Abstract
Planthoppers in the family Cixiidae (Hemiptera: Auchenorrhyncha: Fulgoromorpha) harbor a diverse set of obligate bacterial endosymbionts that provision essential amino acids and vitamins that are missing from their plant-sap diet. “Candidatus Sulcia muelleri” and “Ca. Vidania fulgoroidea” have been associated with cixiid planthoppers since their origin within the Auchenorrhyncha, whereas “Ca. Purcelliella pentastirinorum” is a more recent endosymbiotic acquisition. Hawaiian cixiid planthoppers occupy diverse habitats including lava tube caves and shrubby surface landscapes, which offer different nutritional resources and environmental constraints. Genomic studies have focused on understanding the nutritional provisioning roles of cixiid endosymbionts more broadly, yet it is still unclear how selection pressures on endosymbiont genes might differ between cixiid host species inhabiting such diverse landscapes, or how variation in selection might impact symbiont evolution. In this study, we sequenced the genomes of Sulcia, Vidania, and Purcelliella isolated from both surface and cave-adapted planthopper hosts from the genus Oliarus. We found that nutritional biosynthesis genes were conserved in Sulcia and Vidania genomes in inter- and intra-host species comparisons. In contrast, Purcelliella genomes retain different essential nutritional biosynthesis genes between surface- and cave-adapted planthopper species. Finally, we see the variation in selection pressures on symbiont genes both within and between host species, suggesting that strong coevolution between host and endosymbiont is associated with different patterns of molecular evolution on a fine scale that may be associated with the host diet.
Keywords: troglobite, lava tube caves, genome evolution, Sulcia muelleri, Vidania fulgoroidea, Purcelliella pentastirinorum
Significance.
Cixiid planthoppers harbor three obligate endosymbionts (“Candidatus Sulcia muelleri,” “Ca. Vidania fulgoroidea,” and “Ca. Purcelliella pentastirinorum”) that contribute to essential nutrition provisioning. Current knowledge of cixiid endosymbionts is lacking information on how selection pressures on symbiont functional genes might vary between their planthopper host species and thus impact the functional roles of symbionts. In this study, we find that selection pressures on symbiont genes do vary between two Hawaiian cixiid planthopper host species adapted to surface versus cave environments as well as within an individual cave-adapted host species. We also see that functional gene retention varies among the four Purcelliella genomes sampled as well as between and within the two planthopper host species. These findings indicate that coevolution between cixiid host and their Purcelliella endosymbiont is associated with variation in selection on symbiont genes. This has resulted in variation in Purcelliella functional roles between the two cixiid host species studied.
Introduction
Many species within the Hemiptera insect order rely on obligate endosymbiotic bacteria to provision essential nutrients missing from their plant-sap diets (Buchner 1965). Obligate endosymbionts are vertically transmitted through the matriline (Moran et al. 1993, 2009; Wernegreen 2002). Due to the vertical transmission process, symbionts experience small effective population sizes and significant population bottlenecks (Funk et al. 2001), intensifying the effects of genetic drift and accumulation of deleterious mutations (Moran 1996; Rispe and Moran 2000), in addition to widespread gene loss (McCutcheon and Moran 2010; Bennett and Moran 2013; Moran and Bennett 2014; Chong et al. 2019). Despite rapid genome degradation, selection acts to retain genes related to essential functions and maintenance of the symbiosis, including nutritional biosynthesis for the host (McCutcheon and Moran 2007, 2012; Chong et al. 2019). Presumably, differences in the host environment and nutritional resources will cause variation in selection pressures on endosymbionts. As a result, symbiont genomes will experience differential gene loss and genomic divergence across symbiont strains. However, we do not fully understand how genomic variation arises and persists among closely related symbiont strains, particularly for those with tiny genomes.
Symbiont genome degradation typically happens in two phases (Moran and Mira 2001; Kinjo et al. 2021). In the beginning stages of the transition from a free-living bacterium to a host-restricted endosymbiont, bacterial genomes rapidly lose genes that are nonessential to the symbiotic partnership, as there is no selection pressure to maintain them. Nonessential genes that are often lost from symbiont genomes include genes necessary for adenosine triphosphate (ATP) synthesis, DNA replication and initiation, and cell wall components, among others (Toft and Andersson 2010; Clayton et al. 2012; McCutcheon and Moran 2012; Chong et al. 2019). In the later stages, symbionts continue to lose nonessential genes at a much slower pace, often conserving key biological functions and genomic synteny for long periods of evolutionary time (Latorre and Manzano-Marin 2017; McCutcheon et al. 2019). However, essential genes are still often lost from these tiny genomes even among closely related host species likely resulting from strong genetic drift (Chong et al. 2019; Vasquez and Bennett 2022). Gene functions that are lost can be compensated for by several processes such as horizontal gene transfer of host-encoded homologs (Hansen and Moran 2014; Sloan et al. 2014), the acquisition of new co-endosymbionts (McCutcheon et al. 2009; Sheffer et al. 2020; Dial et al. 2021), or senior symbiont replacement by junior endosymbionts (Clayton et al. 2012; Toenshoff et al. 2012; Koga and Moran 2014; Sudakaran et al. 2017; Chong and Moran 2018; Mao and Bennett 2020).
In the planthopper family Cixiidae (Hemiptera: Fulgoromorpha), hosts harbor three obligate endosymbionts, “Candidatus Sulcia muelleri,” “Ca. Vidania fulgoroidea,” and “Ca. Purcelliella pentastirinorum” (hereafter referred to as Sulcia, Vidania, and Purcelliella) that live in specialized host organs known as bacteriomes to provision the ten essential amino acids (EAAs), one non-EAA, and essential vitamins not readily available through planthoppers’ plant-sap diet (Munson et al. 1991; Moran et al. 2005; Bressan et al. 2009; Bressan and Mulligan 2013; Bennett and Mao 2018; Michalik et al. 2021). Sulcia was most likely acquired by a common ancestor of the suborder Auchenorrhyncha and has been coevolving with this group for >280 million years (Myr) (Moran et al. 2005; Bennett and Moran 2013). Vidania is predicted to have been an obligate partner in the Cixiidae planthoppers since at least their origin of the Fulgoroidea and possibly the Auchenorrhyncha (Koga et al. 2013; Bennett and Mao 2018). In contrast, Purcelliella exhibits genomic characteristics of a more recently acquired endosymbiont, including a comparatively larger genome size and higher GC nucleotide content (Bennett and Mao 2018; Michalik et al. 2021). While Sulcia, Vidania, and Purcelliella have all undergone differing degrees of genome reduction in the cixiid planthoppers, each endosymbiont still retains genes necessary for the biosynthesis of essential nutrients for the host (Bennett and Mao 2018; Michalik et al. 2021). Here, we seek to understand how variation in selection pressures on symbiont genomes might correlate with variation in functional gene retention within symbiont strains that are associated with multiple host species inhabiting dramatically different ecological niches.
To address this question, we isolated and comparatively analyzed multiple Sulcia, Vidania, and Purcelliella genomes from two planthopper host species (Cixiidae: Oliarus polyphemus and Oliarus filicicola) collected from distinct habitats on Hawaiʻi Island, Hawaiʻi, USA. Hawaiian cixiids inhabit a range of unique environmental niches throughout the archipelago (Asche 1997; Hoch and Howarth 1999; Hoch 2006). For example, Oliarus polyphemus is a fully troglobitic (cave-adapted) planthopper species that resides within a network of lava tube caves across Hawaiʻi Island (Howarth 1986; Hoch and Howarth 1993, 1999; Wessel et al. 2013). It is hypothesized that O. polyphemus transitioned from an epigean lifestyle to an obligately subterranean one in order to exploit plant-sap (phloem) from the roots of native Hawaiian ʻŌhiʻa lehua trees (Myrtaceae family) that penetrate the cave ceilings (Howarth et al. 2007). Presumably, their association with nutritional endosymbionts played an essential role in the ability of O. polyphemus to colonize a novel habitat with limited nutritional resources. ʻŌhiʻa tree roots are the predominant source of plant-sap underground, leaving O. polyphemus restricted to a single source of nutrition. Conversely, Oliarus filicicola are epigean (surface-adapted) planthoppers that can feed on a variety of host plants including native tree ferns (Cibotiaceae family). The amino acid content of phloem can vary between host plants and has been shown to impact host health and performance (Sandstrom and Pettersson 1994; Karley et al. 2002), which could influence selection pressures on endosymbiont nutritional provisioning genes. In order to persist in nutrient-limited cave environments, endosymbionts of troglobitic species may need to retain a suite of nutritional provisioning genes that would not be required by surface-dwelling species. For this reason, we predict to see increased positive selection pressure on nutritional provisioning genes and also higher retention of nutritional biosynthesis pathways in cave endosymbiont genomes. We also hypothesize that homogenized gene pools within host species and strong host–symbiont coevolution will result in similarities in the rates of molecular evolution across the symbiont genomes within hosts of the same species. However, between species, we expect to see distinct rates of gene loss and rates of molecular evolution due to ongoing genetic drift, which can be disentangled from selection operating on ecologically important traits (e.g., nutrition synthesis).
To test our hypotheses, Oliarus polyphemus samples were collected from different elevations within the same lava flow in the Kaʻu district of Hawaiʻi Island, and Oliarus filicicola samples were collected from tree ferns found in the Puna district of Hawaiʻi Island. Planthoppers have evolved through patterns of migration and diversification across the island chain; however, both O. polyphemus and O. filicicola are singularly endemic to Hawaiʻi Island. As a fully cave-adapted species, O. polyphemus likely diverged from a surface ancestor following colonization of Hawai‘i Island, since there is a low probability that a fully cave-adapted species would have the ability to migrate across the surface to other islands. As a single island endemic, we also presume that O. filicicola diverged from a surface ancestor following the colonization of cixiid planthoppers on Hawaiʻi Island. Therefore, while there is no formal estimate, both species have likely speciated from Hawaiʻi Island surface ancestors, which would constrain the time of divergence between the two species to the age of Hawaiʻi Island, which is 500,000 years old (Price and Clague 2002).
Based on symbiont genome comparisons between and within hosts, our results show that the different bacterial endosymbiont species exhibit dramatic differences in their patterns of gene retention, rates of molecular evolution, and levels of selection pressure on genes. While Sulcia and Vidania show highly conserved nutritional roles, the tertiary symbiont, Purcelliella, varies in its ability to synthesize vitamins between surface and cave hosts. These results suggest that variation in host resource availability and differential selection pressures on endosymbiont functional roles have ultimately resulted in variation in symbiont genome evolution.
Results and Discussion
Host Cytochrome Oxidase 1 (CO1) Barcoding and Phylogeny
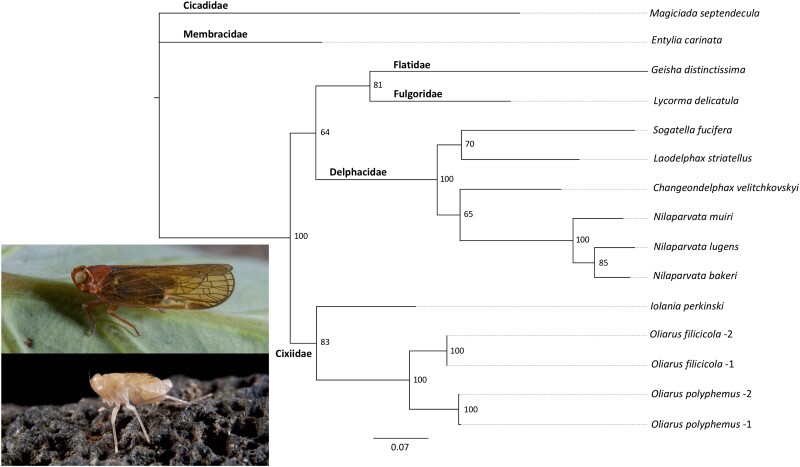
To determine phylogenetic relationships between planthopper host samples, we used a maximum likelihood estimation to reconstruct the host phylogeny based on the mitochondrial gene cytochrome oxidase 1 (CO1). Oliarus filicicola planthopper hosts will hereinafter be refered to as OFIL(1 or 2), and Oliarus polyphemus hosts will be refered to as OPOL(1or 2). Endosymbiont genomes isolated from either O. filicicola host will be referred to as Sulcia-OFIL-(1 or 2), Vidania-OFIL-(1 or 2), and Purcelliella-OFIL-(1 or 2). Endosymbionts isolated from either O. polyphemus host will be referred to as Sulcia-OPOL-(1 or 2), Vidania-OPOL-(1 or 2), and Purcelliella-OPOL-(1 or 2). CO1 gene sequences of both OPOL-1 & OPOL-2 and OFIL-1 & OFIL-2 samples were searched against the NCBI BLASTn database to confirm species identity. The host phylogeny was reconstructed using the complete mitochondrial CO1 sequences of four Oliarus samples (OPOL-1 & OPOL-2 and OFIL-1 & OFIL-2) in addition to the CO1 sequences of other sap-feeding hosts within the Auchenorrhyncha (supplementary table S1, Supplementary Material online). The resulting phylogeny has strong bootstrap support for all nodes (fig. 1) and confirms that O. polyphemus and O. filicicola are closely related species. These results suggest that O. polyphemus and O. filicicola are likely to be independently coevolving with their symbionts, which we predict will result in variation in genome evolution between the respective endosymbionts (Bennett and Moran 2015; Chong and Moran 2016).
Fig. 1.
Host phylogeny based on a maximum likelihood analysis of 13 complete mitochondrial CO1 gene sequences using a translational alignment (supplementary table S1, Supplementary Material online). Tree was rooted using Magiciada septendecula as an outgroup. Numbers at nodes represent bootstrap support values with 1,000 replicates. Inset image of surface species Oliarus filicicola (top) and cave-adapted species Oliarus polyphemus (bottom) adults collected from Hawai’i Island. Photo credit: Michael E. Slay.
Symbiont Genometrics
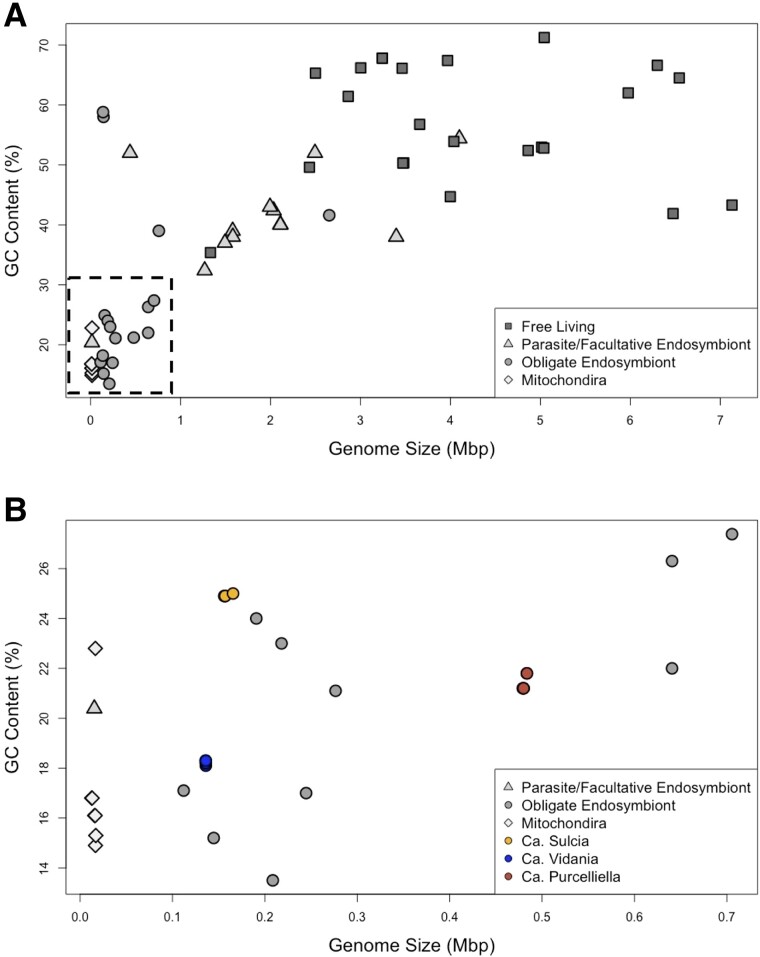
The genomes for Sulcia, Vidania, and Purcelliella were all highly reduced and had low GC content (<25%) compared with free living and facultative bacterial strains (McCutcheon et al. 2019; Perreau and Moran 2022) (fig. 2). Sulcia (156 kb average length) and Vidania (136 kb average length) retain the smallest genomes of the three partners (table 1). As the most recently acquired endosymbiont, Purcelliella has the largest genome (480 kb average length) and the most genes, 445–461 coding sequences (CDS) out of the three partners (table 1). The majority tBLASTx searches on host assemblies revealed a single contig for each endosymbiont genome. With the exceptions being Sulcia genomes from OPOL-1 and OFIL-2 (2 and 4 contigs, respectively) and the Purcelliella genome from OFIL-2 (8 contigs). All contigs for each endosymbiont were circularized into complete genomes (table 1).
Fig. 2.
(A) Relationship between genome sizes (Mbp) and GC content (%) for 72 complete bacterial and mitochondrial genomes, including free-living bacteria, parasites, facultative endosymbionts, and obligate endosymbionts (listed in supplementary table S2, Supplementary Material online) (dashed line box represents the subset of fig. 2A i.e. displayed in fig. 2B). (B) Subset of figure 2A, comparing genome sizes (Mbp) and GC content (%) of the Oliarus filicicola (OFIL-1 and OFIL-2) and Oliarus polyphemus (OPOL-1, OPOL-2) endosymbionts, Ca. Sulcia, Ca. Vidania, and Ca. Purcelliella from this analysis. Oliarus endosymbionts (Sulcia, Vidania, and Purcelliella) are depicted in yellow, blue, and red, respectively.
Table 1.
Genome sizes and features for three endosymbionts, Candidatus Sulcia muelleri, Candidatus Vidania fulgoroidea, and Candidatus Purcelliella pentastirinorum, isolated from two species of cixiid host, epigean Oliarus filicicola (OFIL), and cave-adapted Oliarus polyphemus (OPOL)
| Symbiont | Host ID | Genome Size (bp) | GC (%) | CDS | tRNAs | rRNAs | Coding Density | Coverage | # Contigs |
|---|---|---|---|---|---|---|---|---|---|
| Ca. Sulcia | OFIL-1 | 155,965 | 24.9 | 155 | 29 | 3 | 90.1 | 30X | 1 |
| OFIL-2 | 156,628 | 24.9 | 155 | 29 | 3 | 89.3 | 30X | 4 | |
| OPOL-1 | 157,002 | 25.0 | 155 | 29 | 3 | 90.3 | 13X | 2 | |
| OPOL-2 | 156,525 | 24.9 | 154 | 29 | 3 | 90.2 | 113X | 1 | |
| Ca. Vidania | OFIL-1 | 136,076 | 18.2 | 150 | 30 | 2 | 90.5 | 17X | 1 |
| OFIL-2 | 136,065 | 18.1 | 149 | 30 | 2 | 91.0 | 60X | 1 | |
| OPOL-1 | 135,790 | 18.2 | 150 | 29 | 2 | 89.3 | 60X | 1 | |
| OPOL-2 | 136,060 | 18.3 | 148 | 29 | 2 | 90.2 | 346X | 1 | |
| Ca. Purcelliella | OFIL-1 | 479,076 | 21.2 | 445 | 32 | 3 | 86.6 | 25X | 1 |
| OFIL-2 | 479,868 | 21.2 | 448 | 32 | 3 | 86.7 | 21X | 8 | |
| OPOL-1 | 483,618 | 21.8 | 461 | 31 | 3 | 87.3 | 17X | 1 | |
| OPOL-2 | 483,447 | 21.8 | 445 | 31 | 3 | 87.6 | 156X | 1 |
Based on local tBLASTx searches of host assemblies against the 16S Ribosomal RNA BLAST database, we identified multiple fragments (20 contigs, 6,000–30,000 bp, 10–20 × coverage) within the OPOL-2 assembly matching several “Candidatus Wolbachia sp.” genome sequences from a variety of hosts, including several species of wasp, fruit fly, and psyllid (86–96% identities, 90–100% query cover). Wolbachia is one of the most widespread endosymbionts commonly found in arthropod and nematode species worldwide, and often impacts host reproductive functions such as male-killing and parthenogenesis (Jeyaprakash and Hoy 2000; Jiggins et al. 2001; Werren et al. 2008). Wolbachia infections have been documented in native Hawaiian insects such as Drosophila and Nesophrosyne (leafhoppers) (Dobson et al. 1999; Bennett et al. 2012). The Wolbachia genome in OPOL-2 was highly fragmented (6,000–30,000 bp per fragment), with overall low coverage (10–20×) throughout the host assembly and relatively low percent identities to many of the sequence hits, suggesting that although at least one native Hawaiian planthopper may harbor Wolbachia, it is not an obligate endosymbiont of this individual.
Genomic Comparisons Reveal Variation in Symbiont Gene Retention
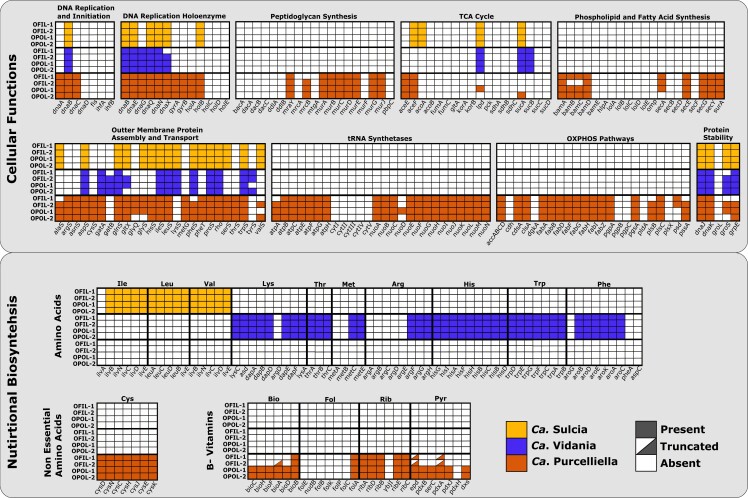
To quantify individual symbiont nutritional provisioning capabilities and functional roles, we annotated complete symbiont genomes and recorded genes retained or lost in each nutritional biosynthesis pathway. From all host samples, we found that genomic synteny was retained between symbiont genomes of the same strain (supplementary fig. S1, Supplementary Material online). Complete genome annotations for each endosymbiont revealed that Sulcia and Vidania retain combined biosynthesis capabilities to provision all ten EAAs and have lost all other nutritional provisioning genes (fig. 3). Similarly, Purcelliella has lost genes necessary for provisioning amino acids and vitamins other than four B vitamins and the non-EAA cysteine (fig. 3). Additionally, all Sulcia and Vidania genomes have lost a majority of the genes necessary for basic bacterial cellular function, including all of the genes necessary for oxidative phosphorylation, peptidoglycan synthesis, phospholipid and fatty acid synthesis, and outer membrane protein assembly and transport (fig. 3). Many of the genes necessary for DNA replication initiation (e.g., dnaACD, fis, and ifhAB) have also been lost from these genomes. A loss of DNA replication genes is associated with enhanced genome degradation and may be contributing to functional gene loss in Sulcia and Vidania (Sloan and Moran 2012).
Fig. 3.
Variation in gene retention for nutritional biosynthesis and bacterial cellular function pathways for Ca. Sulcia, Ca. Vidania, and Ca. Purcelliella, isolated from Oliarus filicicola (OFIL-1, OFIL-2) and Oliarus polyphemus (OPOL-1, OPOL-2) hosts. Essential amino acid biosynthesis pathways include isoleucine (Ile), leucine (Leu), valine (Val), lysine (Lys), threonine (Thr), methionine (Met), arginine (Arg), histidine (his), tryptophan (Trp), phenylalanine (Phe), and non-EAA cysteine (Cys). B-vitamin pathways include biotin (Bio), folate (Fol), riboflavin (Rib), and pyridoxine (Pyr). Each square represents a single gene. Colored squares represent genes that are retained, whereas white squares represent genes that are absent. Half-colored squares represent genes that are truncated.
Purcelliella genomes exhibit a lesser degree of gene loss relative to Sulcia and Vidania genomes. The Purcelliella genomes have lost a number of outer membrane protein assembly and transport genes (e.g., bamE, hlpA, lolABCDE, omp, and secBDF). Purcelliella genes that are involved in oxidative phosphorylation, peptidoglycan synthesis, and phospholipid and fatty acid synthesis have also been lost. However, the degree of loss of genes responsible for cellular functions in Purcelliella genomes is much less extensive than the degree of gene loss in Sulcia and Vidania genomes. For example, while Sulcia and Vidania have lost a majority of the genes necessary for DNA replication and DNA replication holoenzyme, Purcelliella retains all genes in this category except for dnaD, fis, ifhAB, and holCDE. A higher level of gene retention in Purcelliella is expected as it is the most recently acquired endosymbiont and it is likely in the earlier stages of genome degradation (Urban and Cryan 2012).
“Ca. Sulcia muelleri” Retains Minimal Functional Capabilities
Most Sulcia genomes in the Auchenorrhynchan families other than Cixiidae have expanded nutritional provisioning roles (7–8 EAAs), but all Sulcia retain the pathways for the branched-chain amino acids—leucine, valine, and isoleucine—regardless of the host or the capabilities of their partner endosymbionts (McCutcheon and Moran 2007, 2010; McCutcheon et al. 2009; Bennett and Moran 2013). Additionally, Sulcia's reduced functional role of synthesizing only the branhced-chain amino acids in planthoppers is extremely conserved across all four Oliarus hosts specimens (fig. 3), suggesting that the provisioning of these EAAs might be the minimal essential functional role of Sulcia in sap-feeding Auchenorrhyncha. While the biosynthesis pathways for leucine and valine are complete in all Sulcia genomes, the first step in the isoleucine pathway, threonine dehydratase (ilvA) is absent in all four Oliarus Sulcia genomes sampled. Most endosymbionts of sap-feeding insects have also lost ilvA, which converts threonine to alpha-ketobutyrate and ammonia, suggesting that it could be complemented by a host-encoded homolog (Hansen and Moran 2014; Sloan et al. 2014). We performed a tBLASTx search on host assemblies for an ilvA homolog and did not find evidence that ilvA is retained by any of the host genomes, nor in Vidania or Purcelliella isolated from any of the hosts. This result suggests that this precursor metabolite must either be compensated by existing planthoppers’ metabolism or from some other exogenous source like the insect's food.
“Ca. Vidania fulgoroidea” Retains Expanded Functional Capabilities
All Vidania genomes retain a majority of the genes necessary to synthesize the remaining seven EAAs (fig. 3). Complete biosynthesis pathways for threonine, histidine, and tryptophan are retained by each of the four Vidania genomes in this study. The preliminary steps of the methionine biosynthesis pathway, homoserine O-succinyltransferase (metA) and O-succinylhomoserine lyase (metB), have been lost in each Vidania genome. Losses of metA and metB have been reported in other symbiont strains, including the obligate primary endosymbiont Buchnera in pea aphids (McCutcheon and von Dohlen 2011; Chong et al. 2019). The symbiont Buchnera from pea aphids is known to compensate for this loss by utilizing metE, the terminal gene in the methionine biosynthesis pathway, to produce homocysteine from exogenous cystathionine provided by the host (Russell et al. 2013). The metE gene is indeed retained by all Vidania strains sequenced from Oliarus so far, so it is possible that the loss of metA and metB in the methionine pathway is compensated for by a similar mechanism.
Within the Vidania phenylalanine biosynthesis pathway, the preliminary gene phospho-2-dehydro-3-deoxyheptonate aldolase (aroG) and the two terminal genes, aspartate aminotransferase (aspC) and prephenate dehydratase (pheA), have each been lost from all genomes. Transcriptome data from the pea aphid-Buchnera symbiosis have shown that an insect–genome-encoded aspartate aminotransferase gene (EC 2.6.11) is upregulated in host bacteriomes and is likely able to complement the incomplete phenylalanine biosynthesis pathway in Buchnera (Hansen and Moran 2011). Since phenylalanine biosynthesis pathways are often incomplete in symbionts of phloem feeders (Hansen and Moran 2014; Sloan et al. 2014; Mao et al. 2018), it is also likely that planthopper host genes complement this incomplete pathway. Additionally, the first five genes in the arginine biosynthesis pathway, argABCDE, are lost in all of the Vidania genomes. Largely incomplete biosynthesis pathways, such as this one, can be complemented by other obligate symbiont partners (Dial et al. 2021). However, while all Purcelliella genomes do indeed retain argE, we did not find evidence that any of the Purcelliella or Sulcia genomes complement the remainder of the incomplete arginine pathway. If planthoppers receive an adequate source of arginine in their diet, relaxed selection on this pathway in Vidania genomes could result in gene loss. Despite its functional gene losses, Vidania has conserved nutritional biosynthesis pathways across Oliarus host species, suggesting that, despite variation in their ecology and resource limitations, both hosts require these essential functions from Vidania to survive.
In most phloem-feeding hosts, co-symbionts (Nasuia, Zinderia, and Baumannia) complement Sulcia's expanded provisioning role of 7–8 EAAs by provisioning the remaining 2–3 amino acids (Wu et al. 2006; McCutcheon and Moran 2010; Bennett and Moran 2013). In accordance with their reduced nutritional provisioning roles, co-symbiont genomes are typically smaller and have experienced more genome degradation than Sulcia genomes. In Oliarus, despite a more robust functional role than Sulcia, Vidania has a smaller genome and lower GC content overall, which is consistent with previous results (Bennett and Mao 2018). These genomic differences are likely a result of relaxed selection on Sulcia related to the acquisition of Vidania with a robust set of nutritional provisioning genes (Bennett and Mao 2018; Vasquez and Bennett 2022).
“Ca. Purcelliella pentastirinorum” Functional Capabilities Differ
All Purcelliella genomes retain at least a portion of the genes necessary to synthesize four B vitamins, including biotin, folate, riboflavin, pyridoxine, and a semi-essential amino acid, cysteine (fig. 3). The cysteine biosynthesis pathway is complete in all four Purcelliella genomes. Vidania requires cysteine in addition to sulfide in order to complete methionine biosynthesis. The close proximity between the bacteriomes harboring both Purcelliella and Vidania might aid in the provisioning of methionine through direct metabolite transfer (Bressan et al. 2009; Bressan and Mulligan 2013; Bennett and Mao 2018).
The abilities of Purcelliella to synthesize B vitamins are more complicated. The folate biosynthesis pathway is missing all of the necessary genes aside from the final catabolic step, folA, in all genomes. Both Purcelliella-OPOL genomes retain complete biotin biosynthesis pathways. However, Purcelliella-OFIL-1 retains a single gene for biotin biosynthesis, bioB, and Purcelliella-OFIL-2 retains bioB and a truncated bioA gene (213 bp, 16% of predicted length), which we predict might render it nonfunctional and will be eventually lost from the genome. Purcelliella genomes from the four Oliarus retain all of the genes necessary to complete riboflavin biosynthesis except ybjI, which is consistent with riboflavin pathway gene retention in endosymbionts of other sap-feeding hosts (Manzano-Marin et al. 2016; Renoz et al. 2022). The highly conserved riboflavin pathway suggests that neither OPOL nor OFIL species are able to obtain riboflavin from their respective diet sources and thus depend on Purcelliella to provision it for them.
For pyridoxine biosynthesis, both Purcelliella-OPOL genomes have complete pathways with the exception of pdxH. Both Purcelliella-OFIL genomes are missing pdxB, serC, pdxJ, pdxH, and dxs from the pyridoxine pathway. The two genes remaining in the pathway, epd (216 bp, 21% of predicted length) and pdxA (286 bp, 29% of predicted length), are truncated in both genomes, again potentially indicating that these genes are nonfunctional and in the process of being lost from its genome; however, transcriptome and proteome data from endosymbiont genomes would be required to confirm gene expression and functionality in truncated genes. Neither Sulcia-OFIL nor Vidania-OFIL retain these genes, so the source of metabolites for the host may be derived from the host's plant-sap diet. Selection has maintained a majority of the pyridoxine biosynthesis pathway in Purcelliella-OPOL, suggesting that cave-adapted planthoppers may not receive pyridoxine from ʻŌhiʻa tree roots, thus intensifying selection on these endosymbiont genes to be maintained. None of the B-vitamin pathways are complemented by any of the Sulcia or Vidania genomes, so it is unclear if or how Purcelliella-OFIL can provision these vitamins for their host, or even if the host requires them. In some cases, hosts may obtain B vitamins from their diets and, therefore, these genes may be lost (McCutcheon et al. 2009; Douglas 2017).
Purcelliella is not found throughout the rest of the Fulgoroidea and is probably a recent acquisition by an ancestor of the cixiid planthoppers (Bennett and Mao 2018). It is, however, thought to be widely shared among the species in the family (Buchner 1965; Urban and Cryan 2012). For this reason, we expected functional genes to be conserved across cixiid host lineages, especially for closely related species such as O. polyphemus and O. filicicola. Instead, we find that Purcelliella-OFIL have lost genes in both the biotin and pyridoxine pathways that are maintained in Purcelliella-OPOL genomes, suggesting that significant genomic differences in nutritional biosynthesis capabilities of endosymbionts can occur relatively rapidly. Many functional symbiont genes, regardless of being maintained for millions of years, are subject to being purged from their genomes by drift and accumulated deleterious mutations. Selection to avoid dependence on genes that have lost their functions as a result of these processes might result in the host acquiring a new pathway to obtaining the nutrient, such as a new endosymbiotic partner or through its environment (Douglas 2017). Epigean planthoppers feed on a wider variety of host plants than do cave-adapted planthoppers. We predict that this breadth of nutritional resources may provide O. filicicola with B vitamins, allowing selection for maintaining these genes to become relaxed. Although the evolutionary history of Oliarus has yet to be resolved, both O. filicicola and O. polyphemus are endemic to Hawaiʻi Island, with the latter likely to have diverged from a surface ancestor within the last 0.5 Myr. In the case of Purcelliella, our results show evidence of strong coevolution resulting in symbiont functional variation between closely related host species on a relatively short evolutionary time scale.
Selection Analyses Reveal Inter- and Intra-host Variation
To estimate rates of molecular evolution and selection dynamics between symbiont genes, we performed pairwise selection analyses between orthologous coding sequences within and across O. polyphemus and O. filicicola host species. Overall, our results show that Sulcia, Vidania, and Purcelliella all exhibit variation in patterns of molecular evolution within and between host species, with individual genes undergoing both positive and purifying selection. We performed a Fisher's exact test in each genomic comparison to test if any Clusters of Orthologous Genes (COG) category was significantly associated with an increase in positive or purifying selection. Results show that no particular COG category is more likely to be under either form of selection (P >> 0.05).
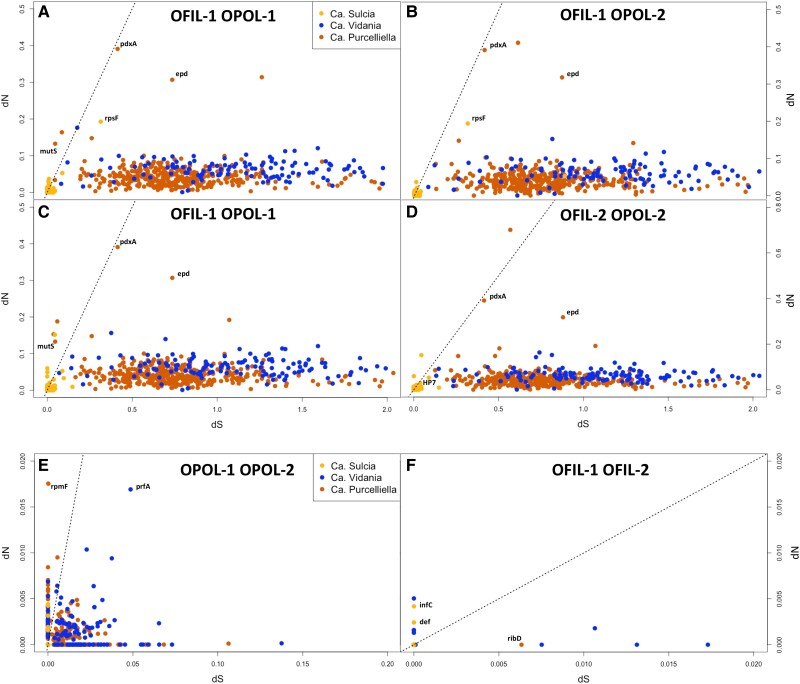
Sulcia genomes have the lowest substitution rates of the three planthopper endosymbionts, indicating highly conserved rates of molecular evolution between and within host species. Within a host species, Sulcia exhibits extremely low rates of molecular evolution. Between the surface hosts, it exhibits no substitutions with the exception of two genes: translation initiation factor IF-3 (infC; dN = 0.0042, dS = 0.0) and peptide deformylase (def; dN = 0.0024, dS = 0.0; fig. 4F). Both of these genes have slightly elevated dN values and, therefore, are under weak positive selection (dN > dS). Sulcia genome comparisons between cave-adapted hosts show six genes that are under positive selection with increased rates of dN, five ribosomal protein genes (rpsACFLN) and an ATP-dependent transporter gene (sufC) (fig. 4E). Between different host species, most Sulcia genes have similarly low, yet nonzero rates of molecular evolution. Only one gene, a ribosomal protein (rpsF), is under strong purifying selection (fig. 4A).
Fig. 4.
(A)–(D) Pairwise intraspecific comparisons of symbiont orthologs showing rates of synonymous (dS) and nonsynonymous (dN) substitutions between host species. Points represent individual orthologous genes from each symbiont comparison. The dashed line represents the point at which dN = dS, or ω = 1. Genes below the line are considered to be under purifying selection (dS > dN), and genes above the line are considered to be under positive selection (dN > dS). (E) and (F) Pairwise interspecific comparisons of symbiont orthologs.
Many studies have shown genomic stability in terms of gene retention and rates of molecular evolution in Sulcia associated with other Auchenorrhyncha hosts (McCutcheon and Moran 2007, 2010; McCutcheon et al. 2009; Bennett and Moran 2013), which has been hypothesized to be linked to its expanded nutritional provisioning role of 7–8 EAAs. Here, we find that, despite Sulcia's more limited functional role in cixiid planthoppers, it still retains depressed rates of molecular evolution between host species. This retained pattern suggests that some feature in Sulcia's DNA replication or repair mechanisms that lead to suppression of substitutions evolved early and has been retained in most, if not all, of its descendant lineages (Waneka et al. 2021). This pattern also indicates that genomic stability in Sulcia may not be directly linked to nutritional provisioning roles as previously predicted.
In contrast, Vidania genomes show higher genome-wide substitution rates than Sulcia between surface and cave-adapted hosts. In comparisons between surface hosts, Vidania exhibit relatively low rates of molecular evolution, with only a few genes experiencing substitution rates greater than zero (fig. 4F). In comparisons between cave-adapted hosts, a majority of the Vidania genes are under weak purifying selection (dS > dN) and a small number of genes are under weak positive selection (dN > dS) with an extremely low rate of nonsynonymous substitutions (dN < 0.0075) (fig. 4E). Vidania has the largest functional role of all of the cixiid endosymbionts, retaining partially complete biosynthesis pathways for 7 out of 10 EAAs. Since its genome is tiny, a large proportion of its genome is dedicated to nutritional provisioning for the hosts. Thus, we see that between host species, increased substitution rates and increased purifying selection in Vidania indicate increased molecular evolution in Vidania, with most genes being selected to be maintained in the genome. It is possible that genetic divergence between O. polyphemus hosts could be associated with the observed increase in rates of molecular evolution between Vidania from cave-adapted planthoppers.
Similar to Vidania, in comparisons between surface and cave host species, most genes in Purcelliella's genome are under purifying selection. However, a few genes are under positive selection. In all pairwise comparisons between surface and cave hosts, pyridoxine biosynthesis genes (epd and pdxA) have increased dN and dS (fig. 4A–D). These genes are retained by both Purcelliella isolated from cave hosts but are truncated and nonfunctional in both Purcelliella isolated from surface hosts. The pyridoxine pathway in Purcelliella-OFIL-1 and Purcelliella-OFIL-2 is extremely degraded. Potentially, surface planthoppers obtain pyridoxine from their environment and increased substitution rates on the retained genes will eventually lead to the pathway being lost all together in Purcelliella-OFIL. Additionally, in both interspecies comparisons involving Purcelliella-OPOL-1, the DNA mismatch repair protein, mutS, is under positive selection (fig. 4A and C). Bacterial strains that have lost mutS have been shown to have a 50-fold higher mutation rate than bacteria that retain mutS (Nilsson et al. 2005). If positive selection between OPOL- and OFIL-Purcelliella leads to an eventual loss of mutS, the mutation rate may accelerate in Purcelliella, leading to increased gene loss and causing the endosymbiont to enter the terminal stages of endosymbiosis.
Within host species comparisons, Purcelliella exhibits contrasting patterns of molecular evolution. Genes compared between Purcelliella from surface hosts have no substitutions between them with the exception of ribD, an essential gene in the riboflavin biosynthesis pathway that is under weak purifying selection (fig. 4F). The surface hosts studied—O. filicicola from the Puna district—are likely to have larger effective population sizes and be more genetically homogeneous because they are not limited by the physical boundaries such as lava tube caves that isolate cave-host populations. In symbionts of surface species, larger effective population sizes may have reduced the number of substitutions among them because symbiont genomes of lower fitness will not have been preserved by genetic drift in small isolated populations.
Within cave hosts, however, Purcelliella, genes have increased rates of substitutions, with many genes under purifying selection and several genes under positive selection (fig. 4E). This contradicts the expectation that symbionts isolated from the same host species should retain low rates of molecular evolution between them. Previous work has shown that there is genetic and phenotypic differentiation between O. polyphemus populations across the island (Hoch and Howarth 1993; Wessel et al. 2013). Oliarus polyphemus planthoppers live within fragmented lava tube cave systems, congregating in areas where ʻŌhiʻa roots project into the cave, creating a suitable habitat for planthoppers to live and feed. Root patches are not continuous throughout the caves, and as a result, planthopper aggregations may become isolated from one another based on the fragmentation of their essential diet source in addition to the natural fragmentation of subterranean lava tubes. Oliarus polyphemus samples were collected from different caves within the same lava flow system giving rise to our hypothesis that host-level reproductive isolation and genetic variation could be associated with differences in rates of molecular evolution between Purcelliella as well as Vidania isolated from cave-adapted planthoppers.
Conclusion
Cixiid planthoppers harbor three obligate endosymbionts, each with distinct functional roles. On Hawaiʻi Island, planthoppers inhabit diverse ecological niches on a geologically young island that is approximately 500,000 years old (Price and Clague 2002). Several planthopper species are cave-adapted having split from their surface ancestors at an unknown point within this time frame. This feature allows us to compare symbiont genomes in hosts that have evolved over a relatively short evolutionary time scale. Comparing symbiont strains across surface and cave-adapted planthopper hosts further enabled us to test for functional gene losses and rates of molecular evolution between symbionts associated with hosts that have variation in their access to nutritional resources. The obligate nature of nutritional endosymbionts leads to ultimate codependency and coevolution between the host and bacteria (Thao et al. 2002; Bennett and Moran 2015). As host species-specific symbiont associations coevolve, symbiont strains may become more genetically divergent between different host species (Bennett and Moran 2015; Chong and Moran 2016), as is reflected in symbiont functional roles and rates of molecular evolution within and between Oliarus species.
We predicted that variation in nutritional resource availability between surface and cave-adapted hosts would result in an increase selection pressures on the endosymbionts of O. polyphemus to retain nutritional biosynthesis pathways in the face of a niche shift to a resource-limited environment. Between cave-adapted O. polyphemus and epigean O. filicicola, we found that Sulcia and Vidania genomes are highly conserved, which is a common feature among co-obligate endosymbiotic partners of Sulcia within other members of the Auchenorrhyncha (Wu et al. 2006; McCutcheon and Moran 2010; Bennett and Moran 2013). In contrast, Purcelliella exhibits remarkable variation in its retention and loss of functional genes and biosynthesis pathways between cave and surface hosts. despite having a highly reduced genome (∼481 kb). Our results indeed indicate that the ecological shift in habitat and restriction on nutritional resources in cave-adapted planthoppers may have increased selection pressures on the retention of nutritional provisioning genes in the relatively young endosymbiont, Purcelliella. Though Purcelliella as a whole shows variation in selection pressure on nutritional biosynthesis genes between surface and cave hosts, substitutions on non-nutritional biosynthesis genes have continued to accumulate between Purcelliella-OPOL1 and Purcelliella-OPOL2. This further indicates that selection is actively maintaining necessary nutritional biosynthesis genes in Purcelliella of cave-adapted Oliarus, whereas nonfunctional genes have increased substitution rates and may be subject to being lost from the genome through the process of genome streamlining. Alternatively, genetic drift due to reduced island and cave insect populations may contribute to the loss of symbiont genes between Oliarus host species. To test these hypotheses, future studies should expand sampling across surface and cave host species and populations to disentangle the effects of adaptation and drift.
We also expected that an increase in divergence between endosymbionts associated with epigean versus cave-adapted host species due to tight coevolution. While Sulcia exhibits extremely low substitution rates between the two host species, its partner endosymbionts Vidania and Purcelliella have much higher substitution rates. This pattern appears to be highly conserved among Auchenorrhyncha hosts that harbor Sulcia and one or two of its partner endosymbionts (Vasquez and Bennett 2022). Additionally, for Purcelliella, we see variation in the rates of molecular evolution between cave and surface host cixiid species. Looking within cave-adapted O. polyphemus hosts, we recorded increased rates of molecular evolution for both Vidania (average dN/dS = 0.11) and Purcelliella (average dN/dS = 0.09). Taken together, our results show that all three endosymbionts exhibit tightly coupled coevolution with their hosts, but that they vary among themselves at inter- and intra-species biological scales.
Previous works on planthopper hosts on Hawaiʻi Island predict that cave-adapted planthoppers, O. polyphemus, are likely members of a divergent species complex with individual lava tube populations that have limited to no gene flow (Hoch and Howarth 1993; Wessel et al. 2013). Our O. polyphemus host samples were collected from the southern Kaʻu district of Hawaiʻi Island; however, samples were collected from different lava tube caves that may represent reproductively isolated subterranean populations. If so, this isolation could explain the variation between functional roles and rates of molecular evolution in O. polyphemus endosymbionts observed in our study (e.g., variation in retention of bacterial cellular function genes and increased rates of substitutions between Vidania and Purcelliella in OPOL-1 and OPOL-2). Coevolution between host and endosymbiont coupled with isolated planthopper populations could result in differential fitness and selective pressures between symbionts isolated from different host populations, despite the hosts being of the same species (Chong and Moran 2016). To fully address these kinds of questions, and the mechanisms driving divergence in cixiid hosts and their symbionts, future work should sample across host populations and their potential symbiont strains. Such work would elucidate how the structure of lava tube cave systems drive host–symbiont coevolution in contrast to epigean species with potentially larger population sizes and expanded nutritional resources.
Materials and Methods
Sample Collection and Sequencing
Four Oliarus specimens were collected from the southern Ka’u and Puna districts of Hawaiʻi Island, Hawaiʻi, USA. Two Ka’u cave-adapted planthoppers were identified in the field as Oliarus polyphemus (OPOL-1 and OPOL-2) and two Puna surface planthoppers were identified as Oliarus filicicola (OFIL-1 and OFIL-2). Specimens were stored in 100% ethanol on site. Whole planthopper specimens were used for DNA extraction using a Qiagen DNeasy Blood and Tissue extraction kit. Genomic libraries were prepared using NEB Next® UltraII DNA Library Prep Kit with average insert sizes of approximately 500 bp and were sequenced using Illumina Technology at NovoGene Corporation (New Jersey, USA).
Host Phylogeny
We used a maximum likelihood approach to estimate phylogenetic relationships of the four Oliarus samples based on the full-length mitochondrial CO1 sequences. Complete CO1 sequences were extracted from each host assembly and aligned with CO1 sequences from a Hawaiian cixiid planthopper Iolania perkinsi (Chong et al. 2022) and other Auchenorrhyncha hosts that were obtained from GenBank (supplementary table S2, Supplementary Material online). A translational alignment was performed on all sequences using MUSCLE (Edgar 2004) in Geneious Prime 2020.1.4. Using IQ tree v1.6.12 (Nguyen et al. 2015), we determined that the best fitting model of nucleotide substitution was GTR + F + I + G4 (Trifinopoulos et al. 2016) based on the Akaike Information Criteria (AIC) (Akaike 1973). The maximum likelihood phylogeny was reconstructed with 10,000 bootstrap replicates using UFBoot2 (Hoang et al. 2018) and visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Symbiont Genome Assembly and Annotation
Raw sequencing reads were filtered for quality and adapter removal using FASTQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads from OFIL-1, OFIL-2, and OPOL-2 samples were trimmed using Trimmomatic v0.39 (Bolger et al. 2014). Reads from OPOL-1 sample were trimmed using Trim Galore! V0.6.4 (phred – 33, paired-end setting, no singletons) (https://github.com/FelixKrueger/TrimGalore). Genomic sequences were assembled using SPAdes v. 3.14.1 (kmer-127) (Nurk et al. 2013). In the case of OFIL-2, raw sequence files were split into two files and assembled in SPAdes v.3.14.0 (kmer-100). Endosymbiont genomes typically have an AT mutational bias that results in lower GC content than the rest of the host genome (Wernegreen 2015; Chong et al. 2019). This allowed us to identify bacterial genomes by their GC percentage in addition to NCBI-tBLASTx searches of the host assemblies against the reference genomes for Ca. Sulcia (CP028359), Ca. Purcelliella (CP028374), and Ca. Vidania (CP028360). We identified contigs in the assemblies matching all three endosymbionts from each of the four host specimens (table 1). Using open-reading frames, symbiont genomes were confirmed against the BLASTP database to ensure sequence validity and then further assembled in Geneious Prime 2020.1.4 (https://www.geneious.com). To identify any contigs present that were associated with other microbial species, we performed a general 16S rRNA gene search with tBLASTx against the 16S ribosomal RNA sequences (Bacteria and Archaea) database.
For each endosymbiont, assembled contigs were circularized into complete genomes using the de novo assemble feature in Geneious Prime 2020.1.4. To verify genome and assembly quality, we aligned reads to the published reference genomes with Bowtie2 to determine consistent coverage, including circularization of linear scaffolds ends (Langmead and Salzberg 2012). Circular bacterial genomes were annotated with RAST v.2.0 (Aziz et al. 2008; Overbeek et al. 2014; Brettin et al. 2015). Circularized genomes were aligned using Mauve (Darling et al. 2004) to estimate genomic synteny. To calculate the coding density for each symbiont genome, we divided the number of combined base pairs for all coding sequences (CDS) in the genome by the total number of base pairs in the genome. Symbiont genes necessary for bacterial cellular function and nutritional biosynthesis were identified using RAST v.2.0 annotations. We searched for nutritional biosynthesis genes for amino acids and vitamins aside from the ten EAAs, one non-EAA, and four B vitamins but none were identified. Genes that were truncated from their predicted full length were aligned to full-length versions of those genes using MUSCLE in Geneious Prime 2020.1.4 (Edgar 2004). Genes were assumed to be nonfunctional if the alignment revealed that they were broken into multiple small pieces (<20 bp) in addition to being truncated. Sequence data for endosymbiont genomes are available on the NCBI GenBank database under the accession numbers CP110504–CP110507 (Ca. Sulcia), CP110500–CP110503 (Ca. Vidania), and CP110496–CP110499 (Ca. Purcelliella).
Rates of Molecular Evolution
To estimate rates of molecular evolution, we performed pairwise comparisons of substitution rates among symbiont genes of the same lineage, across each of the four host specimens, two O. polyphemus and two O. filicicola. One-to-one orthologous protein-coding sequences from each symbiont genome were identified for each pairwise comparison. Orthologous symbiont genes of the same lineage were aligned with MUSCLE (Edgar 2004) in Geneious Prime 2020.1.4 with the bacterial genetic code. A range of CDS were aligned for each symbiont comparison: 128–133 CDS for Sulcia, 131–140 CDS for Vidania, and 363–398 CDS for Purcelliella.
To determine how the selection is shaping symbiont genomic variation between and within host species, we quantified rates of synonymous (dS) and nonsynonymous (dN) substitutions across symbiont genomes. Pairwise gene comparisons of substitution rates were analyzed with the codeML package from PAML v13.14 (Yang 2007), (run mode −2 for pairwise comparisons). Using codeML, we performed maximum likelihood estimation (M0 one-ratio model) to quantify dS and dN between orthologous CDS and estimate the overall selection pressure (ω = dN/dS) for each CDS comparison. Positive selection on an individual gene is indicated by ω > 1 whereby the rate of nonsynonymous substitutions is greater than the rate of synonymous substitutions (dN > dS). Negative purifying selection is indicated by ω << 1 (dN << dS). Synonymous and nonsynonymous substitution rates of each symbiont gene in all comparisons were visualized using the package ggplot2 (Wickham 2016) in Rstudio v1.2.5001 (http://www.rstudio.com). Lastly, to determine if any particular functional category was more likely to be under positive or purifying selection than by random chance, we performed Fisher’s exact test on each Clusters of Orthologous Genes (COG) category for each genome comparison.
Supplementary Material
Acknowledgments
As settlers and guests of Hawaiʻi, we acknowledge that the land on which we conduct research is the home of indigenous Hawaiians, whose home is now illegally occupied by the United States. We recognize the indigenous knowledge of the Hawaiian people as the original caretakers of the ‘āina, past and present. We respect the kapu of the lava caves and are grateful for the opportunity to access these spaces with much care and consideration for the land and the native species that inhabit them.
We would like to thank Dr. Chris Simon, the associate editor, and two anonymous reviewers for their helpful suggestions that benefitted this manuscript. We thank Mireille Steck (UH Mānoa) for providing advice and assistance with data analysis and bioinformatics. We also thank Allen Kalampukattussery for help with selection analyses. Finally, we thank the members of the HICAVES team who collected the planthopper samples that were analyzed in this study. This work was supported by funding from the National Science Foundation DEB-2204670 (to R.A.C. and M.L.P.), Jacqueline Maly ARCS Award in Life Sciences (Achievement Rewards for College Students-Honolulu Chapter), Hampton and Meredith Carson Fellowship (Ecology, Evolution, and Conservation Biology graduate program – UH Mānoa), E. Allison Kay Endowed Award (Zoology graduate program – UH Mānoa), and the Graduate Student Organization Research Award (UH Mānoa).
Contributor Information
Jordan M Gossett, School of Life Sciences, University of Hawaiʻi at Mānoa.
Megan L Porter, School of Life Sciences, University of Hawaiʻi at Mānoa.
Yumary M Vasquez, Life and Environmental Sciences Unit, University of California.
Gordon M Bennett, Life and Environmental Sciences Unit, University of California.
Rebecca A Chong, School of Life Sciences, University of Hawaiʻi at Mānoa.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
The data underlying this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/nucleotide, and can be accessed with BioProject: PRJNA896244 and BioSample: SAMN31357385–SAMN31357396 and SAMN31323236–SAMN31323239. Endosymbiont genomes are available under the accession numbers CP110504–CP110507 (Ca. Sulcia), CP110500–CP110503 (Ca. Vidania), and CP110496–CP110499 (Ca. Purcelliella).
Literature Cited
- Akaike H. 1973. Information theory and an extension of the maximum likelihood principle. Selected papers of hirotugu akaike. 199–213.
- Asche M. 1997. A review of the systematics of Hawaiian planthoppers (Hemiptera:Fulgoroidea). Vol. 51. Honolulu HI: Pacific Science, University of Hawaii Press. p. 366–376. [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:Article 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Mao M. 2018. Comparative genomics of a quadripartite symbiosis in a planthopper host reveals the origins and rearranged nutritional responsibilities of anciently diverged bacterial lineages. Environ Microbiol. 20(12):4461–4472. [DOI] [PubMed] [Google Scholar]
- Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 5(9):1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci U S A. 112(33):10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Pantoja NA, O’Grady PM. 2012. Diversity and phylogenetic relationships of Wolbachia in Drosophila and other native Hawaiian insects. Fly (Austin). 6(4):273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan A, Arneodo J, Simonato M, Haines WP, Boudon-Padieu E. 2009. Characterization and evolution of two bacteriome-inhabiting symbionts in cixiid planthoppers (Hemiptera: Fulgoromorpha: Pentastirini). Environ Microbiol. 11(12):3265–3279. [DOI] [PubMed] [Google Scholar]
- Bressan A, Mulligan KL. 2013. Localization and morphological variation of three bacteriome-inhabiting symbionts within a planthopper of the genus Oliarus (Hemiptera: Cixiidae). Environ Microbiol Rep. 5(4):499–505. [DOI] [PubMed] [Google Scholar]
- Brettin T, et al. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 5:Article 8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. New York: John Wiley & Sons. [Google Scholar]
- Chong RA, Moran NA. 2016. Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proc Natl Acad Sci U S A. 113(46):13114–13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong RA, Moran NA. 2018. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J. 12(3):898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong RA, Park H, Moran NA. 2019. Genome evolution of the obligate endosymbiont Buchnera aphidicola. Mol Biol Evol. 36(7):1481–1489. [DOI] [PubMed] [Google Scholar]
- Chong RA, Steck M, Porter ML. 2022. Complete mitochondrial genomes and phylogenetic analysis of the Hawaiian planthoppers Iolania perkinsi and Oliarus cf. filicicola (Hemiptera: Cixiidae). Mitochondrial DNA Part B Resour. 7(6):1015–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, et al. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 8(11):e1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dial DT, et al. 2021. Transitional genomes and nutritional role reversals identified for dual symbionts of adelgids (Aphidoidea: Adelgidae). ISME J. 12: 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson SL, et al. 1999. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol Biol. 29(2):153–160. [DOI] [PubMed] [Google Scholar]
- Douglas AE. 2017. The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr Opin Insect Sci. 23:65–69. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Wernegreen JJ, Moran NA. 2001. Intraspecific variation in symbiont genomes: bottlenecks and the aphid-buchnera association. Genetics 157(2):477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 108(7):2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2014. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol. 23(6):1473–1496. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch H. 2006. Systematics and evolution of Iolania (Hemiptera: Fulgoromorpha: Cixiidae) from Hawai’i. Syst Entomol. 31(2):302–320. [Google Scholar]
- Hoch H, Howarth FG. 1993. Evolutionary dynamics of behavioral divergence among populations of the Hawaiian cave-dwelling planthopper Oliarus polyphemus. In: Homopters: Fulgoroidea: Cixiidae. Vol. 47. Honolulu HI: Pacific Science, University of Hawaii Press. p. 303–318. [Google Scholar]
- Hoch H, Howarth FG. 1999. Multiple cave invasions by species of the planthopper genus Oliarus in Hawaii (Homoptera: Fulgoroidea: Cixiidae). Zool J Linn Soc. 127(4):453–475. [Google Scholar]
- Howarth FG. 1986. The tropical cave environment and the elocution of troglobites. Proceedings of the 9th Congress Internation Speleol, Barcelona Spain, 1–7 August, Vol. 2. Le Bourget-du-Lac, France: Proceedings of the International Congress of Speleology. p. 153–155.
- Howarth FG, James SA, McDowell W, Preston DJ, Imada CT. 2007. Identification of roots in lava tube caves using molecular techniques: implications for conservation of cave arthropod faunas. J Insect Conserv. 11(3):251–261. [Google Scholar]
- Jeyaprakash A, Hoy MA. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 9(4):393–405. [DOI] [PubMed] [Google Scholar]
- Jiggins FM, Bentley JK, Majerus MEN, Hurst GDD. 2001. How many species are infected with Wolbachia? Cryptic sex ratio distorters revealed to be common by intensive sampling. Proc Biol Sci. 268(1472):1123–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karley AJ, Douglas AE, Parker WE. 2002. Amino acid composition and nutritional quality of potato leaf phloem sap for aphids. J Exp Biol. 205(19):3009–3018. [DOI] [PubMed] [Google Scholar]
- Kinjo Y, et al. 2021. Enhanced mutation rate, relaxed selection, and the “domino effect” are associated with gene loss in Blattabacterium, a cockroach endosymbiont. Mol Biol Evol. 38(9):3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 15(7):2073–2081. [DOI] [PubMed] [Google Scholar]
- Koga R, Moran NA. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8(6):1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre A, Manzano-Marin A. 2017. Dissecting genome reduction and trait loss in insect endosymbionts. Ann N Y Acad Sci. 1389(1):52–75. [DOI] [PubMed] [Google Scholar]
- Manzano-Marin A, Simon JC, Latorre A. 2016. Reinventing the wheel and making it round again: evolutionary convergence in Buchnera-Serratia symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol Evol. 8(5):1440–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Bennett GM. 2020. Symbiont replacements reset the co-evolutionary relationship between insects and their heritable bacteria. ISME J. 14(6):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Yang XS, Bennett GM. 2018. Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in a leafhopper host. Proc Natl Acad Sci U S A. 115(50):E11691–E11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Boyd BM, Dale C. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol. 29(11):R485–R495. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 106(36):15394–15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 104(49):19392–19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 my of evolution. Genome Biol Evol. 2:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 21(16):1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik A, et al. 2021. Alternative transmission patterns in independently acquired nutritional cosymbionts of Dictyopharidae planthoppers. mBio. 12(4):Article e01228-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller's Rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93(7):2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM. 2014. The tiniest tiny genomes. In: Gottesman S, editor. Annual review of microbiology. Vol. 68. San Mateo, CA: Annual Reviews. p. 195–215. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic Bacteria. Science 323(5912):379–382. [DOI] [PubMed] [Google Scholar]
- Moran NA, Mira A. 2001. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2(12):research0054.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc Biol Sci. 253(1337):167–171. [Google Scholar]
- Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 71(12):8802–8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson MA, Baumann P, Kinsey MG. 1991. Buchnera gen. Nov. and Buchnera aphidicola sp. Nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 41(4):566–568. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating Maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AI, et al. 2005. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci U S A. 102(34):12112–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, et al. 2013. Assembling single-cell genomes and Mini-metagenomes from chimeric MDA products. J Comput Biol. 20(10):714–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42(D1):D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau J, Moran NA. 2022. Genetic innovations in animal-microbe symbioses. Nat Rev Genet. 23(1):23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JP, Clague DA. 2002. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc Biol Sci. 269(1508):2429–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoz F, et al. 2022. The di-symbiotic systems in the aphids Sipha maydis and Periphyllus lyropictus provide a contrasting picture of recent co-obligate nutritional endosymbiosis in aphids. Microorganisms 10(7):1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispe C, Moran NA. 2000. Accumulation of deleterious mutations in endosymbionts: Muller’s Ratchet with two levels of selection. Am Nat. 156(4):425–441. [DOI] [PubMed] [Google Scholar]
- Russell CW, Bouvaine S, Newell PD, Douglas AE. 2013. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 79(19):6117–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom J, Pettersson J. 1994. Amino-Acid-composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J Insect Physiol. 40(11):947–955. [Google Scholar]
- Sheffer MM, et al. 2020. Tissue- and population-level microbiome analysis of the wasp spider Argiope bruennichi identified a novel dominant bacterial symbiont. Microorganisms 8(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 31(4):857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Moran NA. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 29(12):3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Kost C, Kaltenpoth M. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 25(5):375–390. [DOI] [PubMed] [Google Scholar]
- Thao ML, Gullan PJ, Baumann P. 2002. Secondary (γ-Proteobacteria) endosymbionts infect the primary (β-Proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl Environ Microbiol. 68(7):3190–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toenshoff ER, Gruber D, Horn M. 2012. Co-evolution and symbiont replacement shaped the symbiosis between adelgids (Hemiptera: Adelgidae) and their bacterial symbionts. Environ Microbiol. 14(5):1284–1295. [DOI] [PubMed] [Google Scholar]
- Toft C, Andersson SGE. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 11(7):465–475. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JM, Cryan JR. 2012. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol Biol. 12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez YM, Bennett GM. 2022. A complex interplay of evolutionary forces continues to shape ancient co-occurring symbiont genomes. Vol. 25(8). Amsterdam, Netherlands: Elsevier. p. 104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waneka G, Vasquez YM, Bennett GM, Sloan DB. 2021. Mutational pressure drives differential genome conservation in two bacterial endosymbionts of sap-feeding insects. Genome Biol Evol. 13(3):Article evaa254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ. 2002. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 3(11):850–861. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. 2015. Endosymbiont evolution: predictions from theory and surprises from genomes. Ann N Y Acad Sci. 1360:16–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6(10):741–751. [DOI] [PubMed] [Google Scholar]
- Wessel A, et al. 2013. Founder effects initiated rapid species radiation in Hawaiian cave planthoppers. Proc Natl Acad Sci U S A. 110(23):9391–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. Ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. p. 160–167. [Google Scholar]
- Wu D, et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4(6):e188–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/nucleotide, and can be accessed with BioProject: PRJNA896244 and BioSample: SAMN31357385–SAMN31357396 and SAMN31323236–SAMN31323239. Endosymbiont genomes are available under the accession numbers CP110504–CP110507 (Ca. Sulcia), CP110500–CP110503 (Ca. Vidania), and CP110496–CP110499 (Ca. Purcelliella).