Abstract
The structure of the complex formed between d(CGTACG)2 and 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide, an inactive derivative of the antitumour agents N-[2-(dimethylamino)ethyl]acridine-4-carboxamide (DACA) and 9-amino-DACA, has been solved to a resolution of 1.8 Å using X-ray crystallography. The complex crystallises in the space group P64 and the final structure has an overall R factor of 21.9%. A drug molecule intercalates between each of the CpG dinucleotide steps with its side chain lying in the major groove, and its protonated morpholino nitrogen partially occupying positions close to the N7 and O6 atoms of guanine G2. The morpholino group is disordered, the major conformer adopting a twisted boat conformation that makes van der Waals contact with the O4 oxygen of thymine T3. A water molecule forms bridging hydrogen bonds between the 4-carboxamide NH and the phosphate group of guanine G2. Sugar rings are found in alternating C3′-exo/C2′-endo conformations except for cytosine C1 which is C3′-endo. Intercalation perturbs helix winding throughout the hexanucleotide compared with B-DNA, steps 1 and 2 being unwound by 10 and 8°, respectively, while the central TpA step is overwound by 11°. An additional drug molecule lies at the end of each DNA helix linking it to the next duplex to form a continuously stacked structure. The protonated morpholino nitrogen of this ‘end-stacked’ drug hydrogen bonds to the N7 atom of guanine G6, and its conformationally disordered morpholino ring forms a C–H···O hydrogen bond with the guanine O6 oxygen. In both drug molecules the 4-carboxamide group is internally hydrogen bonded to the protonated N10 atom of the acridine ring. We discuss our findings with respect to the potential role played by the interaction of the drug side chain and the topoisomerase II protein in the poisoning of topoisomerase activity by the acridinecarboxamides.
INTRODUCTION
Drugs that poison topoisomerases by trapping the ‘cleavable complex’ of these essential topology-manipulating enzymes in a ternary complex with DNA are important in the treatment of cancer (1,2). The acridine-4-carboxamides are a series of DNA intercalating topoisomerase poisons, developed by Denny and colleagues at the Auckland Cancer Society Research Centre, that show remarkable variation in antitumour activity for minimal changes in molecular structure. Some are potent topoisomerase poisons with widespread antitumour efficacy, an example being N-[2-(dimethylamino)ethyl]acridine-4-carboxamide (DACA), which has had extensive clinical trials (3–5), whereas others show no biological activity despite maintaining high DNA-binding affinity (6). For the 9-aminoacridine class of these compounds, the parent of which is 9-amino-DACA, there are tight correlations between ligand structure, cytotoxicity and DNA-binding kinetics (6–10). The most significant features of the structure–activity relationships are that the side chain must be in the acridine 4 position, that the carboxamide must have an unsubstituted NH group, and that there must be two methylene groups between the carboxamide NH and the terminal protonated N,N-dimethylamino group (7). These findings, coupled with the dependence of the dissociation kinetics profile on ligand structure (6), imply that the side chain makes specific interactions with the DNA that are sensed by topoisomerase II in the ternary complex.
To provide insight into the nature of these molecular interactions, we have recently solved the crystal structures of several 9-aminoacridine-4-carboxamides complexed to the DNA hexanucleotide d(CGTACG)2 (11–14). Three of these structures provide valuable information about the interaction of the chromophore and side chain with the DNA, revealing that a drug molecule intercalates between each CpG dinucleotide step, its side chain lying in the major groove, with the protonated dimethylamino group making hydrogen-bonding interactions with the O6 and N7 atoms of guanine (11–13). A water molecule forms bridging hydrogen bonds between the 4-carboxamide NH and the phosphate group of the same guanine, and the 4-carboxamide group is additionally internally hydrogen bonded to the protonated N10 atom of the acridine ring. The compounds investigated in these three studies are all biologically active agents and the solution of the structures allowed a rationalisation of the known structure–activity relationships for the poisoning of topoisomerase II activity, cytotoxicity and DNA-binding kinetics for the 9-aminoacridine-4-carboxamides (11,12). In particular, they permitted the critical step in the dissociation kinetics profile that correlates with cytotoxicity and antitumour activity to be identified with the side chain–guanine interaction (6,11).
However, one anomaly stood out in the correlations between biological activity and kinetics, namely the derivative in which the N,N-dimethyl group of the side chain is replaced with a morpholino moiety. This compound is biologically inactive, but its DNA dissociation kinetics profile indicates that its morpholino side chain is still able to make hydrogen-bonding interactions with guanine bases (6,11,12). Indeed, the affinities and kinetics profiles of 9-amino-DACA and the morpholino derivative are indistinguishable at pH 7, which not only reveals that the latter binds as a dication under these conditions, but suggests that its DNA complex is structurally very similar to that of the parent compound (6). These findings imply that the inactivity of the morpholino analogue results from differences in the way topoisomerase II interacts with the drug–DNA complex, rather than from the fact that the morpholino and dimethylammonium groups interact differently with DNA per se. However, they do not resolve whether the origin of this difference lies in modifications to perturbations of DNA structure caused by binding of the two drugs, and sensed by topoisomerase II, or whether inactivity is more likely a consequence of differences in direct molecular contacts between the protein and the ligand in the DNA–ligand–enzyme ternary complex. As an alternative explanation, Rewcastle et al. (8) have suggested that the morpholino derivative may be inactive because of the lower basicity of its side chain compared with the side chain of 9-amino-DACA.
To help resolve the origins of the lack of activity of the morpholino derivative, we now present the crystal structure of the complex formed between 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide (morpholino-9-amino-DACA) (see Fig. 1) and d(CGTACG)2 in a buffer of pH 6. Our findings confirm that it is, indeed, structurally very similar to the 9-amino-DACA–DNA complex, with the morpholino side chain hydrogen bonding to guanine O6 and N7 atoms, and differences in perturbations to DNA structure being minimal for the two ligands. Thus, we conclude, assuming that the morpholino side chain is protonated in vivo, that the origins of the loss of biological activity probably lie in the consequences of direct interactions between topoisomerase II and the morpholino group.
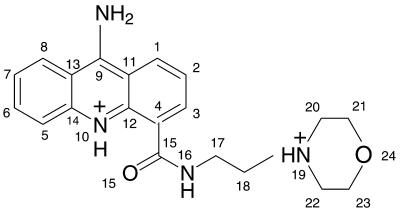
Figure 1.
Morpholino-9-amino-DACA.
MATERIALS AND METHODS
Crystallisation and data collection
The HPLC-purified self-complementary deoxyribonucleotide CGTACG was purchased from Oswel DNA Service (University of Southampton, UK). Morpholino-9-amino-DACA was synthesised as the dihydrochloride salt as previously described by Atwell et al. (7). The DNA and the ligand were both dissolved in water and stored frozen at 253 K. Crystals were grown at 285 K by vapour diffusion in sitting drops in Cryschem 24-well crystallisation plates using the Hampton Nucleic Acid Mini Screen (Hampton Research, Laguna Hills, CA). The drop initially contained 20 mM sodium cacodylate buffer pH 6.0, 0.6 mM DNA (duplex concentration), 1.2 mM ligand, 6 mM spermine tetrahydrochloride, 40 mM KCl, 10 mM MgCl2 and 5% 2-methyl-2,4-pentanediol (MPD) equilibrated against a 1 ml reservoir of 60% MPD. Yellow rods measuring 0.3 × 0.1 × 0.1 mm appeared in 5 weeks at which time a crystal was removed from the drop, placed in Riedel de Haen perfluoropolyether RS 3000 oil, mounted in a cryoloop and frozen at 110 K in a N2 cryostream. Diffraction intensities were recorded on a Rigaku R-axis II image plate system mounted on a Rigaku RU-200 rotating anode generator with focusing mirror optics (CuKα, 1.5418 Å) at a crystal-to-detector distance of 65 mm. The data were processed to 1.8 Å resolution with the DENZO and SCALEPACK programs (15), which indicated this crystal form to be in the space group P64 (or P62) with a = b = 30.2 Å and c = 39.3 Å (Table 1). Eight percent of the reflections were separated into a reference set to monitor Rfree.
Table 1. Crystal details, data collection and final refinement parameters.
| Unit cell dimensions (Å) | a = b = 30.2, c = 39.3 |
| Space group | P64 |
| No. of observations | 30 670 |
| No. of unique reflections | 1955 |
| Resolution range (Å) | 40–1.8 |
| Rmerge (%) | 9.4 (35.1)a |
| Completeness (%) | 99.8 (100)a |
| I/σI | 21.5 (5.0)a |
| No. of water molecules | 17 full occupancy and three partial occupancy |
| R (%) | 21.9 |
| Rfree (%) | 29.1 |
| RMSD from ideal geometry of final model distances | |
| Bonds (Å) | 0.004 (0.03)b |
| Angles (Å) | 0.012 (0.05)b |
| Planes (Å) | 0.027 (0.10)b |
| Average B values (Å2) | |
| Bases | 25 |
| Sugars | 28 |
| Phosphates | 32 |
| Intercalated ligand (100% occupancy) | 29 |
| Intercalated ligand side chain (66% occupancy) | 49 |
| End-stacked ligand (50% occupancy) | 26 |
| Water | |
| Full occupancy | 34 |
| Partial occupancy | 43 |
aValues in parentheses are for the last shell, 1.86–1.80 Å.
bTarget values are in parentheses.
Solution and refinement
The data are isomorphous with our previously solved structure of the 9-amino-DACA/d(CGTACG)2 complex (11), NDB accession number DD0015, with an Rmerge (= ∑ │ I – 〈I〉 │ / ∑ 〈I〉) = 12.6% and therefore we commenced refinement with the 9-amino-DACA structure as a starting model, having first removed all water molecules and ligands. After 150 cycles of conjugate-gradient least-squares refinement, using SHELX97-2 (16) in the resolution range 40–1.8 Å, where the resolution was increased in 0.01 Å steps from 3.0 to 1.8 Å, the R factor was 0.40 and the Rfree was 0.47. An iterative refinement procedure was then conducted interspersed with inspection of electron density maps and manual model rebuilding with the program O (17). Bond lengths and bond angles within the DNA bases were restrained to target values as specified in the SHELX 2000 DNA dictionary. All other chemically equivalent DNA bond lengths and bond angles were restrained by similarity distance restraints but without specification of an actual target value. For morpholino-9-amino-DACA bond lengths and bond angles were restrained to specified target values generated from an idealised structure built with INSIGHT II (18) and optimised with an AMBER force field using the DISCOVER program (18). The DNA bases, the acridine ring and the carboxamide group of morpholino-9-amino-DACA were restrained to be planar whereas all other torsion angles remained unrestrained.
3Fo–2Fc and Fo–Fc maps were examined after the initial rounds of refinement. The positions of the drug chromophores were clearly visible as well as several water molecules, and these were included in the refinement. As the remaining water molecules were added, a few at a time, the side chain positions of the two drug molecules became apparent in the Fo–Fc maps. Regions of diffuse solvent were modelled using Babinet’s principle as implemented in the SWAT command in SHELX97-2 (16). The side chain of the intercalated ligand from C18 onwards was found to be disordered, having high B factors (≈60). We assigned this portion of the side chain 66% occupancy but could not find a position for the remaining 33%. There was evidence in the density maps that the morpholino ring in both the intercalated and end-stacked ligands is conformationally disordered. We have modelled the major, twisted boat, conformer. The final R factor is 0.219 and the final Rfree is 0.291. The final refinement parameters are given in Table 1. Structural parameters for the DNA were analysed using the Curves 5.2 program (19). The coordinates and structure factors have been deposited with the Nucleic Acid Database, accession no. DD048 and the Protein Data Bank, accession no. 1KCI.
RESULTS
Global structure and crystal packing
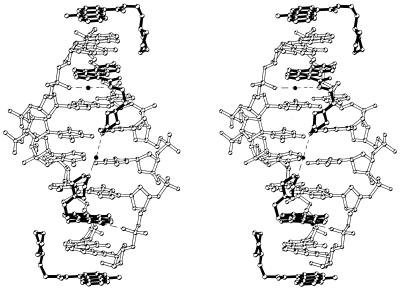
The global structure of the morpholino-9-amino-DACA/d(CGTACG)2 complex is very similar to that of the isomorphous 9-amino-DACA/d(CGTACG)2 and 5-F-9-amino-DACA/d(CGTACG)2 complexes (11,12). An asymmetric unit contains a single strand of DNA hexamer, one intercalated morpholino-9-amino-DACA molecule, 20 water molecules and an additional morpholino-9-amino-DACA. No metal ions or spermine were observed in the density maps. The two strands of the DNA are related by a crystallographic dyad and form a right-handed B-DNA-like duplex with Watson–Crick base pairs. The nucleotides are labelled C1–G6 in the 5′→3′ direction and the numbering scheme used for the drug is given in Figure 1. A morpholino-9-amino-DACA molecule is intercalated between each of the CpG dinucleotide base pair steps with its side chain lying in the major groove pointing towards the centre of the duplex. The protonated nitrogen of the partially disordered morpholino group refines to a position within hydrogen bonding distance of the N7 and O6 atoms of guanine G2. A water molecule on the dyad axis forms bridging hydrogen bonds to the morpholino oxygen atoms (O24) of the two symmetry-related intercalated ligands (Fig. 2). The additional drug molecule, also on a 2-fold axis, stacks on the end of each DNA helix and links it to the next duplex to form a continuous column of duplexes in the ab plane with the polarity of the DNA backbone reversing at this point (see fig. 2 in ref. 11). The protonated morpholino nitrogen of the end-stacked ligand is in hydrogen bonding distance of the N7 atom of guanine G6, and there is good evidence for a C–H···O hydrogen bond between C21 of this ligand and the O6 atom of G6, where the C–O distance, D, is 2.90 Å, and the C–H–O angle, θ, is 150° (20). The stacked columns of duplexes lie with their helical axes in planes related by a 31 axis parallel to c, the columns in neighbouring planes intersecting in projection at the dyad axis.
Figure 2.
Stereo view looking into the major groove of a bis-intercalated d(CGTACG)2 hexanucleotide with an end-stacked ligand bound at each end. The bonds of the ligands are drawn in black. The two water molecules located in the major groove are drawn in black and the hydrogen bonds associated with these water molecules are depicted by dashed lines.
DNA conformation
The DNA main chain and glycosidic torsion angles, as well as the furanose conformations, are listed in the Supplementary Material (Table S1). The P values for the sugar rings are larger than for the isomorphous 9-amino-DACA and 5-F-9-amino-DACA structures by an average of 26 and 23°, respectively. Each alternate sugar pucker now falls outside the range for C2′-endo of 140–180° and becomes the closely related C3′-exo pucker. The corresponding C4′–C3′ (δ) torsion angles, which are known to be correlated with sugar pucker (21), remain in the region associated with C2′-endo puckers of ∼150°. The glycosidic angles, χ, are in the medium anti conformation range for C1 and G2 whereas for all other bases they are high anti as is typically found in B-DNA (21). With the exception of the phosphate P–O5′ and C5′–C4′ torsion angles α and γ at both guanine G2 and G6 residues, the main chain dihedral angles for all six nucleotides fall within ranges characteristic of B-DNA (22). At G2 and G6 the α values have rotated by 116 and 129°, respectively, compared with those of B-DNA and the γ values by 142 and 119°. These coupled α/γ rotations at the guanines are the principal modifications to the backbone torsion angles responsible for opening up the intercalation cavity (11).
The Supplementary Material (Table S2) lists the geometrical properties of the base pairs. For step 1 the rise of 7.1 Å is as expected for an intercalation cavity and its twist angle shows the DNA to be unwound by 10° compared with the standard B-DNA value at the binding site. Base pair C1–G6 has a normal B-DNA propeller twist and a positive buckle of 3.0°, whereas propeller for G2–C5 is flattened to –2° and it has a 7.5° buckle in the opposite direction. The roll angles for all three steps, although different from average B-DNA values, have the sign and magnitude expected for the observed twist angles, given that roll is negatively correlated with twist (23). There is minimal slide and shift at step 1 but the tilt angle is large and negative. The helix winding at step 2, G2–T3, which has slide and shift values increased slightly over step 1 but not appreciably different from B-DNA, is reduced by 8° compared with the canonical structure. In contrast to steps 1 and 2, the central T3–A4 step is overwound by 11° compared with an average B-DNA twist of 36°.
Intercalated ligand conformation and DNA interactions
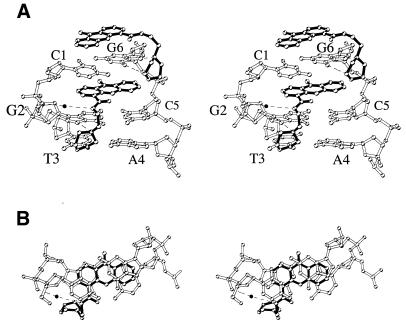
Figures 2 and 3 show stereo views of morpholino-9-amino-DACA intercalated via the major groove of the d(CpG).d(GpC) binding site which illustrate some important features of the complex. The 4-carboxamide group is planar, with the carbonyl oxygen forming an internal hydrogen bond to the protonated acridine ring nitrogen (see Table 2 for side chain dihedral angles). This interaction brings the carboxamide group to lie within 14° of the acridine plane. In this configuration the carboxamide NH group points towards the sugar–phosphate backbone where it hydrogen bonds to a water molecule which in turn hydrogen bonds to the phosphate group of guanine G2. The 9-amino group of the intercalated drug, which lies in the minor groove, forms no interactions with the DNA at the intercalation site, although it hydrogen bonds to the phosphate group of a symmetry-related duplex. The stacking interactions between the chromophore and the DNA bases are shown in projection in Figure 3B, where it can be seen that the acridine ring is fully enveloped by the two base pairs and the three atoms of the 4-carboxamide group are stacked on the C4, O4 and C5 atoms of cytosine C1. In the refined conformation of the partially disordered portion of the side chain, the ethylmorpholino moiety projects at right angles to the plane of the acridine ring, placing its protonated nitrogen within hydrogen bonding distance of the N7 nitrogen (3.0 Å) and the O6 oxygen (2.9 Å) of guanine G2. The conformation of the morpholino ring is disordered. The major conformer, the twisted boat, brings its C23 carbon atom into van der Waals contact (2.8 Å) with the O4 oxygen of thymine T3. The morpholino oxygen is in a position to participate in a bridging-water interaction with its symmetry-related mate intercalated between the C5pG6 dinucleotide (Fig. 2).
Figure 3.
(A) Stereo view looking into the major groove of mopholino-9-amino-DACA intercalated into the d(CpG).d(GpC) site of the d(CpGpT).d(GpCpA). Dashed lines indicate possible hydrogen bonds. (B) Stereo view of the projection down the helix axis of a d(CpG).d(GpC) dinucleotide with sandwiched intercalated ligand (bonds drawn in black).
Table 2. Refined mopholino-9-amino-DACA side chain torsion angles (°) of the major conformer.
| Morpholino-9-amino-DACA | 1a | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercalated | 14 (4) | –169 (4) | –111 (9) | –23 (12) | –74 (13) | 149 (12) | –121 (10) | 178 (8) | –61 (8) | –60 (7) | 40 (13) | 20 (14) |
| End-stacked | 4 (5) | 179 (4) | 117 (6) | –74 (8) | –57 (8) | 171 (6) | 171 (7) | 155 (8) | 35 (9) | 32 (12) | 28 (12) | –64 (8) |
aTorsion angle definition: 1, C3–C4–C15–N16; 2, C3–C4–C15–O15; 3, C15–N16–C17–C18; 4, N16–C17–C18–N19; 5, C17–C18–N19–C20; 6, C17–C18–N19–C22; 7, C18–N19–C20–C21; 8, C18–N19–C22–C23; 9, N19–C22–C23–O24; 10, N19–C20–C21–O24; 11, C20–C21–O24–C23; 12, C22–C23–O24–C21.
End-stacked ligand conformation and DNA interactions
The end-stacked morpholino-9-amino-DACA molecule is sandwiched between the C1–G6 base pairs of two symmetry-related hexanucleotides in the quasi-continuous stack. A second, symmetry related, ligand molecule is superimposed on the first, but flipped over along its major axis. As with the intercalated ligand, the 4-carboxamide group is planar and the carbonyl oxygen forms an internal hydrogen bond to the protonated acridine ring nitrogen (Table 2). The carboxamide group lies within 4° of the acridine plane and the side chain projects at right angles to the plane of the chromophore in an extended conformation (Table 2). A water molecule forms a hydrogen-bonded bridge between the ligand 9-amino group and the phosphate group of guanine G6 in a symmetry-related duplex. The protonated morpholino nitrogen of the end-stacked ligand is in hydrogen bonding distance of the N7 atom of guanine G6 (2.9 Å). The C21 carbon atom of the morpholino ring forms a C–H···O hydrogen bond with the O6 of guanine G6 as described above. The side chain of the end-stacked ligand is not disordered as it is in the intercalated ligand; however, there is evidence once again for conformation disorder of the morpholino ring. In the major, twisted boat, conformation the morpholino oxygen O24 is in close contact (3.2 Å) with the C5 carbon of the intercalated drug.
DISCUSSION
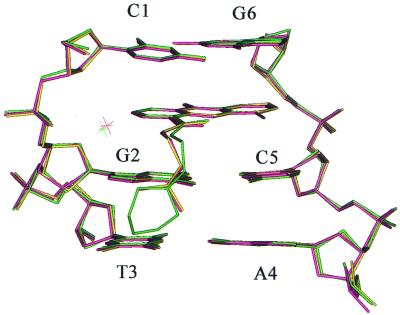
The work presented here is the first example of a crystal structure of an inactive derivative of the acridine-4-carboxamide class of topoisomerase poisons bound to normal duplex DNA. Notwithstanding caveats concerning uncertainties about the protonated state of the morpholino side chain in vivo, discussed in the Introduction, it therefore offers the opportunity for new insights into the molecular origins that underpin trapping of the cleavable complex by these agents. From a molecular pharmacological perspective the most important finding is that morpholino-9-amino-DACA can bind to d(CGTACG)2 in the same manner as its active congeners bearing an N,N-dimethylaminoethyl side chain. The similarity between the intercalated complexes of the active ligands and morpholino-9-amino-DACA is made clear in Figure 4 where the structures of the parent compound (11), its 5-F derivative (12) and morpholino-9-amino-DACA are overlaid for the purposes of comparison. Here it can be seen that the DNA conformation, the chromophore positioning and the ubiquitous phosphate-carboxamide-bridging water molecule are practically superposable. The ethyl linkage of the morpholino derivative is slightly displaced compared with the other two ligands, but in all three cases the terminal nitrogen and its two covalently bonded methyl or methylene carbons once again fall into the same locations, with the protonated nitrogen in each case hydrogen bonding to the guanine O6/N7 atoms. These findings focus attention on the remaining atoms of the morpholino ring, the –CH2–O–CH2– fragment, and their interaction with the DNA as likely sources of the loss of biological activity.
Figure 4.
Superposed line models of 9-amino-DACA (yellow), 5-F-9-amino-DACA (pink) and morpholino-9-amino-DACA (green).
Given the above, the most obvious feature of the morpholino group is that it extends the DNA–ligand interaction to include the next base pair. The twisted boat conformation adopted by the major conformer of the morpholino group is unusual given that the chair conformation is energetically more stable, and is the conformation usually seen in crystal structures of morpholine and its derivatives (24–27). However, Gao and Wang (28) have reported that morpholino groups in substituted derivatives of doxorubicin bound to d(CGATCG)2 and d(CGTACG)2 have flexible geometry that depends on the sequence context. In the structure reported here, in the twisted boat conformation, the C23 methylene group forms a close van der Waals interaction with the O4 oxygen of thymine T3, with the methylene hydrogens ‘embracing’ the thymine oxygen. In this position the morpholino oxygen atom is also able to hydrogen bond to its symmetry-related mate intercalated between C5 and G6 via a bridging water molecule located on the dyad axis. Neither of these interactions would be possible in the chair conformation. Despite this interaction of the morpholino group with T3, there is no evidence that this perturbs the helical parameters of the AT base pair compared with those found in the complexes of the active agents (11,12). Similarly, the helix winding of the three base pair steps for the morpholino complex falls within the range found for the biologically active ligands (11,12). The modified sugar pucker found in the morpholino complex, where some residues have become C3′-exo rather than C2′-endo, is more apparent than real, since this modest change leaves the sugars in the same family of puckers and has few structural consequences (Fig. 4). Thus, in short, it seems that the morpholino group has caused minimal, if any, perturbation to the structure of the DNA/9-aminoacridine-4-carboxamide complex. Similarly, it is apparent that the kinetic and thermodynamic stability of the DNA complex of the morpholino derivative is indistinguishable from that of the parent 9-amino-DACA (6).
It follows from these findings, assuming that the side chain is protonated in vivo, that the most likely causes for inactivity of the morpholino derivative are associated with interactions between the topoisomerase protein and the DNA and/or with the –CH2–O–CH2– fragment of the morpholino group itself. From all the available evidence it appears that active topoisomerase poisons amongst the acridine-4-carboxamides form well defined ternary complexes with the enzyme and DNA. If so, the results presented here suggest that the topoisomerase may be trying to access the base one removed from the intercalated chromophore on its 3′ side, in this case a thymine, and that this interaction is blocked by the morpholino group. Alternatively, it may be that the –CH2–O–CH2– fragment adversely interacts directly with the enzyme itself. Either of these possibilities may prevent formation of a stable ternary ‘cleavable’ complex.
Lastly, we note that the morpholino-9-amino-DACA/d(CGTACG)2 complex contains an interesting feature not commonly seen in structures of DNA or DNA–drug complexes. The morpholino group of the end-stacked ligand makes two distinct hydrogen-bonding interactions with the terminal guanine (see Fig. 3). There is a conventional hydrogen bond from the protonated morpholino nitrogen atom to the guanine N7 nitrogen, but, in addition, there is a second hydrogen bond involving the guanine O6 oxygen and the hydrogen atom of carbon C21. The C–O distance is 2.90 Å and the C–H–O angle is 150°, which, according to Desiraju (20), indicates that this is a strong C–H···O hydrogen bond of almost ideal geometry for a carbonyl oxygen. C–H···O hydrogen bonds have also been described in a DNA triplex involving uridine and hypoxanthine (29), and in intercalated four-stranded poly dC oligomers where there are cytosine–cytosine interactions and such bonding between sugar rings (30). More recently, Ghosh and Bansal (31) have drawn attention to the possibility of C–H···O bonding between AT base pairs in A-track DNA sequences, and Neidle and colleagues (32) have proposed C–H···O bonding between a tris-benzimidazole ligand and a thymine oxygen in a AT-tract minor groove complex. Thus, these occurrences suggest that it may be profitable to consider inclusion of C–H···O, or C–H···N, bonding interactions in the future design of DNA-binding ligands (20).
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
This work was funded by The Australian National Health and Medical Research Council.
Accession nos+ To whom correspondence should be addressed. Tel: +61 2 9351 7817; Fax: +61 2 9351 4726; Email: a.adams@biochem.usyd.edu.au NDB accession no. DD048 and PDB accession no. 1KCI
REFERENCES
- 1.Malonne H. and Atassi,G. (1997) DNA topoisomerase targeting drugs – mechanisms of action and perspectives. Anticancer Drugs, 8, 811–822. [DOI] [PubMed] [Google Scholar]
- 2.Denny W.A. (1997) Dual topoisomerase I/II poisons as anticancer drugs. Expert Opin. Invest. Drugs, 6, 1845–1851. [DOI] [PubMed] [Google Scholar]
- 3.Baguley B.C., Zhuang,L. and Marshall,E. (1995) Experimental solid tumor activity of N-[2-(dimethylamino)ethyl]acridine-4-carboxamide. Cancer Chemother. Pharmacol., 36, 244–248. [DOI] [PubMed] [Google Scholar]
- 4.Kestell P., Dunlop,I.C., McCrystal,M.R., Evans,B.D., Paxton,J.W., Gamage,R.S.K.A. and Baguley,B.C. (1999) Plasma pharmacokinetics of N-[2-(dimethylamino)ethyl]acridine-4-carboxamide in a phase I trial. Cancer Chemother. Pharmacol., 44, 45–50. [DOI] [PubMed] [Google Scholar]
- 5.McCrystal M.R., Evans,B.D., Harvey,V.J., Thompson,P.I., Porter,D.J. and Baguley,B.C. (1999) Phase I study of the cytotoxic agent N-[2-(dimethylamino)ethyl]acridine-4-carboxamide. Cancer Chemother. Pharmacol., 44, 39–44. [DOI] [PubMed] [Google Scholar]
- 6.Wakelin L.P.G., Atwell,G.J., Rewcastle,G.W. and Denny,W.A. (1987) Relationships between DNA-binding kinetics and biological activity for the 9-aminoacridine-4-carboxamide class of antitumor agents. J. Med. Chem., 30, 855–861. [DOI] [PubMed] [Google Scholar]
- 7.Atwell G.J., Cain,B.F., Baguley,B.C., Finlay,G.J. and Denny,W.A. (1984) Potential antitumor agents. Part 43. Synthesis and biological activity of dibasic 9-aminoacridine-4-carboxamides, a new class of antitumor agent. J. Med. Chem., 27, 1481–1485. [DOI] [PubMed] [Google Scholar]
- 8.Rewcastle G.W., Atwell,G.J., Chambers,D., Baguley,B.C. and Denny,W.A. (1986) Potential antitumor agents. 46. Structure–activity relationships for acridine monosubstituted derivatives of the antitumor agent N-[2-(dimethylamino)ethyl]-9-aminoacridine-4-carboxamide. J. Med. Chem., 29, 472–477. [DOI] [PubMed] [Google Scholar]
- 9.Denny W.A., Roos,I.A.G. and Wakelin,L.P.G. (1986) Interrelations between antitumor activity, DNA breakage, and DNA binding kinetics for 9-aminoacridinecarboxamide antitumor agents. Anticancer Drug Des., 1, 141–147. [PubMed] [Google Scholar]
- 10.Denny W.A., Atwell,G.J., Rewcastle,G.W. and Baguley,B.C. (1987) Potential antitumor agents. 49. 5-Substituted derivatives of N-[2-(dimethylamino)ethyl]-9-aminoacridine-4-carboxamide with in vivo solid-tumor activity. J. Med. Chem., 30, 658–663. [DOI] [PubMed] [Google Scholar]
- 11.Adams A., Guss,J.M., Collyer,C.A., Denny,W.A. and Wakelin,L.P.G. (1999) Crystal structure of the topoisomerase II poison 9-amino-[N-(2-dimethylamino)ethyl]acridine-4-carboxamide bound to the DNA hexanucleotide d(CGTACG)2. Biochemistry, 38, 9221–9233. [DOI] [PubMed] [Google Scholar]
- 12.Adams A., Guss,J.M., Collyer,C.A., Denny,W.A., Prakash,A.S. and Wakelin,L.P.G. (2000) Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides. Mol. Pharmacol., 58, 649–658. [DOI] [PubMed] [Google Scholar]
- 13.Todd A.K., Adams,A., Thorpe,J.H., Denny,W.A., Wakelin,L.P.G. and Cardin,C.J. (1999) Major groove binding and ‘DNA-induced’ fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethylamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5-BrU)ACG)2 at 1.3-Å. resolution. J. Med. Chem., 42, 536–540. [DOI] [PubMed] [Google Scholar]
- 14.Adams A., Guss,J.M., Collyer,C.A., Denny,W.A. and Wakelin,L.P.G. (2000) A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)2. Nucleic Acids Res., 28, 4244–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski Z. (1993) Oscillation data reduction program. In Sawyer,L., Isaacs,N. and Bailey,S.S. (eds), Proceedings of the CCP4 Study Weekend: Data Collection and Processing. SERC Daresbury Laboratory, Warrington, UK, pp. 56–62.
- 16.Sheldrick G.M. (1997) The SHELX-97 Manual. University of Gottingen, Germany.
- 17.Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., A47, 110–119. [DOI] [PubMed] [Google Scholar]
- 18. Biosym/MSI (1995) INSIGHTII User Guide. San Diego, CA.
- 19.Lavery R. and Sklenar,H. (1989) Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 20.Desiraju G.R. (1996) The C–H···O hydrogen bond: structural implications and supramolecular design. Acc. Chem. Res., 29, 441–449. [DOI] [PubMed] [Google Scholar]
- 21.Neidle S. (1994) DNA Structure and Recognition, 1st Edn. Oxford University Press, Oxford, UK.
- 22.Schneider B., Neidle,S. and Berman,H.M. (1997) Conformations of the sugar-phosphate backbone in helical DNA crystal structures. Biopolymers, 42, 113–124. [DOI] [PubMed] [Google Scholar]
- 23.Dickerson R.E. (1999) Helix structure and molecular recognition by B-DNA. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford, UK, pp. 145–197.
- 24.Raj S.S.S., Ponnuswamy,M.N., Shanmugam,G. and Kandaswamy,M. (1993) Synthesis and structural characterization of 2,6-bis(morpholinomethyl)-4-tert-butylphenol. J. Crystallogr. Spectrosc. Res., 23, 607–610. [Google Scholar]
- 25.Raj S.S.S., Velmurugan,D., Ponnuswamy,M.N., Shanmugam,G. and Kandaswamy,M. (1994) Structural characterization of 2,6-bis(N-methylenepiperidino)-4-nitrophenol (MPN) and 2,6-bis-(N-methylenemorpholino)-4-nitrophenol (MMN). J. Chem. Crystallogr., 24, 187–191. [Google Scholar]
- 26.Raj S.S.S., Thirumurugan,R., Shanmugam,G., Fun,H.-K., Manonmani,J. and Kandaswamy,M. (1999) {4,4′-Dimethyl-6,6′-bis(1-morpholiniomethyl)-2,2′-[1,2-ethanediylbis(nitrilomethylidene-N)]diphenolato-O,O′}copper(II) diperchlorate monohydrate. Acta Crystallogr., C55, 894–896. [Google Scholar]
- 27.Ertan M., Yesilada,A., Tozkoparan,B., Tarimci,C., Krebs,B. and Lage,M. (1997) (1R,2S,4S,11S)-4-Isopropyl-1-methyl-2-(α-N-morpholino-3-methoxybenzyl)cyclohexan-3-one. Acta Crystallogr., C53, 1466–1468. [Google Scholar]
- 28.Gao Y. and Wang,A.H.J. (1995) Crystal structures of four morpholino-doxorubicin anticancer drugs complexed with d(CGTACG) and d(CGATCG): implications in drug–DNA crosslink. J. Biomol. Struct. Dyn., 13, 103–118. [DOI] [PubMed] [Google Scholar]
- 29.Marfurt J. and Leumann,C. (1998) Evidence for C–H···O hydrogen bond assisted recognition of a pyrimidine base in the parallel DNA triple-helical motif. Angew. Chem. Int. Ed., 37, 175–177. [Google Scholar]
- 30.Berger I., Egli,M. and Rich,A. (1996) Inter-strand C–H···O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl Acad. Sci. USA, 93, 12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh A. and Bansal,M. (1999) C–H···O hydrogen bonds in minor groove of A-tracts in DNA double helices. J. Mol. Biol., 294, 1149–1158. [DOI] [PubMed] [Google Scholar]
- 32.Aymami J., Nunn,C.M. and Neidle,S. (1999) DNA minor groove recognition of a non-self-complementary AT-rich sequence by a tris-benzimidazole ligand. Nucleic Acids Res., 27, 2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.