HindIII, using SP6 RNA polymerase and [α-32P]UTP, and gel purified. Substrate containing a 3′-phosphate was obtained by ligating 32P-pCp to the 3′-end of unlabeled oligo(rA)23 and gel purifying the ligated product. Ascending thin layer chromatography on polyethyleneimine–cellulose plates (Sigma) was conducted in 1 M formic acid and 0.5 M LiCl, as described previously (13). Aliquots of 100 nmol cold 5′-UMP, 3′-UMP and 5′-UDP were added to the sample and visualized by UV shadowing. Exonuclease assays were exactly as described (13). In brief, 100 µl reactions were assembled containing 10 mM Tris–HCl, pH 7.6, 50 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 100 µg/ml BSA, 0.8 U/µl RNasin, 1 pmol substrate RNA and 1–3 pmol protein. Aliquots (15 µl) were withdrawn at the indicated time points, added directly to an equal volume of the sequencing loading mix and snap-frozen in liquid nitrogen.
Protein expression, glycerol gradients, protein interaction and RNA binding assays
To create the N-terminal fragment, C-terminal fragment and full-length AtRrp4p fusions, the respective portions of the cDNA were amplified with oDB501 and oDB590 (5′-ATGGTGATGAGAAAGCTACAGTTA-3′ and 5′-TAACTTCCCGTACTTCTGACTTCT-3′), oDB502 and oDB589 (5′-TCACTTTTTCCTCTTTGTACGTCT-3′ and 5′-AAGTACGGGAAGTTAGAGAAGGGA-3′) and oDB501 and oDB502, respectively, and cloned into the blunted EcoRI site of pGEX-2T (Pharmacia), resulting in pDB539, pDB540 and pDB481, respectively. Both N- and C-terminal GST fusions retained the KYGKL spacer that separates the S1 from the KH domain in the full-length protein. GST–AtRrp4p fusions were expressed and purified on glutathione–agarose as described (13) and additionally purified on a heparin column with 0.05–0.6 M KCl gradient elution.
Glycerol gradient fractionation was as described previously (13). GST pulldown assays were conducted essentially as described (17), except that 10 µg protein was used to load glutathione beads. Gel shift assays were conducted as described (18). Briefly, 25 µl reactions were assembled containing 5 fmol 5′-end-labeled oligo(rA)23 probe, 10 mM Tris, pH 8.0, 70 mM NaCl, 100 ng/µl acetylated BSA, 0.01% NP-40, 100 mM β-mercaptoethanol, 2 µg/ml tRNA and varying concentrations of protein. After 15 min at room temperature, 2.5 µl of 2.5 mg/ml heparin was added. After an additional 10 min incubation, binding reactions were loaded onto 6% polyacrylamide (acrylamide:bisacrylamide ratio 60:1), 1× TBE gels containing 0.1% Triton X-100 and run at 20 V/cm at 4°C.
RESULTS AND DISCUSSION
A BLASTP search of the annotated Arabidopsis genome (mips.gsf.de) using the yeast Rrp4p protein sequence as query yielded a single candidate protein of 322 amino acids and predicted molecular mass of 36.6 kDa, hereafter referred to as AtRrp4p (MIPS gene code At1g03360). The cDNA clone encoding AtRrp4p was reconstructed by RT–PCR amplification and major polyadenylation sites were established by 3′-RACE. The 5′-end amplification by 5′-RACE yielded clones containing in-frame stop codons upstream of the ATG predicted to be the initiation codon. The resulting polypeptide sequence and related sequences identified through PSI-BLAST were used as the input in HMM analysis (see Materials and Methods for details). This analysis revealed that, in addition to the previously described S1 RNA-binding domain, the Rrp4p-related proteins, including a plant member of the Rrp4p family, may possess a KH-type RNA-binding domain located just C-terminal to the S1 domain. A similar prediction has been made independently by others (7), although no experimental evidence that this domain binds RNA has been obtained. Interestingly, the S1 and KH domains in AtRrp4p are separated by a short linker whose core sequence (KYGKL, starting at column 87 in the alignment shown in Fig. 1) is 100% conserved in the Saccharomyces cerevisiae, Schizosaccharomyces pombe, T.brucei, Caenorhabditis elegans, Drosophila and human Rrp4p homologs and differs by only one amino acid in the Rrp4p-like protein from the malarial parasite Plasmodium falciparum. In fact, this linker represents the longest continuous block of absolute sequence identity among these Rrp4p-related proteins. In contrast, the linker is different in the yeast exosome subunit Rrp40p and its homologs, as well as in more distantly related archaeal and prokaryotic proteins that also possess S1 and KH motifs in a tandem arrangement (Fig. 1). Such a remarkable conservation suggests a potential role for this linker in orienting the S1 and KH motifs for optimal RNA binding and/or protein–protein interactions, a possibility consistent with the data presented below. In addition, recent structural analysis of the Thermotoga maritima NusA protein, which contains a S1 domain followed by two KH domains, demonstrates that the tandem arrangement of S1 and KH domains can form an extended, continuous RNA-binding surface (19). Furthermore, a R199A substitution of the conserved residue located at the interface between the S1 and the first of the two KH domains of E.coli NusA leads to defects in mRNA binding in vitro and other NusA functions (20), probably due to destabilization of the relative orientation of the two RNA-binding domains.
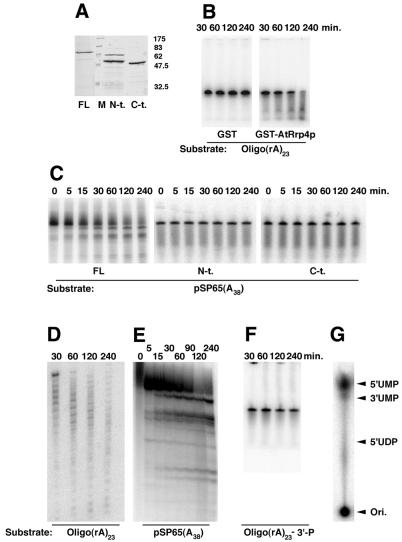
To characterize the enzymatic properties of AtRrp4p, we expressed and purified it from E.coli as a glutathione S-transferase (GST) fusion protein (Fig. 2A). Recombinant GST–AtRrp4p protein, but not GST alone, was able to degrade 5′-end-labeled oligo(rA)23 (Fig. 2B and D), as well as body-labeled RNA transcribed in vitro from the polylinker of plasmid pSP65A38 (16) (Fig. 2C and E), in an apparently distributive manner (the enzyme pause/stop sites are more obvious on the long gels shown in Fig. 2D and E). That incubation with a 5′-end-labeled substrate produced a pattern of partially degraded products (Fig. 2D) demonstrates that the enzyme acts in a 3′→5′, rather than 5′→3′, direction. No RNA degradation was observed when the substrate bore a 3′-phosphate, rather than 3′-hydroxyl, group (Fig. 2F). In order to reveal the nature of RNA cleavage by AtRrp4p, released reaction products were resolved by thin layer chromatography. The reaction products clearly co-migrated with 5′-UMP (Fig. 2G). Therefore, we conclude that AtRrp4p is a hydrolytic, distributive 3′→5′ exonuclease requiring a free 3′-OH on the substrate and releasing nucleoside 5′-monophosphates, much like its yeast counterpart.
Figure 2.
Characterization of AtRrp4p. (A) SDS–PAGE of the purified full-length (labeled FL) and N-terminal (S1 domain, labeled N-t.) and C-terminal (KH domain, labeled C-t.) fragments of AtRrp4p as GST fusions. An ∼60 kDa band in the N-t. lane is an E.coli contaminant that co-purifies with the N-terminal fragments through glutathione and heparin affinity chromatography. (B–F) Exonucleolytic assays using the protein species indicated (reaction time points are shown above the gels) with the 5′-end-labeled oligo(rA)23 ending either with 3′-hydroxyl (B and D) or 3′-phosphate (F) or using an in vitro transcript body-labeled with 32P-UTP made from the polylinker of pSP65(A38) (C and E). Long gels (D and E) are shown to emphasize the distributive nature of the enzyme. (G) Identification of the released product as nucleoside 5′-monophosphates. A TLC plate with the positions of co-chromatographed unlabeled mononucleotide standards marked on the right is shown.
Rrp4p in yeast, the unicellular parasite T.brucei and in humans exists mainly, if not exclusively, as a subunit of the exosome and is indispensable for cell viability, at least in the two unicellular systems. Therefore, it might be expected that the plant homolog is also part of the exosome complex and should be widely expressed and essential. In agreement with this view, a RT–PCR survey of Arabidopsis tissues revealed that AtRRP4 mRNA is expressed in roots, stems, rosette and cauline leaves, flowers and siliques (Fig. 3A). An analysis of the Arabidopsis rrp4 null allele, obtained by T-DNA insertional mutagenesis (J.Dutko, J.Alonso, J.Ecker and D.Belostotsky, in preparation) showed that AtRrp4p is indeed essential. To provide further evidence that Arabidopsis AtRrp4p functions as part of the exosome, rather than as the free protein, its native molecular weight was examined. To simplify this analysis, cauliflower, rather than Arabidopsis, tissue was used. Cauliflower (Brassica oleracea ssp. botrytis) belongs to the same Brassiceaea family and Arabidopsis and Brassica genes share, on average, 87% sequence identity (21). An affinity purified antiserum raised against GST–AtRrp4p fusion protein reacted with a polypeptide of the same size in Arabidopsis and cauliflower extracts (Fig. 3B). When the cauliflower total extract was fractionated by sedimentation through a 10–30% glycerol gradient and subjected to western analysis, the reacting species was found in a fraction corresponding to an ∼500 kDa complex. Moreover, it co-migrated with the cauliflower protein reacting with the previously characterized antibody against Arabidopsis AtRrp41p, another exosome subunit homolog (13) (Fig. 3C). Since we had previously shown that AtRrp41p interacts in vitro with yeast Rrp4p (13), a GST pulldown experiment was performed to confirm that a similar interaction takes place between the two Arabidopsis polypeptides. Glutathione bead-immobilized GST or GST–AtRrp4p fusion was challenged with a maltose-binding protein (MBP)–AtRrp41p fusion protein (13) and after extensive washes, bound proteins were eluted with glutathione and detected by western blotting with anti-AtRrp41p antibodies (Fig. 3D). The results show that Arabidopsis AtRrp41p and AtRrp4p indeed interact. Binding reactions carried out in the presence of RNase A produced identical results (data not shown), indicating that the AtRrp4p–AtRrp41p protein–protein interaction is direct and is RNA-independent. Combined with the above findings, these data strongly suggest that AtRrp4p is a component of the plant exosome.
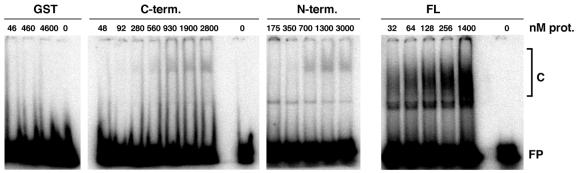
The presence of two distinct RNA-binding motifs in the same protein raises the question of their relative contributions to its properties. In order to address whether both domains are needed for RNA binding, the N- and C-terminal halves of AtRrp4p containing the S1 and KH RNA-binding domains with their respective N- or C-terminal extensions (amino acids 1–179 and 175–322, respectively) were expressed as GST fusions (Fig. 2A) and their ability to bind to oligo(rA)23 was examined by electrophoretic mobility shift assay, using the GST–AtRrp4p full-length fusion protein and GST as controls (Fig. 4) (a magnesium-free buffer was employed to prevent degradation of the substrate by AtRrp4p). The results show that both the N- and C-terminal fragments are individually sufficient for RNA binding, but that binding affinity increases in an apparently synergistic (i.e. non-additive) fashion when both domains are present together in the context of the full-length polypeptide (very high protein concentrations were required to build complete binding curves, thus precluding quantitative determination of Kd values). This is expected if the two domains make independent contacts with RNA, since additivity of the free energies of binding should result in an exponential increase in affinity according to ΔG = –RTlnK, where K is the equilibrium binding constant (the inverse of the equilibrium dissociation constant).
Figure 4.
Electrophoretic mobility shift assay analysis of GST–AtRrp4p and its N-terminal and C-terminal fragments. The protein species used in gel shift assays and their final concentrations in the binding reactions (in nM) are indicated above the respective panels. The protein–RNA complex and the free oligo(rA)23 probe are indicated as C and FP, respectively. The sharp band appearing below the protein–RNA complex in the right two panels is a conformational variant of the oligo(rA)23 probe; its presence and the relative amount does not correlate with the nature or concentration of the protein used in the binding reaction.
The absolute conservation of the five amino acid linker connecting the two domains in all Rrp4p proteins examined so far may indicate that the mutual orientation of the two domains is important to achieve optimal binding. To further examine the potential significance of this tandem arrangement of the S1 and KH domains, the enzymatic activity and ability to interact with AtRrp41p of the N- and C-terminal AtRrp4p domains were also tested. We found that neither GST fusion to the N- or C-terminal domain of AtRrp4p was able to degrade RNA (Fig. 2C, middle and right). To test for possible interactions with AtRrp41p, GST fusions to the C-terminal domain (C-term.) (Fig. 3D, lane 1), to the N-terminal domain (N-term.) (Fig. 3D, lane 2) and to full-length AtRrp4p protein (FL) (Fig. 3D, lane 3) were immobilized on glutathione–agarose beads and challenged with MBP–AtRrp41p fusion protein. After extensive washes, bound species were resolved by SDS–PAGE and detected using antibodies against the C-terminus of AtRrp41p. We found that only GST–full-length AtRrp4p fusion protein was able to interact with the MBP–AtRrp41p fusion (control experiments have shown that the antibody against AtRrp41p does not cross-react with GST–AtRrp4p protein, which apparently co-migrates on SDS–PAGE with MBP–AtRrp41p fusion protein). Notably, the KYGKL linker core was present in both of the GST fusion proteins, suggesting that by itself it was not able to impart the ability to bind RNA or AtRrp41p or to degrade RNA upon the N- or C-terminal halves of AtRrp4p, although a role in binding or catalysis per se in the context of native protein cannot be excluded.
In summary, the results of this report strongly suggest that Arabidopsis AtRrp4p is a component of the plant exosome and is composed of two RNA-binding domains that can bind RNA independently. In addition, both domains are needed for RNA degradation and for interaction with another exosome subunit, AtRrp41p. Absolute conservation of the length and sequence of the short linker connecting the S1 and KH domains suggests that it might play an important role in maintaining the mutual spatial arrangement of the two domains, which is important for protein function. Future work will focus on examining its significance in the context of the whole protein, as well as on identifying the amino acid residues participating in RNA hydrolysis.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank two anonymous reviewers for helpful comments. This research was supported by National Science Foundation grant MCB-9874580 to D.A.B. Work by I.S.M. was supported by the Director, Office of Science, Office of Basic Energy Sciences, US Department of Energy under contract no. DE-AC03-76SF00098.
REFERENCES
- 1.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 2.van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P. and Tollervey,D. (2000) Musing on the structural organization of the exosome complex. Nature Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- 4.Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′→ 5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouwer R., Allmang,C., Raijmakers,R., van Aarssen,Y., Egberts,W.V., Petfalski,E., van Venrooij,W.J., Tollervey,D. and Pruijn,G.J. (2001) Three novel components of the human exosome. J. Biol. Chem., 276, 6177–6184. [DOI] [PubMed] [Google Scholar]
- 6.Estevez A., Kempf,T. and Clayton,C. (2001) The exosome of Trypanosoma brucei. EMBO J., 20, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin E.V., Wolf,Y.I. and Aravind,L. (2001) Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res., 11, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkard K.T. and Butler,J.S. (2000) A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol., 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs M.W., Burkard,K.T.D. and Butler,J.S. (1998) Rrp6, the yeast homologue of the human PM-Scl 100 kDa autoantigen, is essential for efficient 5.8S rRNA 3′-end formation. J. Biol. Chem., 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- 10.Brown J.T., Bai,X. and Johnson,A.W. (2000) The yeast antiviral proteins Ski2p, Ski3p and Ski8p exist as a complex in vivo. RNA, 6, 449–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs Anderson J.S. and Parker,R. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araki Y., Takahashi,S., Kobayashi,T., Kajiho,H., Hoshino,S. and Katada,T. (2001) Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J., 20, 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chekanova J.A., Shaw,R.J., Wills,M.A. and Belostotsky,D.A. (2000) Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem., 275, 33158–33166. [DOI] [PubMed] [Google Scholar]
- 14.Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Logemann J., Schell,J. and Willmitzer,L. (1987) Improved method of isolation of RNA from plant tissues. Anal. Biochem., 163, 16–20. [DOI] [PubMed] [Google Scholar]
- 16.Munroe D. and Jacobson,A. (1990) mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol., 10, 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn U. and Pieler,T. (1996) Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol., 256, 20–30. [DOI] [PubMed] [Google Scholar]
- 19.Worbs M., Bourenkov,G.P., Bartunik,H.D., Huber,R. and Wahl,M.C. (2001) An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell, 7, 1177–1189. [DOI] [PubMed] [Google Scholar]
- 20.Mah T.F., Kuznedelov,K., Mushegian,A., Severinov,K. and Greenblatt,J. (2000) The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev., 14, 2664–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavell A.C., Lydiate,D.J., Parkin,I.A., Dean,C. and Trick,M. (1998) Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome, 41, 62–69. [PubMed] [Google Scholar]