Dear Editor,
Many flowering plants utilize the self-incompatibility (SI) response as a genetic mechanism to prevent self-pollen from establishing on the stigmas, thereby promoting outcrossing and genetic diversity. In Brassica during SI, recognition of the pollen ligand SP11 by S-locus receptor kinase (SRK) results in activation of the E3 ligase ARM-Repeat-Containing protein (ARC1), which leads to proteasomal degradation of compatibility factors required for successful pollen acceptance. ARC1 was originally identified as an interactor of the SRK kinase domain and is highly expressed in mature stigmas (Gu et al., 1998). Antisense suppression of ARC1 resulted in partial breakdown of SI in the self-incompatible Brassica napus W1 line, establishing the role of ARC1 as a positive regulator of SI (Stone et al., 1999). This was further supported by RNAi-mediated suppression of ARC1 in the self-incompatible A. lyrata, which resulted in partial breakdown of the SI pathway (Indriolo et al., 2012). The observed partial compromise of SI in both Brassica and Arabidopsis suggested that either an alternative SI pathway or incomplete suppression of ARC1 could have resulted in the incomplete breakdown of SI. This question has remained unresolved. In this study, we created ARC1 loss-of-function B. napus lines to demonstrate the central role of ARC1 in mediating the SI response.
Despite the partial breakdown of SI shown in two different systems, the role of ARC1 in the Brassicaceae SI response has been questioned for the past several decades. Stable co-expression of Brassica SLG, SRK, and ARC1 was insufficient to confer the SI phenotype in compatible Arabidopsis thaliana (Bi et al., 2000). In another report, when A. thaliana plants were transformed to express SRKb and SCRb genes, a strong SI phenotype was observed in the absence of ARC1 (Kitashiba et al., 2011). A. thaliana plants that exhibit the SI phenotype through transformation of SRK and SCR alone show trends in heritability, developmental regulation, and intensity of SI response similar to those of self-incompatible Brassica cultivars (Nasrallah and Nasrallah, 2014). Although, evolutionarily, Arabidopsis that lacked a bona fide ARC1 ortholog could have re-purposed other E3 ligases to assume the role of ARC1, the question of how complete loss of ARC1 could influence SI in Brassica sp. remained unresolved.
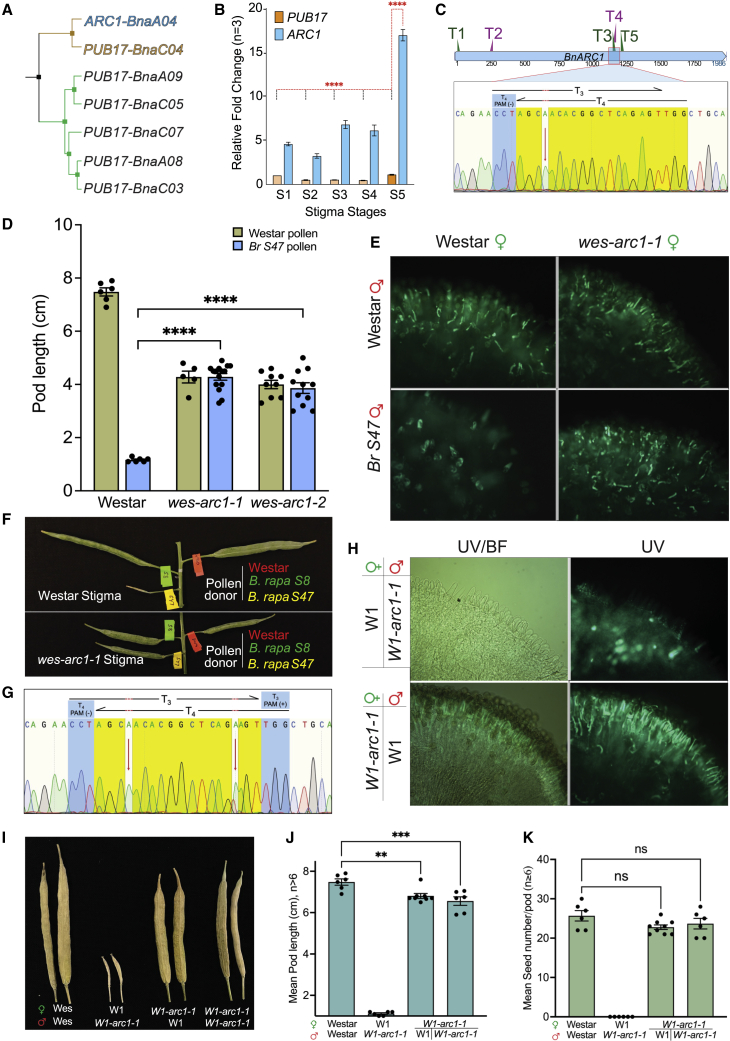
To unequivocally examine the role of ARC1 during SI, we created ARC1 loss-of-function B. napus using the CRISPR–Cas9 platform. In order to efficiently design specific targets for the CRISPR constructs, we retrieved available gene copies of ARC1 or similar genes present in B. napus (allotetraploid from the A and C genomes of B. rapa and B. oleracea) (http://cbi.hzau.edu.cn/bnapus/index.php) (Song et al., 2020). After validation of the sequences across the published NCBI database, TOPO cloning was performed with BnARC1-specific cDNA amplification products obtained from an RNA pool of fully mature B. napus stigmas. Sanger sequencing of the clones revealed that BnARC1 is a single-copy gene derived from Brassica rapa (A genome of rapa), whereas its homolog from the C genome (oleracea) was identified at low frequency and was most similar to PUB17 family genes (Figure 1A). When the expression profiles of these two genes were assessed at various stages of stigma maturity, ARC1 had significantly higher expression than PUB17 and peaked at stigma maturity (Figure 1B).
Figure 1.
CRISPR–CAS9-mediated editing of ARM-Repeat-Containing 1 protein (ARC1) leads to a complete breakdown of the self-incompatibility response in Brassica napus.
(A)ARC1 and related sequences were retrieved from the Brassica napus pangenome information resource database. Highly divergent PUB17 (PUB17-BnaC03a) is not shown in the cladogram.
(B) Relative expression of ARC1 and PUB17 during stigma development was assessed by qPCR. The bars show the fold change in BnPUB17 and BnARC1 during stigma development. Error bars represent the standard error of the mean (±SEM).
(C) Schematic diagram showing the location of the various guideRNA sites along with the chromatogram generated from Sanger sequencing of BnARC1 from wes-arc1-1. Sequence spans the T3 and T4 target sites within the BnARC1 coding region. The red arrow indicates the biallelic insertion of an “A” at precisely 3 bases from the T4 protospacer adjacent motif (PAM) sequence, indicating that the insertion was created during an erroneous endogenous repair of the double stranded break performed by Cas9 at the T4 position.
(D) Bar graph representing the average pod length in wes-arc1 when pollinated with compatible Westar or incompatible S47 pollen. The error bars represent ±SEM.
(E) Aniline blue assay to detect pollen attachment and pollen tube penetration in flowers pollinated with pollen from Westar or S47 haplotypes obtained from B. rapa, 24 h post-anthesis. The images were obtained either with UV (280–390 nm) to visualize aniline blue stain or with a green filter to observe pollen attachment.
(F) Representative picture of pods still attached to inflorescence, pollinated with Westar (self), S47 (incompatible), and S8 (compatible), showing the breakdown of SI in the wes-arc1-1 line.
(G) Chromatogram generated from Sanger sequencing of BnARC1 from W1-arc1-1. The sequence spans the T3 and T4 target sites within the BnARC1 coding region. The red arrows indicate the biallelic insertion of an “A” at precisely 3 bases from the T4 PAM sequence and the biallelic insertions of an “A” or “T” at 3 bases from the T3 PAM sequence.
(H) Aniline blue assay to assess pollen attachment and pollen tube penetration in reciprocal crosses of the CRISPR-edited line W1-arc1-1, showing breakdown of SI in the absence of ARC1.
(I) Representative picture of mature pods from hand-pollinated individual flowers of the combinations shown. The full extension of pods after incompatible pollination indicates compromised SI in W1-arc1-1 lines.
(J and K) Bar graphs representing the average pod length (J) and seed set (K) in W1-arc1-1 pollinated with W1 or self-pollinated compared with self-pollinated Westar. Error bars represent ±SEM.
We next created a multiplex CRISPR–Cas9 ARC1 editing construct and generated several B. napus Westar (SC) transgenic lines harboring the editing system through Agrobacterium-mediated transformation (Ma et al., 2015; Stanic et al., 2021). When ARC1 was amplified from these transgenic lines, several lines, including the wes-arc1-1 line, contained biallelic edits in target 4 that caused a frameshift in the ARC1 sequence (Figure 1C, only target 3/4 [T3/4] shown).
The Westar cultivar is self-compatible due to an insertion in the promoter region of SP11, leading to a lack of expression of SP11, the pollen ligand required for SI response (Okamoto et al., 2007). However, it retains all downstream SI signaling components (SRK and ARC1), as it readily rejects pollen of S47 haplotype origin with a functional SP11 (Okamoto et al., 2007). The T2 generations of two independently edited wes-arc1 plants with biallelic edits were used for pollination assays with compatible Westar or S8 pollen and incompatible S47 pollen. When B. rapa S47 haplotype pollen was applied to control Westar stigmas, a robust SI was observed in the Westar stigmas, which readily rejected pollen of the S47 haplotype (Figure 1D–1F). By contrast, the wes-arc1-1 and wes-arc1-2 plants showed a complete breakdown of the SI response when pollinated with S47 pollen (Figure 1D–1F). When either Westar pollen or B. rapa pollen from an S8 haplotype (SRK8 is absent in Westar) was used as a positive control for stigma receptivity, full acceptance was observed in both Westar controls and wes-arc1 plants, indicating that stigma receptivity was not modified in the edited line (Figure 1D).
We next sought to test whether nullifying ARC1 in the well characterized self-incompatible W1 background would result in complete breakdown of SI. The isogenic, self-incompatible W1 line was created by introgression of the dominant 910 B. rapa haplotype (SRK 910/SP11-910); it displays a very strong SI phenotype when self-pollinated but is compatible when crossed with Westar. We predicted that regardless of the upstream receptor/ligand complex, abolishing ARC1 function should lead to complete breakdown of SI.
Because W1 plants are incompatible and difficult to propagate/regenerate through conventional Agrobacterium-based tissue culture approaches, W1 plants were crossed with the wes-arc1 mutant transgenic lines harboring the ARC1 editing system. In the F1 generation, we identified a W1-arc1-1 line that displayed strong breakdown of SI when self-pollinated. Sequencing of ARC1 revealed edits at both the T3/4 sites (Figure 1G, only T3/4 shown). When pollen from W1-arc1-1 was tested on W1 stigmas, W1 stigmas rejected W1-arc1-1 pollen, indicating that the SP11-910 haplotype was not altered or deleted. On the other hand, ARC1-edited W1-arc1-1 plants readily accepted pollen from self-incompatible W1 plants (Figure 1H). This complete breakdown resulted in full seed set (Figure 1I) comparable to that observed when stigmas were pollinated with compatible Westar pollen (Figure 1J and 1K). This compromise in SI was further confirmed in at least four successive generations of W1-arc1-1 plants that harbored an intact SRK910. Flowers from these lines were also used to pollinate W1 stigmas and confirm the SI reaction, demonstrating that the S-haplotype was unmodified in these lines.
We have convincingly demonstrated that elimination of ARC1 results in complete breakdown of SI in two different S-haplotypes (S47 and SRK910), confirming the essential nature of ARC1 for SI response in Brassica. In both situations, the closely related PUB17 ortholog was unaltered, indicating the exclusive nature of ARC1 for mediating the SI response. Although this study eliminates any doubt as to whether ARC1 is required for SI in Brassica, the fact that ARC1 was shown to be dispensable in certain cases in A. thaliana suggests that there could be an evolutionary significance to this observation. In a self-incompatible A. lyrata ecotype, ARC1 is present and required for SI response (Indriolo et al., 2012; Indriolo and Goring, 2014), whereas in self-compatible Arabidopsis species, ARC1 is often found to have been deleted (Indriolo et al., 2012, 2014). During the switch from SI to compatibility, A. thaliana could have either lost the ARC1 gene and other components of the SI pathway or neo-functionalized them for various other pathways. Alternatively, ARC1 function could be species specific, as shown in a recent report in which A. thaliana SC transgenic lines overexpressing SCR–SRK–ARC1 from A. halleri displayed the SI phenotype, whereas they failed to manifest the SI phenotype when the ARC1 gene was derived from B. napus. These results indicate that the display of incompatibility in SC A. thaliana might have genus-specific preferences (Zhang et al., 2019).
Nonetheless, this investigation has clearly demonstrated that the absence of functional BnARC1 in Westar and W1 plants leads to their inability to mount successful SI, showing that ARC1 is an indispensable downstream effector of SI in Brassica.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada for M.A.S., K.A., and N.M.N.H. X.L. was supported by the National Natural Science Foundation of China (31870300) and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Author contributions
M.A.S. and K.A. conceived and designed the experiments. K.A., N.M.N.H., and X.L. performed the experiments. K.A. and M.A.S. analyzed the data. K.A. and M.A.S. wrote the manuscript. M.A.S., K.A., N.M.N.H., and X.L. revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
No conflict of interest is declared.
Published: December 14, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Bi Y.M., Brugière N., Cui Y., Goring D.R., Rothstein S.J. Transformation of Arabidopsis with a brassica SLG/SRK region and ARC1 gene is not sufficient to transfer the self-incompatibility phenotype. Mol. Gen. Genet. 2000;263:648–654. doi: 10.1007/s004380051213. [DOI] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E., Goring D.R. A conserved role for the ARC1 E3 ligase in Brassicaceae self-incompatibility. Front. Plant Sci. 2014;5:181. doi: 10.3389/fpls.2014.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E., Tharmapalan P., Wright S.I., Goring D.R. The ARC1 E3 ligase gene is frequently deleted in self-compatible brassicaceae species and has a conserved role in arabidopsis lyrata self-pollen rejectionw. Plant Cell. 2012;24:4607–4620. doi: 10.1105/tpc.112.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E., Safavian D., Goring D.R. The ARC1 E3 ligase promotes two different self-pollen avoidance traits in Arabidopsis. Plant Cell. 2014;26:1525–1543. doi: 10.1105/tpc.114.122879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H., Liu P., Nishio T., Nasrallah J.B., Nasrallah M.E. Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2011;108:18173–18178. doi: 10.1073/pnas.1115283108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Nasrallah J.B., Nasrallah M.E. Robust self-incompatibility in the absence of a functional ARC1 gene in Arabidopsis thaliana. Plant Cell. 2014;26:3838–3841. doi: 10.1105/tpc.114.129387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Odashima M., Fujimoto R., Sato Y., Kitashiba H., Nishio T. Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes. Plant J. 2007;50:391–400. doi: 10.1111/j.1365-313X.2007.03058.x. [DOI] [PubMed] [Google Scholar]
- Song J.M., Liu D.X., Xie W.Z., Yang Z., Guo L., Liu K., Yang Q.Y., Chen L.L. BnPIR: Brassica napus Pan-genome Information Resource for 1, 689 accessions. Plant Biotechnol. J. 2020;19:412–414. doi: 10.1111/pbi.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic M., Hickerson N.M.N., Arunraj R., Samuel M.A. Gene-editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola) Plant Biotechnol. J. 2021;19:639–641. doi: 10.1111/pbi.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science. 1999;286:1729–1731. doi: 10.1126/science.286.5445.1729. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhou G., Goring D.R., Liang X., Macgregor S., Dai C., Wen J., Yi B., Shen J., Tu J., et al. Generation of transgenic self-incompatible Arabidopsis thaliana shows a genus-specific preference for self-incompatibility genes. Plants. 2019;8:570–616. doi: 10.3390/plants8120570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.