Abstract
The non-homologous end-joining pathway promotes direct enzymatic rejoining of DNA double-strand breaks (DSBs) and is an important determinant of genome stability in eukaryotic cells. Although previous work has shown that this pathway requires Ku, DNA-PKcs and the DNA ligase IV/XRCC4 complex, we found that these proteins alone did not promote efficient joining of cohesive-ended DNA fragments in a cell-free assay. To identify factors that were missing from the reaction, we screened fractions from HeLa cell extracts for the ability to stimulate the joining of cohesive DNA ends in a complementation assay containing other known proteins required for DNA DSB repair. We identified a factor that restored end-joining activity to the level observed in crude nuclear extracts. Factor activity copurified with Rad50, Mre11 and NBS1, three proteins that have previously been implicated in DSB repair by genetic and cytologic evidence. Factor activity was inhibited by anti-Mre11 antibody. The reconstituted system remained fully dependent on DNL IV/XRCC4 and at least partially dependent on Ku, but the requirement for DNA-PKcs was progressively lost as other components were purified. Results support a model where DNA-PKcs acts early in the DSB repair pathway to regulate progression of the reaction, and where Mre11, Rad50 and NBS1 play a key role in aligning DNA ends in a synaptic complex immediately prior to ligation.
INTRODUCTION
The DNA double-strand break (DSB) end-joining pathway contributes to the maintenance of genome stability in eukaryotic organisms. It has been the subject of intense scrutiny. The pathway is dependent on at least five polypeptides: the two subunits of Ku protein, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), DNA ligase IV (DNL IV) and XRCC4 (reviewed in 1,2). Ku and DNA-PKcs carry out the initial recognition of broken DNA ends (3,4), and a complex of DNL IV and XRCC4 catalyzes the actual step of phosphodiester bond formation (5–9). Mutations affecting any of these proteins in mice lead to hypersensitivity to ionizing radiation and an inability to complete V(D)J recombination, a process that proceeds through a DSB intermediate (10–16). Except for DNA-PKcs, the components of the end-joining pathway are conserved in Saccharomyces cerevisiae, where they play a role similar to that in higher organisms (reviewed in 2). Components of the DNA end-joining machinery also associate with telomeres. They are required for the maintenance of telomere length and structure in S.cerevisiae (17–20) and for the suppression of telomere fusions in higher eukaryotes (21–24).
It is unclear how many other proteins, in addition to Ku, DNA-PKcs and the DNL IV/XRCC4 complex, are essential for the end-joining reaction. A number of proteins are candidates for involvement, based on their biochemical activities, their ability to localize to sites of DSBs in vivo or the phenotypes associated with mutations. For example, the phosphorylated γ-H2AX histone isoform is associated with altered chromatin domains near DNA breaks and helps to recruit additional repair proteins (25,26). The WRN helicase interacts with Ku protein in vitro and has a proposed role in unwinding and processing of DNA ends (27–30). The Mre11/Rad50/NBS1 complex strikingly relocalizes to sites of DSBs in vivo and has a nuclease activity capable of specifically degrading mismatched DNA ends to expose regions of microhomology (31–37). BRCA1 and 53 BP1 localize to sites of DSBs in vivo, but their functions are unknown (38–40). Some of these proteins may participate directly in repair, whereas others may affect DNA damage signaling.
The role of the Mre11/Rad50/NBS1 complex has been particularly hard to understand because of conflicting results obtained in different systems. In S.cerevisiae, Rad50 and Mre11 proteins are essential for DSB end joining (41,42) and are components of the same repair pathway as Ku (18,43). However, in Schizosaccharomyces pombe, these proteins appear to have no role in DSB repair (44). In higher eukaryotes, the role of these proteins has been difficult to assess, because deletion of either Mre11 or NBS1 leads to loss of functions essential for normal cell growth (45–47). Natural mutant alleles of NBS1 and Mre11 exist in the human population, and are associated with Nijmegen breakage syndrome and a variant form of ataxia telangiectasia, respectively (32,48,49). Afflicted individuals are sensitive to radiation but do not appear to have defects in DSB repair itself.
We have used a biochemical fractionation and reconstitution approach to gain insights into the requirements for the mammalian DNA end-joining reaction. Recent developments have facilitated the use of this approach. One of these is the availability of cell-free end-joining systems that show a faithful dependence on Ku, DNA-PKcs and other proteins known to be required for end joining in vivo (50–53). In addition, pure and active preparations of Ku protein, DNA-PKcs and DNL IV/XRCC4 have become available, and these have been shown to synergize with other components in cell extracts to give a high level of in vitro end-joining activity (53–55). The availability of these reagents has allowed us to focus on the identification of additional, unknown proteins required for end joining.
We have carried out a multicolumn fractionation of human cell extracts, using a biochemical complementation assay to identify a fraction that restores efficient end joining in the presence of DNL IV/XRCC4 and Ku. The fraction contains Mre11, Rad50 and NBS1, and its activity is inhibited by anti-Mre11 antibody, suggesting that the Mre11/Rad50/NBS1 complex is an intrinsic participant in the mammalian DNA end-joining pathway under the conditions used. Additionally, we find that the requirement for DNA-PKcs is lost as other components of the end-joining pathway are purified, suggesting that although DNA-PKcs may regulate the reaction, it is not an intrinsic participant in the final end-joining complex.
MATERIALS AND METHODS
HeLa cell nuclear extracts
Extracts were prepared as described (56) with modifications. Cells were swollen in hypotonic buffer and lysed by Dounce homogenization, and nuclei were collected and extracted with 4 packed cell volumes of buffer containing 50 mM Tris–HCl pH 7.9, 420 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 10% sucrose, 2 mM dithiothreitol (DTT), 10 µg/ml phenylmethylsulfonyl fluoride and 1 µg/ml each of pepstatin A, soybean protease inhibitor and leupeptin. After stirring for 30 min at 4°C, nuclei were pelleted at 26 500 g for 30 min, 0.33 g/ml (NH4)2SO4 was added to the supernatant and the precipitate was collected by centrifugation at 20 500 g for 10 min. The pellet was resuspended in 0.25 packed cell volumes of 0.1 M KOAc in DB buffer (20 mM Tris–HCl pH 7.9, 0.5 mM EDTA, 1 mM DTT, 20% glycerol and protease inhibitors). After dialysis, the preparation was clarified by centrifugation at 85 000 g for 60 min and stored at –80°C.
Protein purification
Recombinant DNL IV/XRCC4 complex and non-his-tagged Ku heterodimer were purified as described (54,57). Native DNA-PKcs was purified from HeLa cells through the DEAE–Sephacel step as described (56). Purification was completed by sequential chromatography on single-stranded DNA–agarose and Superdex 200 (Amersham Pharmacia Biotech).
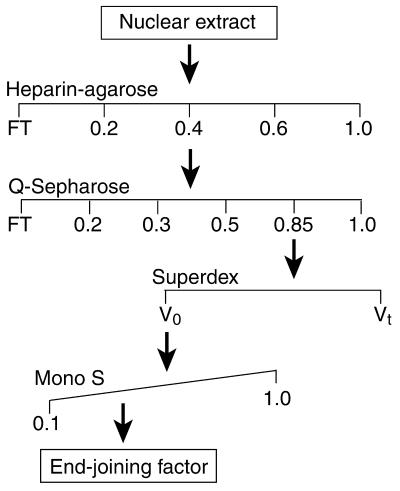
Purification of fractions with end-joining complementation activity was as follows: nuclear extract from 30 l of HeLa cell culture was applied to a 40 ml heparin–agarose column (Sigma) that had been pre-equilibrated with 0.1 M KOAc in DB buffer. The column was eluted stepwise with 0.1, 0.2, 0.4, 0.6 and 1.0 M KOAc in DB buffer. The 0.4 M fraction was dialyzed against 0.1 M KOAc in DB buffer and applied to a 25 ml Q-Sepharose HP column (Amersham Pharmacia Biotech) equilibrated in the same buffer. The column was eluted stepwise with 0.1, 0.2, 0.3, 0.5, 0.85 and 1.0 M KOAc in DB buffer. The 0.85 M fraction was dialyzed against 0.1 M KOAc in DB buffer and subjected to Superdex 200 gel chromatography (HR 16/60 column, Amersham Pharmacia Biotech). Active fractions were pooled and applied to a Mono S column (HR 5/10, Amersham Pharmacia Biotech). The column was eluted with a gradient of 0.1–1.0 M KOAc in DB buffer. Active fractions were aliquotted and stored at –80°C. Protein concentrations were determined by a dye-binding procedure (Bio-Rad). Immunoblotting was performed as described (58), except that membranes were developed using ECL substrate (Amersham Pharmacia Biotech).
DNA end-joining activity assay
Substrate DNA was prepared by digestion of pBluescript II KS+ plasmid (Stratagene) with restriction endonuclease BamHI and was 5′ end-labeled with polynucleotide kinase and [γ-32P]ATP. End-joining assays were performed in a volume of 20 µl, and contained 50 mM triethanolamine, pH 7.5, 10 mM Tris–HCl, pH 7.9, 65 mM KOAc, 0.25 mM EDTA, 0.5 mM DTT, 10% glycerol, 1.0 mM Mg(OAc)2, 100 ng/µl bovine serum albumin, 1 mM ATP, 0.5 ng/µl substrate DNA and proteins as specified in the figure legends. All components except for DNA were mixed, then incubated for 10 min at 37°C. DNA was added, and incubation was continued for 30 min at 37°C. Reactions were terminated by addition of 5 µl of 1% SDS, 30% glycerol, 0.1% bromphenol blue, 0.1% xylene cyanol. Samples were incubated for 15 min at 70°C and subjected to electrophoresis on a 0.6% agarose gel cast and run in buffer containing 40 mM Tris–acetate pH 8.0, 1 mM EDTA and 0.1% SDS. Radiolabeled DNA was visualized by PhosphorImager analysis.
RESULTS
End-joining assay
The substrate DNA used in these studies was a BamHI-digested, 5′-32P-labeled plasmid DNA. We have shown previously that DNL IV/XRCC4-supplemented cell extracts join the ends of this substrate perfectly, without loss or gain of nucleotides, to recreate a BamHI site (54). Because this DNA substrate does not require processing of DNA ends prior to ligation, it allows the identification of the minimal set of proteins required for end joining in the simplest case. The protein extract used in these experiments was from HeLa cell nuclei. When supplemented with DNL IV/XRCC4, this extract has activity equivalent to a whole-cell lymphoblast extract used earlier (54), and is more readily prepared in large quantities.
Resolution of a stimulatory factor from Ku and DNA-PKcs
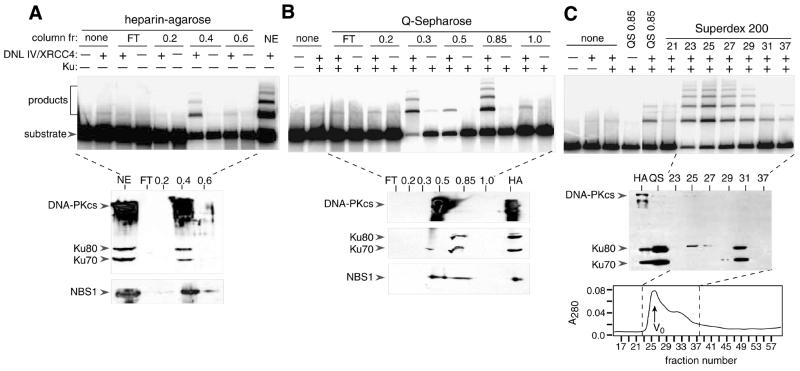
Fractionation of HeLa cell extracts was performed by sequential chromatography on heparin–agarose, Q-Sepharose HP and Superdex 200 columns, as outlined in Figure 1. Results of functional assays are shown in Figure 2. Consistent with previous results, DNL IV/XRCC4 had little or no end-joining activity in the absence of additional column fractions. Figure 2A (upper panel) shows that end-joining activity was present at a high level when DNL IV/XRCC4-containing reactions were supplemented with nuclear extract. Activity was quantitatively removed from the flow-through fraction when nuclear extract was passed over a heparin–agarose column (lanes labeled ‘FT’) and was eluted in a 0.4 M KOAc step fraction. None of the other heparin–agarose fractions showed activity. The activity of the 0.4 M fraction was strictly dependent on recombinant DNL IV/XRCC4.
Figure 1.
Fractionation scheme used in these studies. Material from a HeLa cell nuclear extract was subjected to sequential chromatography using heparin–agarose, Q-Sepharose, Superdex 200 and Mono S columns. Fractions at each stage were assayed for complementation activity in an end-joining assay, pooled, and passed over the next column.
Figure 2.
Characterization of end-joining activity and immunoreactive polypeptides in heparin–agarose, Q-Sepharose and Superdex 200 fractions. (A) Upper panel: end-joining assays using heparin–agarose column fractions. Assays were performed as described in Materials and Methods, and contained 2 µl of flow-through (FT), 2 µl of fractions eluted with buffer containing KOAc (molar concentration indicated) or 2 µl of nuclear extract (NE). Fractions containing >0.1 M KOAc were subjected to dialysis against DB buffer containing 0.1 M KOAc prior to assay. Reactions were performed in the presence or absence of 100 ng of purified, exogenous DNL IV/XRCC4 complex as indicated (+ or –). Products were resolved by SDS–agarose gel electrophoresis and visualized by PhosphorImager analysis. Lower panels: immunoblotting using heparin–agarose column fractions. Fractions are the same as in the upper panel, except that 10 µl was used. Proteins were resolved by 7.5% SDS–PAGE and transferred to nitrocellulose. Blots were probed with a mixture of anti-DNA-PKcs (mAb 18-2), anti-Ku80 (mAb 111) and anti-Ku70 (mAb N3H10), or with anti-NBS1 (rabbit polyclonal serum, Novus Biologicals, Littleton, CO) as indicated. (B) Upper panel: end-joining assays using Q-Sepharose column fractions. Assays were performed as in (A), and contained 2 µl of fractions eluted with buffer containing KOAc (molar concentrations indicated). Some reactions contained Ku heterodimer (25 ng) as indicated. Lower panel: immunoblotting using Q-Sepharose fractions. Fractions are the same as in the upper panel, except that 15 µl was used. Blots were probed with the same antibodies as in (A). (C) Upper panel: end-joining assays using Superdex 200 fractions. Reactions contained 2 µl of pooled Q-Sepharose 0.85 M fraction or 3 µl of individual Superdex 200 fractions as indicated. Middle panels: immunoblotting using 10 µl of 0.4 M heparin–agarose fraction (HA), 15 µl of 0.85 M Q-Sepharose fraction (QS) or 20 µl of Superdex 200 fractions as indicated. Blots were probed with a mixture of antibodies as in (A). Lower panel: Superdex 200 elution profile showing absorbance trace at 280 nm. Dashed lines indicate region of profile analyzed in activity and immunoblotting assays. Column void volume is indicated (V0).
Immunoblotting showed that endogenous Ku and DNA-PKcs co-eluted with activity in the 0.4 M fraction (Fig. 2A, lower panel). Consistent with the abundance of endogenous Ku and DNA-PKcs in this fraction, there was no further increase in activity when exogenous Ku and DNA-PKcs were supplied (not shown). Because of subsequent results showing a requirement for the Mre11/Rad50/NBS1 complex in the reconstituted end-joining system, fractions from the early steps of purification were retrospectively analyzed using anti-NBS1 peptide antibody. Immunoblotting showed that NBS1 was present mostly in the 0.4 M KOAc fraction.
Fractionation of the HeLa cell extract was continued using a Q-Sepharose HP column. From this point on, recombinant Ku protein was included in the end-joining complementation assays. Again, activity was absent from the flow-through (Fig. 2B, upper panel). End-joining activity was detectable in 0.3, 0.5 and 0.85 M KOAc step fractions, and in each case was DNL IV/XRCC4-dependent. Because the 0.85 M KOAc fraction was the most active, it was selected for further characterization. Activities in the other fractions have not yet been characterized.
Immunoblotting showed that most of the Ku eluted in the active 0.85 M KOAc fraction, whereas most of the DNA-PKcs eluted in the less active 0.5 M KOAc fraction (Fig. 2B, lower panel). The high level of end-joining activity in the 0.85 M fraction suggests either that DNA-PKcs is not essential for end joining under these conditions, or that a trace amount of remaining DNA-PKcs is sufficient. NBS1 was detected in both the 0.5 and the 0.85 M KOAc fractions.
Fractionation was continued by passing the Q-Sepharose 0.85 M KOAc fraction over a Superdex 200 gel filtration column. Activity eluted in a sharp peak, which coincided with the void volume (Fig. 2C, upper panel). This implies a native molecular weight of >500 kDa, based on a calibration of the column with standard proteins. In an immunoblotting assay, Superdex fractions 23–29, which have end-joining activity, gave no detectable signal at the positions expected for Ku and DNA-PKcs polypeptides (Fig. 2C, middle panel). Fractions 25 and 27 contained a trace of an unidentified cross-reacting polypeptide, which was slightly larger than Ku80. The bulk of the Ku protein eluted at its expected position, starting in fraction 31, consistent with its native molecular weight of 153 kDa. If present, DNA-PKcs would have eluted from this column slightly after the void peak (56). Taken together, the data in Figure 2 demonstrate that HeLa nuclear extracts contain a factor that is essential for efficient DNA end joining in the presence of DNL IV/XRCC4 and that is separable from Ku and DNA-PKcs.
Stimulatory activity purifies in association with Mre11, Rad50 and NBS1
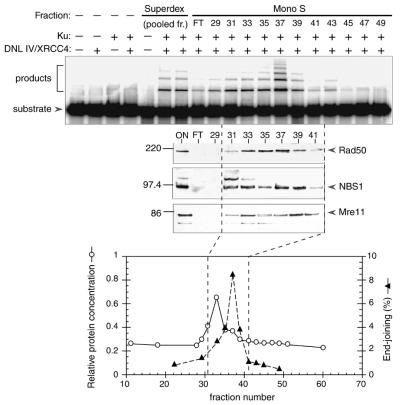
Active fractions from the Superdex 200 column were pooled and subjected to high resolution Mono S chromatography. Activity eluted in a single peak (Fig. 3A). Comparison with the protein profile (Fig. 3A, lower panel) indicated that activity was resolved from the bulk of the protein eluting from the column.
Figure 3.
Characterization of Mono S fractions. Mono S elution profile. Upper panel: end-joining assays were performed as in Figure 2, and contained 2 µl of material from the Superdex 200 column (pooled fractions 25–28) or 3 µl from the Mono S column (flow-through, FT, or undialyzed individual fractions, as indicated). Reactions were performed in the presence or absence of DNL IV/XRCC4 complex (100 ng) and Ku heterodimer (25 ng) as indicated. Middle panels: immunoblotting was performed using the same fractions as in the upper panel, except that 20 µl of the Superdex fraction and 25 µl of the Mono S fractions were used. Three similar blots were probed with rabbit polyclonal sera directed against mouse Rad50, human NBS1 or human Mre11 (Novus Biologicals). The expected position of each protein is indicated by an arrow on the right. Migration of molecular weight markers is indicated on the left. Bottom panel: column profile showing relative protein concentration (determined by the Bradford assay) and end-joining activity in each fraction. Dashed lines indicate the approximate boundaries of the end-joining activity peak.
It was of interest to determine whether the active fractions represented a novel activity, or whether they contained proteins that had previously been implicated in DSB repair by genetic or cytologic criteria. Column fractions were subjected to analysis by immunoblotting using various sera that were purchased commercially or were provided by other laboratories. Positive results were obtained using anti-Rad50, anti-Mre11 and anti-NBS1 sera. These polypeptides co-eluted with end-joining activity in fractions 31–39 (Fig. 3A, center panel). Preliminary attempts to detect other known repair proteins, including BRCA1 and 53 BP1, gave negative results (data not shown). The presence of Mre11, Rad50 and NBS1 in the active fractions suggests that these proteins are direct participants in the mammalian DSB end-joining reaction under the conditions used.
Inhibition by anti-Mre11 antibodies
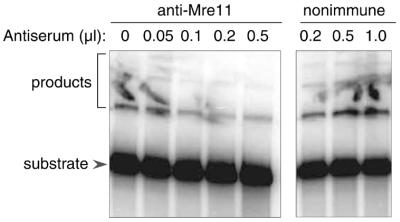
To further investigate the requirements for activity in the fractionated system, we tested the effect of an anti-Mre11 serum in an end-joining assay. As shown in Figure 4, addition of increasing amounts of anti-Mre11 serum resulted in a progressive decrease in end-joining activity. In contrast, addition of non-immune serum resulted in a modest increase in activity, presumably because the higher total protein concentration in the reaction had a protective effect on the repair enzymes. The difference between immune and non-immune serum is clear, and the results strongly suggest that Mre11 is essential for DNA end joining in the reconstituted system.
Figure 4.
Effect of anti-Mre11 serum on end-joining activity. End-joining assays were performed as in Figure 2, and contained 100 ng of DNL IV/XRCC4 complex, 25 ng of Ku heterodimer, and the indicated amount of material from the Mono S column (pooled fractions 35–38). In addition, reactions contained the indicated amounts of anti-Mre11 rabbit serum or non-immune rabbit serum. Non-specific background is seen in some lanes in this experiment and in Figure 5. The background appears to be attributable to contamination of the gel surface with radiolabel and is not correlated with the contents of the reactions.
Lack of dependence on DNA-PKcs
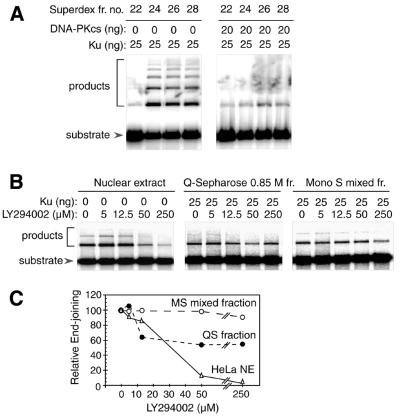
Although Q-Sepharose chromatography removed most of the endogenous DNA-PKcs from the active 0.85 M fraction (Fig. 2B), it is possible that undetected trace levels of DNA-PKcs remained and contributed to the end-joining reaction. To further investigate the requirement for DNA-PKcs, we tested the effect of adding purified, active DNA-PKcs to the fractionated system. As shown in Figure 5A, small amounts of DNA-PKcs inhibited end-joining activity in reactions containing DNL IV/XRCC4 and active Superdex 200 fractions. Thus, not only is the requirement for DNA-PKcs lost as other factors are purified, but DNA-PKcs actually interferes with the reaction. It is unlikely that this interference arises from the presence of non-specific inhibitors in the DNA-PKcs preparation, as similar preparations of DNA-PKcs markedly stimulate end-joining activity in a crude cell extract that has been immunodepleted with anti-DNA-PKcs antibodies (55). Presumably, the difference in the results obtained in the purified system and in the crude extracts reflects the presence, in the latter system, of additional proteins required for productive function of DNA-PKcs. For example, under the conditions used, DNA-PKcs is in stoichiometric excess over DNA ends. It may be that additional factors in the extract are capable of promoting release of DNA-PKcs from these DNA ends, allowing DNL IV and other proteins to gain access.
Figure 5.
End-joining activity in the presence and absence of DNA-PKcs, and in the presence of varying amounts of DNA-PKcs inhibitor. (A) Effect of DNA-PKcs. End-joining assays were performed as in Figure 2. Reactions contained 3 µl of individual Superdex 200 fractions as indicated. Reactions were performed in the presence and absence of DNA-PKcs and Ku, as indicated. All reactions contained DNL IV/XRCC4 complex (100 ng). (B) End-joining activity in the presence of LY294002. Reactions contained HeLa cell nuclear extract (10 µg), Q-Sepharose 0.85 M fraction (2 µl) or Mono S mixed fraction (3 µl) as indicated. Reactions also contained Ku protein (0 or 25 ng) and DNL IV/XRCC4 complex (100 ng). LY294002 was prepared as a 5 mM stock in dimethylsulfoxide (DMSO) and was added to give the final concentrations indicated (0–250 µM). Compensating amounts of DMSO were added to other reactions, such that the final concentration was held constant at 5%. (C) Quantitation of results from (B). End-joining activity was calculated as the ratio of ligated products to total DNA, and is expressed as a percentage of the activity observed with each set of proteins in the absence of LY294002.
To confirm that DNA-PKcs is required for the end-joining reaction in crude extract but not in the purified system, we tested the effect of a small molecule inhibitor, LY294002. This compound inhibits the enzymatic activity of DNA-PKcs and blocks end joining in lymphoblast cell extracts (53). As expected, there was substantial inhibition of activity in reactions containing unfractionated HeLa nuclear extract (Fig. 5B and C). There was only ∼50% inhibition of activity reactions containing the Q-Sepharose 0.85 M fraction, and there was no inhibition in reactions containing Mono S fractions. These results confirm that the requirement for DNA-PKcs activity is lost as other factors are purified.
DISCUSSION
We have carried out biochemical fractionation and in vitro reconstitution studies in order to gain insight into the requirements for the eukaryotic DNA end-joining reaction. Previous work has shown that proteins required for this reaction include Ku, DNA-PKcs and the DNL IV/XRCC4 complex. We show here that these proteins alone are insufficient to reconstitute activity to the levels seen in crude extracts, and that there is a requirement for at least one additional factor. This stimulatory factor copurifies with immunoreactive Mre11, Rad50 and NBS1 polypeptides and is inhibited by anti-Mre11 antibody. A second finding is that the requirement for DNA-PKcs is conditional, and is lost as the other proteins required for the reaction are purified.
Potential role of the Mre11/Rad50/NBS1 complex in the alignment of DNA ends
Presumably, a major function of the additional repair factors is to accelerate the reaction by aligning the DNA ends in the correct position and orientation. Purified DNL IV/XRCC4 complex efficiently uses an oligo(dT)–poly(dA) model substrate, where DNA ends are aligned for ligation by base pairing to a complementary homopolymer, but is much less efficient in its use of natural DNA fragments. One of the key issues in the field is the identity of the proteins that maintain synapsis between the ends of double-stranded DNA fragments in preparation for ligation. Interestingly, there is evidence that each of the known DSB repair proteins contributes to synapsis of the DNA ends in some way. For example, several lines of evidence suggest that Ku protein participates in transient, non-covalent complexes containing two DNA molecules (59–63). The enzymatic activity of DNA-PKcs is modulated in ways that suggest the ability to interact with more than one DNA at once (35,64). The nuclease activity of the Mre11/Rad50/NBS1 complex is arrested when opposing DNAs are base paired, indicating that two DNA ends can be accommodated at the same time within the active site (35). Consistent with this, structures have very recently been observed by electron microscopy and by atomic force microscopy in which Mre11/Rad50 complexes (or yeast Mre11/Rad50/Xrs2 complexes) appear to bridge two DNA ends (65,66). Finally, the presence of two ligase active sites in the tetrameric DNL IV/XRCC4 complex is suggestive of the ability to interact with two DNAs simultaneously (54), and complexes where DNL IV/XRCC4 bridges the ends of two different DNAs have been observed directly (67). Because the DNA termini can make only a limited number of contacts at once, it seems likely that the various repair proteins come into contact with the ends sequentially, rather than simultaneously.
Our data suggest that Ku and the Mre11/Rad50/NBS1 complex, but not DNA-PKcs, participate together in a complex that is formed immediately prior to strand rejoining. The primary evidence for this is that Ku, DNL IV/XRCC4 and the Mre11/Rad50/NBS1-containing fraction, but not DNA-PKcs, have been shown to cooperate directly to reconstitute end joining in the most purified system. DNA-PKcs may act at an earlier step and dissociate prior to the formation of the synaptic complex, or it may participate in an alternative subpathway of repair. Current data do not permit us to distinguish between these alternatives.
Importantly, the requirement for Mre11/Rad50/NBS1 is seen with cohesive-ended DNA fragments, which do not require processing for ligation. Thus, the requirement for the Mre11/Rad50/NBS1 complex cannot be attributed solely to the structure-dependent nuclease activity that is associated with these proteins. We suggest that the nuclease and alignment functions of the Mre11/Rad50/NBS1 complex may be dependent on a single DNA binding site, which is capable of binding the ends of both DNAs simultaneously, remodeling them as needed to remove radiation-damaged and non-cohesive base pairs, and holding them in position for ligation by DNL IV/XRCC4. If this model is correct, then the cohesive DNA substrates used in the present study are able to enter directly into this site, but the ends are not processed because they are already fully base paired. A high-resolution structure of a related archaeal complex is available (68), but does not give insights into the nature of the end-bridging phenomenon in the eukaryotic complexes. Thus, a more critical evaluation of the nature of the DNA binding site awaits further studies.
The proposal that the Mre11/Rad50/NBS1 complex has an alignment function, independent of its nuclease activity, is consistent with the results of genetic studies in S.cerevisiae. In this organism, Mre11 and Rad50 protein are essential for DSB rejoining, but a point mutation affecting only nuclease activity is associated with a phenotype that is milder than that of the null mutant (69,70).
Role of Ku
Further work will be needed to better define the role of Ku protein in the ligation complex. It has been suggested that Ku protects DNA ends against excessive degradation prior to repair (71,72). Consistent with this, we have observed that Ku tends to protect against non-specific loss of radiolabeled phosphate from the DNA ends in the in vitro system, particularly in the presence of crude fractions, where non-specific nucleases and phosphatases are more likely to be present. This complicates efforts to detect specific effects on the end-joining reaction itself. We did observe that Ku stimulated end joining in the most purified system (data not shown) but we have only explored this to a limited extent, and current data are not sufficient to establish that the stimulation occurs by a specific, rather than a non-specific, mechanism.
Prior studies disagree over whether Ku is able to directly affect the activity of eukaryotic DNA ligases, in the absence of other factors (63,67). In the current study, we performed a number of control reactions in which the effect of Ku was tested in the absence of other stimulatory factors (Figs 2, 3 and 5). The amount of ligation under these conditions was barely discernable, and there was no obvious difference in the presence or absence of Ku. The reason for the different results obtained in different studies is unclear.
Regulatory role for DNA-PKcs
Current findings suggest that DNA-PKcs is able to play a dual role, inhibiting end joining in the context of purified system, but promoting it in the context of crude extract or the intact cell. One possible explanation for the dual role of DNA-PKcs is that it controls progression through a reaction checkpoint. For example, several lines of evidence suggest that DNA-PKcs is able to interact with more than one DNA end at once, and the presence of an opposing DNA end may provide a signal that allows the reaction to proceed (64). The lack of a requirement for DNA-PKcs in the purified system could be explained if proper functioning of the checkpoint requires interaction of DNA-PKcs with another, regulatory protein, which is lost as the system is purified.
In vitro studies show that DNA-PKcs is capable of phosphorylating sites in both Ku and XRCC4 (73–75). The finding that DNA-PKcs is not required in a purified system, even though Ku and XRCC4 are present, argues that phosphorylation of these proteins is not intrinsically required for completion of the end-joining reaction. However, phosphorylation could modulate interactions of Ku and XRCC4 with DNA-PKcs itself or with other regulatory proteins not present in the minimal system.
A recent report describes stimulation of end joining by inositol hexakisphosphate (IP6), which apparently works by activating DNA-PKcs (68). We observed no effect when this molecule was added to a partially purified system (data not shown), suggesting that the absence of IP6 does not, in itself, account for the failure of DNA-PKcs to cooperate in end joining.
How many proteins participate in the end-joining pathway?
The results of fractionation studies present a relatively simple picture. In contrast to the multiplicity of factors uncovered when similar approaches have been applied to such processes as eukaryotic transcription and replication, it appears that as few as seven polypeptides could be sufficient for reconstitution of the end-joining pathway: the two subunits of Ku, DNL IV, XRCC4, Mre11, Rad50 and NBS1. The predicted molecular weight of the Mre11/Rad50/NBS1 complex is at least 524 kDa, assuming that two copies of the 80 kDa Mre11 polypeptide, two copies of the 140 kDa Rad50 polypeptide and at least one copy of the 84 kDa NBS1 polypeptide are present (68). Thus, elution of the end-joining factor at a >500 kDa position in the Superdex 200 profile could be explained on the basis of a complex containing Mre11, Rad50 and NBS1 alone. However, a number of additional polypeptides are visible in the most purified fractions after SDS–PAGE and silver staining, and we do not yet know whether these are stably associated with the factor and contribute to its activity.
Another study has recently shown that yeast Mre11/Rad50/Xrs2 complex promotes DNL4-catalyzed DNA end joining in a cell-free system (65). The results are similar to the findings presented here with the mammalian system. Taken together, these studies suggest that the fundamental mechanism by which DNA ends are aligned has been conserved between the yeast and mammalian systems.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Sunghan Yoo (Georgetown University Medical Center) for anti-Mre11 antiserum and Dr Patrick Sung (University of Texas Health Sciences Center-San Antonio) for unpublished information about the purification of the Mre11/Rad50/NBS1 complex. We thank Ms Farlyn Hudson for recombinant Ku and DNL IV/XRCC4, Ms Nancy Miller for consistent cell culture, and the members of the Dynan laboratory and Rhea-Beth Markowitz for helpful comments. This work was supported by US National Science Foundation Grant MCB-9906440. W.S.D. received support as an Eminent Scholar of the Georgia Research Alliance.
REFERENCES
- 1.Smith G.C. and Jackson,S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- 2.Haber J.E. (2000) Partners and pathways repairing a double-strand break. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- 3.Dvir A., Peterson,S.R., Knuth,M.W., Lu,H. and Dynan,W.S. (1992) Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl Acad. Sci. USA, 89, 11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb T.M. and Jackson,S.P. (1993) The DNA-dependent protein kinase: Requirement for DNA ends and association with Ku antigen. Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 5.Critchlow S.E., Bowater,R.P. and Jackson,S.P. (1997) Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- 6.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 7.Schär P., Herrmann,G., Daly,G. and Lindahl,T. (1997) A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev., 11, 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teo S.-H. and Jackson,S.P. (1997) Identification of Saccharomyces cerevisae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J., 16, 4788–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson T.E., Grawunder,U. and Lieber,M.R. (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature, 388, 495–498. [DOI] [PubMed] [Google Scholar]
- 10.Blunt T., Finnie,N.J., Taccioli,G.E., Smith,G.C., Demengeot,J., Gottlieb,T.M., Mizuta,R., Varghese,A.J., Alt,F.W., Jeggo,P.A. and Jackson,S.P. (1995) Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell, 80, 813–823. [DOI] [PubMed] [Google Scholar]
- 11.Kirchgessner C.U., Patil,C.K., Evans,J.W., Cuomo,C.A., Fried,L.M., Carter,T., Oettinger,M.A. and Brown,J.M. (1995) DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science, 267, 1178–1183. [DOI] [PubMed] [Google Scholar]
- 12.Peterson S.R., Kurimasa,A., Oshimura,M., Dynan,W.S., Bradbury,E.M. and Chen,D.J. (1995) Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc. Natl Acad. Sci. USA, 92, 3171–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nussenzweig A., Chen,C., da Costa Soares,V., Sanchez,M., Sokol,K., Nussenzweig,M.C. and Li,G.C. (1996) Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature, 382, 551–555. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C., Bogue,M.A., Lim,D.-S., Hasty,P. and Roth,D.B. (1996) Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell, 86, 379–389. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y., Sun,Y., Frank,K.M., Dikkes,P., Fujiwara,Y., Seidl,K.J., Sekiguchi,J.M., Rathbun,G.A., Swat,W., Wang,J., Bronson,R.T., Malynn,B.A., Bryans,M., Zhu,C., Chaudhuri,J., Davidson,L., Ferrini,R., Stamato,T., Orkin,S.H., Greenberg,M.E. and Alt,F.W. (1998) A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell, 95, 891–902. [DOI] [PubMed] [Google Scholar]
- 16.Barnes D.E., Stamp,G., Rosewell,I., Denzel,A. and Lindahl,T. (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol., 8, 1395–1398. [DOI] [PubMed] [Google Scholar]
- 17.Boulton S.J. and Jackson,S.P. (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulton S.J. and Jackson,S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 20.Nugent C.I., Bosco,G., Ross,L.O., Evans,S.K., Salinger,A.P., Moore,J.K., Haber,J.E. and Lundblad,V. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol., 8, 657–660. [DOI] [PubMed] [Google Scholar]
- 21.Hsu H.L., Gilley,D., Blackburn,E.H. and Chen,D.J. (1999) Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey S.M., Meyne,J., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu H.L., Gilley,D., Galande,S.A., Hande,M.P., Allen,B., Kim,S.H., Li,G.C., Campisi,J., Kohwi-Shigematsu,T. and Chen,D.J. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev., 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samper E., Goytisolo,F.A., Menissier-de Murcia,J., Gonzalez-Suarez,E., Cigudosa,J.C., de Murcia,G. and Blasco,M.A. (2001) Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol., 154, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogakou E.P., Boon,C., Redon,C. and Bonner,W.M. (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol., 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paull T.T., Rogakou,E.P., Yamazaki,V., Kirchgessner,C.U., Gellert,M. and Bonner,W.M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol., 10, 886–895. [DOI] [PubMed] [Google Scholar]
- 27.Cooper M.P., Machwe,A., Orren,D.K., Brosh,R.M., Ramsden,D. and Bohr,V.A. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev., 14, 907–912. [PMC free article] [PubMed] [Google Scholar]
- 28.Li B. and Comai,L. (2000) Functional interaction between Ku and the werner syndrome protein in DNA end processing. J. Biol. Chem., 275, 28349–28352. [DOI] [PubMed] [Google Scholar]
- 29.Orren D.K., Machwe,A., Karmakar,P., Piotrowski,J., Cooper,M.P. and Bohr,V.A. (2001) A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res., 29, 1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yannone S.M., Roy,S., Chan,D.W., Murphy,M.B., Huang,S., Campisi,J. and Chen,D.J. (2001) Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem., 276, 38242–38248. [DOI] [PubMed] [Google Scholar]
- 31.Maser R.S., Monsen,K.J., Nelms,B.E. and Petrini,J.H. (1997) hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol., 17, 6087–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carney J.P., Maser,R.S., Olivares,H., Davis,E.M., Le Beau,M., Yates,J.R.,III, Hays,L., Morgan,W.F. and Petrini,J.H. (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell, 93, 477–486. [DOI] [PubMed] [Google Scholar]
- 33.Paull T.T. and Gellert,M. (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell, 1, 969–979. [DOI] [PubMed] [Google Scholar]
- 34.Trujillo K.M., Yuan,S.S., Lee,E.Y. and Sung,P. (1998) Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11 and p95. J. Biol. Chem., 273, 21447–21450. [DOI] [PubMed] [Google Scholar]
- 35.Paull T.T. and Gellert,M. (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA, 97, 6409–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirzoeva O.K. and Petrini,J.H. (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol., 21, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trujillo K.M. and Sung,P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50/Mre11 complex. J. Biol. Chem., 276, 35458–35464. [DOI] [PubMed] [Google Scholar]
- 38.Scully R., Chen,J., Ochs,R.L., Keegan,K., Hoekstra,M., Feunteun,J. and Livingston,D.M. (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell, 90, 425–435. [DOI] [PubMed] [Google Scholar]
- 39.Schultz L.B., Chehab,N.H., Malikzay,A. and Halazonetis,T.D. (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol., 151, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Q., Chen,C.F., Li,S., Chen,Y., Wang,C.C., Xiao,J., Chen,P.L., Sharp,Z.D. and Lee,W.H. (1999) Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science, 285, 747–750. [DOI] [PubMed] [Google Scholar]
- 41.Moore J.K. and Haber,J.E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsukamoto Y., Kato,J. and Ikeda,H. (1996) Effects of mutations of RAD50, RAD51, RAD52 and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics, 142, 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milne G.T., Jin,S., Shannon,K.B. and Weaver,D.T. (1996) Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisae. Mol. Cell. Biol., 16, 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo G., Yao,M.S., Bender,C.F., Mills,M., Bladl,A.R., Bradley,A. and Petrini,J.H. (1999) Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA, 96, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi-Iwai Y., Sonoda,E., Sasaki,M.S., Morrison,C., Haraguchi,T., Hiraoka,Y., Yamashita,Y.M., Yagi,T., Takata,M., Price,C., Kakazu,N. and Takeda,S. (1999) Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J., 18, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J., Petersen,S., Tessarollo,L. and Nussenzweig,A. (2001) Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol., 11, 105–109. [DOI] [PubMed] [Google Scholar]
- 48.Varon R., Vissinga,C., Platzer,M., Cerosaletti,K.M., Chrzanowska,K.H., Saar,K., Beckmann,G., Seemanova,E., Cooper,P.R., Nowak,N.J., Stumm,M., Weemaes,C.M., Gatti,R.A., Wilson,R.K., Digweed,M., Rosenthal,A., Sperling,K., Concannon,P. and Reis,A. (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell, 93, 467–476. [DOI] [PubMed] [Google Scholar]
- 49.Stewart G.S., Maser,R.S., Stankovic,T., Bressan,D.A., Kaplan,M.I., Jaspers,N.G., Raams,A., Byrd,P.J., Petrini,J.H. and Taylor,A.M. (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell, 99, 577–587. [DOI] [PubMed] [Google Scholar]
- 50.Pfeiffer P. and Vielmetter,W. (1988) Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res., 16, 907–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu X.Y., Weinfeld,M.A. and Povirk,L.F. (1998) Implication of DNA-dependent protein kinase in an early, essential, local phosphorylation event during end-joining of DNA double-strand breaks in vitro. Biochemistry, 37, 9827–9835. [DOI] [PubMed] [Google Scholar]
- 52.Labhart P. (1999) Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol., 19, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumann P. and West,S.C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee K.J., Huang,J., Takeda,Y. and Dynan,W.S. (2000) DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system. J. Biol. Chem., 275, 34787–34796. [DOI] [PubMed] [Google Scholar]
- 55.Woodard R.L., Lee,K., Huang,J. and Dynan,W.S. (2001) Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J. Biol. Chem., 276, 15423–15433. [DOI] [PubMed] [Google Scholar]
- 56.Dvir A., Stein,L.Y., Calore,B.L. and Dynan,W.S. (1993) Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem., 268, 10440–10447. [PubMed] [Google Scholar]
- 57.Yoo S., Kimzey,A. and Dynan,W.S. (1999) Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem., 274, 20034–20039. [DOI] [PubMed] [Google Scholar]
- 58.Nueda A., Hudson,F., Mivechi,N.F. and Dynan,W.S. (1999) DNA-dependent protein kinase protects against heat-induced apoptosis: evidence for a regulatory interaction with HSF1. J. Biol. Chem., 274, 14988–14996. [DOI] [PubMed] [Google Scholar]
- 59.Bliss T.M. and Lane,D.P. (1997) Ku selectively transfers between DNA molecules with homologous ends. J. Biol. Chem., 272, 5765–5773. [DOI] [PubMed] [Google Scholar]
- 60.Pang D., Yoo,S., Dynan,W.S., Jung,M. and Dritschilo,A. (1997) Ku protein joins DNA fragments as shown by atomic force microscopy. Cancer Res., 57, 1412–1415. [PubMed] [Google Scholar]
- 61.Cary R.B., Peterson,S.R., Wang,J., Bear,D.G., Bradbury,E.M. and Chen,D.J. (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl Acad. Sci. USA, 94, 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramsden D.A. and Gellert,M. (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J., 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nick McElhinny S.A., Snowden,C.M., McCarville,J. and Ramsden,D.A. (2000) Ku recruits the XRCC4–ligase IV complex to DNA ends. Mol. Cell. Biol., 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leuther K.K., Hammarsten,O., Kornberg,R.D. and Chu,G. (1999) Structure of DNA-dependent protein kinase: implications for its regulation by DNA. EMBO J., 18, 1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L., Trujillo,K.M., Ramos,W., Sung,P. and Tomkinson,A.E. (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell, 8, 1105–1115. [DOI] [PubMed] [Google Scholar]
- 66.de Jager M., van Noort,J., van Gent,D.C., Dekker,C., Kanaar,R. and Wyman,C. (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell, 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- 67.Chen L., Trujillo,K., Sung,P. and Tomkinson,A.E. (2000) Interactions of the DNA ligase IV/Xrcc4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem., 275, 26196–26205. [DOI] [PubMed] [Google Scholar]
- 68.Hopfner K., Karcher,A., Craig,L., Woo,T.T., Carney,J.P. and Tainer,J.A. (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell, 105, 473–485. [DOI] [PubMed] [Google Scholar]
- 69.Bressan D.A., Olivares,H.A., Nelms,B.E. and Petrini,J.H. (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics, 150, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moreau S., Ferguson,J.R. and Symington,L.S. (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol., 19, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boulton S.J. and Jackson,S.P. (1996) Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J., 15, 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 72.Lee S.E., Moore,J.K., Holmes,A., Umezu,K., Kolodner,R.D. and Haber,J.E. (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell, 94, 399–409. [DOI] [PubMed] [Google Scholar]
- 73.Leber R., Wise,T.W., Mizuta,R. and Meek,K. (1998) The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem., 273, 1794–1801. [DOI] [PubMed] [Google Scholar]
- 74.Chan D.W., Ye,R., Veillette,C.J. and Lees-Miller,S.P. (1999) DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry, 38, 1819–1828. [DOI] [PubMed] [Google Scholar]
- 75.Modesti M., Hesse,J.E. and Gellert,M. (1999) DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J., 18, 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]