Abstract
Proanthocyanidins (PAs) are natural flavan-3-ol polymers that contribute protection to plants under biotic and abiotic stress, benefits to human health, and bitterness and astringency to food products. They are also potential targets for carbon sequestration for climate mitigation. In recent years, from model species to commercial crops, research has moved closer to elucidating the flux control and channeling, subunit biosynthesis and polymerization, transport mechanisms, and regulatory networks involved in plant PA metabolism. This review extends the conventional understanding with recent findings that provide new insights to address lingering questions and focus strategies for manipulating PA traits in plants.
Key words: condensed tannin, proanthocyanidin, carbocation chemistry, metabolic channeling, non-enzymatic polymerization
Proanthocyanidins are flavonoid polymers with biological activities that affect plant and animal health. Unlike most plant natural products, their biosynthetic pathways appear to involve unstable intermediates and enzyme-catalyzed and non-enzymatic reactions. This review summarizes our current understanding of how these complex molecules are assembled from intermediates in the core flavonoid pathway.
Introduction
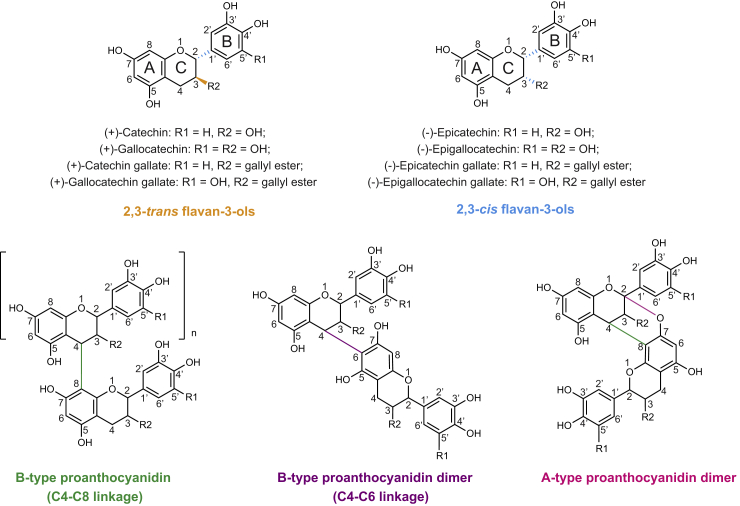
Proanthocyanidins (PAs), also called condensed tannins, are polymeric flavan-3-ols that probably constitute the second most abundant class of polyphenols in the plant kingdom after lignin. Flavan-3-ols possess the characteristic “three-ring system” of flavonoids, with the C2 position of a benzopyran (A and C rings) linked with another aromatic ring (B ring) (Figure 1). The gallate modification of the hydroxyl group at the C3 position, the number of hydroxyl groups on the B ring, and the cis/trans configuration at C2 and C3 contribute to the diversity of flavan-3-ols (Figure 1). (+)-Catechin and (−)-epicatechin represent the basic common PA building blocks with 2,3-trans and 2,3-cis stereochemistry, respectively (Gu et al., 2003; Xie and Dixon, 2005). The adjacent subunits of PAs are usually connected via C4-C8 or, less commonly, C4-C6 bonds. These molecules are collectively referred to as B-type PAs and are present in commonly studied species such as Arabidopsis thaliana, Medicago truncatula, and grapevine (Vitis vinifera) (Lepiniec et al., 2006; Huang et al., 2012; Jun et al., 2018). Besides the C4-C8 linkage, in some cases the upper unit C2 can also link with the hydroxyl group on the A ring of the lower unit (Figure 1), forming A-type PAs in species such as cranberry (Vaccinium macrocarpon) and peanut (Arachis hypogaea) (Lou et al., 1999; Howell et al., 2005).
Figure 1.
Structures of flavan-3-ol monomers and B-type and A-type PAs.
PAs are widely present in fruits, bark, leaves, and seeds and play important roles in resistance to biotic and abiotic stress (Dixon et al., 2005). Depending on their ability to chelate metal ions, PAs reduce the activity of iron-, copper-, or zinc-dependent enzymes in pathogens, inhibiting their infestation or reproduction (Scalbert, 1991; Dixon et al., 2005; Yuan et al., 2012). Induced by environmental stress (e.g., high light and strong ultraviolet B light), PAs can protect plants by scavenging harmful reactive oxygen species (Mellway et al., 2009; Gourlay et al., 2020). The antioxidant properties of PAs also benefit neurological and cardiovascular health in humans (Middleton et al., 2000; Cos et al., 2004). With their protein-binding activity, PAs help protect plants against feeding insects by binding to digestive proteins and causing mid-gut lesions (War et al., 2012). This protein binding also helps maintain ruminal protein nitrogen and prevent lethal pasture bloat in ruminant animals and affects the feeding behavior of livestock and birds (Li et al., 1996; Xie et al., 2019; Xie and Xu, 2019). Plant-derived PAs contribute bitterness and astringency to food products, including red wine and tea (Pang et al., 2013; Ma et al., 2014). A high PA content leads to unwanted drying and puckering in some fruits, such as persimmon (Diospyros kaki) and pomegranate (Punica granatum), and de-astringency processing is needed before marketing (Wu et al., 2022). The bitter and astringent intensities of PAs are influenced by the content, degree of polymerization, 2,3-trans/cis stereochemistry, extent of galloylation, and number of hydroxyl groups on the PA molecules (Thorngate and Noble, 1995; Peleg et al., 1999; Vidal et al., 2003; Suzuki et al., 2005; Hufnagel and Hofmann, 2008; Ma et al., 2014). PAs enhance color stabilization during red wine aging via co-pigmentation and condensation with anthocyanins (Zhang et al., 2022b) and through interactions with proteins and cause hazing in fruit juice, wine, and beer (e.g., Collin et al., 2013). Oxidized PAs possess a brown color and are the major pigment in natural-colored cotton (Gossypium arboretum) fibers, which are an eco-friendly raw material in the textile industry because less dyeing and disposal of toxic waste are needed (Feng et al., 2014). In the terrestrial ecosystem, PA accumulation above ground in plants was recently proposed as a promising carbon sink for greenhouse gases (Dixon and Sarnala, 2020), and PAs from leaf litter and roots contribute to carbon sequestration in soil by forming complexes with necromass and shaping communities and metabolism of rhizosphere microbiomes below ground (Adamczyk et al., 2019a, 2019b, 2020; Zak et al., 2019; McGivern et al., 2021; Ward et al., 2022).
Despite the rapid development of genome editing technology and synthetic genetic circuit design (Gao, 2021; Brophy et al., 2022), deployment of PA-related traits has been limited by the lack of a comprehensive understanding of PA biosynthesis, transport, and regulation. Here we review recent advances in the field that may facilitate precise manipulation of PA traits in commercial crops and exploration of bio-strategies for mitigating global climate change.
Channeling the products upstream of the PA pathway: A matter of protein-protein interactions?
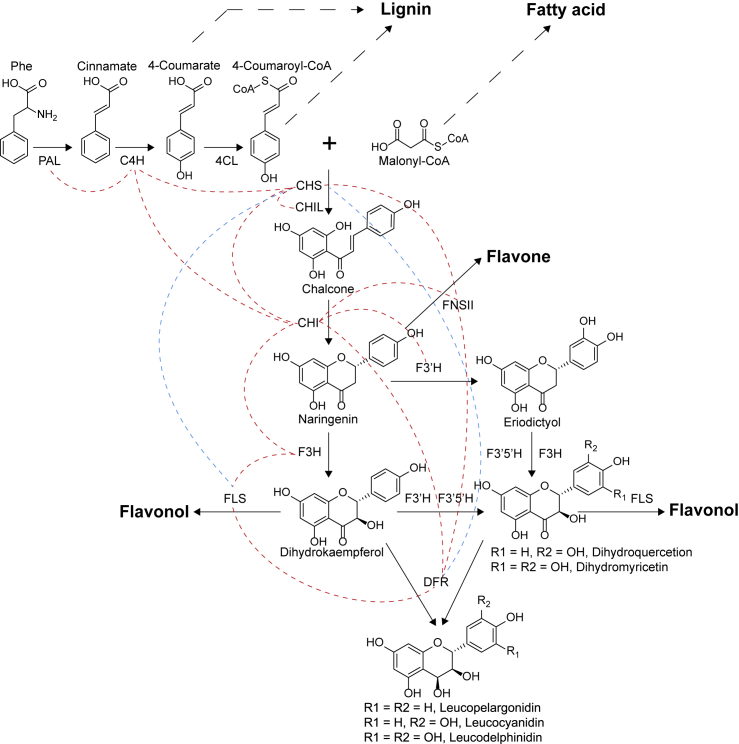
Biosynthesis of PAs is a branch of the flavonoid pathway, located downstream of the core phenylpropanoid metabolic route (Vogt, 2010; Figure 2). L-phenylalanine ammonia-lyase (PAL) is the entry enzyme of the phenylpropanoid pathway, producing cinnamate through deamination of L-phenylalanine (Rasmussen and Dixon, 1999). 4-Coumarate and 4-coumaroyl-coenzyme A (CoA) are sequentially generated from L-phenylalanine in two successive steps catalyzed by cinnamate 4-hydroxylase (C4H) and 4-coumarate CoA-ligase (Schilmiller et al., 2009). Chalcone synthase (CHS) then catalyzes condensation of one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA to generate one molecule of chalcone (Hrazdina et al., 1986), resulting in the C6-C3-C6 skeleton of flavonoids (Figure 2). Because malonyl-CoA is also an essential precursor for fatty acid biosynthesis, enhanced flow of carbon to fatty acids and their derived volatile compounds has been observed when the flavonoid pathway is downregulated (Li et al., 2018; Xuan et al., 2018; Xie et al., 2019; Figure 2). At the same time, 4-coumarate and 4-coumaroyl-CoA are both substrates in the lignin pathway. 4-Coumaroyl-CoA can be converted to the lignin monomer 4-coumaryl alcohol (H unit) through successive reductions by cinnamoyl-CoA reductase and cinnamyl alcohol dehydrogenase or channeled via 4-hydroxycinnamoyl CoA:shikimate hydroxycinnamoyltransferase or (as the free acid) 4-coumarate-3-hydroxylase to generate other forms of lignin monomers (C/G/5H/S units) (Barros et al., 2019). Chalcone isomerase (CHI) catalyzes intramolecular cyclization of chalcone to naringenin (flavanone) (Moustafa and Wong, 1967), with the benzopyran structure and 2R stereochemistry shared by PA building blocks. Accepting naringenin as the substrate, flavanone 3/3′/3′5′-hydroxylases add hydroxyl groups to the C-3, C-3′, and C-5′ of the C6-C3-C6 backbone to generate various substituted flavanols and dihydroflavonols (Forkmann et al., 1980; Britsch and Grisebach, 1986; Menting et al., 1994). It is because of the existence of F3H that dihydroflavonols possess a 2R,3S configuration. Through the activity of dihydroflavonol 4-reductase (DFR), the carbonyl group at the C4 position is reduced to a hydroxyl group, resulting in 2R,3S,4S-leucoanthocyanidins (flavan-3,4-diols) (Fischer et al., 1988).
Figure 2.
Upstream pathway of PA biosynthesis and the interaction network among the enzymes.
PAL, L-phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA-ligase; CHS, chalcone synthase; CHI, chalcone isomerase; CHIL, CHI-like protein; F3H/F3′H/F3′5′H, flavanone 3/3′/3′5′-hydroxylase; FLS, flavonol synthase; FNS II, flavone synthase II; DFR, dihydroflavonol 4-reductase. Red dashed lines indicate a direct interaction between enzymes, and blue dashed lines indicate that FLS competes with DFR to bind CHS. Dotted arrows indicate multiple reactions.
It is believed that the formation of enzyme complexes concentrates substrates and accelerates the exchange of intermediates between adjacent enzymes, thereby enhancing catalytic efficiency (Winkel-Shirley, 1999). In tobacco (Nicotiana tabacum), PAL is closely associated with C4H, although the interaction strength is weak, as shown by fluorescence energy resonance transfer (FRET) studies (Achnine et al., 2004). Yeast two-hybrid (Y2H) studies revealed the three interacting protein pairs CHS-CHI, CHS-DFR, and CHI-DFR in Arabidopsis, and affinity chromatography and immunoprecipitation assays demonstrated interactions between CHS, CHI, and F3H in lysates from 3-day-old Arabidopsis seedlings (Burbulis and Winkel-Shirley, 1999). Among six flavonol synthase (FLS) isoforms of Arabidopsis, FLS1 plays a central role in converting dihydroflavonol to flavonol and interacts with CHS, F3H, and DFR, as shown by Y2H analysis (Owens et al., 2008). FRET detected by fluorescence lifetime imaging microscopy provided in vivo evidence showing that DFR competed with FLS in physically binding to CHS in Arabidopsis, suggesting potentially distinct channels to the flavonol and PA/anthocyanin branches (Crosby et al., 2011). Using bimolecular fluorescence complementation and co-immunoprecipitation approaches, it was shown that soybean (Glycine max) isoflavone synthase interacted with CHS, CHI, and C4H, suggesting the formation of an isoflavone biosynthesis metabolon (Dastmalchi et al., 2016). Y2H and bimolecular fluorescence complementation assays revealed that snapdragon (Antirrhinum majus) flavone synthase II (FNS II) interacted with CHS, CHI, and DFR and that CHI was the shared interaction partner of DFR and F3′H (Fujino et al., 2018). By contrast, similar approaches failed to verify interaction between FNS II and CHS in torenia (Torenia hybrida), and an FNS II–F3H interaction was identified instead (Fujino et al., 2018). In rice (Oryza sativa), a complex has been shown between CHS, F3H, F3′H, DFR, and anthocyanidin synthase (ANS) (Shih et al., 2008). C4H, F3′H, and FNS II, as endoplasmic reticulum membrane-localized proteins, are thought to anchor the phenylpropanoid/flavonoid enzyme complex to the cytosolic side of the endoplasmic reticulum (Winkel-Shirley, 1999, 2001; Fujino et al., 2018).
In addition to forming metabolons, protein-protein interactions can also increase the specific activity of enzymes. Recent studies found that the conserved CHI-like protein could rectify CHS activity to produce chalcone rather than derailment polyketides through protein-protein interactions, facilitating influx of substrates from the phenylpropanoid pathway to the flavonoid pathway (Jiang et al., 2015; Waki et al., 2020; Figure 2).
Biosynthesis and polymerization of PA building blocks: A complex, non-conserved network
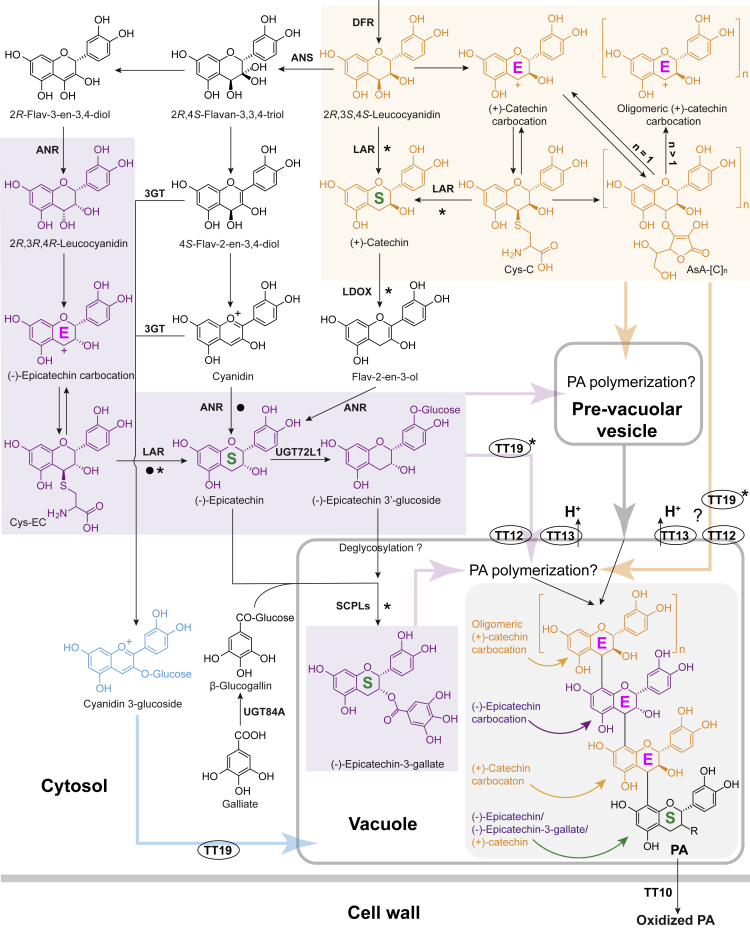
Nearly two decades ago, ANS and leucoanthocyanidin reductase (LAR) were found to use 2R,3S,4S-leucoanthocyanidins as substrates to produce anthocyanidins and 2,3-trans flavan-3-ol monomers (e.g., (+)-catechin), respectively (Tanner and Kristiansen, 1993; Saito et al., 1999; Tanner et al., 2003), and anthocyanidin reductase (ANR) was found to act downstream of ANS to convert anthocyanidins to 2,3-cis flavan-3-ol monomers (e.g., (−)-epicatechin) (Xie et al., 2003; Figure 3). Although publications have stated that flavan-3-ol monomers alone can be polymerized non-enzymatically or through catalysis by laccases, the resulting products are oligomers that are not usually present in nature (Pourcel et al., 2005). Instead, in vitro studies support a more plausible non-enzymatic mechanism of PA polymerization in which flavan-3-ol monomers serve as the starter units for sequential trapping of flavan-3-ol carbocations (extension units) via nucleophilic attack (Wang et al., 2020). 2R,3S,4S-Leucoanthocyanidin was first described as a 2,3-trans extension unit precursor for PA polymerization in vitro in the 1980s (Delcour et al., 1983), whereas the likely precursors of 2,3-cis extension units, 2R,3R,4R-leucocyanidin and 4β-(S-cysteinyl)-(−)-epicatechin (Cys-EC), were only found recently (Liu et al., 2016; Jun et al., 2021). In the past 6 years, new enzymes, novel functions of known enzymes, and new intermediates of the PA pathway have been discovered, providing explanations for the previously contradictory observations concerning in vivo versus in vitro enzyme activity, how PA chain length is regulated, the mechanism of flux channeling into 2,3-trans and 2,3-cis PA subunits, sequential assembly of PA building blocks, and how galloylated PA subunits are produced, as discussed below.
Figure 3.
General pathways for biosynthesis, transport and non-enzymatic polymerization of PA building blocks.
PA starter units are denoted as “S” and extension units as “E.” Black arrows without enzymes indicate non-enzymatic reactions. Bold arrows in purple, yellow, blue, and gray indicate transport routes. Question marks indicate possible pathways that may exist. Black asterisks indicate enzymes or transporters that do not exist or are not functionally conserved in all plants. Black dots indicate the less preferred activities of the corresponding enzymes. DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; LDOX, leucoanthocyanidin dioxygenase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; 3GT, 3-O-glucosyl-transferase; SCPL, serine carboxypeptidase-like protein; Cys-C, 4β-(S-cysteinyl)-(+)-catechin; Cys-EC, 4β-(S-cysteinyl)-(−)-epicatechin; AsA-[C]n, ascorbate conjugates of (+)-catechin. “R” at C3 of the PA structure represents a hydroxyl or galloyl group.
New insights into the in vivo roles of ANS
ANS exists in all PA-producing plants. In the presence of 2-oxoglutarate, ferrous ion, and ascorbate (AsA), recombinant ANSs of beefsteak plant (Perellia frutescens), snapdragon, maize (Zea mays), and torenia were shown to convert 2R,3S,4S-leucoanthocyanidins to corresponding anthocyanidins, with loss of the stereochemistry at C2 and C3 (Saito et al., 1999; Nakajima et al., 2001). On the basis of what was believed to be its primary substrate, ANS is also often called leucoanthocyanidin dioxygenase (LDOX). However, a series of follow-up studies questioned ANS-catalyzed transformation of leucoanthocyanidins as a major source of anthocyanidins. Incubation of Arabidopsis ANS with leucocyanidin substrate gave quercetin as the major product, with small amounts of dihydroquercetin and cyanidin as the minor products (Turnbull et al., 2000, 2003). ANS from grapevine could only produce quercetin using 2R,3S,4S-leucocyanidin as the substrate in vitro (Zhang et al., 2019). ANSs from Arabidopsis, grapevine, and Ginkgo biloba have also been observed to possess FLS activity (conversion of dihydroquercetin to quercetin) in vitro (Turnbull et al., 2000; Xu et al., 2008; Zhang et al., 2022a). Thus, ANS-catalyzed reactions in plants remain to be clarified.
The phenotypes of Arabidopsis tds4 (ans) mutants provide insights into the substrate and products of ANS in vivo. Loss of function of ANS in Arabidopsis results in deficiency of PAs and anthocyanins (Abrahams et al., 2003), supporting the idea that ANS is a part of the PA and anthocyanin pathways. Quercetin and kaempferol levels were the same in 5-day-old seedlings of the Arabidopsis tds4 (ans) mutant and the wild type (Bowerman et al., 2012). 2R,3S,4S-Leucocyanidin does not accumulate in wild-type Arabidopsis but is detectable in Arabidopsis tds4 (ans) mutant seeds (Jun et al., 2021). This evidence suggests that 2R,3S,4S-leucocyanidin is an in vivo substrate forANS but does not participate significantly in flavonol production, at least in Arabidopsis.
Recently, Zhang et al. (2019) trapped 2R,4S-flavan-3,3,4-triol as the initial product of VvANS from grapevine using 2R,3S,4S-leucocyanidin as the substrate, supporting the idea that the generic function of ANS is to catalyze C3-hydroxylation of the flavonoid C-ring (Welford et al., 2001; Wilmouth et al., 2002). In contrast to the full in vitro reaction of VvANS to generate only quercetin from 2R,3S,4S-leucocyanidin, the non-enzymatic degradation products of the purified 2R,4S-flavan-3,3,4-triol included significant amounts of dihydroquercetin and cyanidin but only trace amounts of quercetin (Zhang et al., 2019). 2R-Flav-3-en-3,4-diol and 4S-flav-2-en-3,4-diol are expected to be immediate dehydration products from the unstable 2R,4S-flavan-3,3,4-triol (Welford et al., 2001; Nakajima et al., 2006; Zhang et al., 2019; Figure 3). It is proposed that cyanidin can be obtained via non-enzymatic dehydration of 4S-flav-2-en-3,4-diol (Figure 3) and that the subsequent tautomerization of 2R-flav-3-en-3,4-diol will give dihydroquercetin, which can be further oxidized to quercetin in the presence of ANS in vitro (Wilmouth et al., 2002). On the basis of density functional theory using a molecular dynamics simulation of Arabidopsis ANS under physiological conditions, it was concluded that 4S-flav-2-en-3,4-diol is also likely to be the intermediate for anthocyanin formation via glucosylation and quercetin formation via another catalytic cycle (Nakajima et al., 2006; Figure 3). We therefore reason that, to produce significant levels of anthocyanin and PA in vivo, it is necessary to rapidly separate 2R-flav-3-en-3,4-diol and 4S-flav-2-en-3,4-diol from ANS to avoid the second cycle of enzymatic oxidation. Such a separation mechanism might include glycosylation by flavonoid 3-O-glucosyltransferase, binding with a transporter, or reduction by ANR involving substrate channeling between closely arranged enzymes to prevent back-reaction with ANS (Figure 3).
Apart from participating in the PA and anthocyanin pathways, ANS is also somehow involved in vacuole development; the endothelial cells of two independent Arabidopsis tds4 (ans) mutants failed to form the large central vacuole observed in the wild type, but contained multiple small vacuoles instead (Abrahams et al., 2003). More work is needed to determine whether and how biosynthesis of PAs or anthocyanin is mechanistically linked to formation of the central vacuole.
Models for generating (−)-epicatechin-derived PAs
The downstream PA pathways in Medicago and Arabidopsis have been well characterized by genetic and biochemical approaches, providing the basis for studying PA biosynthesis in non-model crops. Arabidopsis possesses exclusively (−)-epicatechin-type PAs, and ANR, encoded by the BANYLUS gene in Arabidopsis, converts cyanidin to the (−)-epicatechin monomer (Xie et al., 2003; Figure 3). Knockout of ANR in Arabidopsis results in a red–gray seed coat with negative p-dimethylaminocinnamaldehyde staining as a result of loss of PAs and over-accumulation of anthocyanins (Appelhagen et al., 2014). Conversely, ectopic expression of AtANR leads to reduction of anthocyanin levels and accumulation of PAs in tobacco (N. tabacum) petals (Xie et al., 2003). Because of these clearly visible phenotypes, the Arabidopsis banyuls (anr) mutant and tobacco petals have become commonly used systems for testing the biochemical functions of ANRs from non-model species.
Compared with Arabidopsis, Medicago also possesses an LAR that has been found to convert 2R,3S,4S-leucocyanidin to (+)-catechin (Pang et al., 2007; Jun et al., 2018). However, although LAR is expressed at high levels in Medicago pods and seeds, only small amounts of (+)-catechin-based PAs are present in these organs during the immature stage (Pang et al., 2007). Liu et al. (2016) found that Medicago accumulates Cys-EC, an alternative substrate for MtLAR, producing (−)-epicatechin (Liu et al., 2016). Jun et al. (2018) identified a homolog of ANS (LDOX) in Medicago that converted (+)-catechin to the intermediate flav-2-en-3-ol, which is subsequently oxidized to cyanidin, especially under acidic conditions. Time-course enzymatic studies showed that flav-2-en-3-ol, rather than cyanidin, was the preferred substrate of Medicago ANR for synthesis of the (−)-epicatechin monomer (Jun et al., 2021). Using MtDFR–MtANS–ANR-coupled reactions in vitro, it was shown that dicot, but not monocot, ANRs accept 2R-flav-3-en-3,4-diol (a proposed intermediate product of ANS) to produce 2R,3R,4R-leucocyanidin (Jun et al., 2021; Figure 3). Cys-EC and 2R,3R,4R-leucocyanidin could auto-polymerize with flavan-3-ol monomers at neutral pH to yield natural procyanidins with (−)-epicatechin extension units, suggesting that these two newly identified compounds could provide the (−)-epicatechin carbocation to attack the flavan-3-ol monomer (the starter unit) for non-enzymatic PA chain elongation (Liu et al., 2016; Jun et al., 2021; Figure 3). Cys-EC can be formed by a non-enzymatic reaction between Cys and 2R,3R,4R-leucocyanidin in vitro (Jun et al., 2021; Figure 3), consistent with loss of Cys-EC in the Medicago anr mutant (Liu et al., 2016). These findings provide a reasonable explanation for why (−)-epicatechin PA extension units are derived from the ANS–ANR pathway and (−)-epicatechin PA starter units are synthesized by the LAR–LDOX–ANR pathway, as supported by genetic studies with Medicago mutants (Jun et al., 2018). It has been suggested that separation of the starter and extension unit biosynthetic pathways may be the result of protein complex formation (Lu et al., 2022), but more evidence needs to be provided. Loss of function of LAR in Medicago results in decreased levels of soluble PAs but increased levels of high-molecular-weight, insoluble PAs (Liu et al., 2016). Based on the current non-enzymatic PA polymerization model, LAR is a key enzyme in regulating the ratio of starter and extension units (Liu et al., 2016). However, no flavan-3-ol monomer or starter units but only (−)-epicatechin extension units are detected after phloroglucinolysis (a technique that reveals the nature and proportions of PA starter and extension units) of PA extracts from the Medicago lar mutant (Jun et al., 2021). Perhaps alternative non-conventional starter units exist to initiate PA oligomerization (Yu et al., 2022).
How PA chain length is regulated in plants with no LAR remains to be resolved. A possible mechanism for generating (−)-epicatechin starter and extension units in Arabidopsis, which lacks the LAR–LDOX route, is that AtANR uses cyanidin to generate the (−)-epicatechin monomer or, like MtANR, accepts flav-3-en-3,4-diol to synthesize 2R,3R,4R-leucocyanidin (Jun et al., 2018). It has also been reported that recombinant ANR from the tea plant (Camellia sinensis) can convert cyanidin to (−)-epicatechin, a flav-en-3-ol-like compound, and (−)-epicatechin carbocation (Pang et al., 2013; Wang et al., 2020), suggesting another possible route for (−)-epicatechin starter and extension unit synthesis for plants that do not possess LDOX.
The central role of 2R,3S,4S-leucocyanidin in generating (+)-catechin-derived PAs
Many crops that possess LAR, including grapevine, tea, persimmon, and cacao (Theobroma cacao), produce significant levels of (+)-catechin-derived PAs (Suzuki et al., 2005; Huang et al., 2012; Liu et al., 2013; Wang et al., 2018), regardless of whether homologs of MtLDOX are present or functional in these species. It has been shown repeatedly that reconstruction of the (+)-catechin pathway by overexpression of LARs in wild-type Arabidopsis is not possible; (−)-epicatechin is formed, and (+)-catechin is never observed in the seeds (Ferraro et al., 2014; Zhang et al., 2020b). By contrast, introduction of LARs from cacao or tea into the Arabidopsis tds4 (ans) mutant did lead to accumulation of (+)-catechin-derived PAs (Liu et al., 2013; Zhang et al., 2020b). Mass spectrometry analysis showed that the known PA precursors in wild-type Arabidopsis were only Cys-EC and 2R,3R,4R-leucocyanidin (with the (−)-epicatechin 2,3-cis stereochemistry), whereas the Arabidopsis tds4 (ans) mutant lost the (−)-epicatechin PA precursor and instead accumulated 2R,3S,4S-leucocyanidin, 4β-(S-cysteinyl)-(+)-catechin (Cys-C, the 2,3-trans isomer of Cys-EC) and AsA conjugates of (+)-catechin (AsA-[C]n with a (+)-catechin backbone) (Yu et al., 2019, 2022; Wang et al., 2020; Jun et al., 2021). 2R,3S,4S -Leucocyanidin is a known (+)-catechin extension unit for non-enzymatic PA polymerization in vitro (Delcour et al., 1983). Both derived from 2R,3S,4S -leucocyanidin, Cys-C and AsA-[C]n co-exist with Cys-EC in grape berries and provide (+)-catechin extension units for non-enzymatic PA polymerization in vitro (Figure 3; Jun et al., 2018; Yu et al., 2019, 2022). Besides accepting Cys-EC and 2R,3S,4S-leucocyanidin to produce (−)-epicatechin and (+)-catechin, respectively, two grapevine LARs (VvLAR1 and VvLAR2) convert Cys-C to (+)-catechin much more efficiently than they convert Cys-EC to (−)-epicatechin, presumably because of steric hindrance of the 2,3-cis conformation of Cys-EC in the VvLAR active sites (Yu et al., 2019). It can therefore be concluded that (+)-catechin starter and extension units are derived from 2R,3S,4S-leucocyanidin via direct catalysis by LAR or conversion of Cys-C, respectively. As a result, the phenotype observed upon expression of LAR in wild-type Arabidopsis is due to lack of available 2R,3S,4S-leucocyanidin and its derived Cys-C so that the only substrate for the introduced LAR is the less preferred (−)-epicatechin precursor (e.g., Cys-EC) to generate (−)-epicatechin. Such prevention of (+)-catechin precursor accumulation in Arabidopsis may result from strict co-expression of DFR and ANS during seed coat development (Dean et al., 2011) or metabolic channeling analogous to the CHS1–F3H–F3′H–DFR–ANS complex formation observed in rice (Shih et al., 2008). Based on these considerations, it is clear that release of 2R,3S,4S-leucocyanidin from the coupled DFR-ANS flux is a prerequisite for attempts to engineer (+)-catechin-derived PAs. To assess its availability, 2R,3S,4S-leucocyanidin must be measured immediately after metabolite extraction from plant materials because it is highly reactive. Alternative ways to evaluate 2R,3S,4S-leucocyanidin levels in planta are to trap its derived and more stable Cys-C or AsA-C conjugates (Yu et al., 2019, 2022; Jun et al., 2021) or to use acidified methanol/water solution to extract the metabolites and analyze the (+)-catechin-4-O-methyl ether conjugates as the carbocation indicator (Wang et al., 2020). Inability to detect the presence of free 2R,3S,4S-leucocyanidin would suggest that it may be necessary to somehow prevent the coupling between DFR and ANS to allow 2R,3S,4S-leucocyanidin to serve as a substrate for (+)-catechin-derived PA biosynthesis.
Sequential assembly of PAs
Exactly how PA blocks are polymerized in vivo still remains unclear. Given that no evidence to date supports the presence of an enzyme involved in PA polymerization, the nucleophilic/electrophilic attack between flavan-3-ol (starter unit) and its carbocation (extension unit) is the most likely mechanism for the last step of PA block assembly (Wang et al., 2020). By summarizing the results from in vitro auto-polymerization assays, possible donors of flavan-3-ol carbocations have been identified, including leucoanthocyanidins (e.g., 2R,3S,4S/2R,3R,4R-leucocyanidin) (Delcour et al., 1983; Jun et al., 2021) and their derived conjugates with chalcogens between AsA/Cys moieties and C4 of flavan-3-ols (e.g., Cys-C, Cys-EC, and AsA-[C]n) (Liu et al., 2016; Jun et al., 2018; Yu et al., 2019, 2022). Unlike other known monomeric PA extension unit precursors, AsA-[C]n can provide oligomeric extension units for PA polymerization in vitro, suggesting that the starter unit to initiate PA oligomerization does not have to be the flavan-3-ol monomer and that PA elongation does not necessarily proceed by sequential addition of a single extension unit (Yu et al., 2022). It is believed that the reactive leucocyanidins may auto-oxidize to p-quinone methides, which could then transform into (+)-catechin or (−)-epicatechin carbocations, whereas formation of carbocations from the more stable flavan-3-ol conjugates depends on dissociation of the C4-O or C4-S bonds at neutral pH (Liu et al., 2016; Jun et al., 2021; Yu et al., 2022; Figure 3). Considering their reactivity and biosynthetic route, leucoanthocyanidins are more likely to be direct PA extension unit donors than AsA or Cys conjugates of flavan-3-ols. The flavan-3-ol conjugates may function as buffer pools of excess leucoanthocyanidin-derived carbocations for stabilization before participating in PA polymerization (Dixon and Sarnala, 2020; Lu et al., 2022; Yu et al., 2022).
Among plants that possess (−)-epicatechin PA building blocks, Cys-EC is present in Medicago, Arabidopsis, and grapevine, but it is rarely found in the tea plant (Liu et al., 2016; Yu et al., 2019; Zhang et al., 2020b). This indicates that assembly of 2R,3R,4R-leucocyanidin-derived extension units may bypass biosynthesis of Cys-EC in some cases or that nucleophiles other than Cys possibly exist to conjugate with (−)-epicatechin carbocation. As for AsA conjugates of flavan-3-ols, although AsA can trap (−)-epicatechin carbocation in vitro, only AsA-[C]n, but not AsA-[EC]n, is detected in plants (Yu et al., 2022). One possible reason is that AsA may not co-localize with 2R,3R,4R-leucocyanidin or its (−)-epicatechin carbocation in vivo. This implies that the subcellular compartments for storage of (+)-catechin and (−)-epicatechin PA extension units may be separated in plant cells. To incorporate (+)-catechin and (−)-epicatechin subunits into the PA molecules commonly present in crops, the corresponding extension units must reach the same polymerization site as the starter units. Thus, unless there is very tight channeling into the site of polymerization, the relatively stable flavan-3-ol conjugates are more likely candidates for transport than the reactive leucoanthocyanidins or carbocations. To date, the functions of the proposed PA extension unit precursors have only been inferred by in vitro assays and on the basis of their accumulation patterns in PA-producing tissues from a series of developmental stages (Delcour et al., 1983; Liu et al., 2016; Wang et al., 2020; Jun et al., 2021; Yu et al., 2022). Whether these precursors truly participate in PA extension in vivo needs to be addressed. Feeding reactive PA intermediates to plant tissues can be confounded by poor uptake, and there are no criteria for assessing whether these exogenous substrates successfully reach the correct compartment for polymerization or are channeled to detoxification pathways. To overcome these difficulties, the design of specific probes for labeling PAs and their precursors in situ and the development of suitable single-particle tracking methods using high-resolution microscopy technology are possible strategies for monitoring the trafficking and derivation of PA compounds in living cells to provide insights into assembly of PA building blocks (Cui et al., 2018; Hu et al., 2018).
Galloyl decoration of PA building blocks
Galloylation of the 3-hydroxyl group of (−)-epi(gallo)catechin starter and extension units is common in several species, including tea, grapevine, and persimmon, although galloylated (+)-catechin is rarely seen. Although the effects of the galloyl substituent have long been studied with respect to the antioxidant and antimicrobial properties of epigallocatechin gallate (e.g., Taylor et al., 2005), the in planta functions of the galloyl groups on PA oligomers and polymers remain to be determined. The enzymology of the galloylation reaction has proven to be quite complex.
Enzymatic reactions using proteins purified from tea leaves revealed a two-step reaction for (−)-epicatechin galloylation: uridine diphosphate glucose (UDP)-glucose:galloyl-1-O-D-glucosyltransferase first glucosylates gallic acid using UDP-glucose as a glucose donor to generate β-glucogallin (ether-linked gallic acid and glucose), and (−)-epicatechin:1-O-galloyl-D-glucose-O-galloyltransferase (ECGT) then uses β-glucogallin and (−)-epi(gallo)catechin to produce galloylated (−)-epi(gallo)catechins (Liu et al., 2012).
Members of the UGT84A subfamily are responsible for glucosylation of gallic acid (Figure 3). Three genes encoding UGT84A proteins (VvgGT1/2/3) have been identified as the targets of PA-specific activities in grapevine, and these three enzymes individually catalyze formation of β-glucogallin from gallic acid and UDP-glucose in vitro (Khater et al., 2012). Similar biochemical activities have been observed for UGT84A13 from English oak (Quercus robur), UGT84A25/26 from red gum (Eucalyptus camaldulensis), and UGT84A22 from tea (Mittasch et al., 2014; Cui et al., 2016; Tahara et al., 2018). Although UGT84A23/24 of pomegranate (Punica granatum) could also generate β-glucogallin in vitro, simultaneous knockout of the corresponding genes resulted in increased β-glucogallin levels in plants, suggesting that additional enzymes may be responsible for gallic acid glycosylation in this species (Ono et al., 2016).
Enzymatic and SDS-PAGE analysis revealed that ECGT in tea leaf extracts exists in monomeric, dimeric, trimeric, and tetrameric forms, and mass spectrometry suggested that ECGT is a serine carboxypeptidase-like (SCPL) protein (Liu et al., 2012) (Figure 3). Other studies have shown that five SCPLs (SCPL4/5/11/13/14) were involved in transacylation of (-)-epi(gallo)catechin with gallic acid ester in tea (Ahmad et al., 2020; Yao et al., 2022). Recombinant CsSCPL11/13/14 purified from Escherichia coli could generate galloylated epi(gallo)catechin using 1,2,3,4,6-pentagalloylglucose rather than β-glucogallin as an acyl donor, although 1,2,3,4,6-pentagalloylglucose is not commonly found in tea (Ahmad et al., 2020). In a tobacco leaf transient expression system, neither CsSCPL4 nor CsSCPL5 alone could catalyze galloylation of (-)-epi(gallo)catechin, which required co-expression of both proteins; CsSCPL4 plays the role of acyltransferase, whereas CsSCPL5 is a pseudo-enzyme that interacts with CsSCPL4 as a molecular chaperone (Yao et al., 2022). DkSCPL1 and DkSCPL2 are downregulated in PA-deficient persimmon varieties (Ikegami et al., 2007; Akagi et al., 2011), and the acyltransferase activity also relies on co-existence of the two DkSCPLs (Yao et al., 2022). It is believed that the N-terminal signal peptide in SCPLs is essential for post-translational modifications and targeting to the vacuole (Bontpart et al., 2015), consistent with the observation that galloylated (-)-epi(gallo)catechin is localized in the vacuole, as determined by immunohistochemistry (Xu et al., 2016).
VvGAT1 and VvGAT2 (encoding SCPLs) have repeatedly been reported to be co-regulated with PA-related genes in grapevine (Terrier et al., 2008; Carrier et al., 2013; Koyama et al., 2014), but the two VvGATs co-purified from the tobacco leaf expression system could not catalyze formation of galloylated (-)-epigallocatechin from (-)-epigallocatechin and β-glucogallin unless their N-terminal signal peptides were replaced with those from CsSCPL4 and CsSCPL5, respectively (Yao et al., 2022). Because grapevine also possesses significant levels of digalloylglucose (Narduzzi et al., 2015), whether native VvGATs accept digalloylglucose but not β-glucogallin as an acyl donor remains to be tested.
These findings highlight the complexities underlying the biosynthesis of galloylated (-)-epi(gallo)catechin monomer or PA starter units. However, how galloyl decoration proceeds on the PA extension units still remains unclear. A major question is whether galloylation occurs on the (-)-epi(gallo)catechin conjugates or carbocations before their polymerization or on the subsequent PA polymers.
Functions of PA transporters across plant species
In addition to the complexities of the biosynthetic pathways themselves, there are even more uncertainties regarding the mechanisms whereby the PAs arrive at their final destinations in the cell. Vanillin staining of developing wild-type Arabidopsis seeds suggests that PAs are stored in the central vacuole (Debeaujon et al., 2003; Figure 3). It has been proposed that a Phi-family glutathione S-transferase (GST/GSTF/TT19), a tonoplastic multidrug and toxic compound extrusion (MATE/TT12) transporter, and a tonoplast P-type H+-ATPase (AHA10/TT13) form a transporter-mediated pathway for vacuolar loading of PAs or PA precursors from their sites of synthesis in the cytosol (Figure 3). Golgi apparatus-derived TT9 (a peripheral membrane protein localized at the Golgi apparatus) and ECHIDNA on the trans-Golgi network are involved not only in vesicle trafficking of overall flavonoids but also in vacuolar biogenesis and plant growth (Ichino et al., 2014, 2020). Although loss of function of TT19 (GST), TT12 (MATE), or TT13 (AHA10) also disturbs vacuolar integrity, the corresponding effects on plant development are not as significant as those seen in tt9 (gfs9) and echidna mutants (Debeaujon et al., 2001; Kitamura et al., 2004, 2010; Appelhagen et al., 2015).
The Arabidopsis tt19 (gst)-null mutant has lost anthocyanidin accumulation and the brown seed coat color (mainly contributed by oxidized PAs) (Kitamura et al., 2004). Acid butanolysis assays showed that the tt19 (gst) mutant possesses lower levels of soluble PAs but higher levels of insoluble PAs than the wild type (Kitamura et al., 2010). In immature tt19 (gst) seeds, vanillin staining showed that PAs or their derivatives are accumulated in small vacuolar-like structures (Kitamura et al., 2010). A recent study showed that the tt19 (gst) mutant is not affected in the level of Cys-EC extension units but accumulates only trace amounts of the (−)-epicatechin monomer, indicating that TT19 GST is necessary for maintenance of starter units (Lu et al., 2022). Because TT19 GST is localized in the cytosol (Kitamura et al., 2010; Sun et al., 2012), it is still a mystery how its loss of function, like that of ANS, interferes with formation of the large central vacuole.
In Medicago, the glucosyltransferase UGT72L1 is co-regulated with PA biosynthesis and specifically glucosylates epicatechin at the 3′-O- position (Pang et al., 2008). However, whether this enzyme is critical for PA biosynthesis or is instead involved in detoxification of excess epicatechin is still unresolved. In Arabidopsis, the TT12 MATE transporter binds to and transports epicatechin-3′-O-glucoside to vacuoles, dependent on a proton gradient generated by TT13 (AHA10) (Zhao and Dixon, 2009; Aprile et al., 2011; Appelhagen et al., 2015). The tt13 (aha10) mutant phenocopies the tt12 (mate) mutant, and neither is significantly affected in anthocyanin accumulation (Debeaujon et al., 2001; Appelhagen et al., 2015), consistent with the observation that cyanidin-3-O-glucoside is not a preferred ligand for TT12 MATE (Zhao and Dixon, 2009). Compared with the wild type, tt12 (mate) mutant seeds contain higher levels of (−)-epicatechin-3′-O-glucoside (Kitamura et al., 2010). Because epicatechin-3′-O-glucoside is rarely seen as a PA starter unit, and its levels are not as high as those of (−)-epicatechin monomers, a specific glucosidase may exist to hydrolyze (−)-epicatechin-3′-O-glucoside to (−)-epicatechin after transport into the vacuole (Figure 3). Thus, (−)-epicatechin-3′-O-glucoside in the tt12 (mate) mutant might mainly remain in the cytosol in the detoxified conjugate form. The levels of soluble PAs, insoluble PAs, and flavonols are dramatically decreased in immature tt12 (mate) mutant seeds (Debeaujon et al., 2001; Kitamura et al., 2010), indicating that TT12 may have a feedback effect on the upstream flavonoid pathway besides being a functional transporter. Vanillin and toluidine blue O staining assays show that PAs or PA derivatives are at the peripheral regions of vacuoles in tt12 (mate) single mutants and restricted outside the small vacuolar-like structures in tt12 (mate):tt19 (gst) double mutants (Kitamura et al., 2010). These PA-containing small vacuolar-like structures can be outlined with TT12 (MATE) fused with green fluorescent protein in the tt19 (gst) mutant (Kitamura et al., 2010). Thus, TT12 is required to pump PAs or their precursors into vacuolar-like structures in tt19 (gst) and into the central vacuole in the wild-type Arabidopsis seed coat. This is consistent with the hypothesis that PAs are polymerized in pre-vacuolar vesicles that are subsequently fused into the central vacuole (Dixon and Sarnala, 2020) (Figure 3); whether these are truly analogous to pre-vacuolar structures in wild-type tissues is not clear.
In grapevine, VvMATE1 and VvMATE2 are proposed to be the two putative seed-specific TT12-like transporters based on gene co-expression analysis and localization in the tonoplast and Golgi complex, respectively, but their roles in vivo remain to be deciphered (Pérez-Díaz et al., 2014). MdMATE1 and MdMATE2 from apple (Malus × domestica) complement the Arabidopsis tt12 (mate) PA phenotype, and their genes are differentially expressed in fruit flesh, fruit skin, and leaves (Frank et al., 2011). In rapeseed (Brassica napus), the absence of BnTT12-2 (MATE) expression in developing seeds is linked with the L2-type yellow-seed stocks (with reduced seed coat pigmentation levels and increased seed oil content) (Chai et al., 2009). Similarly, significant differential expression of GhTT12 (MATE) is also observed between brown and white cotton. Using a virus-induced gene silencing approach, FaTT12-1 (MATE) was shown to be responsible for PA but not anthocyanin accumulation in strawberry (Fragaria × ananassa) fruit (Chen et al., 2018). Persimmon DkMATE1 complemented the Arabidopsis tt12 (mate) mutant phenotype, and transient overexpression or silencing of DkMATE1 in persimmon leaves led to an increase or decrease in PA content, respectively (Yang et al., 2016). These findings suggest that TT12 (MATE) represents a conserved feature of PA transport among plants.
TT19 (GST) homologs from a large number of species are associated with anthocyanin accumulation but do not always complement the PA phenotype of Arabidopsis tt19 (gst) mutants. Among the three TT19 GST-like proteins (VvGST1/3/4) in grapevine, VvGST3/4 specifically complement the Arabidopsis tt19 (gst) PA phenotype, whereas VvGST4 complements PA and anthocyanin accumulation (Pérez-Díaz et al., 2016). Such dual function is also observed for AcGST1 from kiwifruit (Actinidia chinensis) and CMGSTF12 from the Brassicaceae (Camelina sativa) (Wang et al., 2012; Liu et al., 2019). Rather than restoring the natural B-type PAs, overexpression of pear (Pyrus communis) PcGSTF12 gives rise to A-type PAs in the Arabidopsis tt19 (gst) mutant (Zhang et al., 2020a), the mechanism of which remains unclear. However, litchi (Litchi chinensis) LcGST4, Medicago MtGSTF7, cineraria (Senecio cruentus) ScGST3, cotton GhGSTF12, strawberry Reduced Anthocyanin in PetiolesRAP/GST, tea plant CsGSTF1, peach (Prunus persica) PpGST1, and apple MdGST1/6 can only rescue loss of anthocyanin but not PA in the Arabidopsis tt19 (gst) mutant (Hu et al., 2016; Han et al., 2018; Luo et al., 2018; Jiang et al., 2019; Wei et al., 2019; Zhao et al., 2020; Cui et al., 2021; Shao et al., 2021; Wang et al., 2022). Considering this, it is possible that TT19 GST proteins are not necessarily required for PA transport but are crucial for anthocyanin production through stabilization/protection of anthocyanidins in all plants. However, because the functions of TT19 homologs have mainly been assessed in the Arabidopsis model system, we cannot rule out the possibility that they are functional in the PA pathway in their species of origin. Li et al. (2011) reported that the Trp-to-Leu transition at amino acid 205 of TT19 GST in Arabidopsis disturbs accumulation of PA but not that of anthocyanin, suggesting that the non-conserved C-terminal region is important for PA substrate binding. A better understanding of the function and evolution of the C terminus of TT19 homologs might be helpful for engineering the ratio between soluble and insoluble PAs via rational protein design.
Insights into regulating PA content in plants
Regulators targeting the MYB–bHLH–WD40 complex
PA pathway transcription factors, including MYB, basic helix-loop-helix (bHLH)/MYC, WD40, WRKY, MADS-box, and WIP domain-containingzinc finger family proteins, have been intensively studied across species (He et al., 2008; Yu et al., 2020; Wei et al., 2021; Mora et al., 2022). First discovered in Arabidopsis, the MYB–bHLH–WD40 (MBW) complex, composed of TT2 (MYB123), TT8 (bHLH42), and TTG1 (WD40 family), plays a major role in activating the expression of PA pathway genes, including DFR, ANS, ANR, TT12 (MATE), TT13 (AHA10), and TT19 (GST) (Baudry et al., 2004). Evidence, albeit largely from in vitro experiments, has shown that TT2 and TT8 are mainly responsible for binding downstream target promoters, whereas TTG1 is crucial for maintaining MBW complex activity, possibly via a stabilization effect (Baudry et al., 2004; Xu et al., 2015). TTG1 can also form ternary complexes with coupled MYB5–TT8 and TT2-GLABRA3/ENHANCER OF GLABRA3 to regulate the pathway shared by PA and anthocyanin biosynthesis to a lesser extent (Xu et al., 2014). Apart from its role in regulation of the flavonoid pathway, TTG1 is also involved in regulation of mucilage biosynthesis, trichome and root hair development, storage protein and fatty acid accumulation in seeds, and regulation of the circadian clock (Airoldi et al., 2019; Tian and Wang, 2020).
In sorghum (Sorghum bicolor), mutation of Tannin1 (encoding the homolog of TTG1) results in much lower levels of PAs and anthocyanins but increased levels of fatty acid-derived volatiles, affecting bird feeding behavior in sorghum field production (Xie et al., 2019). Transcription of the Medicago TTG1 homolog MtWD40-1 can be activated by three MYB proteins (MtPAR, MtMYB5, and MtMYB14), of which MtMYB5 and MtMYB14 synergistically promote PA biosynthesis in the presence of MtTT8 and MtWD40 (Verdier et al., 2012; Liu et al., 2014). At the post-transcriptional level, Arabidopsis TTG1 can be phosphorylated by SHAGGY-like kinases 11/12 (SK11/12), which abolishes the interaction of TTG1 with TT2 and compromises activation of downstream GLABRA2 (a negative regulator of fatty acid production), mediating carbon partitioning from flavonoids to fatty acids (Li et al., 2018). When serving as a substrate of the GSK3-like kinase BIN2, phosphorylated TTG1 inhibits the activity of the WEREWOLF-GLABRA3-TTG1 complex to affect root hair development (Cheng et al., 2014). Whether BIN2-mediated phosphorylation of TTG1 is involved in PA regulation remains unknown. Kinases can also regulate MBW complex formation by indirectly targeting the bHLH protein. In apple, the TT8 homolog MdbHLH3 is prevented from forming a complex with MdMYB1/9/11 and MdTTG1 by several jasmonate-zim-domain proteins via direct protein-protein interaction (An et al., 2015; Liu et al., 2017). An SNF1-related kinase (MdSnRK1.1) phosphorylates MdJAZ18 to facilitate its degradation through the 26S proteasome, rendering the released MdbHLH3 functional in activation of the PA and anthocyanin pathways (Liu et al., 2017). More directly, an apple bric-à-brac, tramtrack, and broad complex protein, MdBT2, interacts with MdMYB9 to mediate degradation of the target by the 26S proteasome, reducing production of PAs and anthocyanins (An et al., 2018). Activated by anthocyanin- and PA-related MYB activators, Medicago MtMYB2 and peach PpMYB18 encode R2R3 MYB repressors that compete with MYB activators for binding bHLHs, defining the temporal and spatial pattern of anthocyanin and PA accumulation and preventing over-synthesis of related metabolites (Jun et al., 2015; Zhou et al., 2019). This suggests that the previously characterized PA/anthocyanin repressors VvMYBC2-L1/3 from grapevine (phylogenetically clustered with MtMYB2 and PpMYB18) might also participate in the feedback loop to regulate MBW complex activity in the flavonoid pathway (Huang et al., 2014; Cavallini et al., 2015; Zhou et al., 2019).
Tuning the flux between PA and anthocyanin branches
As indicated above, PA and anthocyanin genes are generally co-regulated through the core MBW regulatory complex, somewhat diminishing the flexibility for balancing levels of PAs and anthocyanins. Without shifting toward lignin, anthocyanin, or flavonol production, VvMYBPA1 and VvMYBPA2 were shown to be positive regulators specific to the PA pathway, activating VvDFR, VvANS, VvANR, and VvLAR1 but not VvLAR2 in transgenic grape tissues (Bogs et al., 2007; Terrier et al., 2008). Similarly, overexpression of persimmon DkMYB4 (encoding the homolog of VvMYBPA1 and VvMYBPA2) promotes PA but not anthocyanin production in kiwifruit, and a reduced PA level is observed in DkMYB4-silenced persimmon callus (Akagi et al., 2009). Ectopic expression of MtPAR in Medicago hairy roots activates PA genes and represses isoflavone genes but has no effect on anthocyanin gene expression, resulting in increased PA levels but decreased anthocyanin levels through redirecting flux (Li et al., 2016). Recently, grape VvMYB86 was found to regulate PA and anthocyanin biosynthesis in different directions by activating PA genes and positive regulators but downregulating anthocyanin genes (Cheng et al., 2021). At the post-transcriptional level, the peptide miPEP164c, encoded by a pre-processed microRNA (miRNA) primary transcript, was shown to target VvMYBPA1 to inhibit PA production and stimulate anthocyanin accumulation in grape (Vale et al., 2021). Under light stress and induced by the B-box protein MdBBX22, the miRNA miR858 targets three PA-positive regulatory genes, MdMYB9/11/12, to downregulate the PA pathway and enhance anthocyanin synthesis in apple peel (Zhang et al., 2022c). Understanding the cross-talk between PA biosynthesis and fruit coloration in the field can suggest optimal conditions for harvest depending on the desired composition of the fruit berries.
Regulators responsible for PA insolubilization
Soluble PAs make more important contributions to bitterness and astringent mouth feel than insoluble PAs, which are believed to be tightly crosslinked with the cell wall complex in raw plant materials (Mattivi et al., 2009; Bordiga et al., 2011). The mechanism for PA insolubilization during plant development has not been fully resolved and may differ among plant species. As indicated in the Arabidopsis tt19 (gst) and Medicago lar mutants, the remarkable shift from soluble PAs to insoluble PAs in the seed coat might be primarily due to the increased ratio between extension and starter units, yielding excess high-molecular-weight PAs with low solubility via non-enzymatic polymerization (Liu et al., 2016; Lu et al., 2022). In Arabidopsis, loss of function of TT10 (encoding a laccase-like polyphenol oxidase) results in more (−)-epicatechin monomers and soluble PAs in seeds compared with the wild type (Pourcel et al., 2005). Given that the major product of TT10 with (−)-epicatechin as a substrate is unnatural dehydrodiepicatechin A (Pourcel et al., 2005), TT10 is more likely to be involved in oxidative polymerization of PAs for insolubilization rather than initial oligomerization.
In the persimmon industry, to remove the unpleasant astringency in fruit flesh, warm water, ethanol, and high-concentration CO2 treatments are widely used to promote accumulation of endogenous acetaldehyde, which can chemically bridge soluble PA oligomers to form insoluble gel-like complexes (Matsuo and Itoo, 1982; Tanaka et al., 1994; Fang et al., 2016). The astringency of Chinese-pollination-constant non-astringent (C-PCNA) persimmon, which naturally diminishes during fruit development on the tree, is regulated at multiple levels. In C-PCNA, the expression patterns of DkLAC1 (encoding a TT10 homolog) are linked with a reduction in soluble PAs during fruit ripening (Hu et al., 2013). Two pyruvate kinases (DkPK7/8) upregulate expression of pyruvate decarboxylase and alcohol dehydrogenase genes (DkPDC and DkADH) in acetaldehyde metabolism, resulting in conversion of soluble PAs to insoluble PAs (Guan et al., 2017). More recently, C-PCNA DkMYB14 was found to be a bifunctional transcription factor that suppresses PA pathway genes and activates acetaldehyde biosynthesis to promote insolubilization of soluble PAs (Chen et al., 2021), providing a new strategy for flexible regulation of soluble and insoluble PA content.
Concluding remarks
Biosynthesis of (+)-catechin and (−)-epicatechin PA subunits is mostly understood, but several aspects still need to be addressed, including exactly how and where PAs are polymerized in vivo, how galloyl decoration of PA extension units is processed, and how PAs are transported from their site(s) of synthesis to the vacuole. For PA engineering, although knockout of key enzymes in the PA pathway can lead to PA compositional changes in some species (Liu et al., 2016; Jun et al., 2018; Robinson et al., 2021), this approach is limited to PA-producing tissues. For broader applications, the currently preferred strategy for engineering PAs in plants is to overexpress transcription factors. However, this often leads to negative growth phenotypes, possibly because of off-target effects or toxicity of ectopically expressed phenolic compounds such as PAs and their chemically active precursors (Lu et al., 2022). Although reactive PA extension units are proposed to be toxic to plant cells, mutants (e.g., Medicago lar and lar:ans; Arabidopsis tds4 [ans]), that over-accumulate PA extension units do not exhibit significant growth inhibition (Liu et al., 2016; Jun et al., 2018; Yu et al., 2022). By contrast, an increased level of stable (+)-catechin can be problematic for seed development, as shown in the Medicago ldox mutant (Jun et al., 2018). The recent discovery of AsA-[C]n in the Arabidopsis tds4 (ans) mutant suggests that initiation of PA oligomerization does not necessarily require (+)-catechin or (−)-epicatechin as the starter unit (Yu et al., 2022), inspiring future attempts to introduce PA-like molecules instead of classic PAs into plants to avoid negative effects on growth. Besides targeting the core MBW complex in the general flavonoid pathway, precise regulation of PA content can also be achieved by tuning the flux between anthocyanin and PA branches or by balancing the portion of soluble and insoluble PAs by manipulating bifunctional transcription factors (e.g., MtPAR, VvMYB86, and DkMYB14). Apart from transcription factors, the kinases, miRNAs, and small peptides that regulate PA pathway activity at the post-transcriptional level are candidates for tailoring PA compositions in crops. Selecting and benchmarking native or synthetic promoters can also facilitate tissue-specific PA engineering (Cui et al., 2022). Root-specific expression will prove to be increasingly important for enhancing belowground PA generation for carbon sequestration and greenhouse gas amelioration.
Funding
This work was supported by the Program of Introducing Talents of Discipline to Universities (111 Project, B13007), the National Natural Science Foundation of China (32030010), and the China Postdoctoral Science Foundation (2020M680404 to K.Y.).
Author contributions
K.Y., Y.S., J.L., and R.A.D. wrote and revised the manuscript.
Acknowledgments
No conflict of interest is declared.
Published: November 26, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Contributor Information
Jinxing Lin, Email: linjx@ibcas.ac.cn.
Richard A. Dixon, Email: richard.dixon@unt.edu.
References
- Abrahams S., Lee E., Walker A.R., Tanner G.J., Larkin P.J., Ashton A.R. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003;35:624–636. doi: 10.1046/j.1365-313x.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Achnine L., Blancaflor E.B., Rasmussen S., Dixon R.A. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell. 2004;16:3098–3109. doi: 10.1105/tpc.104.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk B., Sietiö O.M., Biasi C., Heinonsalo J. Interaction between tannins and fungal necromass stabilizes fungal residues in boreal forest soils. New Phytol. 2019;223:16–21. doi: 10.1111/nph.15729. [DOI] [PubMed] [Google Scholar]
- Adamczyk B., Sietiö O.M., Strakov' P., Prommer J., Wild B., Hagner M., Pihlatie M., Fritze H., Richter A., Heinonsalo J. Plant roots increase both decomposition and stable organic matter formation in boreal forest soil. Nat. Commun. 2019;10:3982. doi: 10.1038/s41467-019-11993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk B., Heinonsalo J., Simon J. Mechanisms of carbon sequestration in highly organic ecosystems – importance of chemical ecology. ChemistryOpen. 2020;9:464–469. doi: 10.1002/open.202000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M.Z., Li P., She G., Xia E., Benedito V.A., Wan X.C., Zhao J. Genome-wide analysis of serine carboxypeptidase-like acyltransferase gene family for evolution and characterization of enzymes involved in the biosynthesis of galloylated catechins in the tea plant (Camellia sinensis) Front. Plant Sci. 2020;11:848–921. doi: 10.3389/fpls.2020.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Ikegami A., Tsujimoto T., Kobayashi S., Sato A., Kono A., Yonemori K. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009;151:2028–2045. doi: 10.1104/pp.109.146985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airoldi C.A., Hearn T.J., Brockington S.F., Webb A.A.R., Glover B.J. TTG1 proteins regulate circadian activity as well as epidermal cell fate and pigmentation. Nat. Plants. 2019;5:1145–1153. doi: 10.1038/s41477-019-0544-3. [DOI] [PubMed] [Google Scholar]
- Akagi T., Katayama-Ikegami A., Yonemori K. Proanthocyanidin biosynthesis of persimmon (Diospyros kaki Thunb.) fruit. Sci. Hortic. (Amst.) 2011;130:373–380. [Google Scholar]
- An X.H., Tian Y., Chen K.Q., Liu X.J., Liu D.D., Xie X.B., Cheng C.G., Cong P.H., Hao Y.J. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015;56:650–662. doi: 10.1093/pcp/pcu205. [DOI] [PubMed] [Google Scholar]
- An J.P., An X.H., Yao J.F., Wang X.N., You C.X., Wang X.F., Hao Y.J. BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiol. 2018;38:1578–1587. doi: 10.1093/treephys/tpy063. [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Thiedig K., Nordholt N., Schmidt N., Huep G., Sagasser M., Weisshaar B. Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta. 2014;240:955–970. doi: 10.1007/s00425-014-2088-0. [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Nordholt N., Seidel T., Spelt K., Koes R., Quattrochio F., Sagasser M., Weisshaar B. TRANSPARENT TESTA 13 is a tonoplast P3A-ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 2015;82:840–849. doi: 10.1111/tpj.12854. [DOI] [PubMed] [Google Scholar]
- Aprile A., Federici C., Close T.J., De Bellis L., Cattivelli L., Roose M.L. Expression of the H +-ATPase AHA10 proton pump is associated with citric acid accumulation in lemon juice sac cells. Funct. Integr. Genomics. 2011;11:551–563. doi: 10.1007/s10142-011-0226-3. [DOI] [PubMed] [Google Scholar]
- Barros J., Escamilla-Trevino L., Song L., Rao X., Serrani-Yarce J.C., Palacios M.D., Engle N., Choudhury F.K., Tschaplinski T.J., Venables B.J., et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019;10:1994–2011. doi: 10.1038/s41467-019-10082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Bogs J., Jaffé F.W., Takos A.M., Walker A.R., Robinson S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007;143:1347–1361. doi: 10.1104/pp.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontpart T., Cheynier V., Ageorges A., Terrier N. BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 2015;208:695–707. doi: 10.1111/nph.13498. [DOI] [PubMed] [Google Scholar]
- Bordiga M., Travaglia F., Locatelli M., Coïsson J.D., Arlorio M. Characterisation of polymeric skin and seed proanthocyanidins during ripening in six Vitis vinifera L. cv. Food Chem. X. 2011;127:180–187. [Google Scholar]
- Bowerman P.A., Ramirez M.V., Price M.B., Helm R.F., Winkel B.S.J. Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia. BMC Res. Notes. 2012;5:485. doi: 10.1186/1756-0500-5-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britsch L., Grisebach H. Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur. J. Biochem. 1986;156:569–577. doi: 10.1111/j.1432-1033.1986.tb09616.x. [DOI] [PubMed] [Google Scholar]
- Brophy J.A.N., Magallon K.J., Duan L., Zhong V., Ramachandran P., Kniazev K., Dinneny J.R. Synthetic genetic circuits as a means of reprogramming plant roots. Science. 2022;377:747–751. doi: 10.1126/science.abo4326. [DOI] [PubMed] [Google Scholar]
- Burbulis I.E., Winkel-Shirley B. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier G., Huang Y.F., Le Cunff L., Fournier-Level A., Vialet S., Souquet J.M., Cheynier V., Terrier N., This P. Selection of candidate genes for grape proanthocyanidin pathway by an integrative approach. Plant Physiol. Biochem. 2013;72:87–95. doi: 10.1016/j.plaphy.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Cavallini E., Matus J.T., Finezzo L., Zenoni S., Loyola R., Guzzo F., Schlechter R., Ageorges A., Arce-Johnson P., Tornielli G.B. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015;167:1448–1470. doi: 10.1104/pp.114.256172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y.R., Lei B., Huang H.L., Li J.N., Yin J.M., Tang Z.L., Wang R., Chen L. TRANSPARENT TESTA 12 genes from Brassica napus and parental species: cloning, evolution, and differential involvement in yellow seed trait. Mol. Genet. Genom. 2009;281:109–123. doi: 10.1007/s00438-008-0399-1. [DOI] [PubMed] [Google Scholar]
- Chen S.Y., Tang Y.M., Hu Y.Y., Wang Y., Sun B., Wang X.R., Tang H.R., Chen Q. FaTT12-1, a multidrug and toxin extrusion (MATE) member involved in proanthocyanidin transport in strawberry fruits. Sci. Hortic. (Amst.) 2018;231:158–165. [Google Scholar]
- Chen W., Zheng Q., Li J., Liu Y., Xu L., Zhang Q., Luo Z. DkMYB14 is a bifunctional transcription factor that regulates the accumulation of proanthocyanidin in persimmon fruit. Plant J. 2021;106:1708–1727. doi: 10.1111/tpj.15266. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Zhu W., Chen Y., Ito S., Asami T., Wang X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. Elife. 2014;3:e02525. doi: 10.7554/eLife.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Yu K., Shi Y., Wang J., Duan C. Transcription factor VviMYB86 oppositely regulates proanthocyanidin and anthocyanin biosynthesis in grape berries. Front. Plant Sci. 2021;11:613677–613716. doi: 10.3389/fpls.2020.613677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin S., Jerkovic V., Bröhan M., Callemien D. In: Natural Products. Ramawat K., Mérillon J.M., editors. Springer; Berlin, Heidelberg: 2013. Polyphenols and beer quality. [DOI] [Google Scholar]
- Cos P., De Bruyne T., Hermans N., Apers S., Berghe D.V., Vlietinck A.J. Proanthocyanidins in health care: current and new trends. Curr. Med. Chem. 2004;11:1345–1359. doi: 10.2174/0929867043365288. [DOI] [PubMed] [Google Scholar]
- Crosby K.C., Pietraszewska-Bogiel A., Gadella T.W.J., Winkel B.S.J. Förster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett. 2011;585:2193–2198. doi: 10.1016/j.febslet.2011.05.066. [DOI] [PubMed] [Google Scholar]
- Cui L., Yao S., Dai X., Yin Q., Liu Y., Jiang X., Wu Y., Qian Y., Pang Y., Gao L., Xia T. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis) J. Exp. Bot. 2016;67:2285–2297. doi: 10.1093/jxb/erw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Yu M., Yao X., Xing J., Lin J., Li X. Single-particle tracking for the quantification of membrane protein dynamics in living plant cells. Mol. Plant. 2018;11:1315–1327. doi: 10.1016/j.molp.2018.09.008. [DOI] [PubMed] [Google Scholar]
- Cui Y., Fan J., Lu C., Ren J., Qi F., Huang H., Dai S. ScGST3 and multiple R2R3-MYB transcription factors function in anthocyanin accumulation in Senecio cruentus. Plant Sci. 2021;313:111094. doi: 10.1016/j.plantsci.2021.111094. [DOI] [PubMed] [Google Scholar]
- Cui X., Jun J.H., Rao X., Bahr C., Chapman E., Temple S., Dixon R.A. Leaf layer-based transcriptome profiling for discovery of epidermal-selective promoters in Medicago truncatula. Planta. 2022;256:31. doi: 10.1007/s00425-022-03920-4. [DOI] [PubMed] [Google Scholar]
- Dastmalchi M., Bernards M.A., Dhaubhadel S. Twin anchors of the soybean isoflavonoid metabolon: evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 2016;85:689–706. doi: 10.1111/tpj.13137. [DOI] [PubMed] [Google Scholar]
- Dean G., Cao Y., Xiang D., Provart N.J., Ramsay L., Ahad A., White R., Selvaraj G., Datla R., Haughn G. Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol. Plant. 2011;4:1074–1091. doi: 10.1093/mp/ssr040. [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Peeters A.J., Léon-Kloosterziel K.M., Koornneef M. The TRANSPARENT TESTA 12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13:853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour J.A., Ferreira D., Roux D.G. Synthesis of condensed tannins. Part 9. The condensation sequence of leucocyanidin with (+)-catechin and with the resultant procyanidins. J. Chem. Soc. Perkin 1. 1983:1711–1717. [Google Scholar]
- Dixon R.A., Sarnala S. Proanthocyanidin biosynthesis- a matter of protection. Plant Physiol. 2020;184:579–591. doi: 10.1104/pp.20.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Xie D.-Y., Sharma S.B. Proanthocyanidins – a final frontier in flavonoid research? New Phytol. 2005;165:9–28. doi: 10.1111/j.1469-8137.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- Fang F., Wang M.M., Zhu Q.G., Min T., Grierson D., Yin X.R., Chen K.S. DkMYB6 is involved in persimmon fruit deastringency, via transcriptional activation on both DkPDC and DkERF. Postharvest Biol. Technol. 2016;111:161–167. [Google Scholar]
- Feng H., Li Y., Wang S., Zhang L., Liu Y., Xue F., Sun Y., Wang Y., Sun J. Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.) J. Exp. Bot. 2014;65:5759–5769. doi: 10.1093/jxb/eru286. [DOI] [PubMed] [Google Scholar]
- Ferraro K., Jin A.L., Nguyen T.-D., Reinecke D.M., Ozga J.A., Ro D.-K. Characterization of proanthocyanidin metabolism in pea (Pisum sativum) seeds. BMC Plant Biol. 2014;14:238. doi: 10.1186/s12870-014-0238-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D., Stich K., Britsch L., Grisebach H. Purification and characterization of (+)dihydroflavonol (3-hydroxyflavanone) 4-reductase from flowers of Dahlia variabilis. Arch. Biochem. Biophys. 1988;264:40–47. doi: 10.1016/0003-9861(88)90567-x. [DOI] [PubMed] [Google Scholar]
- Forkmann G., Heller W., Grisebach H. Anthocyanin biosynthesis in flowers of Matthiola incana flavanone 3-and flavonoid 3′-hydroxylases. Zeitschrift für. Naturforschung C. 1980;35:691–695. [Google Scholar]
- Frank S., Keck M., Sagasser M., Niehaus K., Weisshaar B., Stracke R. Two differentially expressed MATE factor genes from apple complement the Arabidopsis transparent testa 12 mutant. Plant Biol. 2011;13:42–50. doi: 10.1111/j.1438-8677.2010.00350.x. [DOI] [PubMed] [Google Scholar]
- Fujino N., Tenma N., Waki T., Ito K., Komatsuzaki Y., Sugiyama K., Yamazaki T., Yoshida S., Hatayama M., Yamashita S., et al. Physical interactions among flavonoid enzymes in snapdragon and torenia reveal the diversity in the flavonoid metabolon organization of different plant species. Plant J. 2018;94:372–392. doi: 10.1111/tpj.13864. [DOI] [PubMed] [Google Scholar]
- Gao C. Genome engineering for crop improvement and future agriculture. Cell. 2021;184:1621–1635. doi: 10.1016/j.cell.2021.01.005. [DOI] [PubMed] [Google Scholar]
- Gourlay G., Ma D., Schmidt A., Constabel C.P. MYB134-RNAi poplars show reduced tannin synthesis in leaves but not roots, and increased susceptibility to oxidative stress. J. Exp. Bot. 2020;2019:1–11. doi: 10.1093/jxb/eraa371. [DOI] [PubMed] [Google Scholar]
- Gu L., Kelm M.a., Hammerstone J.F., Zhang Z., Beecher G., Holden J., Haytowitz D., Prior R.L. Liquid chromatographic/electrospray ionization mass spectrometric studies of proanthocyanidins in foods. J. Mass Spectrom. 2003;38:1272–1280. doi: 10.1002/jms.541. [DOI] [PubMed] [Google Scholar]
- Guan C., Du X., Zhang Q., Ma F., Luo Z., Yang Y. DkPK genes promote natural deastringency in C-PCNA persimmon by up-regulating DkPDC and DkADH expression. Front. Plant Sci. 2017;08:1–14. doi: 10.3389/fpls.2017.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Zhang C., Tian Y., Gul H., Cong P., Zhang L. Cloning and characterization of MdGST1 from red apple leaves. Can. J. Plant Sci. 2018;98:1150–1158. [Google Scholar]
- He F., Pan Q.H., Shi Y., Duan C.Q. Biosynthesis and genetic regulation of proanthocyanidins in plants. Molecules. 2008;13:2674–2703. doi: 10.3390/molecules13102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A.B., Reed J.D., Krueger C.G., Winterbottom R., Cunningham D.G., Leahy M. A-type cranberry proanthocyanidins and uropathogenic bacterial anti-adhesion activity. Phytochemistry. 2005;66:2281–2291. doi: 10.1016/j.phytochem.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Hrazdina G., Lifson E., Weeden N.F. Isolation and characterization of buckwheat (Fagopyrum esculentum M.) chalcone synthase and its polyclonal antibodies. Arch. Biochem. Biophys. 1986;247:414–419. doi: 10.1016/0003-9861(86)90600-4. [DOI] [PubMed] [Google Scholar]
- Hu Q., Luo C., Zhang Q., Luo Z. Isolation and characterization of a laccase gene potentially involved in proanthocyanidin polymerization in oriental persimmon (Diospyros kaki Thunb.) fruit. Mol. Biol. Rep. 2013;40:2809–2820. doi: 10.1007/s11033-012-2296-2. [DOI] [PubMed] [Google Scholar]
- Hu B., Zhao J., Lai B., Qin Y., Wang H., Hu G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016;35:831–843. doi: 10.1007/s00299-015-1924-4. [DOI] [PubMed] [Google Scholar]
- Hu F., Zeng C., Long R., Miao Y., Wei L., Xu Q., Min W. Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat. Methods. 2018;15:194–200. doi: 10.1038/nmeth.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.F., Vialet S., Guiraud J.L., Torregrosa L., Bertrand Y., Cheynier V., This P., Terrier N. A negative regulator of proanthocyanidin accumulation in grape berry, identified through expression quantitative locus mapping. New Phytol. 2014;201:795–809. doi: 10.1111/nph.12557. [DOI] [PubMed] [Google Scholar]
- Huang Y.-F., Doligez A., Fournier-Level A., Le Cunff L., Bertrand Y., Canaguier A., Morel C., Miralles V., Veran F., Souquet J.-M., et al. Dissecting genetic architecture of grape proanthocyanidin composition through quantitative trait locus mapping. BMC Plant Biol. 2012;12:30. doi: 10.1186/1471-2229-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel J.C., Hofmann T. Quantitative reconstruction of the nonvolatile sensometabolome of a red wine. J. Agric. Food Chem. 2008;56:9190–9199. doi: 10.1021/jf801742w. [DOI] [PubMed] [Google Scholar]
- Ichino T., Fuji K., Ueda H., Takahashi H., Koumoto Y., Takagi J., Tamura K., Sasaki R., Aoki K., Shimada T., Hara-Nishimura I. GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. Plant J. 2014;80:410–423. doi: 10.1111/tpj.12637. [DOI] [PubMed] [Google Scholar]
- Ichino T., Maeda K., Hara-Nishimura I., Shimada T. Arabidopsis ECHIDNA protein is involved in seed coloration, protein trafficking to vacuoles, and vacuolar biogenesis. J. Exp. Bot. 2020;71:3999–4009. doi: 10.1093/jxb/eraa147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami A., Eguchi S., Kitajima A., Inoue K., Yonemori K. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 2007;172:1037–1047. [Google Scholar]
- Jiang W., Yin Q., Wu R., Zheng G., Liu J., Dixon R.A., Pang Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 2015;66:7165–7179. doi: 10.1093/jxb/erv413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Chen M., He N., Chen X., Wang N., Sun Q., Zhang T., Xu H., Fang H., Wang Y., et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019;6:40. doi: 10.1038/s41438-019-0118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.H., Liu C., Xiao X., Dixon R.A. The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell. 2015;27:2860–2879. doi: 10.1105/tpc.15.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.H., Xiao X., Rao X., Dixon R.A. Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat. Plants. 2018;4:1034–1043. doi: 10.1038/s41477-018-0292-9. [DOI] [PubMed] [Google Scholar]
- Jun J.H., Lu N., Docampo-Palacios M., Wang X., Dixon R.A. Dual activity of anthocyanidin reductase supports the dominant plant proanthocyanidin extension unit pathway. Sci. Adv. 2021;7:eabg4682. doi: 10.1126/sciadv.abg4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater F., Fournand D., Vialet S., Meudec E., Cheynier V., Terrier N. Identification and functional characterization of cDNAs coding for hydroxybenzoate/hydroxycinnamate glucosyltransferases co-expressed with genes related to proanthocyanidin biosynthesis. J. Exp. Bot. 2012;63:1201–1214. doi: 10.1093/jxb/err340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S., Shikazono N., Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004;37:104–114. doi: 10.1046/j.1365-313x.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- Kitamura S., Matsuda F., Tohge T., Yonekura-Sakakibara K., Yamazaki M., Saito K., Narumi I. Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J. 2010;62:549–559. doi: 10.1111/j.1365-313X.2010.04174.x. [DOI] [PubMed] [Google Scholar]
- Koyama K., Numata M., Nakajima I., Goto-Yamamoto N., Matsumura H., Tanaka N. Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J. Exp. Bot. 2014;65:4433–4449. doi: 10.1093/jxb/eru213. [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.-M., Baudry A., Pourcel L., Nesi N., Caboche M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- Li Y.-G., Tanner G., Larkin P. The DMACA–HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J. Sci. Food Agric. 1996;70:89–101. [Google Scholar]
- Li X., Gao P., Cui D., Wu L., Parkin I., Saberianfar R., Menassa R., Pan H., Westcott N., Gruber M.Y. The Arabidopsis tt19-4 mutant differentially accumulates proanthocyanidin and anthocyanin through a 3’ amino acid substitution in glutathione S-transferase. Plant Cell Environ. 2011;34:374–388. doi: 10.1111/j.1365-3040.2010.02249.x. [DOI] [PubMed] [Google Scholar]
- Li P., Dong Q., Ge S., He X., Verdier J., Li D., Zhao J. Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanidin precursor flux in legume. Plant Biotechnol. J. 2016;14:1604–1618. doi: 10.1111/pbi.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhang B., Chen B., Ji L., Yu H. Site-specific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat. Commun. 2018;9:571. doi: 10.1038/s41467-018-03013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gao L., Liu L., Yang Q., Lu Z., Nie Z., Wang Y., Xia T. Purification and characterization of a novel galloyltransferase involved in catechin galloylation in the tea plant (Camellia sinensis) J. Biol. Chem. 2012;287:44406–44417. doi: 10.1074/jbc.M112.403071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shi Z., Maximova S., Payne M.J., Guiltinan M.J. Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 2013;13:202. doi: 10.1186/1471-2229-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Jun J.H., Dixon R.A. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 2014;165:1424–1439. doi: 10.1104/pp.114.241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wang X., Shulaev V., Dixon R.A. A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat. Plants. 2016;2:16182. doi: 10.1038/nplants.2016.182. [DOI] [PubMed] [Google Scholar]
- Liu X.-J., An X.-H., Liu X., Hu D.-G., Wang X.-F., You C.-X., Hao Y.-J. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 2017;68:2977–2990. doi: 10.1093/jxb/erx150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Qi Y., Zhang A., Wu H., Liu Z., Ren X. Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis) Plant Mol. Biol. 2019;100:451–465. doi: 10.1007/s11103-019-00870-6. [DOI] [PubMed] [Google Scholar]
- Lou H., Yamazaki Y., Sasaki T., Uchida M., Tanaka H., Oka S. A-type proanthocyanidins from peanut skins. Phytochemistry. 1999;51:297–308. [Google Scholar]
- Lu N., Jun J.H., Liu C., Dixon R.A. The flexibility of proanthocyanidin biosynthesis in plants. Plant Physiol. 2022;190:202–205. doi: 10.1093/plphys/kiac274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Dai C., Li Y., Feng J., Liu Z., Kang C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018;69:2595–2608. doi: 10.1093/jxb/ery096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Guo A., Zhang Y., Wang H., Liu Y., Li H. A review on astringency and bitterness perception of tannins in wine. Trends Food Sci. Technol. 2014;40:6–19. [Google Scholar]
- Matsuo T., Itoo S. A model experiment for de-astringency of persimmon fruit with high carbon dioxide treatment: in vitro gelation of kaki-tannin by reacting with acetaldehyde. Agric. Biol. Chem. 1982;46:683–689. [Google Scholar]
- Mattivi F., Vrhovsek U., Masuero D., Trainotti D. Differences in the amount and structure of extractable skin and seed tannins amongst red grape varieties. Aust. J. Grape Wine Res. 2009;15:27–35. [Google Scholar]
- McGivern B.B., Tfaily M.M., Borton M.A., Kosina S.M., Daly R.A., Nicora C.D., Purvine S.O., Wong A.R., Lipton M.S., Hoyt D.W., et al. Decrypting bacterial polyphenol metabolism in an anoxic wetland soil. Nat. Commun. 2021;12:2466. doi: 10.1038/s41467-021-22765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellway R.D., Tran L.T., Prouse M.B., Campbell M.M., Constabel C.P. The wound-pathogen-and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in Poplar. Plant Physiol. 2009;150:924–941. doi: 10.1104/pp.109.139071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menting J.G.T., Scopes R.K., Stevenson T.W. Characterization of flavonoid 3’, 5’-hydroxylase in microsomal membrane fraction of Petunia hybrida Flowers. Plant Physiol. 1994;106:633–642. doi: 10.1104/pp.106.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E., Kandaswami C., Theoharides T.C. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000;52:673–751. [PubMed] [Google Scholar]
- Mittasch J., Böttcher C., Frolova N., Bönn M., Milkowski C. Identification of UGT84A13 as a candidate enzyme for the first committed step of gallotannin biosynthesis in pedunculate oak (Quercus robur) Phytochemistry. 2014;99:44–51. doi: 10.1016/j.phytochem.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Mora J., Pott D.M., Osorio S., Vallarino J.G. Regulation of plant tannin synthesis in crop species. Front. Genet. 2022;13:870976–871018. doi: 10.3389/fgene.2022.870976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa E., Wong E. Purification and properties of chalcone-flavanone isomerase from soya bean seed. Phytochemistry. 1967;6:625–632. [Google Scholar]
- Nakajima J., Tanaka Y., Yamazaki M., Saito K. Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J. Biol. Chem. 2001;276:25797–25803. doi: 10.1074/jbc.M100744200. [DOI] [PubMed] [Google Scholar]
- Nakajima J.I., Sato Y., Hoshino T., Yamazaki M., Saito K. Mechanistic study on the oxidation of anthocyanidin synthase by quantum mechanical calculation. J. Biol. Chem. 2006;281:21387–21398. doi: 10.1074/jbc.M600303200. [DOI] [PubMed] [Google Scholar]
- Narduzzi L., Stanstrup J., Mattivi F. Comparing wild American grapes with Vitis vinifera: a metabolomics study of grape composition. J. Agric. Food Chem. 2015;63:6823–6834. doi: 10.1021/acs.jafc.5b01999. [DOI] [PubMed] [Google Scholar]
- Ono N.N., Qin X., Wilson A.E., Li G., Tian L. Two UGT84 family glycosyltransferases catalyze a critical reaction of hydrolyzable tannin biosynthesis in pomegranate (Punica granatum) PLoS One. 2016;11:e0156319–e0156325. doi: 10.1371/journal.pone.0156319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens D.K., Alerding A.B., Crosby K.C., Bandara A.B., Westwood J.H., Winkel B.S.J. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 2008;147:1046–1061. doi: 10.1104/pp.108.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Peel G.J., Sharma S.B., Tang Y., Dixon R.A. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of. Proc. Natl. Acad. Sci. USA. 2008;105:14210–14215. doi: 10.1073/pnas.0805954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Peel G.J., Wright E., Wang Z., Dixon R. a. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 2007;145:601–615. doi: 10.1104/pp.107.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Abeysinghe I.S.B., He J., He X., Huhman D., Mewan K.M., Sumner L.W., Yun J., Dixon R.A. Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 2013;161:1103–1116. doi: 10.1104/pp.112.212050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg H., Gacon K., Schlich P., Noble A.C. Bitterness and astringency of flavan-3-ol monomers, dimers and trimers. J. Sci. Food Agric. 1999;79:1123–1128. [Google Scholar]
- Pérez-Díaz R., Ryngajllo M., Pérez-Díaz J., Peña-Cortés H., Casaretto J.A., Gonz'lez-Villanueva E., Ruiz-Lara S. VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Rep. 2014;33:1147–1159. doi: 10.1007/s00299-014-1604-9. [DOI] [PubMed] [Google Scholar]
- Pérez-Díaz R., Madrid-Espinoza J., Salinas-Cornejo J., Gonz'lez-Villanueva E., Ruiz-Lara S. Differential roles for VviGST1, VviGST3, and VviGST4 in proanthocyanidin and anthocyanin transport in Vitis vinífera. Front. Plant Sci. 2016;7:1–13. doi: 10.3389/fpls.2016.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L., Routaboul J.-M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell. 2005;17:2966–2980. doi: 10.1105/tpc.105.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S., Dixon R.A. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell. 1999;11:1537–1552. doi: 10.1105/tpc.11.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.P., Bogs J., McDavid D.A.J., Hooper L.C., Speirs J., Walker A.R. Transgenic grapevines with decreased expression of tannin synthesis genes have altered grape and wine flavonoid composition. Aust. J. Grape Wine Res. 2021;27:106–117. [Google Scholar]
- Saito K., Kobayashi M., Gong Z., Tanaka Y., Yamazaki M. Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 1999;17:181–189. doi: 10.1046/j.1365-313x.1999.00365.x. [DOI] [PubMed] [Google Scholar]
- Scalbert A. Antimicrobial properties of tannins. Phytochemistry. 1991;30:3875–3883. [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.K., Humphreys J., Ruegger M.O., Chapple C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009;60:771–782. doi: 10.1111/j.1365-313X.2009.03996.x. [DOI] [PubMed] [Google Scholar]
- Shao D., Li Y., Zhu Q., Zhang X., Liu F., Xue F., Sun J. GhGSTF12, a glutathione S-transferase gene, is essential for anthocyanin accumulation in cotton (Gossypium hirsutum L.) Plant Sci. 2021;305:110827. doi: 10.1016/j.plantsci.2021.110827. [DOI] [PubMed] [Google Scholar]
- Shih C.H., Chu H., Tang L.K., Sakamoto W., Maekawa M., Chu I.K., Wang M., Lo C. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta. 2008;228:1043–1054. doi: 10.1007/s00425-008-0806-1. [DOI] [PubMed] [Google Scholar]
- Sun Y., Li H., Huang J.R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant. 2012;5:387–400. doi: 10.1093/mp/ssr110. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Someya S., Hu F., Tanokura M. Comparative study of catechin compositions in five Japanese persimmons (Diospyros kaki) Food Chem. X. 2005;93:149–152. [Google Scholar]
- Tahara K., Nishiguchi M., Frolov A., Mittasch J., Milkowski C. Identification of UDP glucosyltransferases from the aluminum-resistant tree Eucalyptus camaldulensis forming β-glucogallin, the precursor of hydrolyzable tannins. Phytochemistry. 2018;152:154–161. doi: 10.1016/j.phytochem.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takahashi R., Kouno I., Nonaka G.i. Chemical evidence for the de-astringency (insolubilization of tannins) of persimmon fruit. J. Chem. Soc. Perkin 1. 1994;20:3013–3022. [Google Scholar]
- Tanner G.J., Kristiansen K.N. Synthesis of 3, 4-cis-[3H]leucocyanidin and enzymatic reduction to catechin. Anal. Biochem. 1993;209:274–277. doi: 10.1006/abio.1993.1119. [DOI] [PubMed] [Google Scholar]
- Tanner G.J., Francki K.T., Abrahams S., Watson J.M., Larkin P.J., Ashton A.R. Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 2003;278:31647–31656. doi: 10.1074/jbc.M302783200. [DOI] [PubMed] [Google Scholar]
- Taylor P.W., Hamilton-Miller J.M.T., Stapleton P.D. Antimicrobial properties of green tea catechins. Food Sci. Technol. Bull. 2005;2:71–81. doi: 10.1616/1476-2137.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N., Torregrosa L., Ageorges A., Vialet S., Verriès C., Cheynier V., Romieu C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2008;149:1028–1041. doi: 10.1104/pp.108.131862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorngate J.H., Noble A.C. Sensory evaluation of bitterness and astringency of 3R(−)-epicatechin and 3S(+)-catechin. J. Sci. Food Agric. 1995;67:531–535. [Google Scholar]
- Tian H., Wang S. TRANSPARENT TESTA GLABRA1, a key regulator in plants with multiple roles and multiple function mechanisms. Int. J. Mol. Sci. 2020;21:4881–4915. doi: 10.3390/ijms21144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J.J., Sobey W.J., Aplin R.T., Hassan A., Schofield C.J., Firmin J.L., Prescott A.G. Are anthocyanidins the immediate products of anthocyanidin synthase? Chem. Commun. 2000;24:2473–2474. [Google Scholar]
- Turnbull J.J., Nagle M.J., Seibel J.F., Welford R.W.D., Grant G.H., Schofield C.J. The C-4 stereochemistry of leucocyanidin substrates for anthocyanidin synthase affects product selectivity. Bioorg. Med. Chem. Lett. 2003;13:3853–3857. doi: 10.1016/s0960-894x(03)00711-x. [DOI] [PubMed] [Google Scholar]
- Vale M., Rodrigues J., Badim H., Gerós H., Conde A. Exogenous application of non-mature miRNA-encoded miPEP164c inhibits proanthocyanidin synthesis and stimulates anthocyanin accumulation in grape berry cells. Front. Plant Sci. 2021;12:706679–706712. doi: 10.3389/fpls.2021.706679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J., Zhao J., Torres-Jerez I., Ge S., Liu C., He X., Mysore K.S., Dixon R.A., Udvardi M.K. MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. USA. 2012;109:1766–1771. doi: 10.1073/pnas.1120916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S., Francis L., Guyot S., Marnet N., Kwiatkowski M., Gawel R., Cheynier V., Waters E.J. The mouth-feel properties of grape and apple proanthocyanidins in a wine-like medium. J. Sci. Food Agric. 2003;83:564–573. [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Waki T., Mameda R., Nakano T., Yamada S., Terashita M., Ito K., Tenma N., Li Y., Fujino N., Uno K., et al. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 2020;11:870. doi: 10.1038/s41467-020-14558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang Y., Zhang M., Cai F., Qin J., Wang Q., Liu C., Wang G., Xu L., Yang L., et al. Molecular cloning and functional characterization of a glutathione S-transferase involved in both anthocyanin and proanthocyanidin accumulation in Camelina sativa (Brassicaceae) Genet. Mol. Res. 2012;11:4711–4719. doi: 10.4238/2012.September.25.4. [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang L., Jiang X., Dai X., Xu L., Li T., Xing D., Li Y., Li M., Gao L., Xia T. Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta. 2018;247:139–154. doi: 10.1007/s00425-017-2771-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu Y., Zhang L., Wang W., Hou H., Zhao Y., Jiang X., Yu J., Tan H., Wang Y., et al. Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J. 2020;101:18–36. doi: 10.1111/tpj.14515. [DOI] [PubMed] [Google Scholar]
- Wang R., Lu N., Liu C., Dixon R.A., Wu Q., Mao Y., Yang Y., Zheng X., He L., Zhao B., et al. MtGSTF7 , a TT19-like GST gene, is essential for accumulation of anthocyanins, but not proanthocyanins in Medicago truncatula. J. Exp. Bot. 2022;73:4129–4146. doi: 10.1093/jxb/erac112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- War A.R., Paulraj M.G., Ahmad T., Buhroo A.A., Hussain B., Ignacimuthu S., Sharma H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012;7:1306–1320. doi: 10.4161/psb.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E.B., Duguid M.C., Kuebbing S.E., Lendemer J.C., Bradford M.A. The functional role of ericoid mycorrhizal plants and fungi on carbon and nitrogen dynamics in forests. New Phytol. 2022;235:1701–1718. doi: 10.1111/nph.18307. [DOI] [PubMed] [Google Scholar]
- Welford R.W.D., Turnbull J.J., Claridge T.D.W., Schofield C.J., Prescott A.G. Evidence for oxidation at C-3 of the flavonoid C-ring during anthocyanin biosynthesis. Chem. Commun. 2001;18:1828–1829. doi: 10.1039/b105576n. [DOI] [PubMed] [Google Scholar]
- Wei K., Wang L., Zhang Y., Ruan L., Li H., Wu L., Xu L., Zhang C., Zhou X., Cheng H., Edwards R. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019;97:825–840. doi: 10.1111/tpj.14161. [DOI] [PubMed] [Google Scholar]
- Wei X., Ju Y., Ma T., Zhang J., Fang Y., Sun X. New perspectives on the biosynthesis, transportation, astringency perception and detection methods of grape proanthocyanidins. Crit. Rev. Food Sci. Nutr. 2021;61:2372–2398. doi: 10.1080/10408398.2020.1777527. [DOI] [PubMed] [Google Scholar]
- Wilmouth R.C., Turnbull J.J., Welford R.W.D., Clifton I.J., Prescott A.G., Schofield C.J. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure. 2002;10:93–103. doi: 10.1016/s0969-2126(01)00695-5. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plantarum. 1999;107:142–149. [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Zhu Q.G., Wang W.Q., Grierson D., Yin X.R. Molecular basis of the formation and removal of fruit astringency. Food Chem. 2022;372:131234. doi: 10.1016/j.foodchem.2021.131234. [DOI] [PubMed] [Google Scholar]
- Xie D.-Y., Dixon R.A. Proanthocyanidin biosynthesis – still more questions than answers? Phytochemistry. 2005;66:2127–2144. doi: 10.1016/j.phytochem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Xie Q., Xu Z. Sustainable agriculture: from sweet sorghum planting and ensiling to ruminant feeding. Mol. Plant. 2019;12:603–606. doi: 10.1016/j.molp.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Xie D.-Y., Sharma S.B., Paiva N.L., Ferreira D., Dixon R. a ( Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science. 2003;299:396–399. doi: 10.1126/science.1078540. [DOI] [PubMed] [Google Scholar]
- Xie P., Shi J., Tang S., Chen C., Khan A., Zhang F., Xiong Y., Li C., He W., Wang G., et al. Control of bird feeding behavior by Tannin1 through modulating the biosynthesis of polyphenols and fatty acid-derived volatiles in Sorghum. Mol. Plant. 2019;12:1315–1324. doi: 10.1016/j.molp.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Xu F., Cheng H., Cai R., et al. Molecular cloning and function analysis of an anthocyanidin synthase gene from Ginko biloba, and its expression in abiotic stress responses. Mol Cells. 2008;26:536.547. [PubMed] [Google Scholar]
- Xu W., Lepiniec L., Dubos C. New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signal. Behav. 2014;9:e28736. doi: 10.4161/psb.28736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang Y., Chen Y., Zhang P., Zhao Y., Huang Y., Wang X., Sheng J. Subcellular localization of galloylated catechins in tea plants [Camellia sinensis (L.) O. Kuntze] assessed via immunohistochemistry. Front. Plant Sci. 2016;7:728–810. doi: 10.3389/fpls.2016.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan L., Zhang C., Yan T., Wu D., Hussain N., Li Z., Chen M., Pan J., Jiang L. TRANSPARENT TESTA 4-mediated flavonoids negatively affect embryonic fatty acid biosynthesis in Arabidopsis. Plant Cell Environ. 2018;41:2773–2790. doi: 10.1111/pce.13402. [DOI] [PubMed] [Google Scholar]
- Yang S., Jiang Y., Xu L., Shiratake K., Luo Z., Zhang Q. Molecular cloning and functional characterization of DkMATE1 involved in proanthocyanidin precursor transport in persimmon (Diospyros kaki Thunb.) fruit. Plant Physiol. Biochem. 2016;108:241–250. doi: 10.1016/j.plaphy.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Yao S., Liu Y., Zhuang J., Zhao Y., Dai X., Jiang C., Wang Z., Jiang X., Zhang S., Qian Y., et al. Insights into acylation mechanisms: co-expression of serine carboxypeptidase-like acyltransferases and their non-catalytic companion paralogs. Plant J. 2022;111:117–133. doi: 10.1111/tpj.15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Jun J.H., Duan C., Dixon R.A. VvLAR1 and VvLAR2 are bifunctional enzymes for proanthocyanidin biosynthesis in grapevine. Plant Physiol. 2019;180:1362–1374. doi: 10.1104/pp.19.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Huang T., Tian B., Zhan J. Advances in biosynthesis and biological functions of proanthocyanidins in horticultural plants. Foods. 2020;9:1774–1822. doi: 10.3390/foods9121774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Dixon R.A., Duan C. A role for ascorbate conjugates of (+)-catechin in proanthocyanidin polymerization. Nat. Commun. 2022;13:3425. doi: 10.1038/s41467-022-31153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Wang L., Han Z., Jiang Y., Zhao L., Liu H., Yang L., Luo K. Molecular cloning and characterization of PtrLAR3, a gene encoding leucoanthocyanidin reductase from Populus trichocarpa, and its constitutive expression enhances fungal resistance in transgenic plants. J. Exp. Bot. 2012;63:2513–2524. doi: 10.1093/jxb/err425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D., Roth C., Unger V., Goldhammer T., Fenner N., Freeman C., Jurasinski G. Unraveling the importance of polyphenols for microbial carbon mineralization in rewetted riparian peatlands. Front. Environ. Sci. 2019;7:1–14. [Google Scholar]
- Zhang J.R., Trossat-Magnin C., Bathany K., Delrot S., Chaudière J. Oxidative transformation of leucocyanidin by anthocyanidin synthase from Vitis vinifera leads only to quercetin. J. Agric. Food Chem. 2019;67:3595–3604. doi: 10.1021/acs.jafc.8b06968. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Tian C., Zhang Y., Li C., Li X., Yu Q., Wang S., Wang X., Chen X., Feng S. Transcriptomic and metabolomic analysis provides insights into anthocyanin and procyanidin accumulation in pear. BMC Plant Biol. 2020;20:129–214. doi: 10.1186/s12870-020-02344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang P., Ma X., Zhao W., Li M., Yao S., Liu Y., Gao L., Xia T. Exploration of the substrate diversity of leucoanthocyanidin reductases. J. Agric. Food Chem. 2020;68:3903–3911. doi: 10.1021/acs.jafc.9b06353. [DOI] [PubMed] [Google Scholar]
- Zhang J.-R., Trossat-Magnin C., Bathany K., Negroni L., Delrot S., Chaudière J. Oxidative transformation of dihydroflavonols and flavan-3-ols by anthocyanidin synthase from Vitis vinifera. Molecules. 2022;27:1047. doi: 10.3390/molecules27031047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.K., Jeffery D.W., Li D.M., Lan Y.B., Zhao X., Duan C.Q. Red wine coloration: a review of pigmented molecules, reactions, and applications. Compr. Rev. Food Sci. Food Saf. 2022;21:3834–3866. doi: 10.1111/1541-4337.13010. [DOI] [PubMed] [Google Scholar]
- Zhang B., Yang H.J., Qu D., Zhu Z.Z., Yang Y.Z., Zhao Z.Y. The MdBBX22–miR858– MdMYB9/11/12 module regulates proanthocyanidin biosynthesis in apple peel. Plant Biotechnol. J. 2022;20:1683–1700. doi: 10.1111/pbi.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Dixon R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell. 2009;21:2323–2340. doi: 10.1105/tpc.109.067819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Dong W., Zhu Y., Allan A.C., Lin-Wang K., Xu C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020;18:1284–1295. doi: 10.1111/pbi.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Wang F., Espley R.V., Ren F., Zhao J., Ogutu C., He H., Jiang Q., Allan A.C., Han Y. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019;221:1919–1934. doi: 10.1111/nph.15486. [DOI] [PubMed] [Google Scholar]