Abstract
The ATPase ISWI is the catalytic core of several nucleosome remodeling complexes, which are able to alter histone–DNA interactions within nucleosomes such that the sliding of histone octamers on DNA is facilitated. Dynamic nucleosome repositioning may be involved in the assembly of chromatin with regularly spaced nucleosomes and accessible regulatory sequence elements. The mechanism that underlies nucleosome sliding is largely unresolved. We recently discovered that the N-terminal ‘tail’ of histone H4 is critical for nucleosome remodeling by ISWI. If deleted, nucleosomes are no longer recognized as substrates and do not stimulate the ATPase activity of ISWI. We show here that the H4 tail is part of a more complex recognition epitope which is destroyed by grafting the H4 N-terminus onto other histones. We mapped the H4 tail requirement to a hydrophilic patch consisting of the amino acids R17H18R19 localized at the base of the tail. These residues have been shown earlier to contact nucleosomal DNA, suggesting that ISWI recognizes an ‘epitope’ consisting of the DNA-bound H4 tail. Consistent with this hypothesis, the ISWI ATPase is stimulated by isolated H4 tail peptides ISWI only in the presence of DNA. Acetylation of the adjacent K12 and K16 residues impairs substrate recognition by ISWI.
INTRODUCTION
Chromatin is not a static structure in which eukaryotic DNA is condensed for storage, but a rather flexible medium that serves to densely package DNA, while at the same time assuring that regulatory sequence elements remain accessible as needed. Central to the establishment of ‘flexibility’ are multiprotein complexes that modify chromatin structure (1–5). While some enzymes alter the properties of chromatin by introducing covalent, post-translational modifications into the histone N-terminal tails, others act in more subtle ways. Energy-dependent nucleosome remodeling machines use the energy released by ATP hydrolysis to ‘remodel’ chromatin by altering histone–DNA interactions within the nucleosome, such that nucleosomal DNA is rendered more accessible, histone octamers begin to slide on DNA and can even be displaced onto competing DNA in trans, if available. While the importance of nucleosome remodeling machines for gene expression has been amply documented in vivo (5), insight into the potential of nucleosome remodeling for a variety of nuclear processes, including chromatin assembly, transcription, DNA repair and site-specific recombination, has mainly come from mechanistic in vitro analysis (6,7).
The main feature of these chromatin remodeling machines is their modularity. The critical core of remodeling activity is a dedicated ATPase subunit with which other subunits associate to regulate and target the activity. According to its domain composition, Drosophila ISWI defines a subfamily of ATPases within the larger family of SWI2/SNF2-type ATPases (for a recent review of ISWI see ref. 8). ISWI contains SANT-like domains in the C-terminal region, but no bromo- or chromodomains, which are hallmarks of SWI2/SNF2- and CHD-type remodeling ATPases, respectively (5). ISWI is a remodeling factor by itself (9–13) but association with other proteins enhances its activity substantially, both quantitatively and qualitatively (14–16). In extracts derived from Drosophila embryos ISWI resides in at least three nucleosome remodeling complexes: the nucleosome remodeling factor (NURF) (16,17), the ATP-utilizing chromatin assembly and remodeling factor (18,19) and the chromatin accessibility complex (15,20). Analogous complexes have been found in other species from yeast to man (for review see 8).
We recently suggested a model that allows us to explain the phenomenology of ISWI-induced nucleosome remodeling in mechanistic terms (8,21,22). The model assumes that the rate-limiting step in nucleosome mobilization is the displacement of a DNA segment from the histone octamer surface by an adjacent segment of DNA, creating a tight loop, or bulge. Propagation of this ‘bulge’ over the histone surface would lead to the translocation of the DNA relative to the histones. The underlying assumption is that interactions of the remodeling enzyme with both DNA and histones are required in order to dislocate the two moieties relative to each other. Both histone and DNA contacts have been highlighted by earlier experiments (11,21). The low, basal ATPase activity of ISWI in the absence of substrate can be stimulated to some degree by the presence of free DNA, but much more profoundly if the DNA is wound around a histone octamer. Since free histones do not trigger ATPase activity, ISWI apparently recognizes a specific feature of the nucleosome that is not present in either free DNA or free histones (9,11,23).
We recently demonstrated an interaction of ISWI with DNA close to the point of entry into the nucleosome (21,24). A requirement for a histone component has been deduced from experiments using nucleosome substrates reconstituted from variant, recombinant histones (11). Nucleosomes lacking the histone H4 tails are not recognized by ISWI and, accordingly, are no longer substrates for nucleosome remodeling and mobilization. This requirement for the H4 tail occurs at a step subsequent to the binding of ISWI to the nucleosome (11,15).
The N-termini of histones protrude from the nucleosome particle and are available for contact by other segments of DNA, histones and non-histone proteins (for review see 25,26). Through these interactions they contribute to the folding of the nucleosomal fiber into higher order structures and hence the efficiency of gene transcription (25,27,28). The importance of the N-terminal domains is particularly well documented for the ‘tails’ of H3 and H4: their amino acid sequences are identical in yeast and human, with the exception of one conservative substitution (25).
The H3 and H4 ‘tail’ domains are also the target for a large number of post-translational modifications (29,30), which are likely to influence their structure and molecular interactions. For example, acetylation of the N-termini can diminish or enhance the affinity for interacting proteins (31–34). Histone acetylation leads to unfolding of the condensed chromatin fiber and permits transcription (35–38).
Since multiple chromatin regulators are able to interact with the H4 N-terminus, and post-translational modifications of the tail are known to affect these interactions, we wished to further characterize the H4 tail requirement for nucleosome remodeling by ISWI (11). Our analysis highlights the importance of a basic patch, consisting of amino acids R17H18R19 at the emergence of the H4 tail from the compact core, as a major determinant of ISWI recognition. Binding a histone H4 tail peptide to DNA reconstituted this recognition epitope, indicating that the tail peptide is likely to be in contact with DNA under physiological conditions. Acetylation of adjacent lysines modulates the recognition of the nucleosomal substrate by ISWI.
MATERIALS AND METHODS
Histone expression and refolding into octamers
The expression and purification of histones were performed essentially as described by Luger et al. (39). Histones were cloned, expressed individually in pET-3b vectors in BL21 pLysS, and purified from inclusion bodies under denaturing conditions as described (11). Octamers were reconstituted by mixing the relevant histones in the appropriate stoichiometry and refolding by slow removal of the denaturant. Octamers were separated from aggregates and subnucleosomal particles by gel filtration (11).
For the H4 deletion mutants [Δ5-H4, Δ10-H4, Δ15-H4 and Δ19-H4 (previously gH4) lacking the first 5, 10, 15 or 19 residues, respectively], we followed protocols developed at LBME-IBCG-CNRS in Toulouse (V. Morales, personal communication). Briefly, the isoleucine codon AUA was inserted after the methionine and the histones were expressed in BL-21-CodonPlus™-RIL (Stratagene), which contains the gene for the ileY tRNA. This approach was also used to generate the chimeric H4 histones, in which a 21 residue peptide corresponding to the Drosophila histone H4 N-terminal tail (residues T1-K20 and an additional isoleucine residue at position 1) was attached to the globular regions of Drosophila histones H2A and H3, as defined by the sites of trypsin digestion (H2A A13-A123; H3, S28-A135). These were constructed by PCR, using large 5′ primers coding for the H4 tail and recipient histone linker sites, and 3′ primers corresponding to the histone C-terminal region, and cloned into the NdeI–BamHI sites of pET3a. The individual histones were subsequently purified and the octamers refolded as described (11). The globular portions of the other histones (39) are: histone H3 (K27 to A135); histone H4 (K20 to G102); histone H2A (K13 to K118) and histone H2B (K27 to K125).
Chromatin assembly, MNase digestion
Chromatin assembly was performed using recombinant yNAP-1 under the conditions previously described (9,11). Briefly, assembly reactions (50 µl) contained 300 ng of plasmid DNA, 450 ng of specified histones, 35 ng of yNAP-1, 120 ng of recombinant ISWI, 3 mM ATP and an ATP-regenerating system in a buffer consisting of 10 mM HEPES–KOH, pH 7.6, 65 mM KCl, 3 mM MgCl2, 0.5 mM EGTA, 10 mM β-glycerophosphate, 10% glycerol, 1 mM DTT, 0.2 mM PMSF and 100 µg/µl chicken egg albumin. The reaction was incubated at 26°C. Micrococcal nuclease (MNase) digestion was performed, after 4 h of incubation, for 20, 50 and 110 s with 0.2 U of MNase at 26°C. DNA was treated with 50 µg proteinase K at 55°C for 1 h, precipitated prior to separation on a 1.3% Tris–glycine agarose gel and visualized with SYBR®Gold staining (Molecular Probes).
ATPase assay
ATPase assays (9,11) were performed under assembly conditions (see above), but in the absence of an ATP regeneration system and in the presence of 0.7 µCi [γ-32P]ATP at 26°C for 1 h. Unhydrolyzed ATP and free phosphate were separated by thin-layer chromatography using PEI Cellulose F foils (Merck). Spots were quantified by PhosphoImager and Tina software.
Histone tail peptide stimulation of ISWI ATPase activity
Indicated amounts of 20mer unmodified or monoacetylated histone tail peptides (SGRGKGGKGLGKGGAKRHRC; Upstate Biotechnology) or 25mer (wild-type, TGRGKGGKGLGKGGAKRHRKVLRDC; mutant, TGRGKGGKGLGKGGAKAHAKV-RDC; Peptide Specialty Laboratories GmbH, Heidelberg, Germany) were added to 300 ng of plasmid DNA in the assembly buffer (see above). ISWI ATPase activity was measured as described above.
RESULTS
The position of the H4 tail on the nucleosome is critical for ISWI ATPase activity
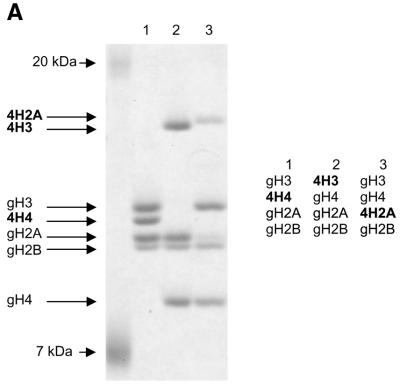
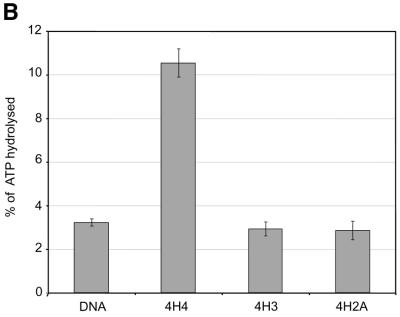
Using recombinant histone technology we recently demonstrated that the histone H4 N-terminal tails are necessary and sufficient to induce ISWI ATPase activity. Nucleosomes containing just an H4 tail stimulate the ATPase of ISWI as strongly as nucleosomes made of wild-type histones (11). To elucidate whether the H4 tail per se is recognized by ISWI, or whether its specific location on the nucleosomal core particle was important, we generated a set of histone octamers that lacked all N-terminal tails, except for the H4 tail which was relocated onto the globular domains of histone H3 and H2A, which emerge from opposite sides of the nucleosome particle (Fig. 1A). This experiment was inspired by earlier work of Grunstein and colleagues who had shown that swapping the H4 and H3 tails in yeast cells did not impair chromatin in general, but rather disrupted the regulation of specific genes as well as the silencing of the yeast mating type cassettes (40). The variant octamers were then used to create nucleosomal arrays by NAP-1 assisted assembly during which we tested for stimulation of ISWI ATPase activity. We found the presence of the H4 tail in its normal context (attached to the globular domain of H4) stimulated the ATPase of ISWI significantly (Fig. 1B, 4H4), whereas particles that contained the tail fused to the globular domains histones H3 or H2A (Fig. 1B, 4H3 and 4H2A) were not recognized by ISWI. As expected, no regular nucleosome arrangement was observable in the absence of ATPase activity (data not shown). These results demonstrate that the location of the H4 tail on the nucleosome is crucial for the activation of ISWI.
Figure 1.
Grafting the histone H4 N-terminus on H3 and H2A prevents substrate recognition by ISWI. (A) Analysis of chimeric histone octamers. Histone octamers were refolded from sets of variant histones as indicated on the right. The globular portions of the histones (39) are indicated with the prefix ‘g’. gH3, histone H3 from K27 to A135; gH4, histone H4 from K20 to G102; gH2A, histone H2A from K13 to K118; and gH2B, histone H2B from K27 to K125. Those histones to which the H4 N-terminus was added (see Materials and Methods) are indicated with the prefix 4. Histones were analyzed by SDS–PAGE in an 18% gel and stained with Coomassie Blue. The migration of the histones and markers are indicated on the left. (B) ATPase assays with variant histone octamers. The histones shown in (A) were reconstituted into nucleosomes by NAP-1 assisted assembly in the presence of ISWI. A control reaction contained only DNA in the absence of histones. The ATPase activity of ISWI during the reaction is displayed as the percentage of ATP hydrolyzed during the assay. The bars represent the average of three independent experiments, and the variability is indicated by the error bars.
The basic amino acids R17H18R19 within the H4 tail are crucial for ISWI remodeling activity
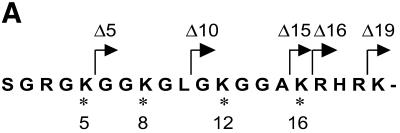
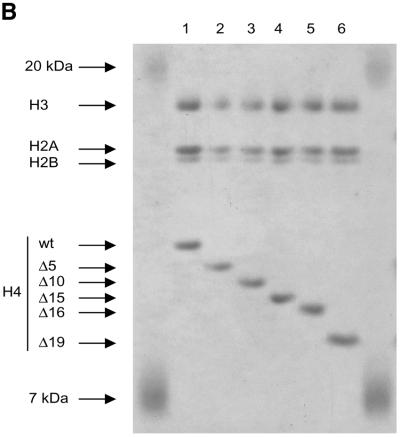
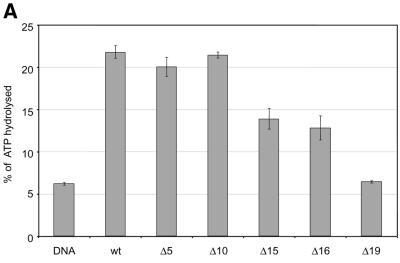
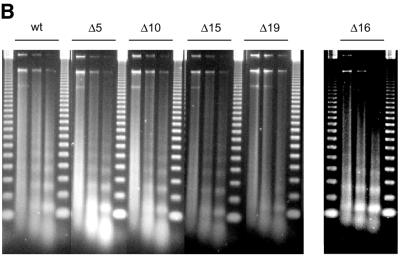
The tail grafting experiments pointed to a more complex ISWI recognition motif, made up from features of the histone tail that required a specific nucleosomal context. In order to delineate this structure more precisely, we generated a set of histone octamers in which the histone H4 tail was sequentially deleted: Δ5-H4, Δ10-H4, Δ15-H4, Δ16-H4, Δ19-H4 lack the first 5, 10, 15, 16 or 19 residues, respectively (Fig. 2). These deletions also progressively removed the lysine residues known to be acetylated in vivo (asterisks in Fig. 2A). Histone octamers were reconstituted from the appropriate histone mixes (Fig. 2B) and used to assemble chromatin in the NAP-1 assisted system (11,18,41), during which we measured the ATPase activity of ISWI. DNA wrapped around a histone octamer containing an intact H4 tail stimulated the ATPase activity ~4-fold compared to the level of DNA alone (Fig. 3A, wt). Histones alone do not stimulate the ATPase at all (9,23). Deleting the first 10 N-terminal amino acids did not influence this level of ATPase hydrolysis. Deletion of the next 5 amino acids led to a partial but significant loss of stimulation (Fig. 3A, and Δ15 and Δ16). Finally, further removal of residues 17–19 resulted in a complete loss of nucleosomal stimulation, such that the ATPase activity did not exceed the level induced by free DNA (Fig. 3A, Δ19). Monitoring the development of nucleosome regularity during the reaction demonstrated that the reduced ATPase levels upon deletion of the H4 N-terminus up to Δ16 still supported the ‘spacing’ reaction (9,11) (Fig. 3B). However, no regularity was obtained if the H4 tail was deleted up to Δ19. Together, these results identify residues R17H18R19 in the H4 tail as a critical motif for ISWI function.
Figure 2.
Characterization of histone octamers harboring systematic deletions of the H4 N-terminus. (A) Amino acid sequence of the histone H4 N-terminal ‘tail’ domain. The endpoints of the deletions are indicated. Asterisks indicate the lysine residues that can be acetylated in vivo. (B) Histone octamers were refolded from histones H3, H2A, H2B and a series of H4 variants from which N-terminal amino acids were sequentially deleted. The histones were analyzed as in Figure 1. The identity of the bands is indicated on the left.
Figure 3.
Identification of crucial determinants for ISWI function in the H4 N-terminus. (A) ATPase activity of ISWI in the presence of the variant nucleosomes shown in Figure 2. ATPase activity is expressed as the percentage of ATP hydrolyzed during the assay. The bars represent the average of three independent experiments, the variability is indicated by the error bars. (B) Nucleosome ‘spacing’ activity of ISWI in the presence of the variant nucleosomes shown in Figure 2. Removal of the amino acids R17H18R19 abolishes the ‘spacing’ capacity of ISWI. The histone octamers were reconstituted into nucleosomes on plasmid DNA by NAP-1 assisted assembly in the presence of ISWI and subjected to MNase digestion. Chromatinized DNA (300 ng) was digested with 0.2 U of MNase for 20, 50 and 110 s. The resulting DNA fragments were purified and visualized by agarose gel electrophoresis and SybrGold“ staining. The marker is a 123 bp DNA ladder.
Interaction of the H4 tail with DNA generates a complex epitope which is recognised by ISWI
The residues that appear critical for ISWI function are part of a highly basic patch, K16R17H18R19K20, at the base of the H4 tail (42). This sequence has earlier been suggested to contact nucleosomal DNA (43–45). Since histone tail peptides alone, tested as fusion proteins to glutathione S-transferase (GST), do not trigger the ATPase activity of ISWI (9,23), we tested whether binding of 20mer peptides corresponding to the first 19 amino acids of the H4 N-terminus (plus a C-terminal cysteine due to the synthesis) to plasmid DNA would reconstitute the critical ISWI recognition motif.
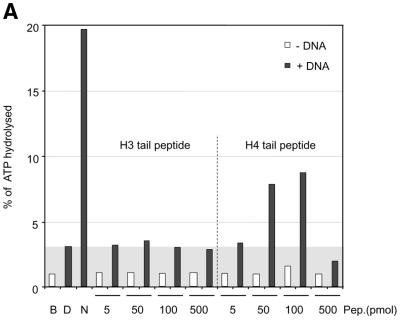
Remarkably, ISWI responded to a mixture of DNA and histone H4 tail peptides by a significant 3-fold increase in ATPase activity (Fig. 4A), although the level of nucleosomal stimulation was not reached. In the absence of DNA, histone tail peptides were not recognized by ISWI. Titration of peptide into the reaction revealed an optimal range of concentrations (50–100 pmol), beyond which the ATPase dropped below the levels of DNA stimulation presumably because excess peptides (500 pmol) led to the formation of inactive aggregates. The response of ISWI to the tail-bound peptide was specific since corresponding H3 tail petides failed to stimulate the ATPase beyond the levels elicited by free DNA (Fig. 4A).
Figure 4.
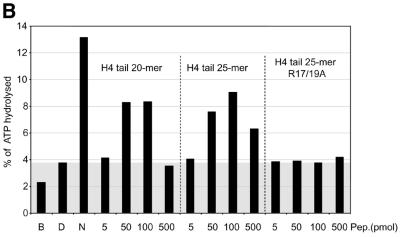
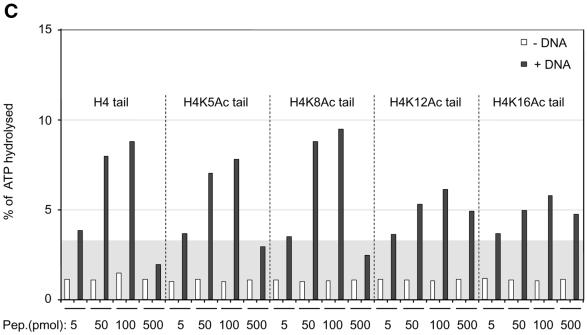
Interaction of H4 tail peptides with DNA reconstitutes a minimal determinant for ISWI response. (A) ISWI ATPase assays in the presence of various substrates. B, buffer; D, DNA; N, nucleosomes; H3 or H4 tail peptides in the indicated amounts (5–500 pmol) in the absence (–) or presence (+) of DNA. The ATPase activity is expressed as the percentage of ATP hydrolyzed during the assay. The ATPase activity level elicited by free DNA is extended through the figure as a gray background, for reference. This result has been obtained consistently in a number of independent experiments under different conditions. (B) R17/R19 within the histone H4 tail are crucial for ISWI ATPase activity. A 25mer H4 tail peptide TGRGKGGKGLGKGGAKRHRKVLRDC induces the ATPase of ISWI as well as the 20mer used in (A). A variant 25mer H4 tail peptide in which R17 and R19 were substituted by alanines does not trigger ATPase acivity. All reactions contained DNA. (C) ISWI ATPase activity is sensitive to site-specific acetylation of histone H4 tail. The data are presented as in (A).
In order to get further support for a role of the R17H18R19 motif in this context, we mutated the two arginines to alanines. These mutations were introduced in the context of a longer peptide, representing the first 25 amino acids of the H4 N-terminus in order to avoid potential end effects. While the longer peptide corresponding to the wild-type sequence stimulated the ISWI ATPase as well as the short peptide, mutation of arginines 17 and 19 to alanines completely abolished the effect of the peptide (Fig. 4B).
ISWI ATPase activity is sensitive to specific acetylation of histone H4 tails
We previously found that acetylation of lysine 16 and 12 in the H4 N-terminus compromised the ability of a tail peptide to compete for nucleosome-stimulated ATPase (D.F.V.Corona, C.R.Clapier, P.B.Becker and J.W.Tamkun, manuscript submitted). We therefore tested whether site-specific acetylation might modify the recognition of the DNA-bound tail by ISWI. Acetylation of the H4 tail peptide at lysines 16 or 12, close to the critical basic patch (K16) or in the region of the tail that contributes to full ISWI stimulation (K12), respectively, severely compromised the potential of the tail peptide to stimulate the ATPase (Fig. 4C). The effect is unlikely to be due to non-specific charge reduction since acetylation of lysine 8 had no effect on ATPase activity. Acetylation of lysine 5 has an intermediate effect, the significance of which is unclear. These data document a direct effect of acetylation within the H4 N-terminus on substrate recognition by the nucleosome remodeling ATPase ISWI.
DISCUSSION
Our present study highlights the importance of the H4 N-terminal residues R17H18R19 for substrate recognition by the nucleosome remodeling ATPase ISWI. Out of context, either as isolated tail peptides or grafted onto histone H3 or H2A, H4 tails containing this sequence element will not trigger the ATPase activity of ISWI. Position-dependent functions of the H4 tail have been observed in yeast: while nucleosome assembly and structure per se is unaffected by swapping the tail domains, the regulation of specific loci through chromatin, however, requires the H4 tail at its native position (40). Our data are consistent with an independent analysis of the histone tail requirements for nucleosome sliding by NURF (46).
The critical amino acids are part of a larger, basic patch, K16R17H18R19K20, where the tail emerges from the compact globular histone domains. Early crosslinking experiments demonstrated a direct contact of histidine 18 with nucleosomal DNA (43). Our finding that combining H4 tail peptides with DNA, but neither of the components alone, elicits an ATPase response in ISWI strongly supports the idea that some part of the H4 tail associates with DNA, generating an ‘epitope’ consisting of DNA and protein that triggers the ATPase activity of ISWI. The fact that mutagenesis of arginines 17 and 19 to alanines abolishes this effect may point to a critical role for these residues in DNA binding. However, we also cannot exclude the alternative possibility that these arginines are directly recognized by ISWI.
The crystal structure of the nucleosome (26,42) yields little further insight. One H4 tail is not at all visible in the structure while the second one is ordered by extensive interactions with an acidic patch on the H2A–H2B dimer surface of an adjacent nucleosome. Since both tails are not equivalent in the crystal and the localization of the visible tail involves interactions that may be dictated by crystal packing forces (mutations in this region prevent crystallization) (42), it is possible that the interaction with a neighboring particle leads to an artificial ‘lifting’ of the H4 tail base off the DNA.
It has previously been suggested that interaction of the H4 tail with the minor groove of DNA may lead to the induction or stabilization of an α-helical conformation of the part in question. On the basis of elegant in vivo experiments using the yeast model, Grunstein and colleagues proposed that residues 16–19 of the H4 tail might adopt an α-helical structure upon charge neutralization by an acidic protein or DNA (44). Later, Baneres et al. (45) concluded on the basis of CD spectroscopy and secondary structure prediction that the H4 tail sequences between 16 and 24 are in an α-helical conformation upon interaction with DNA (45). Residues 16–20, which contain the ISWI recognition epitope, were found to be important for repression of the yeast mating type loci by Johnson et al. (44,47). In a search for extragenic suppressors of mutations in these residues these authors identified SIR3. Direct interaction of SIR3 with this site within the H4 tail induces the heterochromatic state that characterizes the silent mating type cassettes (48). However, SIR3 (and its companion SIR4) are apparently able to interact with the H4 tail in the absence of DNA, although the formation of an α-helix in the critical region was assumed by these authors in order to explain the results of their mutagenesis study (48). In contrast, we have been unable to identify a stable interaction between the GST–H4 tail fusion protein (48) and ISWI in the absence of DNA (unpublished observations).
Several mechanisms, not mutually exclusive, may contribute to repressive function of the DNA-bound basic patch in the H4 N-terminus. Interaction of SIR3/SIR4 proteins may lead to an inaccessible higher order folding of chromatin. At the same time, binding of these proteins may stabilize the contact of the basic patch with DNA, which may restrict the flexibility of the nucleosomal fiber. It is also conceivable that the interaction of ISWI-containing nucleosome remodeling factors with the tail may be modulated by SIR protein binding.
Most interestingly, the yeast homolog of the ATPase ISWI has recently been shown to be involved in the repression of many genes, including some that govern the meiotic programme (49–51), apparently through positioning nucleosomes in ways that are incompatible with transcription. The histone tail requirements for nucleosome remodeling by the ISW1 and ISW2 complexes and their resident ATPases have not yet been determined (41). If the mechanism of action of the ISWI homologs have been as conserved as the H4 tails, our deletion study would predict that the regulatory events that have been ascribed to yeast ISWI function would be compromised in strains that harbor a deletion/mutation of the critical H4 tail residues.
In yeast and higher eukaryotes, active loci are marked by particular histone acetylation patterns whereas repressed domains are generally characterized by hypoacetylation (29,52). The reconstitution of a minimal epitope able to stimulate ISWI ATPase activity allowed us to address whether acetylation of specific lysine residues within the H4 N-terminus affected ISWI function. We found that acetylation of those lysines that reside closest to the ISWI recognition epitope, lysines 16 and 12, interfere significantly with ISWI function. Interestingly, systematic mutational analysis of single amino acids in the H4 N-terminus in yeast suggested that acetylation of lysine 16, but not of any of the other lysines in the H4 tail, interferes with silencing of mating type loci (47). Acetylation of lysine 16 is also a hallmark of the hyperactive, partially decondensed male X chromosome in Drosophila (53). The recent observation that the higher order chromosome structure of the male X chromosome is specifically disrupted in an ISWI homozygous mutant larvae (54), suggested that ISWI-containing nucleosome remodeling complexes counteract the effect of histone acetylation on higher order folding of chromatin, presumably through their nucleosome assembly/spacing functions. Such a scenario is reminiscent of the repressive functions of ISWI-containing complexes in yeast.
Why is the activity of ISWI so sensitive to deletion and modification of the recognition epitope? Early crosslinking studies in nuclei mapped the contact of histidine 18 to a region of DNA 1.5 helical turns off the dyad axis of the nucleosome (43). This site is likely to be in reach for ISWI bound to DNA at the exit point from the nucleosome (21), suggesting that a direct interaction of ISWI may be possible. It is difficult to formulate hypotheses as to how this contact stimulates the ATPase activity of ISWI, since it is not known at which step during the nucleosome remodeling cycle ATP is hydrolyzed. ATP-dependent changes in ISWI conformation may be modulated by contact with the DNA-bound H4 motif. Conversely, disruption of the histone–DNA interactions at that site due to the remodeling process may feed back on the activity of the remodeling enzyme itself.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs V. Morales for helpful suggestions concerning histone expression, D. Corona and J. Tamkun for peptides and R. Aasland for discussions that improved the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through SFB–190, the European Commission through the Human Potential Programme, Contract Number HPRN-CT-2000-00078, and the Wellcome Trust to K.P.N. C.R.C. is a member of the EMBL international Ph.D programme.
REFERENCES
- 1.Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- 2.Peterson C.L. (2000) ATP-dependent chromatin remodeling: going mobile. FEBS Lett., 476, 68–72. [DOI] [PubMed] [Google Scholar]
- 3.Havas K., Whitehouse,I. and Owen-Hughes,T. (2001) ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci., 58, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vignali M., Hassan,A.H., Neely,K.E. and Workman,J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker P.B. and Hörz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., in press. [DOI] [PubMed] [Google Scholar]
- 6.Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- 7.Flaus A. and Owen-Hughes,T. (2001) Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev., 11, 148–154. [DOI] [PubMed] [Google Scholar]
- 8.Längst G. and Becker,P.B. (2001) Nucleosome mobilization and positioning by ISWI-containing chromatin remodeling factors. J. Cell Sci., 114, 2561–2568. [DOI] [PubMed] [Google Scholar]
- 9.Corona D.F.V., Längst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- 10.Längst G., Bonte,E.J., Corona,D.F.V. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- 11.Clapier C.R., Langst,G., Corona,D.F., Becker,P.B. and Nightingale,K.P. (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol., 21, 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guschin D., Wade,P.A., Kikyo,N. and Wolffe,A.P. (2000) ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry, 39, 5238–5245. [DOI] [PubMed] [Google Scholar]
- 13.Havas K., Flaus,A., Phelan,M., Kingston,R., Wade,P.A., Lilley,D.M. and Owen-Hughes,T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [DOI] [PubMed] [Google Scholar]
- 14.Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberharter A., Ferrari,S., Langst,G., Straub,T., Imhof,A., Varga-Weisz,P., Wilm,M. and Becker,P.B. (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J., 20, 3781–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao H., Sandaltzopoulos,R., Wang,H., Hamiche,A., Ranallo,R., Lee,K., Fu,D. and Wu,C. (2001) Dual functions of the largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell, 8, 531–543. [DOI] [PubMed] [Google Scholar]
- 17.Tsukiyama T. and Wu,C. (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell, 83, 1011–1020. [DOI] [PubMed] [Google Scholar]
- 18.Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- 19.Ito T., Tyler,J.K. and Kadonaga,J.T. (1997) Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells, 2, 593–600. [DOI] [PubMed] [Google Scholar]
- 20.Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed] [Google Scholar]
- 21.Längst G. and Becker,P.B. (2001) ISWI induces nucleosome sliding on nicked DNA. Mol. Cell, 8, 1085–1092. [DOI] [PubMed] [Google Scholar]
- 22.Schiessel H., Widom,J., Bruinsma,R.F. and Gelbart,W.M. (2001) Polymer reptation and nucleosome repositioning. Phys. Rev. Lett., 86, 4414–4417. [DOI] [PubMed] [Google Scholar]
- 23.Georgel P.T., Tsukiyama,T. and Wu,C. (1997) Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J., 16, 4717–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brehm A., Langst,G., Kehle,J., Clapier,C.R., Imhof,A., Eberharter,A., Muller,J. and Becker,P.B. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J., 19, 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen J.C., Tse,C. and Wolffe,A.P. (1998) Structure and function of the core histone N-termini: more than meets the eye. Biochemistry, 37, 17637–17641. [DOI] [PubMed] [Google Scholar]
- 26.Luger K. and Richmond,T.J. (1998) The histone tails of the nucleosome. Curr. Opin. Genet. Dev., 8, 140–146. [DOI] [PubMed] [Google Scholar]
- 27.Fletcher T.M. and Hansen,J.C. (1995) Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J. Biol. Chem., 270, 25359–25362. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Ramirez M., Dong,F. and Ausio,J. (1992) Role of the histone “tails” in the folding of oligonucleosomes depleted of histone H1. J. Biol. Chem., 267, 19587–19595. [PubMed] [Google Scholar]
- 29.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 30.Imhof A. and Becker,P.B. (2001) Modifications of the histone N-terminal domains. Evidence for an ‘epigenetic code’? Mol. Biotechnol., 17, 1–13. [DOI] [PubMed] [Google Scholar]
- 31.Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- 32.Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson R.H., Ladurner,A.G., King,D.S. and Tjian,R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- 34.Ornaghi P., Ballario,P., Lena,A.M., Gonzalez,A. and Filetici,P. (1999) The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol., 287, 1–7. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Ramirez M., Rocchini,C. and Ausio,J. (1995) Modulation of chromatin folding by histone acetylation. J. Biol. Chem., 270, 17923–17928. [DOI] [PubMed] [Google Scholar]
- 36.Tse C., Sera,T., Wolffe,A.P. and Hansen,J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nightingale K.P., Wellinger,R.E., Sogo,J.M. and Becker,P.B. (1998) Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J., 17, 2865–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akhtar A. and Becker,P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell., 5, 367–375. [DOI] [PubMed] [Google Scholar]
- 39.Luger K., Rechsteiner,T.J. and Richmond,T.J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol., 304, 3–19. [DOI] [PubMed] [Google Scholar]
- 40.Ling X., Harkness,T.A., Schultz,M.C., Fisher,A.G. and Grunstein,M. (1996) Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev., 10, 686–699. [DOI] [PubMed] [Google Scholar]
- 41.Tsukiyama T., Palmer,J., Landel,C.C., Shiloach,J. and Wu,C. (1999) Characterization of the Imitation Switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev., 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luger K., Mäder,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 43.Ebralidse K.K., Grachev,S.A. and Mirzabekov,A.D. (1988) A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nature, 331, 365–367. [DOI] [PubMed] [Google Scholar]
- 44.Johnson L.M., Fisher,A.G. and Grunstein,M. (1992) Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J., 11, 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baneres J.L., Martin,A. and Parello,J. (1997) The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J. Mol. Biol., 273, 503–508. [DOI] [PubMed] [Google Scholar]
- 46.Hamiche A., Kang,J., Dennis,C., Xiao,H. and Wu,C. (2001) Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl Acad. Sci. USA, 98, 14316–14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson L.M., Kayne,P.S., Kahn,E.S. and Grunstein,M. (1990) Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 87, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hecht A., Laroche,T., Strahl-Bolsinger,S., Gasser,S.M. and Grunstein,M. (1995) Histone H3 and H4 N-termini interact with SIR3 and Sir4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- 49.Goldmark J.P., Fazzio,T.G., Estep,P.W., Church,G.M. and Tsukiyama,T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- 50.Fazzio T.G., Kooperberg,C., Goldmark,J.P., Neal,C., Basom,R., Delrow,J. and Tsukiyama,T. (2001) Widespread collaboration of isw2 and sin3-rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol., 21, 6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kent N.A., Karabetsou,N., Politis,P.K. and Mellor,J. (2001) In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev., 15, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner B.M. (2000) Histone acetylation and an epigenetic code. Bioessays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- 53.Turner B.M., Birley,A.J. and Lavender,J. (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell, 69, 375–384. [DOI] [PubMed] [Google Scholar]
- 54.Deuring R., Fanti,L., Armstrong,J., Sarte,M., Papoulas,O., Prestel,M., Daubresse,G., Verardo,M., Moseley,S., Berloco,M., Tsukiyama,T., Wu,C., Pimpinelli,S. and Tamkun,J.W. (2000) The ISWI chromatin remodeling protein is required for gene expression and the maintenance of higher order chromaitn structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]