Abstract
The prediction system EpiSIX was used to study the COVID-19 epidemic in mainland China between November 2022 and January 2023, based on reported data from December 9, 2022, to January 30, 2023, released by The Chinese Center for Disease Control and Prevention on February 1, 2023. Three kinds of reported data were used for model fitting: the daily numbers of positive nucleic acid tests and deaths, and the daily number of hospital beds taken by COVID-19 patients. It was estimated that the overall infection rate was 87.54% and the overall case fatality rate was 0.078%–0.116% (median 0.100%). Assuming that a new COVID-19 epidemic outbreak would start in March or April of 2023, induced by a slightly more infectious mutant strain, we predicted a possible large rebound between September and October 2023, with a peak demand of between 800,000 and 900,000 inpatient beds. If no such new outbreak was induced by other variants, then the current COVID-19 epidemic course in mainland China would remain under control until the end of 2023. However, it is suggested that the necessary medical resources be prepared to manage possible COVID-19 epidemic emergencies in the near future, especially for the period between September and October 2023.
Keywords: COVID-19, Epidemic, Infection rate, Case fatality rate, Prediction
1. Introduction
On January 30, 2020, the World Health Organization (WHO) announced that the Coronavirus Disease 2019 (COVID-19) was a “Public Health Emergency of International Concern,” and declared it to be a pandemic on March 11, 2020.1 In China, there were several COVID-19 outbreaks before December 7, 2022,1 when the Chinese government issued the “New ten measures” for the prevention and control of COVID-19, following the “Twenty measures” released on November 11, 2022.1 With this policy adjustment, the course of the COVID-19 epidemic in China changed dramatically. On January 30, 2023, the WHO1 confirmed that the COVID-19 pandemic still constituted a public health emergency of international concern and remained a major global health threat.
Since November 2022, the COVID-19 epidemic in mainland China has largely been made up of Omicron variants (mainly strains BA.5.2 and BF.7).2 Two major questions remain. The first one concerns the overall infection rate, and the second one concerns the possible trend in the near future. To answer these questions, the prediction system EpiSIX was used to fit the reported data3 to a general susceptible–exposed–infected–recovered (SEIR) model,4, 5, 6 yielding estimations that the overall infection rate was 87.54%, and the overall case fatality rate was 0.078%–0.116% (median 0.100%).
Several studies7, 8, 9 have attempted to answer the first question. For example, the seroprevalence results to the ORF8 antigen of 1,500 samples from Guangzhou city7 indicated that the infection rate in Guangzhou ranged from 66.7% to 90.7%, and a model study8 using real-time mobility data suggested that the overall infection rate from November 1, 2022, to January 31, 2023, in Beijing would reach 92.3%. Nevertheless, the use of the proportion of individuals who participated in online polls from December 10–22, 2022, as the main indicator for the epidemic course8 is questionable. Finally, a model-based study9 projected an infection fatality rate of 619,549–987,455 from COVID-19 if the entire population of China were infected.
In the present study, concerning the first question stated above, the concept of the Gray Degree of Diagnosis5 was used to measure the gap between the number of cases reported and the number of cases that really existed. Moreover, a general model for estimating the total number of deaths from known inpatient deaths was introduced. Concerning the second question, our modeling revealed a possible optimistic prospect that the current COVID-19 epidemic course in mainland China would remain under control up to the end of 2023 if there were no new COVID-19 outbreaks induced by more transmissible variants. However, our further modeling studies suggested that necessary medical resources should be prepared to manage possible COVID-19 epidemic emergencies in the near future, especially for a possible new outbreak between September and October 2023.
The analysis and prediction method presented in this article could also be used for other countries with similar COVID-19 epidemic patterns.
2. Data sources and methods
2.1. Data sources
The relevant data of the COVID-19 epidemic in mainland China from December 9, 2022, to January 30, 2023, were used as the main data source. These data were released by The Chinese Center for Disease Control and Prevention (CCDC) on February 1, 20233.
2.2. Methods
All estimations were obtained by using EpiSIX to fit the actual data3 to a general SEIR model.4, 5
2.2.1. Estimating numbers of COVID-19 cases and hospitalizations
A recent study10 found that the half-life of the novel coronavirus antibody ranged from 105 to 270 days with a median of 195 days. When using EpiSIX to fit the data to the model, the median value (195 days) was taken as the decay half-time of the hybrid immunity produced by natural infection and vaccination. The daily reported number of cases “diagnosed” as positive with nucleic acid tests of the novel coronavirus infection and the daily reported number of hospitalizations (both from the abovementioned data source) were fitted to the model (see5 for more details) and yielded several estimations of theoretical rates, including the positive rate p of nucleic acid tests and the theoretical outpatient visit rate q of reported cases (persons with positive nucleic acid tests). Finally, the theoretical rate of hospital visits was derived as the product of p and q. The value 1 − p was defined as the Gray Degree of Diagnosis (GDD)5, which measures the errors between the number of cases reported and the theoretical number of cases that exist. One of the important features of EpiSIX was the embedding of GDD into the model settings. Note that the GDD must be considered even under the zero-COVID policy.6
2.2.2. Estimating the number of COVID-19 deaths: A probability model
The following probability model proposed for estimating the number of COVID-19 deaths is generally applicable to other infectious diseases. Let be the probability of hospitalization and be the positive rate of nucleic acid tests. Let and be the in-hospital and out-of-hospital case fatality rates, respectively. Let be the actual total number of COVID-19 infections. Then,
is the total number of deaths in and out of hospitals, respectively. It follows that
The probability of outpatient visits is related to the severity of symptoms, so it can be assumed that the probability of outpatient visits is close to the ratio . It can be assumed that has a 20% deviation around the standard value of 1.0, i.e., the value of varies from 0.80 to 1.20. Correspondingly, the total number of deaths can be calculated as
The value of ranges from to .
Based on the abovementioned CCDC data relating to COVID-19 deaths in hospitals, the range of the number of deaths in hospitals until March 31, 2023, was estimated.
2.2.3. Estimating main epidemiological parameters
The instant transmission rate (called also the effective reproductive number, Reff(t), which can be calculated by an integral4, 5), the latent and infectious periods, the generation time (GT), and GDD were estimated. Here, GT is estimated by the commonly used relation , where and r are the basic reproductive number and the growth rate, respectively.
3. Results
3.1. Overall infection rate
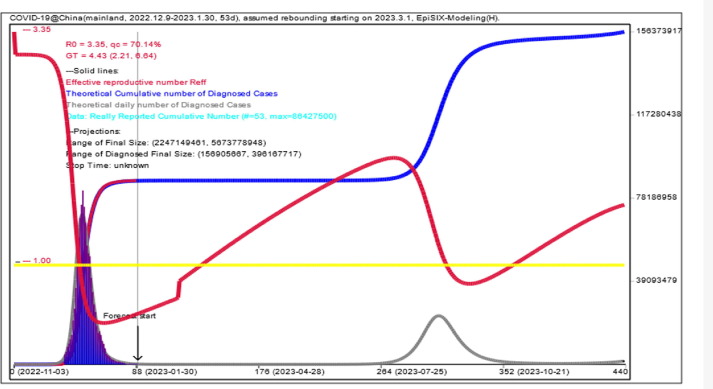
As of January 30, 2023, the theoretical total number of COVID-19 infections in mainland China starting from November 2022 was estimated to be 1.237 billion, yielding a theoretical overall infection rate of 87.54% given the total population of 1.413 billion in China. Moreover, it was predicted that the total number of infections would exceed the total population for the first time on August 21, 2023, and that the corresponding rebound peak of daily infections (27.8 million) would appear on September 5, 2023 (Fig. 1 and Table 1 ), if a new outbreak started in March 2023.
Fig. 1.
Modeling and prediction of the COVID-19 epidemic by EpiSIX from November 2022 in mainland China using data from Dec. 9, 2022, to Jan. 30, 2023 (53 days) released by CCDC. The red line is the instant transmission rate (i.e., effective reproductive number Reff). When its value is less than the threshold 1.0 (yellow line), the epidemic is “under control.” Otherwise, it is “out of control.” The dark blue column is the daily number of reported new cases, and the gray curve is its model fitting. The dark blue solid line is the model fit for the cumulative total number of cases. The abscissa is the date. Assuming that a new outbreak starts on Mar. 1, 2023, the simulation shows that the peak future rebound will appear around Sept. 5, 2023.
Table 1.
Modeling and prediction results of the COVID-19 epidemic in mainland China from Nov. 2022 to Dec. 31, 2023.
| Modeling results and dates | Theoretical total number of infections, unit: 100 million | Theoretical total number (daily number) of cases that should be reported. Unit for total number: 100 million; Unit for daily number: 1 million |
Theoretical total number of positive nucleic acid tests (total number of actual reported cases), unit: 10 thousand |
Theoretical number of inpatient beds required (actual value), unit: 1 k = 1 thousand |
Instant trans- mission rate Rt |
|---|---|---|---|---|---|
| 2022–12-07 | 1.20 | 0.48(10.18) | 42(/) | 3068(/) | 2.83 |
| 2022–12-09 | 1.85 | 0.77(16.04) | 84(40) | 7028(164 k) | 2.67 |
| 2022–12-22 | 9.94 | 7.59(67.85) | 3946(3985) | 470 k(520 k) | 0.79 |
| 2023–01-05 | 12.08 | 11.79(9.19) | 8212(8228) | 1626 k(1625 k) | 0.42 |
| 2023–01-30 | 12.37 | 12.36(0.24) | 8633(8642) | 156 k(144 k) | 0.51 |
| 2023–02-28 | 12.38 | 12.38(67.83) | 8644 | 18 k | 0.66 |
| 2023–03-31 | 12.38 | 12.38(13.36) | 8645 | 6588 | 1.13 |
| 2023–04-30 | 12.38 | 12.38(12.56) | 8645 | 808 | 1.42 |
| 2023–05-31 | 12.38 | 12.38(44.73) | 8646 | 169 | 1.68 |
| 2023–06-30 | 12.39 | 12.38(0.044) | 8649 | 562 | 1.91 |
| 2023–07-31 | 12.56 | 12.38(1.16) | 8715 | 11 k | 2.07 |
| 2023–08-31 | 16.68 | 12.54(25.57) | 10,876 | 340 k | 1.42 |
| 2023–09-30 | 21.23 | 16.94(6.28) | 14,681 | 700 k | 0.82 |
| 2023–10-31 | 21.82 | 21.72(0.92) | 15,220 | 180 k | 1.03 |
| 2023–11-30 | 21.99 | 22.20(0.50) | 15,349 | 44 k | 1.29 |
| 2023–12-31 | 22.21 | 22.18(0.92) | 15,486 | 28 k | 1.52 |
3.2. GDD and positive rate of nucleic acid tests
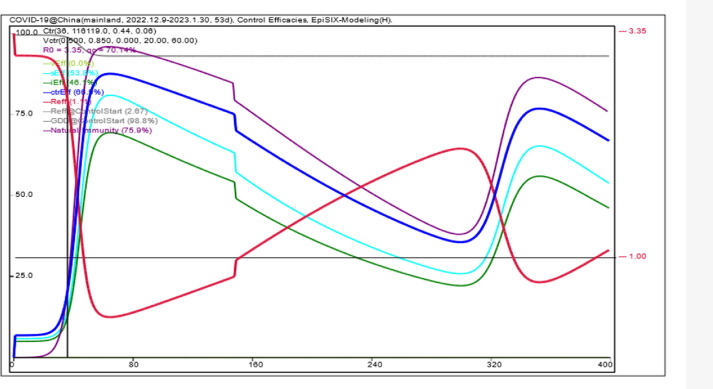
As of January 30, 2023, the reported cumulative number of positive nucleic acid tests was about 86.43 million. The corresponding final GDD was estimated to be approximately 93.0% (Fig. 4). The theoretical ratio of the number of positive nucleic acid tests to the actual number of infections finally converged to about 7.0%.
Fig. 4.
Illustration of control efficacies calculated by EpiSIX for the COVID-19 epidemic course from November 2022 to December 2023 in mainland China (with an assumed new outbreak starting from Mar. 1, 2023). The purple line is the overall control efficacy generated by natural immunity. No vaccination program is assumed to start, and thus the overall vaccine efficacy vEff remains at zero. sEff and iEff are efficacy against susceptibility and infectivity, generated by all control measures including NPIs, vaccination, and natural immunity. All except vEff oscillate according to the loss of antibody protection and the emergence of new outbreaks. The blue line ctrEff is the combined control efficacy of sEff and iEff. The red line is the instant transmission rate Rt (i.e., effective reproductive number Reff). Finally, the gray line is the GDD. For the epidemic course under study, the GDD drops from 100% to 93.0%.
3.3. Hospitalization parameters and rate of outpatient visits
After a positive nucleic acid test, critically ill patients were being transferred to ICU within an average of 13 days and receiving treatment for a maximum of 19 days (average 9.5 days), while moderate and mild patients were being admitted to hospital within an average of 14 days and receiving treatment for a maximum of 17 days (average 8.5 days). Among those with positive nucleic acid tests, the hospitalization rate was 6.873%, of which 6.39% were moderate and mild patients and 0.483% were critically ill patients (accounting for 7.03% of the total hospitalization rate); see Fig. 2 . The theoretical rate of outpatient visits (as the product of this hospitalization rate with the abovementioned positive rate of nucleic acid tests) was 0.481%.
Fig. 2.
Inpatient demand simulated by EpiSIX from Dec. 9, 2022, to Jan. 30, 2023. The blue histogram is the actual daily number of inpatient beds from Dec. 9, 2022, to Jan. 30, 2023, and the red dotted line is its fitting. Estimation results: 0.483% and 6.39% of patients (after testing positive) would be transferred to ICU and general wards, respectively. Assuming a new outbreak starting from Mar. 1, 2023, the simulation shows that the peak demand for inpatient beds will reach 818,000 around Sept. 20, 2023.
3.4. Demand for medical resources from possible new outbreaks
It was assumed that any new mutant strain (except for BA.5.2 and BF.7 strains) would produce a 10% enhancement in the immune escape and pathogenicity compared with previous hybrid immune antibodies, and the half-life of the antibody was shortened from the median of 195 days to the lower bound of 105 days. At the same time, the abovementioned positive rate of nucleic acid tests (7.0%) and hospitalization rate (6.383%) remained unchanged. If the start date was March 1, 2023, the demand for inpatient beds would continue to drop to about 20,000 at the end of February 2023, and a “low epidemic level” period (with a demand of less than 5,000 inpatient beds and less than 500 ICU beds) would continue from April 4 to July 23, 2023 (nearly 4.5 months). However, the demand for inpatient beds would rebound from mid-August 2023 to a certain extent, and would gradually rise from 366,000 on September 1, 2023, to a daily peak of 818,000 beds on September 20, 2023 (including 64,600 ICU beds), approximately 50% of the past peak of 1.625 million hospitalizations on January 5, 2023. The period for which more than 400,000 inpatient beds would be required (approximately 8.0% ICU beds) would appear between September 3 and October 15, 2023. See Fig. 1, Fig. 2, and Table 2 .
Table 2.
Demand for inpatient beds from possible new outbreaks.
| Date | The “low epidemic level” period (less than 5000) | The peak period (more than 400,000) | Peak |
|---|---|---|---|
| 1 March 2023 | 4 Apr.–23 Jul. 2023 (The number of ICU beds required is less than 500) |
3 Sept.-15 Oct. 2023 (The number of ICU beds accounts for about 8%) |
818,000 On 20 September 2023 |
| 1 April 2023 | 4 Apr.-21 Aug. 2023 (The number of ICU beds required is less than 500) |
18 Sept.-8 Nov. 2023 (The number of ICU beds accounts for about 8%) |
887,000 On 13 October 2023 |
Remark. The dates in Table 1 (Dec. 9, 2022, Jan. 30, 2023, Dec. 22, 2022, and Jan. 5, 2023) are the start and end dates of the data released by CCDC on February 1, 2023 [3], and the dates when the number of positive nucleic acid tests and hospitalizations peaked. The total number in Table 1 should be understood as the total number of person-times because some people might be infected multiple times, while others might not be infected at all.
If the start date was postponed to April 1, 2023, the corresponding “low epidemic level” period would extend from April 4 to August 21, 2023 (nearly 4.5 months). The period with more than 400,000 inpatient beds (8.0% ICU beds) would then last from September 28 to November 8, 2023 (the peak value of 887,000 inpatient beds would appear on October 13, 2023); cf. Table 2. For brevity, we have omitted the figures showing all simulation results.
3.5. Case fatality rate of COVID-19
Based on the existing number of deaths caused by COVID-19 infection in hospitals (a cumulative number of about 81,000 cases up to January 30, 2023), it was estimated that the total number of deaths in hospitals would reach 82,806–84,546 up to March 31, 2023 (Fig. 3 ). Combining this with estimated parameters of p = 6.873% and q = 7.0% (Fig. 1, Fig. 2), the method described in Section 2.2.2 yielded a total of 960,000–1,440,000 deaths from COVID-19 from November 2022 to March 31, 2023. The corresponding case fatality rate is 0.078%–0.116% with a median of 0.100%, slightly lower than the reported case fatality rates associated with known Omicron variant strains worldwide.1 Note that the corresponding cumulative number of positive nucleic acid tests was about 86.43 million, yielding an estimated case fatality rate of about 0.094%.
Fig. 3.
Modeling and prediction of in-hospital deaths by EpiSIX, using in-hospital death data from Dec. 9, 2022, to Jan. 30, 2023 (53 days) released by CCDC. Up to Jan. 30, 2023, the total number of in-hospital deaths was about 81,000. The predicted total number of in-hospital deaths ranges from 82,806–84,546 up to Mar. 31, 2023.
3.6. Latent period, infectious period, and GT
The mean latent period was estimated as 1.72 d (95%CI: 1.0–3.9 d), the mean infectious period was estimated as 3.98 d (95%CI: 1.6–11.6 d), and the GT was estimated to average 4.43 d, with a range of 2.2–6.6 d.
3.7. Instant transmission rate
It was estimated that the instant transmission rate had a value of 2.83 on December 7, 2022, 0.79 on December 22, 2022 (the peak of the past biggest wave), and 0.51 on January 30, 2023; see Fig. 1.
3.8. Remarks
Most of the above results had already been obtained in early February 2023. Although the data released by CCDC for the recent period between January 31, 2023, and February 23, 2023, were available, the estimation results and predictions using the data up to January 30, 2023,3 provided a very good match with the real development of the COVID-19 epidemic course under study. For this reason, we did not use the data from the more recent period.
4. Discussion
The prediction system EpiSIX has been improved constantly while our team has been actively working and preparing more than 400 reports (unpublished) on the study and forecasting of the COVID-19 epidemic. Our practical experience combined with published work (e.g.,11, 12) enabled us to determine the main epidemiological parameters of Omicron variants before running EpiSIX, allowing the reported data to be fitted to the model. Compared with other known prediction systems based on the classical SEIR model or its variants, the superiority of EpiSIX lies in the embedding of GDD into the model settings so that the gray degree of reported data can be estimated simultaneously. Note that EpiSIX can be applied to the most infectious epidemic courses worldwide, especially COVID-19.
CRediT authorship contribution statement
Yao Bai: Writing – original draft. Zhihang Peng: Conceptualization. Fengying Wei: Conceptualization. Zhen Jin: Conceptualization. Jinjie Wang: Conceptualization. Ximing Xu: Conceptualization. Xinyan Zhang: Conceptualization. Jun Xu: Conceptualization. Zixiong Ren: Conceptualization. Bulai Lu: Conceptualization. Zhaojun Wang: Conceptualization. Jianguo Xu: Conceptualization. Senzhong Huang: Writing – original draft, Methodology, Software.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by grants from a Consultancy Project of the Chinese Academy of Engineering (CAE, 2022-JB-06), Natural Science Foundation of Shaanxi Province, China (2023-JC-YB-676), Innovation Foundation of Medical Research Project of Xi’an City (2022YXYJ0040), and Natural Science Foundation of Fujian Province of China (2021 J01621). We thank Dr. Luo Huiming and two anonymous reviewers for their many valuable discussions.
References
- 1.National Heath Commission of the People’s Republic of China (https://www.nhc.gov.cn), World Heath Organization (https://www.who.int).
- 2.World Heath Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern[EB/OL]. [2021-12-01][2022-09-21]. https://www.who.int/news/item/ 26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- 3.National Novel Coronavirus Infection Epidemic Situation(in Chinese and English). https://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_13141/202302/t20230201_263576.html.
- 4.Huang S. A new SEIR model with applications to the theory of eradication and control of diseases, and to the calculation of R0. Math Biosci. 2008;215:84–104. doi: 10.1016/j.mbs.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Huang S., Peng Z., Jin Z. Studies of the strategies for controlling the COVID-19 epidemic in China: Estimation of control efficacy and suggestions for policy makers. Sci Sinica Math. 2020;50(6):885–898. doi: 10.1360/SSM-2020-0043. in Chinese. [DOI] [Google Scholar]
- 6.Huang S, Wei F, Peng Z, et al. Assessment method of coronavirus disease 2019 outbreaks under normal prevention and control. Disease Surveill. 2020; 35(08): 679-686 (in Chinese and English). DOI:10.3784/j.issn.1003-9961.2020.08.004.
- 7.Huang J, Zhao S, Chong KC, et al. Infection rate in Guangzhou after easing the zero-COVID policy: seroprevalence results to ORF8 antigen. Lancet infect Dis 2023, published online February 17, 2023. DOI:10.1016/s1473-3099(23)00112-3. [DOI] [PubMed]
- 8.Leung K, Lau EHY, Wong CKH, Leung GM, Wu JT. Estimating the transmission dynamics of SARS-CoV-2 omicron BF.7 in Beijing after adjustment of the zero-COVID policy in November-December 2022. Nat Med 2023; published online January 13, 2023. DOI:10.1038/s41591-023-02212-y. [DOI] [PubMed]
- 9.Ioannidis J., Zonta F., Levitt M. Estimate of COVID-19 deaths in Mainland China after abandoning zero COVID policy. Eur J Clin Invest. 2023 Jan;23:e13956. doi: 10.1111/eci.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bobrovitz N, Ware H, Ma X, et al. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect Dis, Published on January 18, 2023, DOI:10.1016/S1473-3099(22)00801-5. [DOI] [PMC free article] [PubMed]
- 11.Wu Y., Liu M. The incubation period of COVID-19 caused by different SARS-CoV-2 variants. Chinese. Gen Pract. 2022;25(11):1309–1313. doi: 10.12114/j.issn.1007-9572.2022.0078. in Chinese. [DOI] [Google Scholar]
- 12.Liao C.X., Wang B., Lyu J., Li L.M. Progress in research of etiology and epidemiology of 2019-nCoV Omicron variant. Chinese J Epidemiol. 2022;43(11):1691–1698. doi: 10.3760/cma.j.cn112338-20220929-00829. [DOI] [PubMed] [Google Scholar]