Abstract
Non-alcoholic fatty liver disease (NAFLD) is a chronic progressive liver disease with increasing prevalence. Lipophagy is a type of programmed cell death that plays an essential role in maintaining the body’s balance of fatty acid metabolism. However, the livers of NAFLD patients are abnormally dysregulated in lipophagy. mTORC1 is a critical negative regulator of lipophagy, which has been confirmed to participate in the process of lipophagy through various complex mechanisms. Therefore, targeting mTORC1 to restore failed autophagy may be an effective therapeutic strategy for NAFLD. This article reviews the main pathways through which mTORC1 participates in the formation of lipophagy and the intervention effect of mTORC1-regulated lipophagy in NAFLD, providing new therapeutic strategies for the prevention and treatment of NAFLD in the future.
Keywords: NAFLD, lipophagy, mTORC1, hepatic steatosis, insulin resistance, hepatic fibrosis
Introduction
Non-alcoholic fatty liver disease (NAFLD) caused by obesity and excessive/unbalanced diet has become a common liver disease worldwide. According to statistical and epidemiological analysis, 1.9 billion people worldwide suffer from NAFLD, accounting for about 25% of the world’s population (Younossi et al., 2018; Younossi et al., 2019). The etiology of NAFLD is complex and accompanied by multiple complications. Therefore, the development of suitable therapies is slow, and the number of candidate drugs that have successfully entered the late clinical stage is small and challenging to develop. A healthy lifestyle remains the key to prevention and treatment (Romero-Gomez et al., 2017; Sheka et al., 2020). There is an urgency to explore practical strategies for treating NAFLD.
NAFLD is a metabolic stress-induced liver injury disease, mainly manifested by steatosis and lipid accumulation in hepatic parenchymal cells. According to the course of the disease, it is divided into simple fatty liver, steatohepatitis, liver fibrosis, and hepatocellular carcinoma (Marengo et al., 2016; Nassir, 2022). The link between the main disease features of NAFLD and the actual events leading to hepatic cell death—insulin resistance, lipid metabolism imbalance, inflammation, and oxidative stress—is increasingly recognized (Zhao et al., 2021). Programmed cell death is a gene-directed form of cell self-extinction, which is a necessary condition for the normal development of biological tissues, including apoptosis, autophagy, pyroptosis, necroptosis, ferroptosis, and cuproptosis (Lockshin, 2016; Bedoui et al., 2020; Kopeina and Zhivotovsky, 2022). Multiple studies have found that lipophagy, a form of selective autophagy, is significantly inhibited during the development of NAFLD (Chen and Lin, 2022). Alleviating lipid metabolism disorders by restoring lipophagy dysregulation is an effective strategy to prevent and treat NAFLD. Furthermore, the mammalian target of rapamycin (mTOR), an atypical serine/threonine protein kinase and a member of the phosphatidylinositol kinase-related kinase (PIKK) family, plays an important role in NAFLD as Table 1 shows. mTOR exists in two complexes, the target of rapamycin complex 1 (mTORC1) and rapamycin complex 2 (mTORC2). Among them, mTORC1 is a valuable negative core lipophagy regulator (Kim and Guan, 2015; Liu et al., 2022).
TABLE 1.
The effect of and the research development of mTOR in NAFLD.
| mTOR-associated signaling pathway | Compound or Intervention | Main finding in NAFLD | Reference |
|---|---|---|---|
| mTOR/S6K1 | lncRNA-NEAT1 | Reduce lipid accumulation in the liver | Wang (2018) |
| mTORC1/S6K/SREBP1 | PPDPF-LKO | Inhibit mTOR signaling activation, reduce hepatic steatosis | Ma et al. (2021) |
| DEPTOR/mTOR | Neddylation inhibition | Promote fatty acid oxidation, improve lipid denaturation | Serrano-Macia et al. (2021) |
| p-AKT/mTOR/LC-3II | Probiotics and Metformin | Targeting intestinal ecological dysregulation, inflammatory and the autophagy pathway to prevent NAFLD progression and liver injury | Seif El-Din et al. (2021) |
| Nrf2/AMPK/mTOR | Pterostilbene | Alleviate oxidative stress caused by excessive lipid accumulation, promote the metabolism and decomposition of fatty acids | Shen et al. (2023) |
| Geniposide | Reduce lipid accumulation, enhance resistance to oxidative stress and anti-inflammatory capacity | Shen et al. (2020) | |
| AMPK/mTOR | Empagliflozin | Improve liver damage and disorders of lipid metabolism | Meng et al. (2021) |
| Kangtaizhi Granule | Improve lipid accumulation and hepatic steatosis | Zhang et al. (2020) | |
| Dapagliflozin | Activate autophagy, improve liver steatosis | Li et al. (2021); Sun et al. (2015); Porcu et al. (2018) | |
| Hydrogen sulfide | |||
| Oleuropein | |||
| Schisandrin B | Reduce hepatic steatosis and promote fatty acid oxidation | Yan et al. (2022); Lee et al. (2020) | |
| Citrus Peel | |||
| AKT/mTOR | Pueraria flavonoids | Induce autophagy, reduce lipid accumulation and inflammation levels | Sun et al. (2023) |
| Follistatin | Relieve hepatic steatosis, reduce DNL | Tong et al. (2022) | |
| Caveolin-1 | Reduce lipid accumulation, promote autophagy | Xue et al. (2020) | |
| AKT/mTOR/SREBP-1 | Ursodeoxycholic acid | Inhibit hepatic steatosis | Hu et al. (2019) |
| mTORC1/SREBP-1c | Oleuropein | Improve lipid accumulation and insulin resistance | Porcu et al. (2018) |
| AMPK/mTOR/SREBP-1 | Betulinic acid | Improve lipid accumulation in liver cells | Quan et al. (2013) |
| LKB12/AMPK/mTOR | γ-Linolenic Acid | Regulate lipid metabolism and autophagy | Liang et al. (2021) |
| mTORC1 | Korean red ginseng | Inhibit steatosis, liver damage and inflammation | Choi et al. (2019) |
| Acetyl-CoA | Activate autophagy | He et al. (2020) |
This article briefly discusses the role of mTORC1 in the formation of lipophagy, summarizes its intervention effect on the regulation of lipophagy in NAFLD, and preliminarily clarifies the relationship between mTORC1 and lipophagy and NAFLD.
Overview of lipophagy
The term “autophagy” was first introduced by Christian de Duve at the International Conference on Lysosomes in 1963 (Klionsky, 2008; Wen and Klionsky, 2020). More than 20 years later, Yoshinori Ohsumi gradually explored the physiological significance and molecular mechanism of autophagy, laying a solid foundation for the field of autophagy (Tsukada and Ohsumi, 1993; Ohsumi, 2014).
Autophagy is a phenomenon of cellular degradation within lysosomes of defective or aged organelles and proteins, invading pathogens and some cytoplasmic components after receiving induction signals from autophagy-related genes, and is also considered type II programmed cell death (Levine and Klionsky, 2004; Li et al., 2020b). There are three known pathways of cellular autophagy: macroautophagy, microautophagy, and molecular chaperone-mediated autophagy. In general, substrates are classified according to the size and type of the degraded substrate, the scale of their degradation, and the various mechanisms by which the substrate is transported to the lysosome (Parzych and Klionsky, 2014; Bednarczyk et al., 2018; Mizushima and Levine, 2020). Macroautophagy is present in all eukaryotes and is the dominant form of autophagy. Unless otherwise stated, the term “autophagy” generally refers to macroautophagy (Yang and Klionsky, 2009).
Lipophagy is the process in which cells selectively recognize and degrade cholesteryl esters and triglycerides in lipid droplets (LDs) by activating autophagy-associated molecules to form free cholesterol, fatty acids, and glycerol to prevent excessive accumulation of lipids (Schulze et al., 2017). The co-localization of autophagy markers and LDs markers in hepatocytes and the involvement of autophagy in the clearance of lipid droplets and triglycerides identified for the first record the presence of lipophagy (Singh et al., 2009). The series of processes involved in lipophagy is that under the stimulation of internal and external factors, a membrane vesicle with a double-layer membrane structure appears in the cytoplasm, which gradually bends into a cup-like vesicle system through continuous extension and expansion: autophagic precursors. The autophagic precursor can enclose the nearby LDs and form an autophagosome after complete closure. The outer membrane of the autophagosome fuses with the lysosome and enters the lysosome with a monolayer structure. Inside the lysosome, the monolayer structure and contents of the autophagosome can be rapidly degraded by the action of related enzymes and eventually degraded to free fatty acids and glycerol. (Kim et al., 2001; Shin, 2020; Li and Peng, 2022).
Molecular mechanisms of mTORC1 involved in the formation of lipophagy
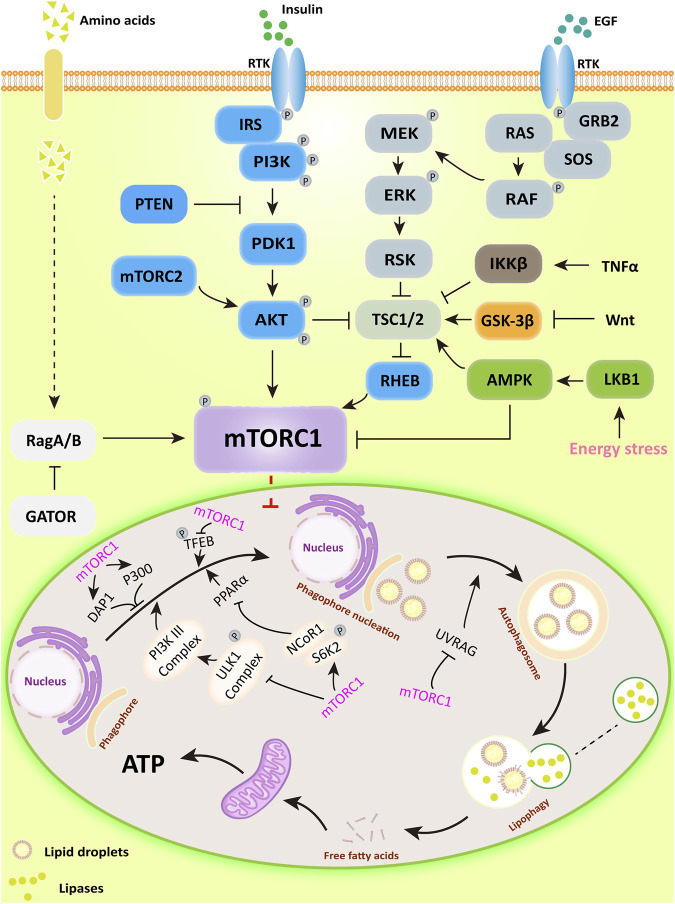
mTORC1 is a negative regulator of lipophagy, the main upstream regulatory pathways (PI3K/AKT, RAS/RAF, AMPK, Wnt, and TNF) and its function in lipophagy are shown in Figure 1.
FIGURE 1.
The major upstream regulatory pathways of mTORC1 and its main mechanisms for regulating lipophagy. PI3K/AKT activates downstream mTORC1 by directly phosphorylating or inhibiting TSC1/2, a potent mTOR inhibitor; AMPK can directly inhibit mTORC1 activity or inactivate mTORC1 in a TSC1/2-dependent manner. Subsequently, mTORC1 regulates lipophagic processes by regulating the ULK1 complex, DAP1, and P300-mediated acetylation.
mTORC1 inhibits lipophagy by affecting autophagosome formation. The ATG kinase complex induces autophagosome formation in yeast macrophages (Kamada et al., 2000). In mammalian cells, the complex consists of the Unc-51 serine/threonine kinase family ULK1/ULK2, ATG13, and 200 kDa adhesion point kinase family interaction proteins (FIP200) (Hara et al., 2008; Ganley et al., 2009; Kim et al., 2011). ULK1/2 is a downstream target of mTORC1, their binding interaction is influenced by nutritional status (Hosokawa et al., 2009b; Nagy et al., 2014). Under nutrient-rich conditions, mTORC1 binds to the complex and dissociates from nutrient-deficient states. When mTORC1 associates with the complex, it can inactivate ULK1/2 and ATG13 by inducing their phosphorylation. When cells are treated with the mTOR inhibitor rapamycin or are undernourished, mTORC1 dissociates from the ULK complex, stimulating the process of lipophagy (Hosokawa et al., 2009a; Jung et al., 2009).
In addition, the Class Ⅲ PI3K complex, a downstream regulator of the ULK1 complex, is also significant for the formation of autophagosomes (Longatti and Tooze, 2009). This complex consists of PIK3C3/VPS34, PIK3R4/p150, and Beclin-1 in mammalian cells (Kihara et al., 2001; Furuya et al., 2005). Beclin-1 binds to Class III PI3K to form the Beclin-1-PIK3C3/VPS34 complex, the effect of which depends mainly on the status and activity of Beclin-1 (Kang et al., 2011). For example, the anti-apoptotic protein Bcl-2 binds Beclin −1 and prevents its interaction with PIK3C3, and the Beclin −1 binding protein Rubicon inhibits PIK3C3 activity in Class III PI3K complex (Pattingre et al., 2005; Matsunaga et al., 2009). In addition, SH3GLB1/Bif-1, a positive regulator of the Class III PI3K complex, promotes autophagy by facilitating the interaction of UVRAG with Beclin-1 (Takahashi et al., 2007). AMBRA1, another positive regulator of the Class III PI3K complex, also promotes autophagosome nucleation directly by facilitating the exchange of Beclin-1 with the kinase VPS34 (Di Bartolomeo et al., 2010). Moreover, mTORC1 can directly phosphorylate AMBRA1 to inhibit lipophagy and also directly phosphorylate ATG14 in the Class III PI3K complex through several sites (Ser3, Ser223, Thr233, Ser383, and Ser440) to inhibit lipophagy (Nazio et al., 2013; Shimobayashi and Hall, 2014). PI3P generated by the Class III PI3K complex is required for autophagy in yeast and mammals (Burman and Ktistakis, 2010). For example, the ATG18 direct homologs WIPI1 and WIPI2 in mammalian cells can participate in autophagy by binding to PI3P under starvation or amino acid deficiency conditions (Polson et al., 2010). Moreover, WIPI2 has been shown to function by binding directly to ATG16-L1 in the ATG12-ATG5-ATG16-L1 complex associated with the ubiquitin-like protein binding system to form autophagosomes. mTORC1 has been shown to phosphorylate the Ser395 site of WIPI2 and induce specific interaction of WIPI2 with the E3 ubiquitin ligase HUWE1 to control autophagosome formation (Dooley et al., 2015; Wan et al., 2018).
mTORC1 also has significant effects on autophagosome and lipophagy gene related transcription factors. It has been found that under nutrient-rich conditions, mTORC1 phosphorylates ribosomal protein S6 kinase (S6K2), forming a complex with nuclear receptor co-repressor 1 (NCoR1). In addition, mTORC1 binds to and phosphorylates transcription factor EB (TFEB), the master transcription factor responsible for ATG and lysosomal genes, remaining in the cytoplasm. In contrast, in the absence of nutrients or inactivation of mTORC1, nuclear translocation of NCoR1 is inhibited. TFEB is subsequently dephosphorylated and enters the nucleus to participate in the transcription of lysosomal genes. ULK1 kinase is activated, which in turn induces lipophagy. Upon activation of lipophagy, on the one hand, peroxisome proliferator-activated receptor α (PPARα) is activated by degrading NCoR1. On the other hand, activated PPARα upregulates lipophagy through AMPK/mTOR signaling, decreasing intrahepatic lipid content and stimulating ß-oxidation in the liver and hepatocytes (Martina et al., 2012; Saito et al., 2019; Tong et al., 2019). Furthermore, studies have shown that PPARα and farnesol-like X receptor (FXR) compete for the same DNA binding site in the ATG gene promoter with opposite transcriptional outputs and that pharmacological activation of PPARα reverses the normal inhibition of lipophagy in the fed state and induces lipid degradation (Lee et al., 2014a).
mTORC1 also has downstream negative regulators of lipophagy. Koren first indicated that death-associated protein 1 (DAP1) is involved in the negative regulation of autophagy. Further studies showed that mTOR is a specific kinase of DAP1 (Koren et al., 2010; Yahiro et al., 2014). In addition, mTORC1 has also been shown to inhibit autophagosome formation through direct phosphorylation and activation of acetyltransferase p300. Acetyltransferase p300 has been identified as a direct phosphorylation substrate for mTORC1. mTOR phosphorylates and activates four serine residues (Ser2271, Ser2279, Ser2291 and Ser2315) in the C-terminal structural domain of p300 (Wan et al., 2017).
mTOR has been found to have a significant role in the terminal phase of lipophagy. mTORC1 phosphorylates the Ser498 site of UVRAG to prevent the maturation of autophagosomes (Kim et al., 2015). And during the sustained starvation response, autophagosomes formed by the fusion of autophagosomes with lysosomes can reactivate mTOR, providing negative feedback to inhibit further autophagosome formation and prevent the breakdown of more components than necessary for survival (Munson and Ganley, 2015).
Targeting mTORC1-dependent lipophagy for the treatment of NAFLD
Lipophagy and hepatic steatosis
Triglycerides stored in LDs, which accumulate in large amounts to initiate hepatocyte steatosis, are essential triggers of NAFLD (Canbay et al., 2007; Scorletti and Carr, 2022). Targeting mTORC1-regulated lipophagy to promote LDs degradation may be an effective therapeutic strategy to improve NAFLD (Filali-Mouncef et al., 2022).
In NAFLD, enhanced lipophagy can directly affect de novo lipogenesis (DNL). For example, perinatal exposure to bisphenol A (BPA) causes NAFLD in both female and male offspring, which is associated with the inhibition of lipophagy via the AKT/mTOR signaling pathway, leading to the upregulation of lipogenic genes (Lin et al., 2019). In addition, extensive studies have found that inhibition of lipophagy exacerbates hepatic lipid accumulation, which is highly correlated with sustained activation of mTORC1 in the liver of high-fat-fed mice (Korsheninnikova et al., 2006). Pharmacological intervention with the autophagy inducers rapamycin or carbamazepine significantly reduced liver, blood triglyceride, glucose, and plasma insulin levels in high-fat diet-fed mice, indicating an improvement in NAFLD by restoring lipophilic flux through inhibition of the mTOR-dependent pathway (Lin et al., 2013). Acyl coenzyme A oxidase 1 (Acox1), an enzyme that catalyzes the first step in peroxisome ß-oxidation, accumulates in the liver and further increases during fasting or a high-fat diet (HFD). Liver-specific Acox1 knockout (Acox1-LKO) protects mice from hepatic steatosis induced by starvation or HFD, which is associated with reduced lysosomal localization of mTOR, leading to impaired activation of mTORC1 and induction of lipophagy (He et al., 2020). In HFD-fed mouse models of early hepatic steatosis and L02 cells with A/O-induced lipid hyperaccumulation, Caveolin1 (CAV1) promotes lipophagy by inhibiting the AKT/mTOR pathway, which attenuates lipid accumulation (Xue et al., 2020). Casein kinase 2-interacting protein 1 (CKIP-1), a positive regulator of autophagy in hepatocytes, activates lipophagy by inhibiting AKT/mTOR signaling, thereby reducing lipid accumulation (Li et al., 2020a).
AMP-dependent protein kinase (AMPK) can inhibit mTOR directly or indirectly, but its activity is reduced in NAFLD (Jacquel et al., 2018; Zhao et al., 2020). Restoring lipophagy by increasing AMPK activity is also an important target for improving NAFLD. For example, the glucagon-like peptide 1 (GLP-1) analog liraglutide (LRG) restores lipophagy by increasing AMPK activity, ultimately improving hepatocyte steatosis (He et al., 2016). Chemotherapeutic isothiocyanate radixanthin (SFN) contributes to lipophagy in adipocytes of mice fed HFD and induces lipophagy via the AMPK-mTOR-ULK1 signaling pathway in differentiated 3T3-L1 cells (Masuda et al., 2022).
The imbalance between lipogenesis and lipolysis may lead to steatosis, and lipophagy is essential in maintaining lipid homeostasis; focusing on the extended side may be the key to NAFLD treatment. Regulation of lipophagy by targeting the mTOR signaling pathway is critical in adipogenesis and lipid metabolism, and is an essential target for the treatment of NAFLD.
Lipophagy and insulin resistance
The development of NAFLD is closely related to insulin resistance. On the one hand, hepatic fat accumulation leads to insulin resistance, and on the other hand, insulin resistance increases serum-free fatty acid levels by promoting lipolysis (Zhang et al., 2018; Watt et al., 2019). In addition, fat accumulation decreases lipophagic flux, leading to defective insulin action (Yang et al., 2010).
It was tentatively suggested that enhancing lipophagy by inhibiting mTOR has an ameliorative effect on insulin resistance: the autophagy inducer rapamycin ameliorated reduced glucose tolerance, decreased insulin sensitivity, dysglycemia, and dyslipidemia, increased hepatic steatosis, enhanced hepatic inflammation, elevated p-mTOR expression and decreased lipophagic flux caused by HFD and STZ-induced T2DM rats, and The autophagy inhibitor 3-MA significantly reversed the effects of rapamycin, which suggested that rapamycin improved insulin resistance and hepatic steatosis in T2DM rats by activating lipophagy, but the upstream molecular mechanism of mTOR was not elucidated (Zhou and Ye, 2018). In addition, bee pollen polysaccharides alleviated hepatic steatosis and insulin resistance in HFD-fed mice by promoting lipophagy through an AMPK/mTOR-dependent signaling pathway. Sitagliptin significantly improved hepatic steatosis and insulin resistance in ob/ob mice by activating lipophagy through the AMPK/mTOR signaling pathway (Li et al., 2017; Zheng et al., 2018). More studies in the future are still needed to elucidate the role of lipophagy in hepatic selective insulin resistance, thus providing new directions and perspectives for the prevention and treatment of NAFLD.
Lipophagy and liver fibrosis
Hepatic fibrosis is a pathological feature of various chronic liver diseases, including NAFLD, characterized by persistent extracellular matrix (ECM) accumulation (Parola and Pinzani, 2019). Substantial evidence indicates that hepatic stellate cells (HSCs) are an essential source of ECM and that their activation is a critical event in the development of liver fibrosis (Elpek, 2014). Therefore, inhibiting the activation of HSCs is considered a vital strategy to reverse liver fibrosis.
Lipophagy has an essential role in liver fibrosis. On the one hand, it has been proposed that lipophagy promotes the activation of hematopoietic stem cells. The main feature of hematopoietic stem cell activation is the release of LDs containing retinol and triglycerides. Meanwhile, lipophagy degrades lipid droplets in hematopoietic stem cells, which hydrolyzes retinol into free fatty acids, further oxidized by mitochondria to generate ATP, providing energy for cell activation (Hernandez-Gea and Friedman, 2012). The insulin-like growth factor binding protein-related protein (IGFBPrP1) has also been shown to upregulate lipophagy by inhibiting the PI3K/AKT/mTOR signaling pathway, thus promoting the activation of hematopoietic stem cells (Huang et al., 2019).
On the other hand, lipophagy has also been suggested to inhibit liver fibrosis. Rutin and curcumin stimulate fatty acid-induced lipophagy in NHSCs (non-chemically induced HSCs) by regulating the PI3K/AKT/mTOR pathway, which ultimately inhibits the activation of NHSCs (Lee et al., 2014b).
The above results suggest the importance of mTOR-regulated lipophagy in liver fibrosis. However, it is unclear why the different effects might be due to varying degrees of lipophagy activation in HSC and thus other effects: moderate activation of lipophagy in HSC cells could provide energy for HSC activation. Conversely, excessive activation of HSC lipophagy might lead to the aging and death of HSC autophagy cells, thus inhibiting HSC activation. Therefore, more studies are still needed to explore the relationship between them.
Conclusion and perspectives
mTORC1, as a central negative regulator of adipose autophagy, exerts an essential regulatory role on the initiation, formation, and end stages of lipophagy and is a highly effective therapeutic target. Moreover, restoration of lipophagy dysregulation based on mTORC1 plays different effects in different stages of NAFLD development. In the early stage of NAFLD progression, the leading role of lipophagy is to degrade LDs in hepatocytes to alleviate steatosis and thus improve NAFLD. Although lipophagy is thought to provide binding energy for the activation of hepatic fibroblasts in the later stages of NAFLD development, enhanced autophagy has also been shown to inhibit the activation of HSCs and thereby ameliorate liver fibrosis. Therefore, accurately assessing the progress stage of NAFLD and targeting the intervention of lipophagic signaling molecules in hepatocytes are more conducive to the treatment of NAFLD. Lipophagy makes essential contributions in regulating hepatic mass metabolism and maintaining energy homeostasis and also provides valuable novel ideas for the therapeutic approach of NAFLD. Multiple molecular mechanisms regulate lipophagy, and in this paper, we discuss mTORC1-centered lipophagy to provide a candidate strategy for the clinical treatment of NAFLD.
Funding Statement
This study was supported by the National Natural Science Foundation of China (Grant No. 82074077). This study was also supported by the Hubei Provincial Natural Science Foundation of China (Grant No. 2022CFB381).
Author contributions
XT, XH, and ZL: writing—original draft, software, and visualization; LC and ZQ: writing—review and editing; XT and ZQ: conceptualization and project administration; JH and ZQ: funding acquisition and conceptualization. All authors have read and agreed to the published version of the manuscript. XT, XH, and ZL have contributed equally to this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Bednarczyk M., Zmarzly N., Grabarek B., Mazurek U., Muc-Wierzgon M. (2018). Genes involved in the regulation of different types of autophagy and their participation in cancer pathogenesis. Oncotarget 9 (76), 34413–34428. 10.18632/oncotarget.26126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S., Herold M. J., Strasser A. (2020). Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 21 (11), 678–695. 10.1038/s41580-020-0270-8 [DOI] [PubMed] [Google Scholar]
- Burman C., Ktistakis N. T. (2010). Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 584 (7), 1302–1312. 10.1016/j.febslet.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Canbay A., Bechmann L., Gerken G. (2007). Lipid metabolism in the liver. Z Gastroenterol. 45 (1), 35–41. 10.1055/s-2006-927368 [DOI] [PubMed] [Google Scholar]
- Chen C. L., Lin Y. C. (2022). Autophagy dysregulation in metabolic associated fatty liver disease: A new therapeutic target. Int. J. Mol. Sci. 23 (17), 10055. 10.3390/ijms231710055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. Y., Park J. S., Shon C. H., Lee C. Y., Ryu J. M., Son D. J., et al. (2019). Fermented Korean red Ginseng Extract enriched in Rd and Rg3 protects against non-alcoholic fatty liver disease through regulation of mTORC1. Nutrients 11 (12), 2963. 10.3390/nu11122963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo S., Corazzari M., Nazio F., Oliverio S., Lisi G., Antonioli M., et al. (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191 (1), 155–168. 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley H. C., Wilson M. I., Tooze S. A. (2015). WIPI2B links PtdIns3P to LC3 lipidation through binding ATG16L1. Autophagy 11 (1), 190–191. 10.1080/15548627.2014.996029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elpek G. O. (2014). Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J. Gastroenterol. 20 (23), 7260–7276. 10.3748/wjg.v20.i23.7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filali-Mouncef Y., Hunter C., Roccio F., Zagkou S., Dupont N., Primard C., et al. (2022). The menage a trois of autophagy, lipid droplets and liver disease. Autophagy 18 (1), 50–72. 10.1080/15548627.2021.1895658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya N., Yu J., Byfield M., Pattingre S., Levine B. (2005). The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 1 (1), 46–52. 10.4161/auto.1.1.1542 [DOI] [PubMed] [Google Scholar]
- Ganley I. G., Lam du H., Wang J., Ding X., Chen S., Jiang X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284 (18), 12297–12305. 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., et al. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181 (3), 497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A., Chen X., Tan M., Chen Y., Lu D., Zhang X., et al. (2020). Acetyl-CoA derived from hepatic peroxisomal beta-oxidation inhibits autophagy and promotes steatosis via mTORC1 activation. Mol. Cell 79 (1), 30–42. 10.1016/j.molcel.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Sha S., Sun L., Zhang J., Dong M. (2016). GLP-1 analogue improves hepatic lipid accumulation by inducing autophagy via AMPK/mTOR pathway. Biochem. Biophys. Res. Commun. 476 (4), 196–203. 10.1016/j.bbrc.2016.05.086 [DOI] [PubMed] [Google Scholar]
- Hernandez-Gea V., Friedman S. L. (2012). Autophagy fuels tissue fibrogenesis. Autophagy 8 (5), 849–850. 10.4161/auto.19947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., et al. (2009a). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20 (7), 1981–1991. 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. (2009b). Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5 (7), 973–979. 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- Hu J., Hong W., Yao K. N., Zhu X. H., Chen Z. Y., Ye L. (2019). Ursodeoxycholic acid ameliorates hepatic lipid metabolism in LO2 cells by regulating the AKT/mTOR/SREBP-1 signaling pathway. World J. Gastroenterol. 25 (12), 1492–1501. 10.3748/wjg.v25.i12.1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. J., Ren J. J., Zhang Q. Q., Kong Y. Y., Zhang H. Y., Guo X. H., et al. (2019). IGFBPrP1 accelerates autophagy and activation of hepatic stellate cells via mutual regulation between H19 and PI3K/AKT/mTOR pathway. Biomed. Pharmacother. 116, 109034. 10.1016/j.biopha.2019.109034 [DOI] [PubMed] [Google Scholar]
- Jacquel A., Luciano F., Robert G., Auberger P. (2018). Implication and regulation of AMPK during physiological and pathological myeloid differentiation. Int. J. Mol. Sci. 19 (10), 2991. 10.3390/ijms19102991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., et al. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20 (7), 1992–2003. 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150 (6), 1507–1513. 10.1083/jcb.150.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Zeh H. J., Lotze M. T., Tang D. (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18 (4), 571–580. 10.1038/cdd.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae . J. Cell Biol. 152 (3), 519–530. 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W. P., Klionsky D. J. (2001). Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152 (1), 51–64. 10.1083/jcb.152.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., Guan K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13 (2), 132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Guan K. L. (2015). mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 125 (1), 25–32. 10.1172/JCI73939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. M., Jung C. H., Seo M., Kim E. K., Park J. M., Bae S. S., et al. (2015). mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell 57 (2), 207–218. 10.1016/j.molcel.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. (2008). Autophagy revisited: A conversation with christian de Duve. Autophagy 4 (6), 740–743. 10.4161/auto.6398 [DOI] [PubMed] [Google Scholar]
- Kopeina G. S., Zhivotovsky B. (2022). Programmed cell death: Past, present and future. Biochem. Biophys. Res. Commun. 633, 55–58. 10.1016/j.bbrc.2022.09.022 [DOI] [PubMed] [Google Scholar]
- Koren I., Reem E., Kimchi A. (2010). DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20 (12), 1093–1098. 10.1016/j.cub.2010.04.041 [DOI] [PubMed] [Google Scholar]
- Korsheninnikova E., van der Zon G. C., Voshol P. J., Janssen G. M., Havekes L. M., Grefhorst A., et al. (2006). Sustained activation of the mammalian target of rapamycin nutrient sensing pathway is associated with hepatic insulin resistance, but not with steatosis, in mice. Diabetologia 49 (12), 3049–3057. 10.1007/s00125-006-0439-5 [DOI] [PubMed] [Google Scholar]
- Lee G. H., Peng C., Park S. A., Hoang T. H., Lee H. Y., Kim J., et al. (2020). Citrus peel Extract ameliorates high-fat diet-induced NAFLD via activation of AMPK signaling. Nutrients 12 (3), 673. 10.3390/nu12030673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Thiyagarajan V., Sie H. W., Cheng M. F., Tsai M. J., Chia Y. C., et al. (2014b). Synergistic effect of natural compounds on the fatty acid-induced autophagy of activated hepatic stellate cells. J. Nutr. Biochem. 25 (9), 903–913. 10.1016/j.jnutbio.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Lee J. M., Wagner M., Xiao R., Kim K. H., Feng D., Lazar M. A., et al. (2014a). Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516 (7529), 112–115. 10.1038/nature13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. (2004). Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 6 (4), 463–477. 10.1016/s1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Li H. Y., Peng Z. G. (2022). Targeting lipophagy as a potential therapeutic strategy for nonalcoholic fatty liver disease. Biochem. Pharmacol. 197, 114933. 10.1016/j.bcp.2022.114933 [DOI] [PubMed] [Google Scholar]
- Li L., He S., Ma B. (2020b). Autophagy and autophagy-related proteins in cancer. Mol. Cancer 19 (1), 12. 10.1186/s12943-020-1138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li Q., Huang W., Han Y., Tan H., An M., et al. (2021). Dapagliflozin alleviates hepatic steatosis by restoring autophagy via the AMPK-mTOR pathway. Front. Pharmacol. 12, 589273. 10.3389/fphar.2021.589273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xie P., Lin W., Liu J., Chen J., Guo Z., et al. (2020a). CKIP-1 augments autophagy in steatotic hepatocytes by inhibiting Akt/mTOR signal pathway. Exp. Cell Res. 397 (1), 112341. 10.1016/j.yexcr.2020.112341 [DOI] [PubMed] [Google Scholar]
- Li X., Gong H., Yang S., Yang L., Fan Y., Zhou Y. (2017). Pectic bee pollen polysaccharide from rosa rugosa alleviates diet-induced hepatic steatosis and insulin resistance via induction of AMPK/mTOR-Mediated autophagy. Molecules 22 (5), 699. 10.3390/molecules22050699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Zhang Z., Tu J., Wang Z., Gao X., Deng K., et al. (2021). γ-Linolenic acid prevents lipid metabolism disorder in palmitic acid-treated Alpha mouse liver-12 cells by balancing autophagy and apoptosis via the LKB1-AMPK-mTOR pathway. J. Agric. Food Chem. 69 (29), 8257–8267. 10.1021/acs.jafc.1c02596 [DOI] [PubMed] [Google Scholar]
- Lin C. W., Zhang H., Li M., Xiong X., Chen X., Chen X., et al. (2013). Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J. Hepatol. 58 (5), 993–999. 10.1016/j.jhep.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Wu D., Wu F. J., Meng Y., Zhang J. H., Wang X. G., et al. (2019). Non-alcoholic fatty liver disease induced by perinatal exposure to bisphenol a is associated with activated mTOR and TLR4/NF-κB signaling pathways in offspring rats. Front. Endocrinol. (Lausanne) 10, 620. 10.3389/fendo.2019.00620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Qiu D., Liang X., Huang Y., Wang Y., Jia X., et al. (2022). Lipotoxicity-induced STING1 activation stimulates MTORC1 and restricts hepatic lipophagy. Autophagy 18 (4), 860–876. 10.1080/15548627.2021.1961072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin R. A. (2016). Programmed cell death 50 (and beyond). Cell Death Differ. 23 (1), 10–17. 10.1038/cdd.2015.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A., Tooze S. A. (2009). Vesicular trafficking and autophagosome formation. Cell Death Differ. 16 (7), 956–965. 10.1038/cdd.2009.39 [DOI] [PubMed] [Google Scholar]
- Ma N., Wang Y. K., Xu S., Ni Q. Z., Zheng Q. W., Zhu B., et al. (2021). PPDPF alleviates hepatic steatosis through inhibition of mTOR signaling. Nat. Commun. 12 (1), 3059. 10.1038/s41467-021-23285-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marengo A., Rosso C., Bugianesi E. (2016). Liver cancer: Connections with obesity, fatty liver, and cirrhosis. Annu. Rev. Med. 67, 103–117. 10.1146/annurev-med-090514-013832 [DOI] [PubMed] [Google Scholar]
- Martina J. A., Chen Y., Gucek M., Puertollano R. (2012). MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8 (6), 903–914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M., Yoshida-Shimizu R., Mori Y., Ohnishi K., Adachi Y., Sakai M., et al. (2022). Sulforaphane induces lipophagy through the activation of AMPK-mTOR-ULK1 pathway signaling in adipocytes. J. Nutr. Biochem. 106, 109017. 10.1016/j.jnutbio.2022.109017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11 (4), 385–396. 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Meng Z., Liu X., Li T., Fang T., Cheng Y., Han L., et al. (2021). The SGLT2 inhibitor empagliflozin negatively regulates IL-17/IL-23 axis-mediated inflammatory responses in T2DM with NAFLD via the AMPK/mTOR/autophagy pathway. Int. Immunopharmacol. 94, 107492. 10.1016/j.intimp.2021.107492 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B. (2020). Autophagy in human diseases. N. Engl. J. Med. 383 (16), 1564–1576. 10.1056/NEJMra2022774 [DOI] [PubMed] [Google Scholar]
- Munson M. J., Ganley I. G. (2015). MTOR, PIK3C3, and autophagy: Signaling the beginning from the end. Autophagy 11 (12), 2375–2376. 10.1080/15548627.2015.1106668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P., Karpati M., Varga A., Pircs K., Venkei Z., Takats S., et al. (2014). Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy 10 (3), 453–467. 10.4161/auto.27442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassir F. (2022). Nafld: Mechanisms, treatments, and biomarkers. Biomolecules 12 (6), 824. 10.3390/biom12060824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M., et al. (2013). mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15 (4), 406–416. 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2014). Historical landmarks of autophagy research. Cell Res. 24 (1), 9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola M., Pinzani M. (2019). Liver fibrosis: Pathophysiology, pathogenetic targets and clinical issues. Mol. Asp. Med. 65, 37–55. 10.1016/j.mam.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Parzych K. R., Klionsky D. J. (2014). An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal 20 (3), 460–473. 10.1089/ars.2013.5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., et al. (2005). Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122 (6), 927–939. 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Polson H. E., de Lartigue J., Rigden D. J., Reedijk M., Urbe S., Clague M. J., et al. (2010). Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6 (4), 506–522. 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- Porcu C., Sideri S., Martini M., Cocomazzi A., Galli A., Tarantino G., et al. (2018). Oleuropein induces AMPK-dependent autophagy in NAFLD mice, regardless of the gender. Int. J. Mol. Sci. 19 (12), 3948. 10.3390/ijms19123948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan H. Y., Kim D. Y., Kim S. J., Jo H. K., Kim G. W., Chung S. H. (2013). Betulinic acid alleviates non-alcoholic fatty liver by inhibiting SREBP1 activity via the AMPK-mTOR-SREBP signaling pathway. Biochem. Pharmacol. 85 (9), 1330–1340. 10.1016/j.bcp.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Romero-Gomez M., Zelber-Sagi S., Trenell M. (2017). Treatment of NAFLD with diet, physical activity and exercise. J. Hepatol. 67 (4), 829–846. 10.1016/j.jhep.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Saito T., Kuma A., Sugiura Y., Ichimura Y., Obata M., Kitamura H., et al. (2019). Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat. Commun. 10 (1), 1567. 10.1038/s41467-019-08829-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R. J., Sathyanarayan A., Mashek D. G. (2017). Breaking fat: The regulation and mechanisms of lipophagy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862 (10), 1178–1187. 10.1016/j.bbalip.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorletti E., Carr R. M. (2022). A new perspective on NAFLD: Focusing on lipid droplets. J. Hepatol. 76 (4), 934–945. 10.1016/j.jhep.2021.11.009 [DOI] [PubMed] [Google Scholar]
- Seif El-Din S. H., Salem M. B., El-Lakkany N. M., Hammam O. A., Nasr S. M., Okasha H., et al. (2021). Early intervention with probiotics and metformin alleviates liver injury in NAFLD rats via targeting gut microbiota dysbiosis and p-AKT/mTOR/LC-3II pathways. Hum. Exp. Toxicol. 40 (9), 1496–1509. 10.1177/0960327121999445 [DOI] [PubMed] [Google Scholar]
- Serrano-Macia M., Simon J., Gonzalez-Rellan M. J., Azkargorta M., Goikoetxea-Usandizaga N., Lopitz-Otsoa F., et al. (2021). Neddylation inhibition ameliorates steatosis in NAFLD by boosting hepatic fatty acid oxidation via the DEPTOR-mTOR axis. Mol. Metab. 53, 101275. 10.1016/j.molmet.2021.101275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheka A. C., Adeyi O., Thompson J., Hameed B., Crawford P. A., Ikramuddin S. (2020). Nonalcoholic steatohepatitis: A review. JAMA 323 (12), 1175–1183. 10.1001/jama.2020.2298 [DOI] [PubMed] [Google Scholar]
- Shen B., Feng H., Cheng J., Li Z., Jin M., Zhao L., et al. (2020). Geniposide alleviates non-alcohol fatty liver disease via regulating Nrf2/AMPK/mTOR signalling pathways. J. Cell Mol. Med. 24 (9), 5097–5108. 10.1111/jcmm.15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Wang Y., Cheng J., Peng Y., Zhang Q., Li Z., et al. (2023). Pterostilbene alleviated NAFLD via AMPK/mTOR signaling pathways and autophagy by promoting Nrf2. Phytomedicine 109, 154561. 10.1016/j.phymed.2022.154561 [DOI] [PubMed] [Google Scholar]
- Shimobayashi M., Hall M. N. (2014). Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15 (3), 155–162. 10.1038/nrm3757 [DOI] [PubMed] [Google Scholar]
- Shin D. W. (2020). Lipophagy: Molecular mechanisms and implications in metabolic disorders. Mol. Cells 43 (8), 686–693. 10.14348/molcells.2020.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., et al. (2009). Autophagy regulates lipid metabolism. Nature 458 (7242), 1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zhang J., Hou J., Hui M., Qi H., Lei T., et al. (2023). Induction of autophagy via the PI3K/Akt/mTOR signaling pathway by Pueraria flavonoids improves non-alcoholic fatty liver disease in obese mice. Biomed. Pharmacother. 157, 114005. 10.1016/j.biopha.2022.114005 [DOI] [PubMed] [Google Scholar]
- Sun L., Zhang S., Yu C., Pan Z., Liu Y., Zhao J., et al. (2015). Hydrogen sulfide reduces serum triglyceride by activating liver autophagy via the AMPK-mTOR pathway. Am. J. Physiol. Endocrinol. Metab. 309 (11), E925–E935. 10.1152/ajpendo.00294.2015 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., et al. (2007). Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9 (10), 1142–1151. 10.1038/ncb1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Cong L., Jia Y., He B. L., Guo Y., He J., et al. (2022). Follistatin alleviates hepatic steatosis in NAFLD via the mTOR dependent pathway. Diabetes Metab. Syndr. Obes. 15, 3285–3301. 10.2147/DMSO.S380053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L., Wang L., Yao S., Jin L., Yang J., Zhang Y., et al. (2019). PPARδ attenuates hepatic steatosis through autophagy-mediated fatty acid oxidation. Cell Death Dis. 10 (3), 197. 10.1038/s41419-019-1458-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae . FEBS Lett. 333 (1-2), 169–174. 10.1016/0014-5793(93)80398-e [DOI] [PubMed] [Google Scholar]
- Wan W., You Z., Xu Y., Zhou L., Guan Z., Peng C., et al. (2017). mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol. Cell 68 (2), 323–335. 10.1016/j.molcel.2017.09.020 [DOI] [PubMed] [Google Scholar]
- Wan W., You Z., Zhou L., Xu Y., Peng C., Zhou T., et al. (2018). mTORC1-Regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Mol. Cell 72 (2), 303–315. 10.1016/j.molcel.2018.09.017 [DOI] [PubMed] [Google Scholar]
- Wang X. (2018). Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell Biochem. 119 (2), 1567–1574. 10.1002/jcb.26317 [DOI] [PubMed] [Google Scholar]
- Watt M. J., Miotto P. M., De Nardo W., Montgomery M. K. (2019). The liver as an endocrine organ-linking NAFLD and insulin resistance. Endocr. Rev. 40 (5), 1367–1393. 10.1210/er.2019-00034 [DOI] [PubMed] [Google Scholar]
- Wen X., Klionsky D. J. (2020). At a glance: A history of autophagy and cancer. Semin. Cancer Biol. 66, 3–11. 10.1016/j.semcancer.2019.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Wang J., Jiang W., Shi C., Wang X., Huang Y., et al. (2020). Caveolin-1 alleviates lipid accumulation in NAFLD associated with promoting autophagy by inhibiting the Akt/mTOR pathway. Eur. J. Pharmacol. 871, 172910. 10.1016/j.ejphar.2020.172910 [DOI] [PubMed] [Google Scholar]
- Yahiro K., Tsutsuki H., Ogura K., Nagasawa S., Moss J., Noda M. (2014). DAP1, a negative regulator of autophagy, controls SubAB-mediated apoptosis and autophagy. Infect. Immun. 82 (11), 4899–4908. 10.1128/IAI.02213-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. S., Zhang S. F., Luo G., Cheng B. C., Zhang C., Wang Y. W., et al. (2022). Schisandrin B mitigates hepatic steatosis and promotes fatty acid oxidation by inducing autophagy through AMPK/mTOR signaling pathway. Metabolism 131, 155200. 10.1016/j.metabol.2022.155200 [DOI] [PubMed] [Google Scholar]
- Yang L., Li P., Fu S., Calay E. S., Hotamisligil G. S. (2010). Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11 (6), 467–478. 10.1016/j.cmet.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J. (2009). An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335, 1–32. 10.1007/978-3-642-00302-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z., Anstee Q. M., Marietti M., Hardy T., Henry L., Eslam M., et al. (2018). Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15 (1), 11–20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., et al. (2019). Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 69 (6), 2672–2682. 10.1002/hep.30251 [DOI] [PubMed] [Google Scholar]
- Zhang F., Hu Z., Li G., Huo S., Ma F., Cui A., et al. (2018). Hepatic CREBZF couples insulin to lipogenesis by inhibiting insig activity and contributes to hepatic steatosis in diet-induced insulin-resistant mice. Hepatology 68 (4), 1361–1375. 10.1002/hep.29926 [DOI] [PubMed] [Google Scholar]
- Zhang J., Du H., Shen M., Zhao Z., Ye X. (2020). Kangtaizhi granule alleviated nonalcoholic fatty liver disease in high-fat diet-fed rats and HepG2 cells via AMPK/mTOR signaling pathway. J. Immunol. Res. 2020, 3413186. 10.1155/2020/3413186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hu Y., Peng J. (2021). Targeting programmed cell death in metabolic dysfunction-associated fatty liver disease (MAFLD): A promising new therapy. Cell Mol. Biol. Lett. 26 (1), 17. 10.1186/s11658-021-00254-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Sun X., Chaggan C., Liao Z., Wong K., He F., et al. (2020). An AMPK-caspase-6 axis controls liver damage in nonalcoholic steatohepatitis. Science 367 (6478), 652–660. 10.1126/science.aay0542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zhou J., Song S., Kong W., Xia W., Chen L., et al. (2018). Dipeptidyl-peptidase 4 inhibitor sitagliptin ameliorates hepatic insulin resistance by modulating inflammation and autophagy in ob/ob mice. Int. J. Endocrinol. 2018, 8309723. 10.1155/2018/8309723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ye S. (2018). Rapamycin improves insulin resistance and hepatic steatosis in type 2 diabetes rats through activation of autophagy. Cell Biol. Int. 42 (10), 1282–1291. 10.1002/cbin.11015 [DOI] [PubMed] [Google Scholar]