Abstract
Genomic imbalance refers to the more severe phenotypic consequences of changing part of a chromosome compared with the whole genome set. Previous genome imbalance studies in maize have identified prevalent inverse modulation of genes on the unvaried chromosomes (trans) with both the addition or subtraction of chromosome arms. Transposable elements (TEs) comprise a substantial fraction of the genome, and their reaction to genomic imbalance is therefore of interest. Here, we analyzed TE expression using RNA-seq data of aneuploidy and ploidy series and found that most aneuploidies showed an inverse modulation of TEs, but reductions in monosomy and increases in disomy and trisomy were also common. By contrast, the ploidy series showed little TE modulation. The modulation of TEs and genes in the same experimental group were compared, and TEs showed greater modulation than genes, especially in disomy. Class I and II TEs were differentially modulated in most aneuploidies, and some superfamilies in each TE class also showed differential modulation. Finally, the significantly upregulated TEs in three disomies (TB-7Lb, TB9Lc, and TB-10L19) did not increase the proportion of adjacent gene expression when compared with non-differentially expressed TEs, indicating that modulations of TEs do not compound the effect on genes. These results suggest that the prevalent inverse TE modulation in aneuploidy results from stoichiometric upset of the regulatory machinery used by TEs, similar to the response of core genes to genomic imbalance.
Key words: genome imbalance, transposable element expression, aneuploidy, polyploidy, Class I and II TEs
Investigation of transposable element (TE) expression in aneuploidy and ploidy series showed that TEs are predominantly inversely modulated in aneuploidies but display little modulation in the ploidy series. This result suggests that the prevalent inverse TE modulation in aneuploidy results from a stoichiometric upset of the regulatory machinery used by TEs, similar to the response of core genes to genomic imbalance.
Introduction
Genome imbalance is a phenomenon in which changing the dosage of an individual chromosome (aneuploidy) has a more detrimental effect on the phenotype than changing the whole set (ploidy). Such a phenomenon has been recapitulated across eukaryotic species, such as Datura stramonium (Blakeslee et al., 1920; Blakeslee, 1921, 1934; Blakeslee and Belling, 1924), Drosophila melanogaster (Bridges, 1925), wheat (Sears, 1953), and human (Schinzel, 2001).
In maize, the study of aneuploidy is greatly facilitated by translocations with the B chromosome. The supernumerary B chromosome is nonessential but is capable of transcribing active genes and impacting A-chromosome gene expression (Shi et al., 2022). The B chromosome can maintain itself in populations by nondisjunction at the second pollen mitosis, which produces the two sperm, followed by preferential fertilization of the egg by the B-containing sperm during the process of double fertilization (Blavet et al., 2021). Utilization of translocations between various normal chromosome (A chromosome) arms and the supernumerary B chromosome has allowed the dosage of the former to be varied in one (monosomy, hypoploid heterozygote), two (euploid heterozygote), and three doses (trisomy, hyperploid heterozygote) (Birchler and Yang, 2021; Shi et al., 2021). Furthermore, hyperploid heterozygotes of each translocation can be crossed as females by a haploid inducer line to obtain disomic haploids (Yang et al., 2021). Although maize is functionally a diploid organism, it behaves like an ancient allotetraploid (Messing, 2009) that is tolerant of producing a wider spectrum of aneuploids and ploidy series (haploid, triploid, and tetraploid), thereby making maize a highly suitable organism for systematic and comprehensive study of genome imbalance.
Once molecular studies were initiated with aneuploids, monitoring the RNA or protein level of genes on the varied chromosome arms (defined as cis) showed gene dosage effects (i.e., gene expression was proportional to the chromosomal dose changes). However, it was also found that a subset of genes in the same varied region often displayed no change in levels of RNA or protein products, a phenomenon called dosage compensation. One classic case that provided insight into the basis of dosage compensation involved testing the enzyme level of maize alcohol dehydrogenase 1 (adh1) in a dosage series of the long arm of chromosome 1 (1L). This structural gene was dosage-compensated in a 1–4 dosage series of 1L (Birchler, 1979). After dissecting 1L into smaller segmental dosage series, adh1 showed a dosage effect in one region that included this gene but was modulated inversely (i.e., gene expression decreased as this chromosomal dose increased) by another portion of 1L, suggesting that the basis of dosage compensation of adh1 is the cancellation of a dosage effect by an inverse effect (Birchler, 1981).
For genes on the unvaried chromosomes (trans genes), global transcriptome analysis of disomy (two doses at the haploid level), monosomy, and trisomy in maize showed that an inverse modulation (or inverse effect) was the most common effect across the genome (Shi et al., 2021; Yang et al., 2021). In other words, in monosomies, reduced dosage of a chromosome arm causes an increase in the expression of most of the trans genes, and in disomies and trisomies, the addition of a chromosomal segment causes reductions in gene expression (Shi et al., 2021; Yang et al., 2021). In addition to the inverse effect, reductions in monosomy or increases in trisomy have also been found (Shi et al., 2021), referred to as direct effects or direct modulation. Compared with trisomic diploids, disomic haploids produce greater inverse modulation owing to their greater genome imbalance (2-fold dosage change in disomy vs. 1.5-fold dosage change in trisomy). There is also a more detrimental and severe phenotype in disomic seedlings (Yang et al., 2021). The magnitude of the inverse effects in disomies was correlated with the size of the cis region across all examined regions. When a greater number of genes showed an inverse effect in trans, more cis genes trended toward dosage compensation, showing a relationship between the inverse dosage effect and dosage compensation for genes in the varied chromosomal regions on a global scale (Birchler and Veitia, 2021; Yang et al., 2021).
The Gene Balance Hypothesis was formulated to explain the mechanism of genome imbalance; it posits that changing the stoichiometry of subunits involved in multicomponent interactions will affect the kinetics and mode of assembly, and can thus disrupt the function of whole macromolecular complexes or multicomponent interactions (Birchler et al., 2005; Birchler and Veitia, 2007, 2010, 2012, 2021). Thus, genes involved in multicomponent interactions are typically dosage sensitive and overrepresented among transcription factors, signal transduction pathway genes, and chromatin modifiers (Birchler et al., 2001). The effects of regulatory pathways depend not only on the quantity of gene expression of any regulatory gene affecting its targets but also on its quantitative relationship with its interactors. Modeling the assembly of functional multisubunit complexes (Bray and Lay, 1997; Veitia, 2002) suggests that by monitoring the level of bridge components that interact with multiple other subunits from low to high concentration, the production of functional complexes proceeds through a peak and then declines at higher concentrations because partial complexes (non-functional) are produced that cannot proceed to completion of the whole (functional). Thus, both under- and overrepresentation of individual subunits can be detrimental. These models parallel typical aneuploid effects at the monosomic and trisomic levels (Lee et al., 1996a, 1996b; Shi et al., 2021).
Evolutionary genomics provides support for this concept. Many eukaryotes have experienced multiple rounds of whole-genome duplication (WGD) (Wolfe and Shields, 1997; Simillion et al., 2002; Maere et al., 2005; Aury et al., 2006; Blomme et al., 2006; Thomas et al., 2006; Freeling, 2009; Jiao et al., 2011). Following WGD, genomes are fractionated so that most genes return to the singleton state, with exceptions that depend on the type of gene function. Dosage-sensitive genes that are involved with multicomponent interactions are overrepresented in the exceptions. On the other hand, gene pairs from small-scale duplications show underrepresentation of genes involved in multicomponent interactions, thus complementing the effects of WGD (Veitia, 2004; Hakes et al., 2007; Freeling et al., 2008; Korbel et al., 2008; Ionita-Laza et al., 2009; Rodgers-Melnick et al., 2012; Tasdighian et al., 2017). Small-scale duplication would also mimic the detrimental effects of aneuploidy, in that changing the copy numbers of interacting genes would have an adverse effect.
In contrast to genes, transposable elements (TEs) are fluid over evolutionary time and constitute the majority of the genome. They are often silenced by the host genome but utilize the host machinery when they are expressed. Modulation of TEs in aneuploidy has not been systematically studied in any organism. TEs can be grouped into two classes, class I and class II, based on their transposition mechanism. Class I elements are also called retrotransposons. They “copy and paste” to increase their copies in the genome. An element’s RNA is used as a template to synthesize DNA molecules that insert into new chromosomal sites. Class I elements are generally divided into two groups on the basis of transposition mechanism and structure: LTR and non-LTR retrotransposons. Class II elements, generally referred to as DNA transposons, often contain terminal inverted repeats (TIRs) and employ “cut and paste” transposition in which an element is cut out of one site in a chromosome and pasted into a new site. In addition to TIR elements, the class II transposons also include Helitron elements that usually contain a DNA helicase. These elements lack TIRs. In plant genomes, DNA transposons are usually present at much lower levels than retrotransposons. Usually, the majority of TEs in the genome remain silenced, but under some conditions (e.g., after treatment with a demethylating agent, after tissue culture, or after chromosomal breakage), TEs can be activated (Rhoades and Dempsey, 1982; Pouteau et al., 1991; Hirochika et al., 1996; Griffin et al., 2016). Repression of TE transposition involves a variety of mechanisms, including histone modifications, DNA methylation, and other factors such as chromatin-remodeling enzymes. Repression of TE transposition relies on RNA-directed DNA methylation (RdDM) (de novo DNA methylation formation) and maintenance of methylation through DNA methyltransferases, especially in euchromatic regions (Sigman and Slotkin, 2016). For example, TEs near genes require RdDM to establish initial methylation, and later the DNA methylation is retained and enhanced through cell division by DNA methyltransferases (Sigman and Slotkin, 2016).
Here, we systematically analyzed the expression of TEs in both haploid and diploid aneuploidy together with the ploidy series and found that most aneuploidies showed inverse modulation of TEs as the major response, whereas the ploidy series showed little TE modulation. Furthermore, there was a trend of stronger modulation of TEs than genes in the same experimental group. Different classes of TEs were differentially modulated in most aneuploidies, and some superfamilies of each TE class also showed differential modulation. These results suggest that prevalent inverse TE modulation in aneuploidy results from stoichiometric upset of the regulatory machinery used by TEs (e.g., for transcription or repression), similar to the response of core genes to genomic imbalance.
Results
Widespread modulation of individual TEs in aneuploidy
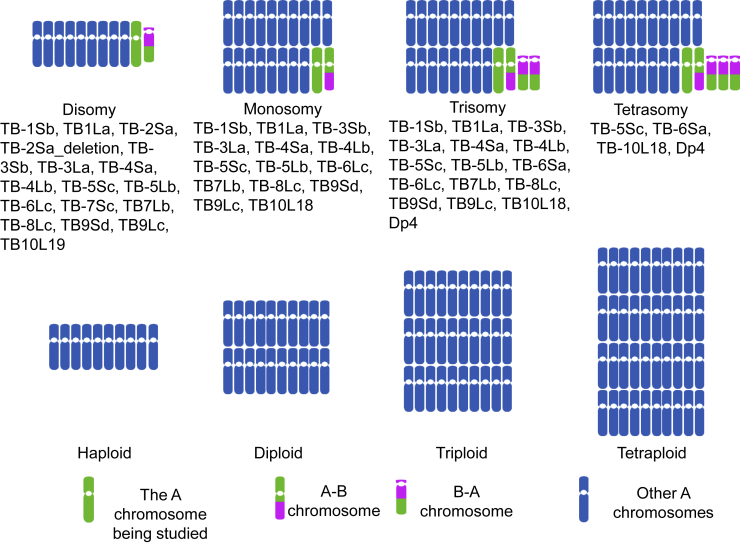
Previous genome balance studies in maize have found greater modulations in aneuploidies compared with the ploidy series (Shi et al., 2021; Yang et al., 2021). To test changes in the expression of TEs, we downloaded RNA-seq data for the aneuploidy and ploidy series from NCBI (Shi et al., 2021; Yang et al., 2021). The aneuploidy data included 17 disomies that have an extra chromosomal segment at the haploid level and diploid aneuploidies, including 14 monosomies, 16 trisomies, and 4 tetrasomies. The ploidy series included haploids, diploids, triploids, and tetraploids. For convenience, disomies and their haploid control were named h2D and h1D, respectively, and monosomies, diploids, trisomies, and tetrasomies in each aneuploid were referred to as 1D, 2D, 3D, and 4D (D designates dosage of the chromosomal arm), whereas haploids, diploids, triploids, and tetraploids in the ploidy series were named 1X, 2X, 3X, and 4X, respectively (X designates the basic chromosome set). Duplication 4 (Dp4), which consists of the A-B chromosomes of TB-4Lb and TB-4Sa, namely 4Lb-B and 4Sa-B, produces duplicated proximal chromosomal segments (centromere 4 and surrounding regions). In addition, whereas both the cis regions of 2Sa and 2Sa_deletion contain two noncontinuous segments, the 2Sa_deletion has a deleted part in the first segment in 2S (Yang et al., 2021). All the RNA-seq data were from the W22 background except for those of the triploid (3X), which originated from the Mo17 inbred. The B-A translocations used to generate aneuploids are described in Figure 1.
Figure 1.
Diagram of aneuploid and ploidy series data used in this study.
The disomy (10A + 1B-A) has an extra A chromosomal segment on the B-A chromosome. The monosomy has 19A chromosomes plus 1A-B chromosome except for TB-5Sc (18A + 2A-B + 1B-A). Trisomy has a chromosome constitution of 19A, 1A-B, and 2B-A except for TB-5Sc (18A + 2A-B + 3B-A) and TB-6Sa (20A + 1B-A). Tetrasomy has 19A, 1A-B, and 3B-A chromosomes except for TB-6Sa (20A + 2B-A). The A refers to chromosomes 1 to 10. Trisomy for Dp4 has 19 copies of the A chromosomes plus one copy each of 4Lb-B and 4Sa-B, whereas tetrasomy has 18 A chromosomes and two copies each of 4Lb-B and 4Sa-B. The chromosome that was studied is depicted in green compared with the rest of the genome, which is depicted in blue. For the B-A translocation, for example, in TB-1La, the number “1” refers to chromosome 1. The uppercase letter “L” denotes the long arm, and the lowercase “a” represents the first B-A translocation produced for that chromosome. The B chromosome portion is shown in magenta. More disomy and diploid aneuploid information, including genomically defined breakpoints, has been reported previously (Shi et al., 2021; Yang et al., 2021). The B chromosome breakpoints have been defined (Blavet et al., 2021).
Because there are many truncated TEs from the same family in maize (Hufford et al., 2021), instead of aligning the data to the W22 genome directly, we mapped the RNA-seq data to W22 TE exemplar sequences that consist of 57 955 intact and non-overlapping TEs (Shi et al., 2022) after reads per kilobase of transcript per million mapped reads (RPKM) normalization to obtain the relative expression level. Principal component analysis (PCA) of the biological replicates in each category using normalized counts showed that most of the aneuploidy and ploidy groups were separated from the respective control, revealing that TE expression showed a distinctive expression pattern in each group. However, in some groups, such as disomy TB-1Sb (1S), two biological replicates did not cluster with the remainder, distinct from the PCA of protein-coding gene expression levels from the same plants, in which all disomic plants were clustered together (Supplemental Figure 1; Yang et al., 2021). To identify possible outliers, we calculated the mean and standard deviation (SD) of the principal components among biological replicates and examined whether the values of PC1 and PC2 for each replicate were within two SDs of the mean. The results indicated that no replicates could be considered outliers (Supplemental Data Set 1).
To further examine how genome imbalance affects the global TE expression trend, ratio distributions were obtained to determine TE expression in the aneuploidy and ploidy series. All of the original B-A translocations used in this study, except TB-10L18 and TB-10L19, were not in the W22 background when originally generated (Beckett, 1991). During backcrossing to W22, because B-A chromosomes have a dominant anthocyanin gene, hyperploid heterozygotes (trisomy) were chosen based on having a colorless endosperm and colored embryo (two copies of the B-A chromosomes in the embryo) (Birchler and Yang, 2021). Both genes and TEs on the A-chromosome portion were maintained as in the original stock. Unlike genes that can display good syntenic locations in different maize lines, the TEs in different maize genomes show high levels of variation (Hufford et al., 2021). Therefore, instead of plotting the individual TEs from the varied and unvaried chromosome arms separately, we plotted all individual TEs for each comparison together. Read counts were averaged across biological replicates. Expression of most of the TEs mapped to the exemplar sequences in both experimental and control groups was suppressed. After filtering out TEs with low expression, the ratios of each expressed TE for each group comparison were generated and then plotted as a ratio distribution. The results showed that most of the TEs (∼90%) remained silenced in both aneuploids and controls, as only ∼10% of the TEs remained after filtering (mean (control) + mean (experimental group)) ≥ 1 (RPKM) (Supplemental Data Set 2).
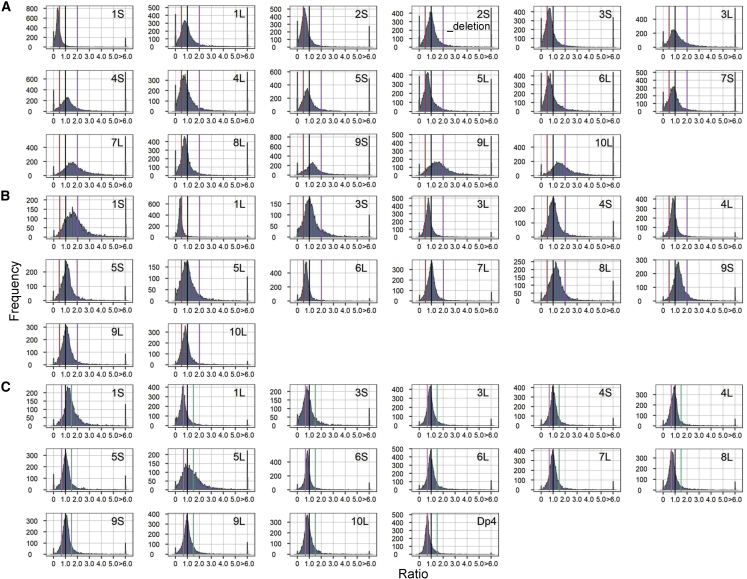
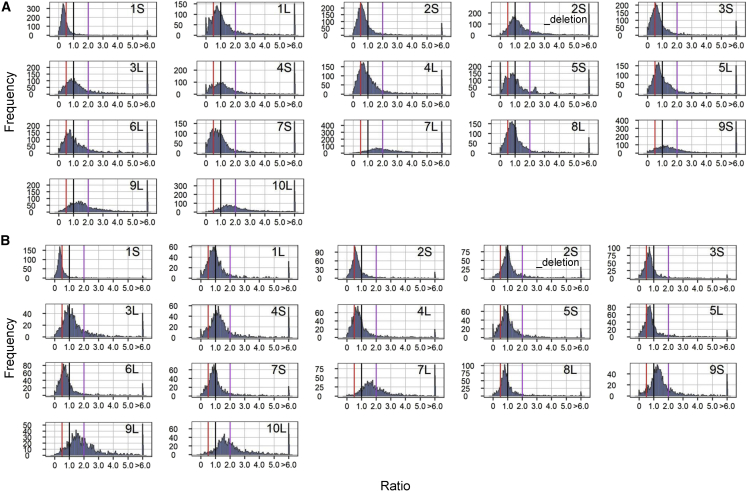
There is a generalized inverse effect in both haploid and diploid aneuploidies. In other words, in h2D/h1D (disomy/haploid control), 3D/2D (trisomy/diploid control), and 4D/2D (tetrasomy/diploid control), a greater number of genotypes show decreased TE expression rather than increased expression. In 1D/2D (monosomy/diploid control), the removal of a chromosome arm causes a trend of increased TE expression in most monosomies (Figure 2; Supplemental Figure 2). For h2D/h1D, 11 out of 17 (1S, 1L, 2S, 3S, 3L, 4L, 5S, 5L, 6L, 7S, 8L) show an inverse effect (major peak below 1.0, which is defined as no modulation), but in some cases, the ratios of some TEs within the same disomy trend above 1.0, indicating direct modulation. For example, 2S_deletion has a major peak around 1.0 but also shows a clear trend of both inverse and direct modulation. TE expression in the remainder of the h2D/h1D comparisons (4S, 7L, 9S, 9L, and 10L) is directly modulated, with a major peak above 1.0 (Figure 2A). TEs have slightly greater modulation in h2D/h1D than in 1D/2D, as shown by a broad range of median TE ratios in h2D/h1D (Supplemental Figure 2). The removal of a chromosomal arm (1D/2D) tends to cause a greater modulation than the addition of a chromosomal arm (3D/2D). In monosomy, TE expression in most genotypes is modulated in both inverse and direct directions, as shown in 1S, 3S, 4S, 5S, 5L, 7L, 8L, 9S, and 9L. The remainder of the 1D/2D comparisons show direct modulation (major peak below 1.0), with 1L being strongly directly modulated (Figure 2B; Supplemental Figure 2). Most of the 3D/2D comparisons (11 out of 16) have a tighter distribution with a peak around 1.0, suggesting that TE expression is less modulated than in 1D/2D (Figure 2C; Supplemental Figure 2). In 3D/2D for 1S, 3S, and 5L, the ratio distributions show both up and down modulations. The TE expression in tetrasomic 1L and Dp4 (Figure 1) is strongly inversely modulated (major peak is 0.5), whereas TEs in 5S tetrasomies were both up and down modulated (Supplemental Figure 3). The other two tetrasomies do not show much modulation in TE expression, with a sharp peak around 1.0. Comparing tetrasomy to trisomy (4D/3D) suggests a progressive effect of TE expression. As in the cases in 4D/3D, 5S does not exhibit much modulation (sharp peak at 1.0), and Dp4 has a major peak close to 1.0. In 6S and 10L, a minor effect (peak below but very close to 1) could be found in the 4D/2D comparison, and the major peak is at 1.0 in the 4D/3D comparison (Supplemental Figure 3).
Figure 2.
Ratio distributions of h2D/h1D, 1D/2D, and 3D/2D.
(A) h2D/h1D.
(B) 1D/2D.
(C) 3D/2D. The normalized TE counts from RNA-seq data were averaged for the biological replicates. TEs with a sum of averaged counts of experimental and control <1 were regarded as having low expression and were filtered out. For each expressed TE, we calculated the ratio of the averaged normalized value in the experimental group relative to the normalized counts in the control. These ratios were plotted in bins of 0.05. The x axis indicates the value for each bin, and the y axis indicates the number of TEs per bin (frequency). A ratio of 1.0 (black line) represents no difference between the experimental genotype and the control. These ratio values are demarcated with labeled vertical lines in purple (2.0) and red (0.5) in h2D/h1D and 1D/2D. In 3D/2D, vertical lines are depicted in green (1.5) and pink (0.67). To avoid the extended nomenclature of B-A translocations, only information on the cis region is shown in the figures; e.g., 1S refers to TB-1Sb, 1L designates TB-1La, and so forth. More B-A translocation information can be found in Figure 1.
A flattened ratio distribution is also found in nine disomies in h2D/h1D (1L, 3L, 4S, 5S, 7S, 7L, 9S, 9L, and 10L). We examined whether this effect is caused by the presence of a greater proportion of TE outliers (ratios ≥ 6 or ≤ 1/6). To this end, the outliers were excluded, and there was still a widespread TE modulation in these disomies (Supplemental Figure 4). In 1D/2D and 3D/2D, there is less modulation (a relatively tighter peak) of TEs in these nine B-A translocation lines. These results indicate that a greater genome imbalance may cause stronger TE modulation in disomies or that the effect of aneuploidy between haploid and diploid levels differs for other reasons.
Scatter plots were used to test the significance of the differential expression of TEs. For the TEs that passed our filtering criteria (mean (control) + mean (experimental group) ≥ 1 (RPKM)), most (∼90%) showed no significant change in aneuploids, except for 7L, 9L, and 10L, in which ∼40% of TEs were significantly upregulated or downregulated (Supplemental Data Set 2). In general, h2D/h1D have more significantly upregulated and downregulated TEs than 1D/2D, 3D/2D, and 4D/2D, a result that complements the finding of greater TE modulation in h2D/h1D from the ratio distribution analysis (Supplemental Figure 5; Supplemental Data Set 2).
Minor modulation of individual TEs in the ploidy series
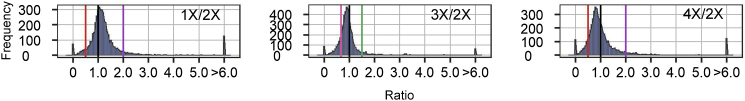
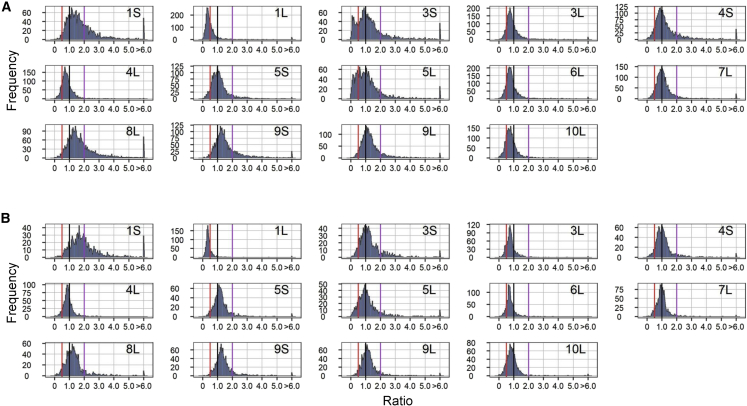
To compare modulation in the ploidy series, expression of individual TEs (same as above) in haploids (1X), triploids (3X), and tetraploids (4X) was compared with that of the diploid controls (2X). In contrast to aneuploidies, the ratio distributions in 1X/2X, 3X/2X, and 4X/2X show much less spread, as illustrated by a tighter peak around 1.0 in each comparison (Figure 3). The number of differentially expressed TEs (DETEs) is 15 in 1X/2X, 1 in 3X/2X, and 0 in 4X/2X, suggesting that changing the whole chromosome set has less effect on TE expression than changing part of the genome (Supplemental Figure 5; Supplemental Data Set 2). Kolmogorov–Smirnov (K-S) tests for distribution differences and Bartlett’s tests for distribution variances confirmed significant differences between the aneuploidy and ploidy series (Supplemental Data Set 3), indicating a greater level of disruption of global TE expression in aneuploidy than in ploidy.
Figure 3.
Ratio distributions of the ploidy series.
Ratio distributions of TE expression in 1X/2X, 3X/2X, and 4X/2X. Ratio distributions were plotted as described in Figure 2. The vertical lines are depicted in purple (2.0) and red (0.5) in 1X/2X and 4X/2X. In 3X/2X, ratio values are demarcated with labeled vertical lines in green (1.5) and pink (0.67).
The TEs on the A chromosomes may be modulated by the presence of the B chromosome, as most of the materials used in this study bear one or two copies of the B chromosome or portions of this supernumerary chromosome. A previous study systematically analyzed the effect of varied B-chromosome copy numbers on A-chromosome genes; there may be a slight effect of the presence of B chromosomes on A-chromosome TEs (Shi et al., 2022).
Greater modulation of individual TEs than genes in aneuploidy
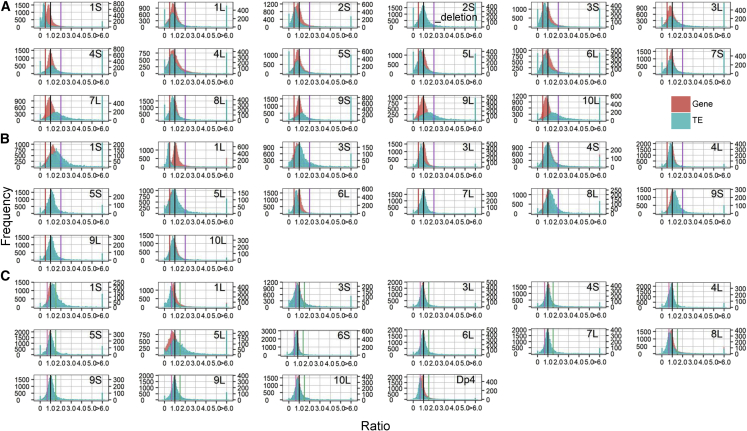
Previous studies have identified a prevalent trend of an inverse effect of core protein-coding genes with either a deletion or addition of a chromosomal segment in cis (Shi et al., 2021; Yang et al., 2021). To test the difference in modulation between TEs and genes in the same aneuploid, the ratios of expression of genes in trans and individual TEs were distributed on the same plot. In general, the ratios of the levels of expression of TEs in h2D/h1D, 1D/2D, 3D/2D, and 4D/2D show clearly different distributions from those of genes (Figure 4; Supplemental Figure 6), as confirmed by K-S and Bartlett’s tests (P value < 0.05, V). The difference between TEs and genes is more evident in disomy. For example, the peak of TE ratios in 1S approaches 0.5, whereas in 7L, 9L, and 10L, the major peak is above 1.5. By contrast, the genes show a trend of inverse modulation (peak below 1.0), suggesting a different response of TEs and genes under genome imbalance in these specific aneuploids (Figure 4A).
Figure 4.
Overlapping plots for genes and TEs.
Ratio distributions were plotted as described in Figure 2.
(A) h2D/h1D.
(B) 1D/2D.
(C) 3D/2D. The ratio distributions for genes and TEs are depicted in red and cyan. The left y axis is the number of genes in each bin, and the right y axis is the TE number.
It must be noted that the trans genes in monosomy showed both inverse and direct modulation, in which medians of ratios across 14 chromosomal arms range from 0.8 to 1.3 (Supplemental Figure 7). However, with aneuploidy involving the addition of a chromosomal segment (disomy, trisomy, and tetrasomy), gene expression was generally inversely modulated (median of ratio below 1.0) (Supplemental Figure 7). For TEs, the modulation varied depending on the cis region being varied, as the major peak of TEs ranged from 0.4 to 1.7 in 1D/2D, 0.7 to 1.3 in 3D/2D, and 0.4 to 1.7 in h2D/h1D. Nevertheless, there is a trend of inverse modulation of TEs in all aneuploids, i.e., increased TE expression (median of ratio >1) in monosomy or decreased TE expression (median of ratio <1) in disomy, trisomy, and tetrasomy. Pearson correlation coefficients (PCCs) were calculated to investigate whether the expression of TEs and genes is correlated in different genotypic comparisons, and the results showed that there is a trend but no significant correlation; PCCs equal 0.44, 0.50, and 0.37 in disomy, monosomy, and trisomy, respectively (Supplemental Figure 8). In parallel, a ploidy series of haploids, triploids, and tetraploids showed many fewer disturbances (a sharp peak near ratio 1.0) in both gene and TE expression, illustrating that gene and TE expression in balanced genomes are much less disrupted globally (Supplemental Figure 6C).
Differential modulation of TE families from two TE classes in aneuploidies
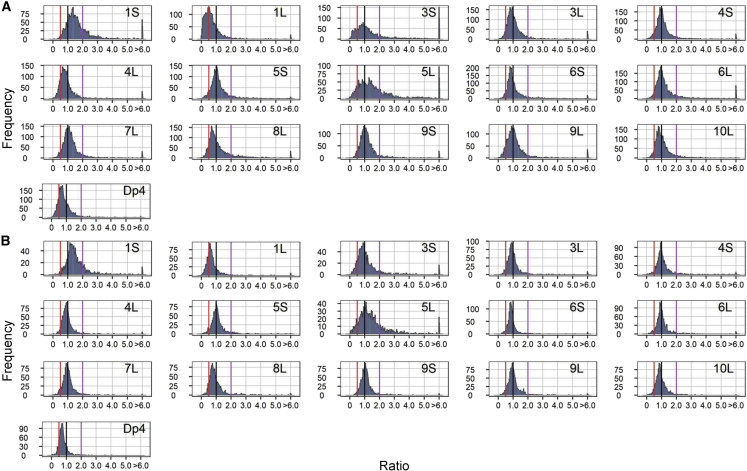
We next tested the responses of two classes of TEs in the aneuploidy and ploidy series. Because TE sequences are highly repetitive, some reads from the RNA-seq data cannot be uniquely mapped to the TE exemplar. However, TE families are connected by lineage and sequence similarity, and we therefore adapted the method from Anderson et al. (2019) to assess per-family expression in this study. Per-family TE transcript abundance was determined using reads that mapped uniquely to a specific TE as well as reads that mapped to multiple locations that were all annotated as members of the same TE family. The read counts were summarized per TE family and normalized by the reads per million (RPM). Ratios were obtained by comparing the experimental groups to the controls and plotted separately for different TE classes. The results showed that class I and class II TEs have distinct ratio distributions in all 1D/2D, 4D/2D, h2D/h1D, and most 3D/2D comparisons (15 out of 16) (Figures 5, 6, and 7), as confirmed by K-S tests (P value < 0.05, Supplemental Data Set 3A). Ratios from the two classes of TEs were tested for variances, and all aneuploids showed significant differences (Bartlett’s test) (Supplemental Data Set 3B). K-S and Bartlett’s tests of the ploidy series also suggested significant differences between the two classes of TEs, even though the expression of few TE families was significantly changed in 1X/2X, 3X/2X, and 4X/2X (Supplemental Data Set 3; Supplemental Figures 9 and 10).
Figure 5.
Ratio distributions of two classes of TEs in h2D/h1D.
Ratio distributions were plotted as described in Figure 2.
(A) The ratio distribution of class I TEs.
(B) The ratio distribution of class II TEs.
Figure 6.
Ratio distributions of the two classes of TEs in 1D/2D.
Ratio distributions were plotted as described in Figure 2.
(A) The ratio distribution of class I TEs.
(B) The ratio distribution of class II TEs.
Figure 7.
Ratio distributions of the two classes of TEs in 3D/2D.
Ratio distributions were plotted as described in Figure 2.
(A) The ratio distribution of class I TEs.
(B) The ratio distribution of class II TEs.
The h2D/h1D comparisons generally have more outlier ratios both in class I and II TEs than in diploid aneuploidies; by contrast, there are few extreme values in the comparisons of haploid to diploid control (1X/2X) in both TE classes. Further analyses were performed to investigate the possibility of activation or repression of the two TE classes in h2D/h1D among the outliers; i.e., whether TEs were derepressed or repressed in disomy in those families with high (≥6) or low (≤1/6) extreme ratios, respectively. In terms of TE activation in disomy, we divided the controls based on expression, RPM ≥ 1 and RPM < 1, and calculated the proportion of the two groups. The results showed that most of the TE families with outlier ratios ≥6 have an expression (RPM) less than 1 in the haploid controls, suggesting the TEs in the control are in a suppressed state, whereas the TEs were activated in disomy (Supplemental Data Set 4). For example, in disomy TB-10L19 (10L), there are 289 class I TE outlier ratios (≥6). About 85.5% (247/289) of them have a mean (RPM) <1 in the haploid control, suggesting that most are still repressed in the control. However, this result does not suggest that expression of these class I families from these outliers in disomy TB-10L19 changes dramatically; most of them (85.5%, 227/289) only have an expression level between 1 and 10 (RPM) in the disomy. For those TE families with a low extreme ratio (≤1/6), most showed low to medium expression in the control (1 ≤ RPM < 10) and even further suppression in disomy (Supplemental Data Set 4).
As the two classes of TEs have significantly different ratio distributions in aneuploids, we next tested whether class I TEs were modulated to a greater extent than class II TEs or vice versa. The proportion of differentially expressed TE families (DETEFs) was calculated for each TE class. If one of the two TE classes was more modulated than the other, there might be a trend of a higher percentage of DETEFs in that class. We calculated the percentage of DETEFs from the outlier (ratio ≤ 1/6 or ratio ≥ 6) and non-outlier ratios (ratio > 1/6 and < 6). The results showed that class I in h2D/h1D generally has a lower proportion of DETEFs than class II for the non-outlier families. For the families that produced outlier ratios, a higher proportion of DETEFs was found in class I (Supplemental Data Set 5). This result suggests that class I is modulated to a greater extent in outliers and less modulated in non-outliers than class II. Nevertheless, this trend is not obvious in 1D/2D and 3D/2D comparisons (Supplemental Data Set 5). This difference might be due to a generalized trend of less modulation in diploid aneuploidies, as fewer outlier ratios were found. For those non-outlier ratios, a greater number of comparisons had no more than 2% DETEFs (5 out of 14 comparisons in 1D/2D, and 10 out of 16 comparisons in 3D/2D) (Supplemental Data Set 5). These results also confirm the findings from the ratio distributions that h2D/h1D have a greater TE modulation than the diploid aneuploidies.
TE superfamilies are differentially modulated in aneuploidies
Because the two classes of TEs were modulated differently in aneuploidies (Figures 5, 6, and 7), we investigated how the TE superfamilies in each TE class were modulated. The expression of each TE family was normalized by RPM. We further divided class I into Ty1/Copia (RLC), Gypsy (RLG), Unknown LTR (RLX), RTE (RIT), L1 (RIL), and SINE (RST) and class II into Helitron (DHH), hAT (DTA), CACTA (DTC), Pif/Harbinger (DTH), Mutator (DTM), Tc1/Mariner (DTT), and Unknown TIR (DTX). We then calculated the proportion of the family members in each superfamily in the DETEFs and compared it to the proportion of the family members in each superfamily in the TE annotation file. The multinomial fit test was performed to calculate the difference. The results showed that there are generally more disomies in h2D/h1D showing a differential modulation of superfamilies in both class I and class II TEs than in diploid aneuploidies (Supplemental Data Set 6). RLG, RLX, and RST are the three most common superfamilies that were differentially modulated in class I. In class II, DTA, DTH, and DHH were the superfamilies that often showed differential modulation (Supplemental Data Set 6).
TEs have minimal effect on adjacent gene expression in aneuploidy
Previous studies have found that individual TEs can contribute to the expression of nearby genes (Makarevitch et al., 2015) or that nearby gene expression is negatively correlated with the density of methylated TEs (Hollister and Gaut, 2009). These results suggest that TEs may provide local enhancer activities that stimulate gene expression. In this study, we showed that the expression of individual TEs in h2D/h1D tends to have more DETEs, especially in TB-7Lb (7L), TB9Lc (9L), and TB-10L19 (10L), in which there are more than 2000 upregulated individual TEs in each of the comparisons. To investigate whether increased TE expression upregulated adjacent gene expression, we first identified TEs located in gene promoter regions and separated the TEs into two groups, significantly upregulated TEs and non-differentially expressed TEs in h2D/h1D. Then we compared the proportion of upregulated genes in these two groups. From the results of 7L, 9L, and 10L, there is no increase in the proportion of upregulated genes with an upregulated TE in the promoter region than that of upregulated genes whose promoter region contains a non-differentially expressed TE (Supplemental Data Set 7). For the remainder of the disomies, there are insufficient upregulated TEs (less than 20) for suitable comparisons.
Discussion
Despite many recent advances in comprehensive gene expression studies on aneuploidy and polyploidy, changes in expression of TEs under genomic imbalance have not been investigated in any organism to our knowledge. In this study, we examined global TE modulation in haploid and diploid maize aneuploids with varied dosages of multiple chromosomal segments, as well as a whole-genome ploidy series including haploids, diploids, triploids, and tetraploids. This analysis enabled us to address several questions about genomic balance. (1) How do aneuploidy and polyploidy impact the expression of TEs globally? (2) Does genomic imbalance have a similar effect on TE expression and protein-coding gene expression? (3) How are the two classes of TEs modulated in aneuploidies? (4) Do the differentially expressed TEs affect adjacent gene expression, which would compound the effect of genomic imbalance? This study provides insights into these questions.
Most TEs remained silenced in both the aneuploidy and ploidy series, but aneuploidy showed greater modulation of those that were expressed
In the genome, repression of TEs is initiated by DNA methylation through the RdDM pathway followed by maintenance of methylation by DNA methyltransferases (Sigman and Slotkin, 2016). TE activation can occur under some stress conditions, such as protoplasting, tissue culture, and chromosomal breakage (Rhoades and Dempsey, 1982; Pouteau et al., 1991; Hirochika et al., 1996). In this study, nearly 90% of TEs failed to pass our filtering criteria for TE expression, suggesting that most remained silenced or were expressed at a very low level. However, for those elements that were expressed, greater TE modulation in aneuploidy than in the ploidy series suggests that changing parts of the genome (aneuploidy) has a greater effect on the regulatory machinery that affects TE expression than changing whole chromosome sets (ploidy series). The prevalent inverse TE modulation in aneuploidy suggests a relationship between the response of TEs to genomic imbalance and that of core genes.
Greater modulation of TEs than of genes
Previous studies have found that different functional groups of genes exhibit distinct modulations in aneuploidies (Hou et al., 2018; Shi et al., 2021; Yang et al., 2021). Typically, transcription factors and signal pathway genes show slight inverse modulation, but some functional groups like structural components of the ribosome are modulated to a greater extent. Here, comparison of the modulation of TEs and core genes under the same experimental conditions shows that TEs are misregulated to a greater extent than core genes.
Differential modulation of two classes of TEs in aneuploids
Nearly 80% of the maize genome consists of two classes of TEs, class I and class II. Although the two TE families utilize different mechanisms to perpetuate themselves, the first principle of TE silencing in plant genomes is the importance of the chromosomal position where the elements are located (Sigman and Slotkin, 2016). Small nonautonomous DNA transposons, particularly Mutator, hAT, Helitron, and miniature TE derivatives (MITEs), prefer to insert into or near a gene (Jiang and Wessler, 2001). The expression of TEs near or within genes is regulated by RdDM and maintenance TE-silencing mechanisms. Retrotransposons are frequently found in heterochromatin, mainly in nested clusters, intermingled with other TEs (SanMiguel and Vitte, 2009). Unlike euchromatic regions, in which RdDM is responsible for TE repression, heterochromatic regions require the chromatin-remodeling protein DECREASED DNA METHYLATION1 (DDM1) for silencing (Fu et al., 2018; Sigman and Slotkin, 2016; Zemach et al., 2013). Because the regulation of class I and class II TEs involves different mechanisms, genomic imbalance of various portions of the genome is likely to affect their expression differently. This is indeed the case in our results, as class I and class II TEs were differentially modulated in most of the aneuploids.
Effect of TEs on adjacent gene expression
TEs can affect gene expression in different ways. For example, TEs might contribute to the activation of maize genes in response to abiotic stress, which suggests that TEs may provide local enhancer activities that stimulate stress-responsive gene expression (Makarevitch et al., 2015). Also, TEs regularly capture fragments of genes. When the host silences these TEs, small interfering RNAs homologous to the captured regions may also target the genes. Such an intragenomic conflict may not ultimately affect important genes but may lead to the pseudogenization of less-constrained genes (Hollister and Gaut, 2009; Bousios and Gaut, 2016; Muyle et al., 2020). In our results, we found that three disomies (TB-7Lb, TB-9Lc, and TB-10L19) showed significant upregulation of more than 2000 TEs. By comparing the proportion of significantly upregulated genes that have a nearby unchanged TE in the promoter region to those with neighboring TEs that are significantly upregulated, we did not find a trend of increase for the significantly upregulated TE group in these three disomies. The results indicate that an interaction of TEs and genes in the context of aneuploidy does not compound expression changes in response to genomic imbalance.
Methods
RNA-seq data and PCA plot analysis
The RNA-seq data were downloaded from NCBI using GEO accession numbers GSE149186 and GSE156986. RNA-seq and PCA plot analyses were described previously (Shi et al., 2021, 2022; Yang et al., 2021). Low-quality reads were filtered out using fastq quality_filter (-Q 33 -q 20 -p 80); the remaining reads were mapped to External RNA Controls Consortium (ERCC) sequences using bowtie2 --phred33 --no-unal so that reads that mapped to ERCC were removed. To deplete reads derived from organelle genes, reads that uniquely mapped to maize organelle genome sequences with no more than two mismatches (mitochondria and chloroplast) using tophat2 were excluded from further analysis. The remaining reads were then mapped to maize W22 TE exemplar sequences that contain intact and non-overlapping TEs (Springer et al., 2018; Shi et al., 2022) based on the TE GFF3 file using bowtie2 (--phred33 -N 0 –no-unal -k 10) (Langmead and Salzberg, 2012).
For individual TE expression, uniquely mapped reads with zero mismatches were selected and normalized by RPKM. For per TE family expression, reads that were uniquely mapped to a TE family or multi-mapped but annotated as members of the same TE family were summarized and quantified by RPM using a Perl script. TEs with low expression (mean of the control group + mean of the experimental group < 1 (RPKM for individual TE and RPM for TE family)) were excluded from further study.
For gene expression, after filtering out ERCC and organelle reads, the remaining reads were mapped to the maize reference genome W22v2 using TopHat2 with default parameters (Kim et al., 2013). Normalized read counts were generated by Cuffdiff (raw-mapped-norm) for each comparison (Trapnell et al., 2013). Genes with low expression (mean of the control group + mean of the experimental group < 1 (RPKM)) were excluded from further study. PCA plots were generated in R using normalized read counts (Supplemental Figure 1). To determine whether there were any outliers, the mean of PC1 or PC2 was subtracted from the first principal component (PC1) or second principal component (PC2), respectively, of each biological replicate. If the absolute value of the subtraction was greater than or equal to the standard deviation (|PC1−PC1mean|≥2×SD or |PC2−PC2mean|≥2×SD), the sample was regarded as an outlier (Supplemental Data Set 1).
Ratio distributions and scatterplots
Ratio distributions and scatterplots were generated using the same methods described in a previous study (Shi et al., 2021, 2022; Yang et al., 2021). In brief, the ratio was generated by dividing the mean of experimental counts (RPKM values) by the mean of control counts. The ratios were plotted as a histogram. For the overlapping plots, the left y axis is the number of genes in each bin, and the right y axis is the TE number. For scatterplots, we performed differential TE expression analysis with edgeR (empirical analysis of DGE [digital gene expression] in R) using raw counts to test the significance of differential expression levels between each treatment group and the control group (Robinson et al., 2010; McCarthy et al., 2012). The FDR value and log2(fold change) were used for the scatterplots. The log2 fold change (logFC) between treatment and control was plotted on the x axis, and the mean of log2 of RPKMof the treatment and control group was plotted on the y axis. Data points with an FDR <0.05 and a corresponding logFC >0 were depicted in magenta, and points with an FDR <0.05 and a corresponding logFC <0 were depicted in green. Otherwise, points were shown in black.
K-S and Bartlett’s test
K-S and Bartlett’s tests were performed using R with extreme ratio values (ratio ≥ 6 or ≤ 1/6) excluded. Details of the statistical tests were described in a previous study (Shi et al., 2021). For an individual TE or TE family, ratios of the expression of the individual TE or TE family were generated for aneuploidy and ploidy series followed by the K-S test to compare the two ratio lists and determine whether there was a significant difference (P value < 0.05). Bartlett’s test was used to examine whether the variances of the ratios were equal across different groups (Supplemental Data Set 3).
Differential modulation analysis of TE superfamilies
To investigate the differential modulation of different TE superfamilies (e.g., RLG, retrotransposon, LTR, Gypsy, RLG00001), we first filtered out TE families with low expression. The number of each superfamily was compared with the number of total families in class I or II and then multiplied by the number of significantly upregulated, significantly downregulated, and non-differentially expressed TE families in order to calculate the expected numbers of significantly upregulated, significantly downregulated, and non-differentially expressed TE superfamilies. With the number of significantly upregulated, significantly downregulated, and non-differentially expressed for each superfamily in the expressed TE family, a multinomial fit test was performed. The null hypothesis expects that there is no differential modulation. This hypothesis would be rejected if the P value of the subsequent Chi-squared test was less than 0.05, and thus differential modulation would be accepted.
TE and adjacent gene expression
TEs were identified in the promoter regions of genes using the same method described in a previous study (Makarevitch et al., 2015). In general, for each gene, transposons located within 1 kb of the transcription start site (TSS) were identified using the W22 reference gene and TE annotations (Springer et al., 2018). The distance of a TE from the TSS of each gene was determined using the closest tool from the BEDTools suite (Quinlan and Hall, 2010); TEs upstream were given a positive distance value, and TEs downstream were given a negative distance value. To determine whether TEs affect nearby gene expression, the TEs in the promoter regions of genes were divided into two groups, non-differentially expressed TEs (FDR ≥ 0.05) or significantly upregulated TEs (FDR < 0.05 and log fold change with base 2 (log2fold_change, or logFC) > 0). Then, the proportion of significantly upregulated genes (corrected P value [q value] < 0.05 and logFC > 0) in these two groups was computed and compared.
Funding
This research was supported by National Science Foundation USA grant IOS-1545780. Computation for this work was performed on the high-performance computing infrastructure provided by Research Computing Support Services and in part by the National Science Foundation USA under grant number CNS-1429294 at the University of Missouri, Columbia, MO, https://doi.org/10.32469/10355/69802.
Author contributions
H.Y., X.S., C.C., J.H., T.J., J.C., and J.A.B. designed the research; H.Y., X.S., C.C., and J.A.B. analyzed the data; and H.Y. and J.A.B. wrote the paper.
Acknowledgments
No conflict of interest is declared.
Published: October 28, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
(A) Disomy. (B) Monosomy, trisomy, tetrasomy, and ploidy series.
If more than 100 TEs are significantly upregulated or significantly downregulated, they are marked in magenta or green, respectively.
K-S test in (A) and Bartlett’s test in (B). Ratio distributions were compared between aneuploidy and ploidy (disomy vs. haploid, monosomy vs. haploid, trisomy vs. triploid, or tetrasomy vs. tetraploid). P values were computed with extreme values (ratio >6 or <1/6) excluded and are listed in the order of mention in the text. The P values are listed for the probability that a pair of compared distributions are the same. P values less than 0.05 are highlighted by pink shading.
(A) Class I outliers with ratios ≥ 6. (B) Class II outliers with ratios ≥ 6. (C) Class I outliers that have ratios ≤ 1/6.
(A and B) Percentage of differentially expressed TE (DETE) families in the non-outlier (ratio <6 & ratio >1/6) (A) or outlier ratios (ratio ≥6 & ratio ≤1/6) (B) in disomy. (C and D) Proportion of DETE families in the non-outlier (C) or outlier ratios (D) in monosomy. (E) Proportion of DETE families in the non-outlier ratios in trisomy.
Class I superfamilies: RLC, Ty1/Copia, RLG, Gypsy. RLX, unknown LTR, RIT, RTE. RIL, L1. RST, SINE. Class II superfamilies: DHH, Helitron. DTA, hAT. DTC, CACTA. DTH, Pif/Harbinger. DTM, Mutator. DTT, Tc1/Mariner. DTX, unknown TIR.
Data availability
The RNA-seq data were downloaded from NCBI using GEO accession numbers GSE149186 and GSE156986.
References
- Anderson S.N., Stitzer M.C., Zhou P., Ross-Ibarra J., Hirsch C.D., Springer N.M. Dynamic patterns of transcript abundance of transposable element families in maize. G3 (Bethesda) 2019;9:3673–3682. doi: 10.1534/g3.119.400431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J.M., Jaillon O., Duret L., Noel B., Jubin C., Porcel B.M., Ségurens B., Daubin V., Anthouard V., Aiach N., et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Beckett J.B. In: Chromosome Engineering in Plants: Genetics, Breeding, Evolution (Part A) Gupta P.K., Tsuchiya T., editors. Elsevier; Amsterdam: 1991. Cytogenetic, genetic and plant breeding applications of B–A translocations in maize; pp. 493–529. [Google Scholar]
- Birchler J.A. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics. 1979;92:1211–1229. doi: 10.1093/genetics/92.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A. The genetic basis of dosage compensation of alcohol dehydrogenase-1 in maize. Genetics. 1981;97:625–637. doi: 10.1093/genetics/97.3-4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Bhadra U., Bhadra M.P., Auger D.L. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Riddle N.C., Auger D.L., Veitia R.A. Dosage balance in gene regulation: biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Yang H. The supernumerary B chromosome of maize: drive and genomic conflict. Open Biol. 2021;11 doi: 10.1098/rsob.210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 2010;186:54–62. doi: 10.1111/j.1469-8137.2009.03087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. One hundred years of gene balance: how stoichiometric issues affect gene expression, genome evolution, and quantitative traits. Cytogenet. Genome Res. 2021;161:529–550. doi: 10.1159/000519592. [DOI] [PubMed] [Google Scholar]
- Blakeslee A.F. Types of mutations and their possible significance in evolution. Am. Nat. 1921;55:254–267. [Google Scholar]
- Blakeslee A.F. New jimson weeds from old chromosomes. J. Hered. 1934;25:81–108. [Google Scholar]
- Blakeslee A.F., Belling J. Chromosomal mutations in the Jimson weed, Datura stramonium. J. Hered. 1924;15:195–206. [Google Scholar]
- Blakeslee A.F., Belling J., Farnham M.E. Chromosomal duplication and Mendelian phenomena in Datura mutants. Science. 1920;52:388–390. doi: 10.1126/science.52.1347.388. [DOI] [PubMed] [Google Scholar]
- Blavet N., Yang H., Su H., Solanský P., Douglas R.N., Karafiátová M., Šimková L., Zhang J., Liu Y., Hou J., et al. Sequence of the supernumerary B chromosome of maize provides insight into its drive mechanism and evolution. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2104254118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T., Vandepoele K., De Bodt S., Simillion C., Maere S., Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousios A., Gaut B.S. Mechanistic and evolutionary questions about epigenetic conflicts between transposable elements and their plant hosts. Curr. Opin. Plant Biol. 2016;30:123–133. doi: 10.1016/j.pbi.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Bray D., Lay S. Computer-based analysis of the binding steps in protein complex formation. Proc. Natl. Acad. Sci. USA. 1997;94:13493–13498. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C.B. Sex in relation to chromosomes and genes. Am. Nat. 1925;59:127–137. [Google Scholar]
- Freeling M., Lyons E., Pedersen B., Alam M., Ming R., Lisch D. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Fu F.F., Dawe R.K., Gent J.I. Loss of RNA-directed DNA methylation in maize chromomethylase and DDM1-Type nucleosome remodeler mutants. Plant Cell. 2018;30:1617–1627. doi: 10.1105/tpc.18.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin P.T., Niederhuth C.E., Schmitz R.J. A comparative analysis of 5-Azacytidine- and zebularine-induced DNA demethylation. G3 (Bethesda) 2016;6:2773–2780. doi: 10.1534/g3.116.030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes L., Pinney J.W., Lovell S.C., Oliver S.G., Robertson D.L. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M.B., Seetharam A.S., Woodhouse M.R., Chougule K.M., Ou S., Liu J., Ricci W.A., Guo T., Olson A., Qiu Y., et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science. 2021;373:655–662. doi: 10.1126/science.abg5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Gaut B.S. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Shi X., Chen C., Islam M.S., Johnson A.F., Kanno T., Huettel B., Yen M.R., Hsu F.M., Ji T., et al. Global impacts of chromosomal imbalance on gene expression in Arabidopsis and other taxa. Proc. Natl. Acad. Sci. USA. 2018;115:E11321–E11330. doi: 10.1073/pnas.1807796115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I., Rogers A.J., Lange C., Raby B.A., Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Wessler S.R. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell. 2001;13:2553–2564. doi: 10.1105/tpc.010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J.O., Kim P.M., Chen X., Urban A.E., Weissman S., Snyder M., Gerstein M.B. The current excitement about copy-number variation: how it relates to gene duplications and protein families. Curr. Opin. Struct. Biol. 2008;18:366–374. doi: 10.1016/j.sbi.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.A., Coe E.H., Darrah L.L. Genetic variation in dosage effects in maize aneuploids. Genome. 1996;39:711–721. doi: 10.1139/g96-090. [DOI] [PubMed] [Google Scholar]
- Lee E.A., Darrah L.L., Coe E.H. Dosage effects on morphological and quantitative traits in maize aneuploids. Genome. 1996;39:898–908. doi: 10.1139/g96-113. [DOI] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Waters A.J., West P.T., Stitzer M., Hirsch C.N., Ross-Ibarra J., Springer N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. In: Handbook of Maize. Bennetzen J.L., Hake S., editors. Springer; New York, NY: 2009. The polyploid origin of maize. [DOI] [Google Scholar]
- Muyle A., Seymour D., Darzentas N., Primetis E., Gaut B.S., Bousios A. Gene capture by transposable elements leads to epigenetic conflictin maize. Mol. Plant. 2020;14:237–252. doi: 10.1016/j.molp.2020.11.003. [DOI] [PubMed] [Google Scholar]
- Pouteau S., Huttner E., Grandbastien M.A., Caboche M. Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 1991;10:1911–1918. doi: 10.1002/j.1460-2075.1991.tb07717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.M., Dempsey E. The induction of mutable systems in plants with the high-loss mechanism. Maize News Lett. 1982;56:21–26. [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Mane S.P., Dharmawardhana P., Slavov G.T., Crasta O.R., Strauss S.H., Brunner A.M., Difazio S.P. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 2012;22:95–105. doi: 10.1101/gr.125146.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P., Vitte C. In: Handbook of Maize. Bennetzen J.L., Hake S., editors. Springer; New York, NY: 2009. The LTR-retrotransposons of maize. [DOI] [Google Scholar]
- Schinzel A. Walter de Gruyter; Berlin, New York: 2001. Catalogue of Unbalanced Chromosome Aberrations in Man. [Google Scholar]
- Sears E.R. Nullisomic analysis in common wheat. Am. Nat. 1953;87:245–252. [Google Scholar]
- Sigman M.J., Slotkin R.K. The first rule of plant transposable element silencing: location, location, location. Plant Cell. 2016;28:304–313. doi: 10.1105/tpc.15.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C., Vandepoele K., Van Montagu M.C.E., Zabeau M., Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Yang H., Chen C., Hou J., Hanson K.M., Albert P.S., Ji T., Cheng J., Birchler J.A. Genomic imbalance determines positive and negative modulation of gene expression in diploid maize. Plant Cell. 2021;33:917–939. doi: 10.1093/plcell/koab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Yang H., Chen C., Hou J., Ji T., Cheng J., Birchler J.A. Effect of aneuploidy of a nonessential chromosome on gene expression in maize. Plant J. 2022;110:193–211. doi: 10.1111/tpj.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N.M., Anderson S.N., Andorf C.M., Ahern K.R., Bai F., Barad O., Barbazuk W.B., Bass H.W., Baruch K., Ben-Zvi G., et al. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet. 2018;50:1282–1288. doi: 10.1038/s41588-018-0158-0. [DOI] [PubMed] [Google Scholar]
- Tasdighian S., Van Bel M., Li Z., Van de Peer Y., Carretero-Paulet L., Maere S. Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell. 2017;29:2766–2785. doi: 10.1105/tpc.17.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia R.A. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Veitia R.A. Gene dosage balance in cellular pathways: implications for dominance and gene duplicability. Genetics. 2004;168:569–574. doi: 10.1534/genetics.104.029785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yang H., Shi X., Chen C., Hou J., Ji T., Cheng J., Birchler J.A. Predominantly inverse modulation of gene expression in genomically unbalanced disomic haploid maize. Plant Cell. 2021;33:901–916. doi: 10.1093/plcell/koab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.-H., et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Disomy. (B) Monosomy, trisomy, tetrasomy, and ploidy series.
If more than 100 TEs are significantly upregulated or significantly downregulated, they are marked in magenta or green, respectively.
K-S test in (A) and Bartlett’s test in (B). Ratio distributions were compared between aneuploidy and ploidy (disomy vs. haploid, monosomy vs. haploid, trisomy vs. triploid, or tetrasomy vs. tetraploid). P values were computed with extreme values (ratio >6 or <1/6) excluded and are listed in the order of mention in the text. The P values are listed for the probability that a pair of compared distributions are the same. P values less than 0.05 are highlighted by pink shading.
(A) Class I outliers with ratios ≥ 6. (B) Class II outliers with ratios ≥ 6. (C) Class I outliers that have ratios ≤ 1/6.
(A and B) Percentage of differentially expressed TE (DETE) families in the non-outlier (ratio <6 & ratio >1/6) (A) or outlier ratios (ratio ≥6 & ratio ≤1/6) (B) in disomy. (C and D) Proportion of DETE families in the non-outlier (C) or outlier ratios (D) in monosomy. (E) Proportion of DETE families in the non-outlier ratios in trisomy.
Class I superfamilies: RLC, Ty1/Copia, RLG, Gypsy. RLX, unknown LTR, RIT, RTE. RIL, L1. RST, SINE. Class II superfamilies: DHH, Helitron. DTA, hAT. DTC, CACTA. DTH, Pif/Harbinger. DTM, Mutator. DTT, Tc1/Mariner. DTX, unknown TIR.
Data Availability Statement
The RNA-seq data were downloaded from NCBI using GEO accession numbers GSE149186 and GSE156986.