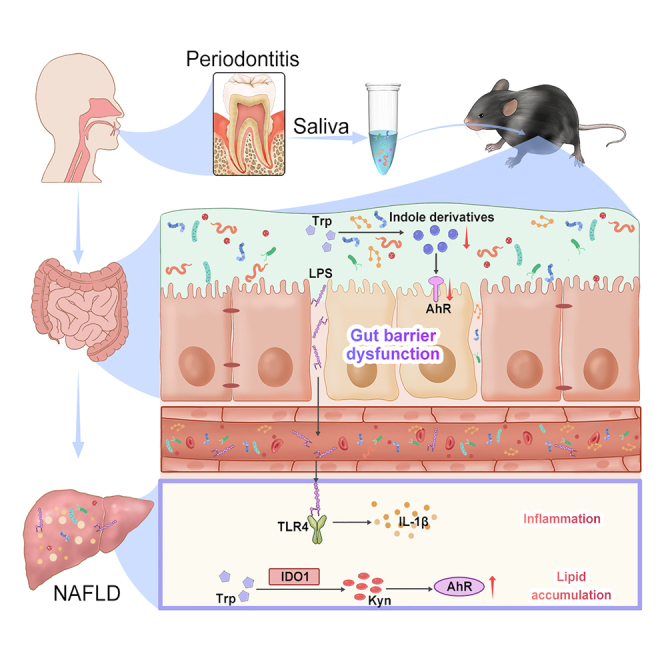
Summary
Periodontitis may aggravate the development of nonalcoholic fatty liver disease (NAFLD); however, the precise mechanism is unknown. In this study, salivary microbiota collected from patients with periodontitis was transferred intragastrically to obese mice induced by high-fat diet. Microbiomics and metabolomics analysis were performed to assess the influence of periodontitis salivary microbiota on gut microbiome and liver metabolism. Periodontitis salivary microbiota altered gut microbiota composition and exacerbated intestinal barrier dysfunction in obese mice. Subsequently, the bacterial lipopolysaccharide transported to liver may activate the toll-like receptor 4 signaling and cause the release of pro-inflammatory factors. Moreover, the tryptophan-kynurenine-AhR signal axis was upregulated in liver, which may be related to aggravated hepatic steatosis and glucolipid metabolism dysregulation during NAFLD development. This study indicated that in the context of obesity, periodontitis salivary microbiota may aggravate the pathological progression of NAFLD, in which the tryptophan-AhR pathway may play a key role.
Subject areas: Hepatology, Microbiome
Graphical abstract
Highlights
-
•
Periodontitis salivary microbiota exacerbates gut barrier dysfunction in obese mice
-
•
The bacterial LPS may transport to the liver and activate the TLR4 signaling pathway
-
•
The upregulated kynurenine-AhR axis may be related to aggravated hepatic steatosis
Hepatology; Microbiome
Introduction
Nonalcoholic fatty liver disease (NAFLD) is characterized by the pathologic hepatocellular accumulation of lipids and encompasses a continuum of liver conditions ranging from nonalcoholic fatty liver to nonalcoholic steatohepatitis, cirrhosis, and hepatocellular carcinoma.1 NAFLD is believed to be related to common diseases such as obesity, metabolic syndrome, and type 2 diabetes mellitus.2 Paralleling the high prevalence of obesity, NAFLD has become the most frequent chronic liver disease worldwide, with a global prevalence of approximately 25–40%.3 A recent cohort study demonstrated that all histological stages of NAFLD are associated with a higher risk of overall mortality, and this risk increases with worsening NAFLD histology.4 Given the growing burden caused by NAFLD, it is important to explore the risk factors for its occurrence and development.
Periodontitis is a chronic inflammatory disease induced by plaque biofilms containing diverse periodontal pathogens that lead to the destruction of tissues supporting the teeth.5 Epidemiological studies have demonstrated that periodontitis is a potential risk factor for NAFLD.6 A recent investigation suggested that higher clinical attachment loss is associated with an increased incidence of liver fibrosis in obese adults with NAFLD.7 It was also demonstrated that periodontal pathogens can aggravate the progression of NAFLD accompanied by enhanced steatosis.8 Furthermore, nonsurgical periodontal treatment can ameliorate liver abnormalities in NAFLD patients with periodontitis.9
The mechanism connecting periodontitis with NAFLD has long been explained by the hematogenous diffusion of periodontal bacteria and inflammatory mediators through ulcerous periodontal pocket.10 However, the precise mechanism remains unclear. Our recent studies demonstrate that periodontitis can induce gut dysbiosis through the influx of salivary microbe, which may mediate the effects of periodontitis on systemic diseases.11,12,13 There are significant differences in the composition of salivary microbiota between periodontitis patients and healthy individuals, with an increase in the relative abundance of Porphyromonas gingivalis, Treponema denticola, and Prevotella intermedia,14 which may enter the intestine and affect the state of gut microbiome.11 It has been also reported that oral pathobionts, including Klebsiella and Enterobacter species, expand due to periodontitis and translocate to the gut, where they trigger inflammation and aggravate colitis.15 Interestingly, mounting evidence suggests that unbalanced gut microbiota and their metabolites impair intestinal barrier function, leading to enhanced bacterial translocation from the gut to the body circulation, which disturbs the gut-liver axis, resulting in the progression of NAFLD.16,17 Thus, intestinal dysbiosis induced by swallowing oral bacteria may be a potential route linking periodontitis and NAFLD.
Therefore, we hypothesized that periodontitis salivary microbiota may cause intestinal microbiota disorders, which may influence the development of NAFLD. To better simulate actual clinical conditions, we used salivary microbiota from patients with severe periodontitis as a model of mixed bacterial dysbiosis to investigate its influence on NAFLD and possible underlying mechanisms.
Results
HFD impaired intestinal barrier and altered tryptophan metabolism
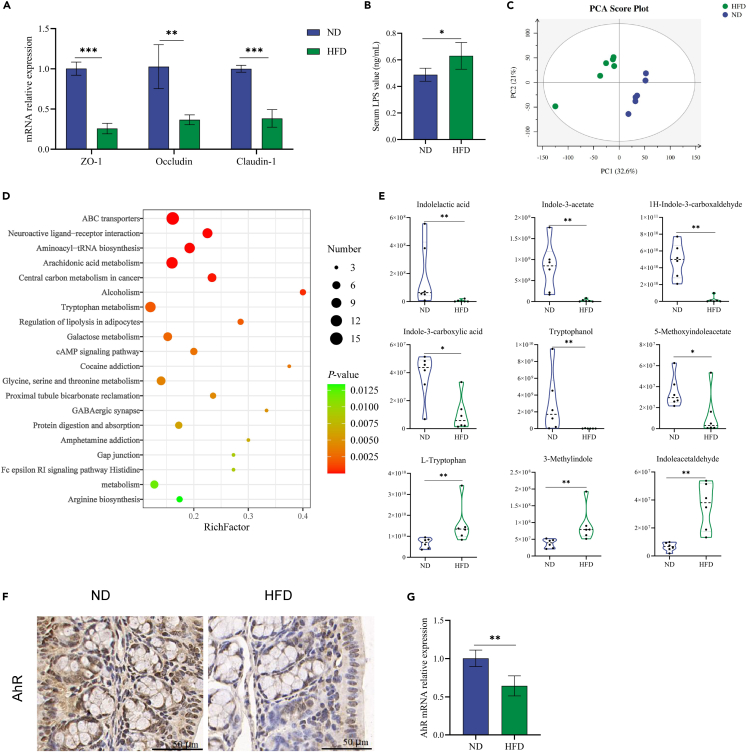
HFD consumption markedly decreased the mRNA expression levels of the tight junction proteins zonula occludens-1 (ZO-1), occludin, and claudin-1 in the colon (Figure 1A), and HFD significantly increased serum lipopolysaccharide (LPS) concentrations (Figure 1B), indicating that the integrity of the intestinal barrier was compromised in HFD mice.
Figure 1.
HFD disrupted intestinal barrier function and altered tryptophan metabolism
(A) Relative mRNA levels of tight junction proteins in colon tissues.
(B) Serum level of LPS.
(C) PCA plot on cecal contents metabolomics.
(D) Bubble diagram of the KEGG enrichment pathway analysis comparing HFD and ND groups.
(E) Relative concentrations of tryptophan metabolites-indole derivatives in cecal contents.
(F and G) IHC stainings and relative mRNA levels of AhR in colon tissues. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. AhR, aryl hydrocarbon receptor; HFD, high-fat diet; IHC, immunohistochemical; LPS, lipopolysaccharide; ND, normal diet; PCA, principal component analysis.
To further study the possible mechanisms underlying the HFD-induced disruption of the intestinal barrier, cecal contents were extracted and analyzed using liquid chromatography-mass spectrometry (LC-MS) strategy. The principal component analysis (PCA) plot showed that HFD induced a specific metabolomic profile of the cecal contents (Figure 1C). The top 20 enrichment pathways with significant differences between HFD and ND mice included arachidonic acid metabolism, tryptophan metabolism, and histidine metabolism (Figure 1D). Tryptophan metabolism is closely related to intestinal barrier function. Although the relative concentration of L-tryptophan was upregulated by the HFD, the levels of intestinal microbiota-derived tryptophan metabolites including indolelactic acid, indole-3-acetate, 1H-indole-3-carboxaldehyde, indole-3-carboxylic acid, tryptophanol, and 5-methoxyindoleacetate were significantly reduced in the HFD group (Figure 1E). These indole derivatives are ligands for aryl hydrocarbon receptor (AhR), and the mRNA and protein expression levels of AhR in the HFD group were significantly decreased in colon tissues (Figures 1F and 1G). Taken together, these data indicate that HFD impaired intestinal barrier and altered tryptophan metabolism in mice.
Periodontitis salivary microbiota worsens liver dysfunction in HFD-induced obese mice
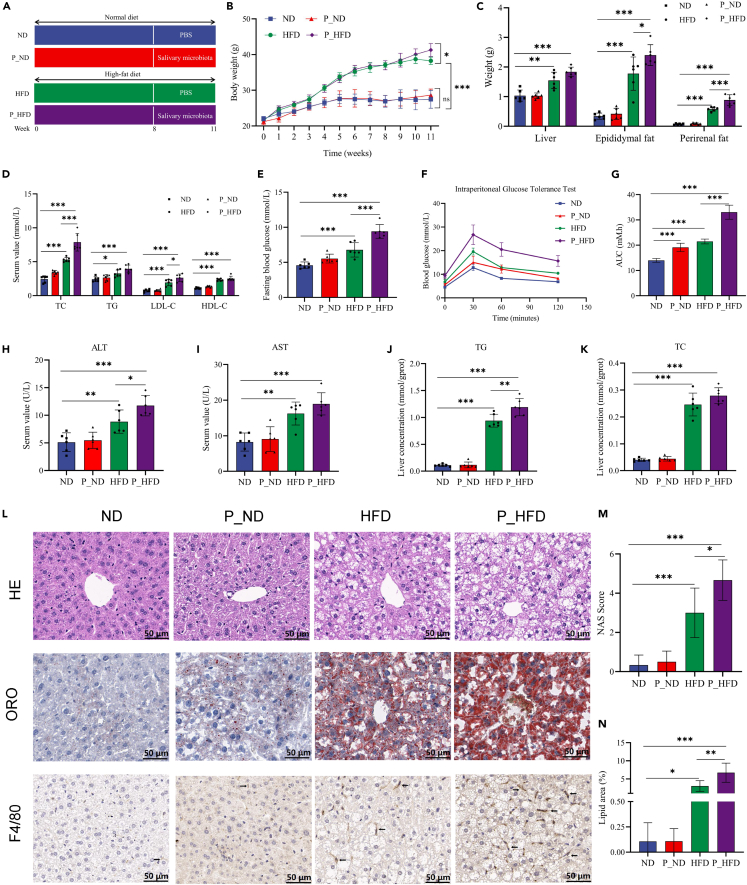
The experimental mice were fed with HFD for 8 months to induce obesity and intragastrically administered with salivary microbiota from the patients with periodontitis or PBS for 3 weeks (Figure 2A). We observed that HFD-fed mice showed obese symptoms as indicated by increased body weight compared to ND group, and periodontitis salivary microbiota further aggravated HFD-induced obesity (Figure 2B). The weight gains were mainly manifested by an increase in epididymal fat and perirenal fat weight (Figure 2C). Additionally, a more serious lipid metabolism disorder in the P_HFD group compared to that in the HFD group was confirmed by significantly higher levels of serum total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-C) (Figure 2D). In terms of glucose metabolism, HFD mice exhibited elevated fasting blood glucose (FBG) levels and increased areas under the curve of the intraperitoneal glucose tolerance test (IGTT), which were further increased in the P_HFD group (Figures 2E–2G).
Figure 2.
Periodontitis salivary microbiota worsens liver dysfunction in HFD-induced obese mice
(A) Schematic diagram of the experiment.
(B)Body weight from 0 to 11 weeks.
(C) Liver, epididymal fat, and perirenal fat weight of mice at 11 weeks.
(D) Serum levels of TC, TG, LDL-C, and HDL-C.
(E) Fasting blood glucose levels.
(F) Curves of the intraperitoneal glucose tolerance test.
(G) Areas under the curve.
(H and I) Serum levels of ALT and AST.
(J and K) TG and TC concentrations in the liver.
(L) HE, ORO, and F4/80 IHC stainings of liver tissues.
(M) NAS values.
(N) Positive area rates in ORO stainings. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ALT, alanine aminotransferase; AST, aspartate aminotransferase; AUC, area under the curve; HDL-C, high-density lipoprotein cholesterol; HE, hematoxylin and eosin; HFD, high-fat diet; IHC, immunohistochemical; LDL-C, low-density lipoprotein cholesterol; NAS, NAFLD activity score; ORO, oil red O; TC, total cholesterol; TG, total triglyceride.
The liver, as the main organ of glucolipid metabolism, impaired liver function is another major abnormality in obesity. Therefore, our study verified whether the periodontitis salivary microbiota affects hepatic pathology in HFD-induced obese mice. We found that P_HFD mice exhibited higher serum alanine aminotransferase (ALT) levels, although aspartate aminotransferase (AST) levels did not differ significantly between P_HFD and HFD mice, confirming the existence of liver dysfunction in the P_HFD group (Figures 2H and 2I). Interestingly, hepatic total triglyceride (TG) levels were significantly increased in the P_HFD group (Figures 2J and 2K), further indicating hepatic lipid metabolism disorders. Periodontitis salivary microbiota promoted the substantial accumulation of lipid droplets with more hepatocyte ballooning degeneration in HE stainings and more infiltration of F4/80-positive macrophages in IHC stainings of liver tissues compared to the HFD group (Figure 2L). According to the NAS histological system, the NAS of the P_HFD group was 4.67 ± 0.94, which was significantly higher than that of the HFD group (3 ± 0.15) (Figure 2M). ORO staining revealed more red-stained lipids in the HFD group, and this number was further increased in the P_HFD group, along with an increase in oil red-positive areas (Figure 2N). In short, the above results indicate that periodontitis salivary microbiota worsened the dysfunction of liver, including hepatic lipid accumulation and impaired glucolipid metabolism.
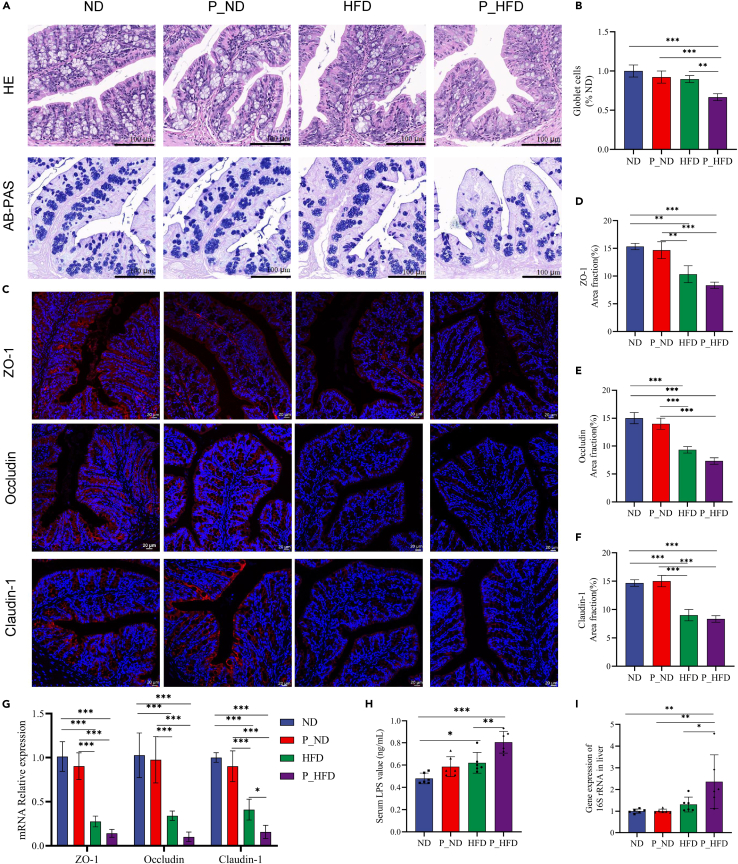
Periodontitis salivary microbiota exacerbates intestinal barrier dysfunction and bacterial translocation in HFD-induced obese mice
HE stainings of colon tissues showed that the arrangement of crypts tended to be disordered in the HFD and P_HFD groups, and the morphology of some crypts was uncharacteristic (Figure 3A). Alcian blue periodic acid Schiff (AB-PAS) stainings showed that the number of goblet cells substantially reduced in the P_HFD group (Figures 3A and 3B). In terms of intestinal permeability, we observed that the protein and mRNA levels of tight junction proteins were markedly reduced in the HFD and P_HFD groups; in particular, the expression of claudin-1 mRNA was further decreased in the P_HFD group compared to that in the HFD group (Figures 3C–3G). Moreover, the serum LPS concentration in the P_HFD group was significantly increased (Figure 3H). Consistent with endotoxemia, the gene expression level of bacterial 16S rRNA in liver was markedly elevated in the P_HFD group (Figure 3I), indicating that periodontitis salivary microbiota increased the translocation of bacteria to the liver tissues in obese mice. Similarly, the relative concentrations of indole derivatives and AhR mRNA expression levels tended to decrease, but the differences between HFD and P_HFD mice were not statistically significant (Figure S1). All these results support that periodontitis salivary microbiota promote LPS leakage into the bloodstream and bacterial translocation to the liver due to HFD-induced disruption of intestinal barrier function.
Figure 3.
Periodontitis salivary microbiota exacerbates intestinal barrier dysfunction and bacterial translocation in HFD-induced obese mice
(A) HE and AB-PAS stainings of colon tissues.
(B) The goblet cells per crypt that normalized to the ND group.
(C) IF stainings of tight junction proteins in colon tissues.
(D–F) Quantification of area fractions of tight junction proteins in colon tissues.
(G) Relative mRNA levels of tight junction proteins in colon tissues.
(H) Serum LPS levels.
(I) Relative quantification of bacterial loads in liver tissues. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. AB-PAS, Alcian blue periodic acid Schiff; HE, hematoxylin and eosin; HFD, high-fat diet; IF, immunofluorescence; LPS, lipopolysaccharide.
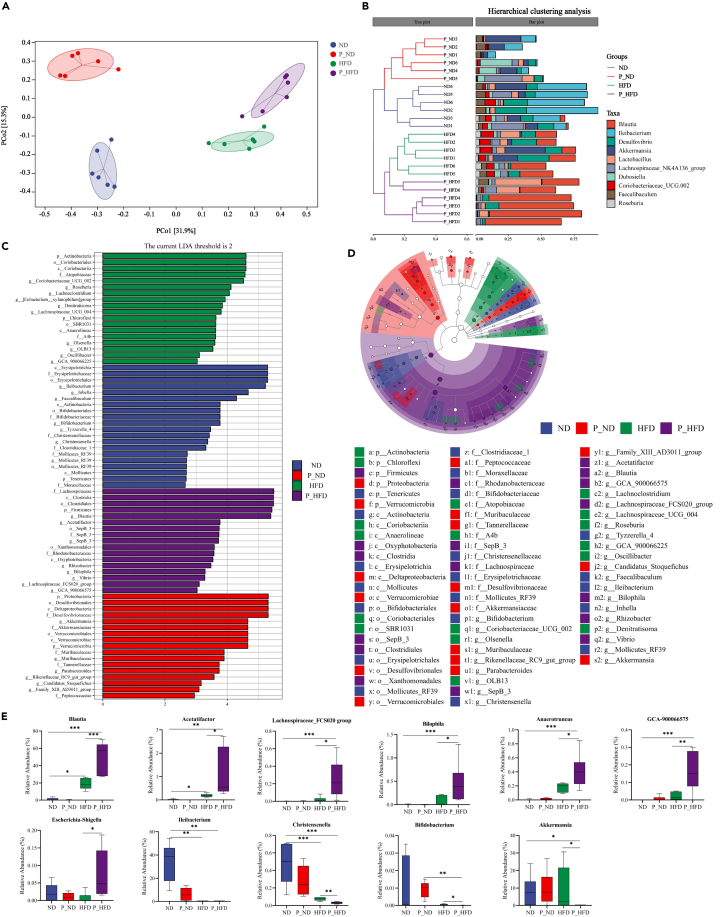
Periodontitis salivary microbiota alters the gut microbiota composition in HFD-induced obese mice
The gut microbiota may be a critical contributor to obesity-related diseases. We used high-throughput sequencing of the 16S rRNA gene to investigate the effects of the periodontitis salivary microbiota on the gut microbiome. The results showed that HFD increased the Shannon index and Chao 1 index, whereas periodontitis salivary microbiota did not affect bacterial α-diversity (Figure S2A). The principal coordinate analysis (PCoA) based on Bray-Curtis analysis indicated that periodontitis salivary microbiota regulated the β-diversity of P_ND and P_HFD mice (Figure 4A). Similarly, hierarchical clustering analysis showed that the gut bacterial composition of the four study groups was distinctly separated into four different clusters (Figure 4B).
Figure 4.
Periodontitis salivary microbiota alters the gut microbiota composition in HFD-induced obese mice
(A) PCoA of Bray-Curtis analysis (R = 0.898, p = 0.001, Anosim test).
(B) Hierarchical clustering analysis based on Bray-Curtis similarity.
(C) LefSe analysis indicating discriminative biomarkers of the four groups (LDA >2).
(D) Cladogram representing the distribution of biomarkers at taxonomic levels from phylum to genus levels. The sizes of nodes indicate the relative abundance of taxa.
(E) Representative genera screened by LefSe and random forest analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. HFD, high-fat diet; LDA, linear discriminant analysis; LefSe, linear discriminant analysis effect size; PcoA, principal coordinate analysis.
To further explore the specific changes in the microbiota community, the relative abundances of the top five phyla were compared among the four groups. The relative abundance of Proteobacteria was significantly increased in the P_ND group, whereas Firmicutes was significantly increased in the P_HFD group (Figure S2B). Random forest analysis at the genus level and Linear discriminant analysis effect size (LefSe) analysis at different levels were used to identify the dominant taxa enriched in the experimental groups (Figures 4C and 4D). In the ND group, Erysipelotrichaceae, Ileibacterium, Faecalibaculum, Bifidobacterium, and Christensenella were significantly enriched. Proteobacteria, Desulfovibrionaceae, and Akkermansia were enriched in the P_ND group. In the HFD group, the enriched taxa included Actinobacteria, Coriobacteriaceae, and Roseburia. Additionally, Firmicutes, Lachnospiraceae, Clostridiales, Blautia, Acetatifactor, and Bilophila were dominant taxa in the P_HFD group. A random forest heatmap showed that Ileibacterium and Blautia were biomarker species for the study groups (Figure S2C).
Next, we performed a statistical analysis of the relative abundance of the differential genera screened by LefSe analysis among the four groups. In the P_HFD group, the relative abundances of Blautia, Acetatifactor, Lachnospiraceae_FCS020 group, Bilophila, Anaerotruncus, GCA-900066575, and Escherichia-Shigella were significantly increased, whereas Akkermansia, Christensenella, and Ileibacterium were almost absent (Figure 4E). Interestingly, Spearman’s correlation heatmap showed that Blautia was positively correlated with the enriched taxa in the HFD group and negatively correlated with the dominant genus taxa in the ND and P_ND groups (Figure S2D). These data provide direct evidence that periodontitis salivary microbiota can alter the gut microbiota composition in HFD-induced obese mice.
Periodontitis salivary microbiota-induced alterations in gut microbiota and tryptophan metabolites are associated with worse obesity-related symptoms
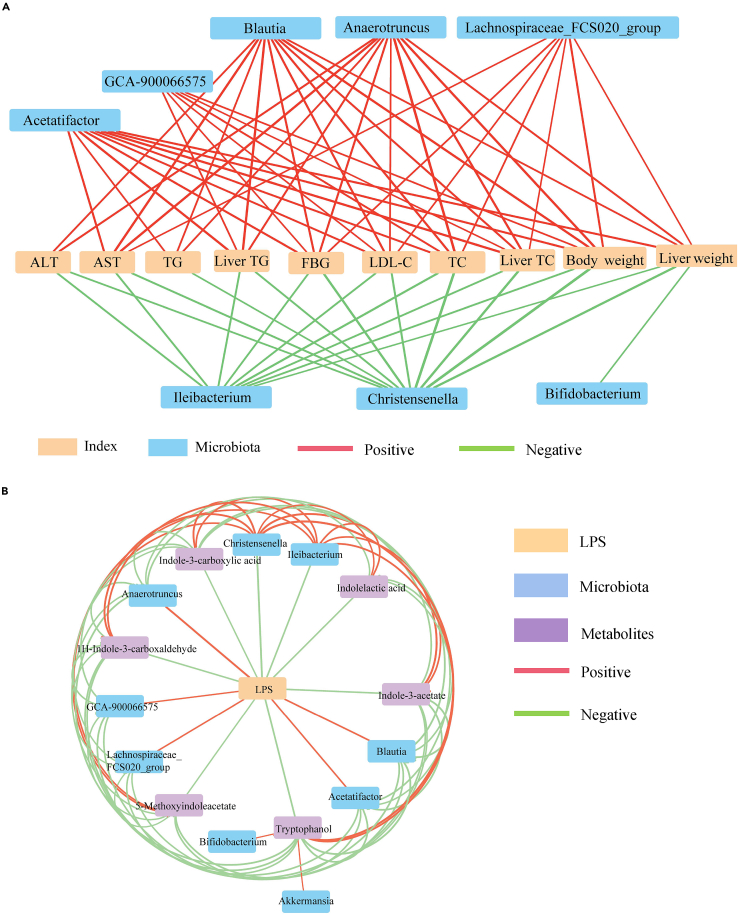
Next, we explored the correlation between the altered gut microbiota and obesity-related symptoms. Spearman’s correlation analysis showed that Lachnospiraceae and unclassified Clostridiales were positively correlated with obesity-related indicators, whereas Erysipelotrichaceae and Christensenellaceae were negatively correlated with obesity-related indicators (Figure S3 and Table S2). At the genus level, Blautia, Acetatifactor, Lachnospiraceae_FCS020 group, and Anerotruncus enriched in the P_HFD group were positively correlated with obesity-related indicators; however, beneficial bacteria such as Ileibacterium and Christensenella were negatively correlated with obesity-related indicators (Figure 5A and Table S3). Altogether, these results indicate that the gut microbiota altered by periodontitis salivary microbiota may be associated with obesity-related symptoms.
Figure 5.
Gut microbiota and tryptophan metabolites with intestinal barrier function in HFD-induced obese mice
(A) Spearman’s correlation between representative genera and obesity-related parameters. Significant associations with p < 0.05 and |r| > 0.7 are shown.
(B) Correlation analysis of representative genera, indole derivatives, and serum LPS. Significant associations with p < 0.05 and |r| > 0.4 are shown (Spearman’s correlation analysis). Red lines indicate positive relationships between nodes, and green lines indicate negative correlations. The thickness of the lines indicates the strength of the correlation. LPS, lipopolysaccharide.
Increasing evidence has shown that indole and its derivatives can enhance intestinal epithelial barrier function and that the serum LPS concentration is an important index for assessing intestinal permeability. To further clarify the role of the alterations in tryptophan metabolite levels, we conducted an association network analysis of the interaction matrix, including representative genera among the four groups selected by the LefSe analysis, indole derivatives, and serum LPS (Table S4). The results showed that LPS was located in the center of the association network, and LPS was negatively correlated with Ileibacterium, Christensenella, and the six indole derivatives, whereas Ileibacterium and Christensenella were positively associated with the six indole derivatives. By contrast, Blautia, Acetatifactor, Lachnospiraceae_FCS020 group, and Anerotruncus were positively correlated with LPS, and these genera showed significant negative correlations with the six indole derivatives (Figure 5B). Thus, these data reveal that alterations in gut microbiota and tryptophan metabolites induced by HFD and periodontitis salivary microbiota play important roles in the deterioration of intestinal barrier function and the promotion of bacterial translocation.
Periodontitis salivary microbiota upregulates the tryptophan-kynurenine-AhR axis in the liver
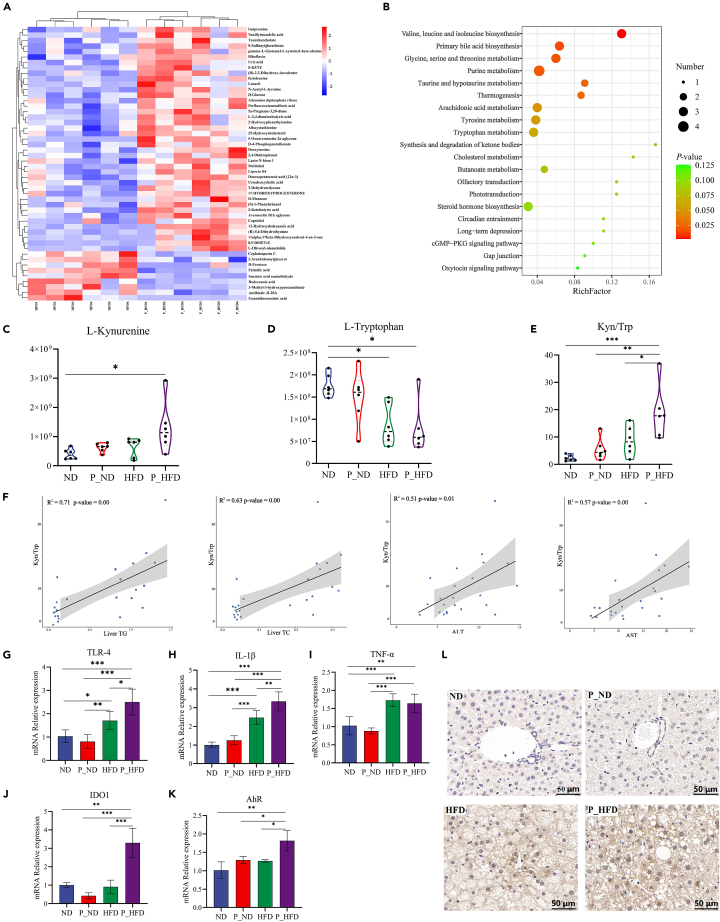
The metabolic profiles of the liver tissues from the four groups were measured using LC-MS. The PLS-DA model showed that samples in the ND and P_ND groups were clearly separated, whereas the samples in the HFD and P_HFD groups were partially clustered together (Figure S4A). However, the OPLS-DA model demonstrated that the P_ND, ND, P_HFD, and HFD groups could be completely separated (Figures S4C and S4D). The metabolites differentially present in the study groups were selected according to a VIP ≥ 1 and p ≤ 0.05. The clustering heatmap showed the top 50 differential metabolites based on the VIP-value (Figure 6A). Subsequently, a KEGG pathway analysis was performed. As shown in Figure 6B, pathways significantly enriched in the P_HFD and HFD groups included valine, leucine, and isoleucine biosynthesis; primary bile acid biosynthesis; glycine, serine, and threonine metabolism; purine metabolism; taurine and hypotaurine metabolism; arachidonic acid metabolism; and tryptophan metabolism. Interestingly, all of these pathways were significantly upregulated. Additionally, the pathways significantly enriched in the P_ND and ND groups were mainly related to amino acid metabolism (Figure S5).
Figure 6.
Periodontitis salivary microbiota stimulates the tryptophan-kynurenine-AhR axis in the liver
(A) Heatmap of the top 50 differential metabolites in VIP-value between P_HFD and HFD groups.
(B) KEGG pathway analysis of the top 20 enriched metabolic pathways in the P_HFD and HFD groups.
(C–E) Relative concentrations of L-kynurenine, L-tryptophan, and the Kyn/Trp ratio in liver tissues.
(F) Spearman’s correlation analysis between Kyn/Trp and liver TG, TC, serum ALT, AST.
(G–K) Relative mRNA levels of TLR4, IL-1β, TNF-α, IDO1, and AhR.
(L) IHC staining of AhR in liver tissues. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. AhR, aryl hydrocarbon receptor; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HFD, high-fat diet; IDO1, indoleamine 2,3-dioxygenase 1; IHC, immunohistochemical; IL, interleukin; Kyn/Trp, ratio of L-kynurenine to L-tryptophan; P_HFD, high-fat diet + periodontitis salivary microbiota; TC, total cholesterol; TG, total triglyceride; TLR, toll-like receptor; TNF, tumor necrosis factor; VIP, variable importance for the projection.
To clarify the specific effects of periodontitis salivary microbiota on tryptophan metabolism in the liver, we further analyzed the metabolites that lead to changes in tryptophan metabolic pathways. The results showed that the ratio of L-kynurenine to L-tryptophan (Kyn/Trp) in the P_HFD group was significantly higher than that in the HFD group, indicating that tryptophan-kynurenine was significantly upregulated in the P_HFD group (Figures 6C–6E). According to the results of Spearman’s correlation analysis, the changes in Kynurenine-AhR axis were positively correlated with hepatic indicators and glucolipid metabolism (Figures 6F and S6).
Indoleamine 2,3-dioxygenase 1 (IDO1) is a key enzyme in the metabolism of L-tryptophan to kynurenine. It has been reported that oxidized LDL (oxLDL) stimulation can activate the toll-like receptor 2/4 (TLR2/4) pathway to produce inflammatory factors and activate IDO1. This promotes the conversion of tryptophan to kynurenine, which can activate the AhR signaling pathway leading to obesity and hepatic steatosis.18 Therefore, we examined the mRNA expression levels of factors related to the LPS-TLR4 signaling pathway, and the results showed that the mRNA expression levels of TLR4, interleukin (IL)-1β, IDO1, and AhR were significantly higher in the P_HFD group (Figures 6G–6K). Moreover, IHC stainings confirmed that the protein levels of AhR were increased in the P_HFD group (Figure 6L). These results suggest that gut bacteria-derived LPS may activate the hepatic TLR4 signaling pathway and release the pro-inflammatory factor IL-1β. On the other hand, IDO1-mediated tryptophan catabolism to kynurenine is promoted and the increased kynurenine may aggravate NAFLD lesions through the AhR signaling pathway.
Discussion
Increasing evidence indicates that gut dysbiosis affects various phenotypic changes and the progression of HFD-associated diseases, including NAFLD.19 Disturbance of the oral microbiota may lead to gut dysbiosis, which is a possible route of oral bacterial dissemination via enteral spreading.20 Numerous oral bacteria in saliva can enter the intestinal tract after swallowing.11 In the present study, the salivary microbiota of patients with severe periodontitis was intragastrically administered to mice. We aimed to explore whether the swallowed periodontitis salivary microbiota affects the pathological progression of NAFLD and examine possible underlying mechanisms.
HFD can directly interact with intestinal epithelial cells or indirectly modulate intestinal immune responses to enhance intestinal permeability.21 In this study, ZO-1, occludin, and claudin-1, biomarkers of intestinal tight junctions, were significantly reduced by HFD, indicating increased intestinal permeability in obese mice. Metabolites produced by the gut microbiota are increasingly appreciated for maintaining intestinal barrier homeostasis.22 We found that HFD altered tryptophan metabolism, consistent with previous observations.23,24 Among the tryptophan metabolites, several indole derivatives were nearly depleted by HFD including indolelactic acid, indole-3-acetate, 1H-Indole-3-carboxaldehyde, indole-3-carboxylic acid, tryptophanol, and 5-methoxyindoleacetate, which are AhR ligands. The AhR signaling, a key component of the intestinal barrier immune response, may be closely related to the pathogenesis of NAFLD. A recent study showed that artificial sugar intake aggravated NAFLD symptoms in mice by reducing the production of AhR ligands.25 Additionally, AhR may be involved in the pathogenesis of metabolic syndrome. A decrease in intestinal AhR ligands was observed in patients with metabolic syndrome and corresponding mouse models, whereas supplementation with Lactobacillus can compensate for impaired AhR signal transduction, thus restoring disrupted intestinal barrier function and improving metabolic disorder-related symptoms.26 Consistently, our findings showed that the expression of AhR in HFD-induced obese mice was significantly reduced. Overall, our data suggest that HFD may disrupt the intestinal barrier by affecting tryptophan metabolism and downregulating the indole derivative-AhR axis.
We further explored whether the periodontitis salivary microbiota could aggravate the progression of NAFLD. In this study, the salivary microbiota of patients with severe periodontitis was used as a model for mixed bacterial dysbiosis. Previous animal studies have mainly focused on the effects of a single periodontal pathogen on NAFLD.8,27,28 However, the key pathogenic hypothesis may not fully reveal the complexity of the pathogenesis and interactions between periodontitis and NAFLD. Consequently, a mixed bacterial dysbiosis model related to periodontitis may provide better scientific support than a single-species model and can better simulate actual clinical conditions.29 Interestingly, periodontitis salivary microbiota promoted the progression of liver pathology in NAFLD mice, accompanied by aggravated disorders of glucolipid metabolism. This is consistent with the latest findings that metabolic dysfunctions, such as impaired glucose tolerance, dyslipidemia, and obesity, are associated with hepatic steatosis.30
The underlying mechanisms by which the periodontitis salivary microbiota affected NAFLD were further investigated. In the present study, periodontitis salivary microbiota caused a significant decrease in the number of goblet cells in the colons of NAFLD mice. Goblet cells are distributed on the surface of the crypt and are vital for maintaining mucus barrier function and protecting intestinal epithelial cells from exogenous substances and microorganisms.31 The decrease in the number of goblet cells leads to a disruption of intestinal mucosal barrier function. Additionally, although periodontitis salivary microbiota did not significantly affect tight junction proteins in NAFLD mice, it increased LPS concentrations in the serum and bacterial 16S rRNA gene expression levels in the liver. Systemic LPS concentration in NAFLD patients is higher compared to controls.32 This is consistent with our results that periodontitis salivary microbiota promoted bacterial translocation to the liver. The translocation of bacteria or bacterial products from the gut, even in minute quantities, has been reported to severely affect Kupffer cells in the liver.33,34 Bacterial LPS and their products translocated into the liver via portal vein blood flow, which are recognized by TLRs on hepatocytes, Kupffer cells, and hepatic stellate cells.35 The signaling ultimately activates NF-κB and the subsequent inflammasome activation. In addition, LPS could directly damage hepatocytes and activate Kupffer cells to produce inflammatory cytokines, followed by the waterfall effect.36 Our results showed that microbiota-derived bacterial LPS may activate the TLR4 signaling pathway and cause the release of pro-inflammatory factors, which may exacerbate the development of NAFLD.
To explore the specific role of gut microbes in the progression of NAFLD exacerbated by periodontitis salivary microbiota, we performed 16S rRNA sequencing of cecal contents. On the background of HFD-induced obesity, periodontitis salivary microbiota significantly increased the relative abundance of Blautia, Acetatifactor, Lachnospiraceae_FCS020 group, Bilophila, Anaerotruncus, GCA-900066575, and Escherichia-Shigella, whereas Christensenella, Ileibacterium, Akkermansia, and Bifidobacterium were nearly absent. These latter bacteria with reduced numbers have great potential as probiotics for the treatment of metabolic diseases related to obesity.37 Studies have shown that the numbers of Blautia, Lachnospiraceae_FCS020 group, and Escherichia-Shigella were increased in the feces of patients with obesity and NAFLD.38 Additionally, Acetatifactor and Bilophila are involved in the production and transformation of bile acids associated with intestinal functional metabolic disorders.39 Anaerotruncus is positively correlated with levels of inflammatory factors such as LPS and IL-6.40 The above studies suggest that Blautia, Acetatifactor, Lachnospiraceae_FCS020 group, Bilophila, Anaerotruncus, GCA-900066575, and Escherichia-Shigella may be the predominant species promoting the progression of NAFLD. These key bacteria were positively correlated with NAFLD indices and LPS and negatively correlated with indole derivatives. Overall, our data demonstrated that the periodontitis salivary microbiota leads to gut microbiome disorders in obese mice, which are associated with increased bacterial translocation from the gut to the liver.
Additionally, we observed that Proteobacteria, Desulfovibrionace, and Akkermansia were enriched by periodontitis salivary microbiota on the background of an ND. Proteobacteria represent an unstable state of the intestine,41 whereas Akkermansia often maintain intestinal homeostasis by binding to TLRs on the intestinal epithelium or by regulating the expression of tight junction proteins.42 This suggests that, on a healthy background, although periodontitis salivary microbiota disrupts intestinal microbiome homeostasis, the intestinal microecology is still in a state of compensatory regulation, manifested by an increase in pathogenic bacteria accompanied by a compensatory increase in beneficial bacteria.
Growing evidence has pointed to the influx of bacteria or bacterial products to the liver via the portal system, worsening the metabolic abnormalities of NAFLD.43,44 The results of liver metabolomics showed that the periodontitis salivary microbiota upregulated the hepatic tryptophan-kynurenine metabolic pathway in obese mice, with a significant increase in the ratio of Kyn/Trp and IDO1 levels. IDO1 is a key enzyme in the metabolism of the L-tryptophan-L-kynurenine pathway. After feeding an HFD for 26 weeks, Ido1−/− mice showed a significantly lower plasma Kyn/Trp ratio and less hepatic steatosis, TG deposition, and weight gain than wild-type mice.45 Moreover, IDO1 expression and activity in fat and liver tissues, plasma kynurenine levels, and Kyn/Trp ratios were higher in obese subjects than in non-obese controls.46 These studies suggest that IDO1 may be a key regulator of the obesity-related kynurenine metabolic pathway.
Besides indole and indole derivatives, kynurenine also acts as an AhR agonist and activates AhR signaling in a variety of cell types, including hepatocytes.47 A recent study reported that kynurenine stimulates AhR expression in adipocytes, which mediates a systemic effect on the development of obesity and insulin resistance.48 The hepatic AhR signaling pathway is activated by the addition of kynurenine to a low-fat diet in mice, inducing the expression of hepatic cytochrome P450 1B1 and stearoyl-CoA desaturase 1, leading to hepatic steatosis.49 Similarly, we found that AhR mRNA expression increased in the P_HFD group, and the hepatic arachidonic acid metabolic pathway was upregulated. Interestingly, several AhR ligands have been reported to modulate arachidonic acid metabolism and higher levels of arachidonic acid metabolites were able to promote hepatic steatosis.50,51 Thus, we speculated that the activation of the kynurenine-AhR signaling pathway in the liver may be related to NAFLD progression.
Conclusions
Our results demonstrate that HFD damages intestinal barrier function via the indole derivatives-AhR signaling pathway. In this context, periodontitis salivary microbiota aggravates gut microbiome disorders and promote bacterial translocation to the liver. The bacterial LPS may activate the TLR4 pathway, leading to hepatic inflammation. Simultaneously, the hepatic tryptophan-kynurenine-AhR signaling axis is upregulated, which may be related to serious hepatic steatosis. The present results provide novel insights into the mechanisms by which periodontitis affects NAFLD pathology. Moreover, these results suggest that poorly managed or untreated severe periodontitis may exacerbate NAFLD progression. Periodontal treatment may positively influence clinical outcomes in patients with NAFLD and periodontitis.
Limitations of the study
This study has potential limitations. First, we observed that periodontitis salivary microbiota aggravated intestinal barrier dysfunction and bacterial translocation in NAFLD mice. However, which bacteria ectopically colonized is involved in the pathological events, remains unclear in our present study. The second limitation concerns the mechanism by which kynurenine deteriorates hepatic steatosis is not fully clarified. Further investigations are required to overcome the limitations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| F4/80 antibody | Proteintech | Cat# 28463-1-AP; RRID: AB_2881149 |
| ZO-1 antibody | Proteintech | Cat# 21773-1-AP; RRID: AB_10733242 |
| Occludin antibody | Proteintech | Cat# 27260-1-AP; RRID: AB_2880820 |
| Claudin-1 antibody | Proteintech | Cat# 13050-1-AP; RRID: AB_2079881 |
| AhR antibody | Invitrogen | Cat# MA1-513; RRID: AB_2223958 |
| Biological samples | ||
| Human saliva samples | Nanjing Stomatological Hospital (Ethical approvement:2019NL008(KS)) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 2-chlorophenylalanine | Aladdin | Cat# 103616-89-3 |
| methanol | Thermo Fisher Scientific | Cat# 67-56-1 |
| chloroform | Thermo Fisher Scientific | Cat# 67-66-3 |
| methanoic acid | Thermo Fisher Scientific | Cat# 64-18-6 |
| acetonitrile | Thermo Fisher Scientific | Cat# 75-05-8 |
| Critical commercial assays | ||
| TG | Jiancheng Bioengineering Institute | Cat# A110-1-1 |
| TC | Jiancheng Bioengineering Institute | Cat# A111-1-1 |
| HDL-C | Jiancheng Bioengineering Institute | Cat# A112-1-1 |
| LDL-C | Jiancheng Bioengineering Institute | Cat# A113-1-1 |
| ALT | Jiancheng Bioengineering Institute | Cat# C009-2-1 |
| AST | Jiancheng Bioengineering Institute | Cat# C010-2-1 |
| Lipopolysaccharide ELISA kit | Cloud-Clone | Cat# SEB526 |
| RNA Easy Fast Tissue Kit | TIANGEN | Cat# DP451 |
| HiScript III RT SuperMix for qPCR | Vazyme | Cat# R323-01 |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A25742 |
| Quant-iT PicoGreen dsDNA Assay Kit | Invitrogen | Cat# P7589 |
| TruSeq Nano DNA Library Prep Kit | Illumina Inc. | Cat# FC-121-4001 |
| MagPure Stool DNA LQ Kit | Magen | Cat# D6364 |
| NovaSeq 6000 SP Reagent Kit | Illumina Inc. | Cat# 20027464 |
| Deposited data | ||
| Raw data | https://www.ncbi.nlm.nih.gov/ | PRJNA857856 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Vital River Laboratory Animal Technology Co., Ltd. | Cat# 219 |
| Oligonucleotides | ||
| Primer sequences. See Table S1 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism v8.0 | GraphPad Software | RRID:SCR_002798 |
| QIIME 2 | https://qiime2.org/ | RRID:SCR_018074 |
| R packages v4.0.0 | https://www.r-project.org/ | RRID: SCR_001905 |
| Silva database Release132 | http://www.arb-silva.de | RRID:SCR_006423 |
| Other | ||
| ND diet | Jiangsu Xietong Bioengineering Co., Ltd | D12450J |
| HFD diet | Jiangsu Xietong Bioengineering Co., Ltd | D12492 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Fuhua Yan (yanfh@nju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study population
Saliva samples were obtained from 20 patients with severe chronic periodontitis. The inclusion criteria were as follows: 1) age ≥ 18 years; 2) ≥ 14 remaining teeth; 3) ≥ 30% teeth with the following three characteristics: I) probing depth ≥ 6 mm; II) clinical attachment loss at the interdental site of greatest loss ≥ 5 mm; III) radiology bone loss exceeding to middle or apical third of the root. The exclusion criteria were as follows: 1) history of metabolic disease, cardiovascular diseases, immune deficiency, chronic gastrointestinal diseases, abnormal gastrointestinal activity, and other systemic diseases; 2) history of periodontal treatment within the last 6 months; 3) use of antimicrobials or anti-inflammatory drugs in the last 6 months; 4) presence of other serious oral diseases; 5) smokers (> 10 PCS/day); 6) pregnancy or lactation; and 7) body mass index ≥ 28 kg/m2. The clinical data regarding the periodontal status of the study population was showed in the Table S5. The procedure was approved by the Ethics Committee of Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, China [No.2019NL008(KS)]. And informed consent was obtained from all patients.
Collection and disposition of periodontitis saliva samples
After fully informed the nature of the study, the risks and benefits, each participant completed a questionnaire and signed an informed consent form. Then, they were given a 50 mL sterile centrifuge tube, the unstimulated saliva was slowly spat into a 50 mL sterile centrifuge tube until the volume of saliva was more than 5 mL. This process could usually be completed within 15 min. The donors were informed not to suck during collection to avoid stimulating gingival bleeding. After collection, the saliva samples were centrifuged at 1000 rpm for 10 min at 4°C to remove impurities, and the supernatants were suspended in an equal volume (w/v) of phosphate buffer saline (PBS) containing 20% glycerol/PBS, snap-frozen in liquid nitrogen, and stored at -80°C until use.
The amount of saliva given to mice according to the weight ratio of human and mouse. Specifically, an adult (60 kg) secretes 1–1.5 L saliva every day, each mouse (20 g) was approximately gavaged with 0.5 mL saliva per day. Therefore, 1 ml of frozen samples was used per mouse per day. After collecting all the required samples, the saliva samples were pooled, separated evenly, and stored at -80°C. Before gavaged in mice, the sample was thawed and centrifuged at 3300×g for 10 min at 4°C to receive microbiota,52 and suspended in PBS (200 μL per mouse per day). Mixed saliva was used to avoid manipulation errors.
Animal models
C57BL/6J mice (male, 6 weeks old) were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The animals were housed in a specific pathogen-free environment with a 12-h light/dark cycle. After adapting to the environment for 1 week, 24 mice were randomly divided into four groups: normal diet (ND), ND + periodontitis salivary microbiota (P_ND), HFD, and HFD + periodontitis salivary microbiota (P_HFD). Mice in the ND and P_ND groups were fed an ND (D12450J, Table S6), and those in the HFD and P_HFD groups were fed an HFD (D12492, Table S6). Periodontitis salivary microbiota was intragastrically administered to mice via a feeding needle in the P_ND and P_HFD groups from the 8th week for 3 weeks, whereas mice in the ND and HFD groups received the same amount of PBS. Animal procedures were conducted according to the “Guide for the care and use of laboratory animals” and approved by the Animal Ethics Committee of Nanjing Agriculture University, Nanjing, China (No. PZW2022014).
Method details
Measurement of glucose metabolic parameters
An intraperitoneal glucose tolerance test (IGTT) was performed 2 days before the end of the experiment. All mice received fresh bedding and were fasted overnight. Blood was collected by cutting the tail tip, and glucose levels were measured as fasting blood glucose using OneTouch Ultra Easy (LifeScan Inc., USA). The mice were then injected with a 25% glucose solution (1 g/kg), and glucose levels were measured at 30, 60, and 120 min after injection.
Measurement of serum and hepatic biochemistry
TC and total triglyceride TG levels in liver tissues and serum concentrations of TC, TG, LDL-C, HDL-C, ALT, and AST were measured using assay kits (Jiancheng Bioengineering Institute, China), according to the manufacturer’s protocols. The serum LPS level was determined using an ELISA kit (Cloud-Clone, China).
Histological analyses
Liver and colon tissues were fixed in 4% paraformaldehyde for 48 h, embedded in paraffin, and sliced into 4-μm sections. The tissue sections were subjected to HE staining, F4/80 and AhR IHC stainings, and ZO-1, Occludin, and Claudin-1 IF stainings. An NAS system was used to evaluate the histological features of the liver.53 Additionally, frozen liver tissues (-20°C) were cut into 10-μm sections and stained with oil red to assess lipid accumulation in the liver. Images were obtained using a Pannoramic Slide Scanner (3D HISTECH, Hungary).
Quantitative real-time polymerase chain reaction (qPCR) of gene expression in colon and liver tissues
Total RNA of colon and liver tissues was extracted using the RNA Easy Fast Tissue Kit (TIANGEN, China). RNA concentrations were measured using Nanodrop One (Thermo Fisher Scientific, USA), followed by a reverse transcription reaction using a HiScript III RT SuperMix Kit (Vazyme, China). PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, USA) was used for qPCR on a ViiA V7Dx qPCR-384 system (Thermo Fisher Scientific, USA). The primer sequences used are listed in Table S1.
qPCR of 16S ribosomal RNA (rRNA) expression in liver tissues
Genomic DNA was extracted from equal liver tissues using the TIANamp Genomic DNA Kit (TIANGEN, China). DNA concentrations were measured using Nanodrop One, and PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, USA) was used for qPCR. The 16S rRNA sequences were 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’).
16S rRNA sequencing of cecal contents microbiota
Total microbial DNA of cecal contents was extracted using the MagPure Stool DNA LQ Kit (Magen, China). The amplified products were purified using Agencourt AMPure beads (Beckman Coulter, USA) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, USA) according to the manufacturer’s instructions. After quality inspection of the sequencing library, sequencing was performed using a NovaSeq 6000 SP Reagent Kit (Illumina Inc., USA). Raw amplification sequencing data were analyzed using QIIME 2 software and R packages (v3.3.2).54 The primer fragments of the paired-end Illumina NovaSeq data were demultiplexed, and the unmatched primer sequences were discarded using the QIIME cutadapt trim pairs. Amplicon sequence variants were determined using DADA2 (version 1.16) with quality filtering, denoising, merging, and chimeras removal.55 Each feature sequence of the amplicon sequence variants was compared with the Silva database (Release132, https://www.arb-silva.de) using the Naïve Bayes Classifier in the feature-classifier plugin. Shannon, Simpson, Chao 1, Goods_coverage, and observed_species indices were used to estimate the alpha (α)-diversity. p < 0.05 in the Kruskal–Wallis test was considered statistically significant. PCoA of the Bray–Curtis analysis was used to demonstrate beta (β)-diversity among different groups. LefSe was used to identify differentially abundant taxa across groups.56 The random forest algorithm was used for sample classification of microbial communities to demonstrate the importance ranking of species.
LC-MS analysis
For cecal contents metabolism, 100 mg samples were mixed with 2-chlorophenylalanine (4 ppm) methanol (0.6 mL, -20°C), vortexed for 30 s, ground for 90 s at 60 Hz, ultrasonicated for 10 min, centrifuged for 10 min at 12000 rpm (4°C), and filtered to obtain samples for LC-MS.57 For liver metabolism, 100 mg samples were added to 1 mL tissue extract (75% 9:1 methanol: chloroform, 25% ddH2O) (-20°C), ground for 60 s at 50 Hz (twice), ultrasonicated for 30 min, settled on ice for 30 min, and centrifuged for 10 min at 12000 rpm (4°C). The supernatant samples (650 μL) were concentrated to dryness in a vacuum and dissolved in 2-chlorobenzalanine (4 ppm) 50% acetonitrile solution (200 μL, -20°C). The supernatants were then filtered through a 0.22-μm membrane to obtain samples for LC-MS.58 Quality control samples were constructed with 20 μL of each prepared sample to monitor deviations and errors.59 For chromatographic separation, an ACQUITY UPLC® HSS T3 (150×2.1 mm, 1.8 μm, Waters) column was used on the Ultimate 3000 system (Thermo Fisher Scientific, USA). Electrospray ionization-tandem mass spectrometry tests were performed with spray voltages of 3.5 kV (positive mode) and −2.5 kV (negative mode) on a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, USA). Raw data were assessed using Proteowizard software (v3.0.8789) and the XCMS package in R (v3.3.2). After the peak intensity was normalized in batches, mass-to-charge ratio (m/z) and retention time (rt) were used for relative quantification analysis. To profile the metabolic variousness, PCA and OPLS-DA were performed using the ropls package in R (v3.3.2). Differential metabolites with biological significance were detected according to p-values in two-tailed Student’s t-tests and VIP scores of the OPLS-DA model. Group differences in metabolites were screened using the standard of p-value ≤ 0.05 and VIP ≥ 1. Thereafter, these metabolites were identified using the Human Metabolome Database (HMDB), Metlin, Massbank, and LipidMap databases. Cluster analyses using the pheatmap package in R and KEGG pathway analysis were conducted to further explore the functions of differential metabolites.60
Quantification and statistical analysis
All data are presented as the mean ± standard deviation. GraphPad Prism version 8.0 (GraphPad Software) was used for data analysis. The independent sample t-test or Mann–Whitney test was used for comparison between two groups, one-way ANOVA or Kruskal–Wallis test was used for comparison among multiple groups, and correlations were evaluated by Spearman's rank correlation analysis. p < 0.05 was considered statistically significant.
Acknowledgments
The present study was supported by National Natural Science Foundation of China (No. 82270979 and 81970939), Jiangsu Province Key Research and Development Program (No. BE2022670), Nanjing Science and Technology Development Plan (No. 2019060009).
We thank Dr. Bin Chen and Junjie Zhao, Department of Periodontology, Nanjing Stomatological Hospital, Medical School of Nanjing University (Nanjing, China), for their help on the collection of saliva samples. We thank Personal Biotechnology Company (Shanghai, China) for providing 16S rRNA and LC-MS sequencing.
Author contributions
M. W. and L.L. contributed to conception, design, data acquisition, analysis, and interpretation, drafted and revised the manuscript; J.Q. and N.W. contributed to data acquisition and interpretation, revised the manuscript; J.B., J.L., Y.L., and F.C. contributed to data acquisition and analysis, revised the manuscript; Y.Z. and F.Y. contributed to conception, design, data interpretation, and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106346.
Contributor Information
Yangheng Zhang, Email: zhangyhdent@nju.edu.cn.
Fuhua Yan, Email: yanfh@nju.edu.cn.
Supplemental information
Data and code availability
-
•
The datasets supporting the conclusions of this article have been deposited at the Sequence Read Archive (SRA) repository (https://www.ncbi.nlm.nih.gov/) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
No new code was generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Powell E.E., Wong V.W.S., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–2224. doi: 10.1016/s0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 2.Polyzos S.A., Kountouras J., Mantzoros C.S. Obesity and nonalcoholic fatty liver disease: from pathophysiology to therapeutics. Metabolism. 2019;92:82–97. doi: 10.1016/j.metabol.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z., Tacke F., Arrese M., Chander Sharma B., Mostafa I., Bugianesi E., Wai-Sun Wong V., Yilmaz Y., George J., Fan J., Vos M.B. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 4.Simon T.G., Roelstraete B., Khalili H., Hagström H., Ludvigsson J.F. Mortality in biopsy-confirmed nonalcoholic fatty liver disease: results from a nationwide cohort. Gut. 2021;70:1375–1382. doi: 10.1136/gutjnl-2020-322786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slots J. Periodontitis: facts, fallacies and the future. Periodontol. 2000. 2017;75:7–23. doi: 10.1111/prd.12221. [DOI] [PubMed] [Google Scholar]
- 6.Shin H.S., Hong M.H., Moon J.Y., Sim S.J. Periodontal disease could be a potential risk factor for non-alcoholic fatty liver disease: an 11-year retrospective follow-up study. Clin. Oral Investig. 2022;26:5503–5514. doi: 10.1007/s00784-022-04518-6. [DOI] [PubMed] [Google Scholar]
- 7.Kuroe K., Furuta M., Takeuchi K., Takeshita T., Suma S., Shinagawa T., Shimazaki Y., Yamashita Y. Association between periodontitis and fibrotic progression of non-alcoholic fatty liver among Japanese adults. J. Clin. Periodontol. 2021;48:368–377. doi: 10.1111/jcpe.13415. [DOI] [PubMed] [Google Scholar]
- 8.Nakahara T., Hyogo H., Ono A., Nagaoki Y., Kawaoka T., Miki D., Tsuge M., Hiraga N., Hayes C.N., Hiramatsu A., et al. Involvement of Porphyromonas gingivalis in the progression of non-alcoholic fatty liver disease. J. Gastroenterol. 2018;53:269–280. doi: 10.1007/s00535-017-1368-4. [DOI] [PubMed] [Google Scholar]
- 9.Yoneda M., Naka S., Nakano K., Wada K., Endo H., Mawatari H., Imajo K., Nomura R., Hokamura K., Ono M., et al. Involvement of a periodontal pathogen, Porphyromonas gingivalis on the pathogenesis of non-alcoholic fatty liver disease. BMC Gastroenterol. 2012;12:16. doi: 10.1186/1471-230x-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuraji R., Sekino S., Kapila Y., Numabe Y. Periodontal disease-related nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: an emerging concept of oral-liver axis. Periodontol. 2000. 2021;87:204–240. doi: 10.1111/prd.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao J., Li L., Zhang Y., Wang M., Chen F., Ge S., Chen B., Yan F. Periodontitis may induce gut microbiota dysbiosis via salivary microbiota. Int. J. Oral Sci. 2022;14:32. doi: 10.1038/s41368-022-00183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Bao J., Chang Y., Wang M., Chen B., Yan F. Gut microbiota may mediate the influence of periodontitis on prediabetes. J. Dent. Res. 2021;100:1387–1396. doi: 10.1177/00220345211009449. [DOI] [PubMed] [Google Scholar]
- 13.Qian J., Lu J., Huang Y., Wang M., Chen B., Bao J., Wang L., Cui D., Luo B., Yan F. Periodontitis salivary microbiota worsens colitis. J. Dent. Res. 2022;101:559–568. doi: 10.1177/00220345211049781. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita T., Kageyama S., Furuta M., Tsuboi H., Takeuchi K., Shibata Y., Shimazaki Y., Akifusa S., Ninomiya T., Kiyohara Y., Yamashita Y. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Sci. Rep. 2016;6 doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamoto S., Nagao-Kitamoto H., Jiao Y., Gillilland M.G., 3rd, Hayashi A., Imai J., Sugihara K., Miyoshi M., Brazil J.C., Kuffa P., et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182:447–462.e14. doi: 10.1016/j.cell.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behary J., Amorim N., Jiang X.T., Raposo A., Gong L., McGovern E., Ibrahim R., Chu F., Stephens C., Jebeili H., et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 2021;12:187. doi: 10.1038/s41467-020-20422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han H., Jiang Y., Wang M., Melaku M., Liu L., Zhao Y., Everaert N., Yi B., Zhang H. Intestinal dysbiosis in nonalcoholic fatty liver disease (NAFLD): focusing on the gut-liver axis. Crit. Rev. Food Sci. Nutr. 2021:1–18. doi: 10.1080/10408398.2021.1966738. [DOI] [PubMed] [Google Scholar]
- 18.Moyer B.J., Rojas I.Y., Kerley-Hamilton J.S., Hazlett H.F., Nemani K.V., Trask H.W., West R.J., Lupien L.E., Collins A.J., Ringelberg C.S., et al. Inhibition of the aryl hydrocarbon receptor prevents Western diet-induced obesity. Model for AHR activation by kynurenine via oxidized-LDL, TLR2/4, TGFβ, and Ido1. Toxicol. Appl. Pharmacol. 2016;300:13–24. doi: 10.1016/j.taap.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aron-Wisnewsky J., Vigliotti C., Witjes J., Le P., Holleboom A.G., Verheij J., Nieuwdorp M., Clément K. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020;17:279–297. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 20.Kitamoto S., Nagao-Kitamoto H., Hein R., Schmidt T.M., Kamada N. The bacterial connection between the oral cavity and the gut diseases. J. Dent. Res. 2020;99:1021–1029. doi: 10.1177/0022034520924633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X.Y., Chen J., Yi K., Peng L., Xie J., Gou X., Peng T., Tang L. Phlorizin ameliorates obesity-associated endotoxemia and insulin resistance in high-fat diet-fed mice by targeting the gut microbiota and intestinal barrier integrity. Gut Microb. 2020;12:1–18. doi: 10.1080/19490976.2020.1842990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postler T.S., Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metabol. 2017;26:110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan S., Ding Y., Saedi N., Choi M., Sridharan G.V., Sherr D.H., Yarmush M.L., Alaniz R.C., Jayaraman A., Lee K. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 2018;23:1099–1111. doi: 10.1016/j.celrep.2018.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondanelli G., Albini E., Orecchini E., Pallotta M.T., Belladonna M.L., Ricci G., Grohmann U., Orabona C. Pathogenetic interplay between IL-6 and tryptophan metabolism in an experimental model of obesity. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.713989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z., Lei H., Chen G., Yuan P., Cao Z., Ser H.L., Zhu X., Wu F., Liu C., Dong M., et al. Impaired intestinal Akkermansia muciniphila and aryl hydrocarbon receptor ligands contribute to nonalcoholic fatty liver disease in mice. mSystems. 2021;6 doi: 10.1128/mSystems.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natividad J.M., Agus A., Planchais J., Lamas B., Jarry A.C., Martin R., Michel M.L., Chong-Nguyen C., Roussel R., Straube M., et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metabol. 2018;28:737–749.e4. doi: 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Ahn J.S., Yang J.W., Oh S.J., Shin Y.Y., Kang M.J., Park H.R., Seo Y., Kim H.S. Porphyromonas gingivalis exacerbates the progression of fatty liver disease via CD36-PPARγ pathway. BMB Rep. 2021;54:323–328. doi: 10.5483/BMBRep.2021.54.6.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuraji R., Ito H., Fujita M., Ishiguro H., Hashimoto S., Numabe Y. Porphyromonas gingivalis induced periodontitis exacerbates progression of non-alcoholic steatohepatitis in rats. Clin. Exp. Dent. Res. 2016;2:216–225. doi: 10.1002/cre2.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freire M., Nelson K.E., Edlund A. The oral host-microbial interactome: an ecological chronometer of health? Trends Microbiol. 2021;29:551–561. doi: 10.1016/j.tim.2020.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Ipsen D.H., Lykkesfeldt J., Tveden-Nyborg P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018;75:3313–3327. doi: 10.1007/s00018-018-2860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyström E.E.L., Martinez-Abad B., Arike L., Birchenough G.M.H., Nonnecke E.B., Castillo P.A., Svensson F., Bevins C.L., Hansson G.C., Johansson M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science. 2021;372 doi: 10.1126/science.abb1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpino G., Del Ben M., Pastori D., Carnevale R., Baratta F., Overi D., Francis H., Cardinale V., Onori P., Safarikia S., et al. Increased liver localization of lipopolysaccharides in human and experimental NAFLD. Hepatology. 2020;72:470–485. doi: 10.1002/hep.31056. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S.S., Wang J., Yannie P.J., Ghosh S. Intestinal barrier dysfunction, LPS translocation, and disease development. J. Endocr. Soc. 2020;4:bvz039. doi: 10.1210/jendso/bvz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazankov K., Jørgensen S.M.D., Thomsen K.L., Møller H.J., Vilstrup H., George J., Schuppan D., Grønbæk H. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 2019;16:145–159. doi: 10.1038/s41575-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 35.Chopyk D.M., Grakoui A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology. 2020;159:849–863. doi: 10.1053/j.gastro.2020.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferro D., Baratta F., Pastori D., Cocomello N., Colantoni A., Angelico F., Del Ben M. New insights into the pathogenesis of non-alcoholic fatty liver disease: gut-derived lipopolysaccharides and oxidative stress. Nutrients. 2020;12 doi: 10.3390/nu12092762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazier W., Le Corf K., Martinez C., Tudela H., Kissi D., Kropp C., Coubard C., Soto M., Elustondo F., Rawadi G., Claus S.P. A new strain of Christensenella minuta as a potential biotherapy for obesity and associated metabolic diseases. Cells. 2021;10 doi: 10.3390/cells10040823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen F., Zheng R.D., Sun X.Q., Ding W.J., Wang X.Y., Fan J.G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2017;16:375–381. doi: 10.1016/s1499-3872(17)60019-5. [DOI] [PubMed] [Google Scholar]
- 39.Wang K., Lv L., Yan R., Wang Q., Jiang H., Wu W., Li Y., Ye J., Wu J., Yang L., et al. Bifidobacterium longum R0175 protects rats against d-galactosamine-induced acute liver failure. mSphere. 2020;5 doi: 10.1128/mSphere.00791-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y., Wu D., Zeng W., Chen Y., Guo M., Lu B., Li H., Sun C., Yang L., Jiang X., Gao Q. The role of intestinal dysbacteriosis induced arachidonic acid metabolism disorder in inflammaging in atherosclerosis. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.618265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Derrien M., Belzer C., de Vos W.M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Mouries J., Brescia P., Silvestri A., Spadoni I., Sorribas M., Wiest R., Mileti E., Galbiati M., Invernizzi P., Adorini L., et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 2019;71:1216–1228. doi: 10.1016/j.jhep.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder B.O., Birchenough G.M.H., Ståhlman M., Arike L., Johansson M.E.V., Hansson G.C., Bäckhed F. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 2018;23:27–40.e7. doi: 10.1016/j.chom.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagano J., Shimizu M., Hara T., Shirakami Y., Kochi T., Nakamura N., Ohtaki H., Ito H., Tanaka T., Tsurumi H., et al. Effects of indoleamine 2,3-dioxygenase deficiency on high-fat diet-induced hepatic inflammation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cussotto S., Delgado I., Anesi A., Dexpert S., Aubert A., Beau C., Forestier D., Ledaguenel P., Magne E., Mattivi F., Capuron L. Tryptophan metabolic pathways are altered in obesity and are associated with systemic inflammation. Front. Immunol. 2020;11:557. doi: 10.3389/fimmu.2020.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun M., Ma N., He T., Johnston L.J., Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR) Crit. Rev. Food Sci. Nutr. 2020;60:1760–1768. doi: 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- 48.Huang T., Song J., Gao J., Cheng J., Xie H., Zhang L., Wang Y.H., Gao Z., Wang Y., Wang X., et al. Adipocyte-derived kynurenine promotes obesity and insulin resistance by activating the AhR/STAT3/IL-6 signaling. Nat. Commun. 2022;13:3489. doi: 10.1038/s41467-022-31126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas I.Y., Moyer B.J., Ringelberg C.S., Wilkins O.M., Pooler D.B., Ness D.B., Coker S., Tosteson T.D., Lewis L.D., Chamberlin M.D., Tomlinson C.R. Kynurenine-induced aryl hydrocarbon receptor signaling in mice causes body mass gain, liver steatosis, and hyperglycemia. Obesity. 2021;29:337–349. doi: 10.1002/oby.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossner P., Jr., Libalova H., Vrbova K., Cervena T., Rossnerova A., Elzeinova F., Milcova A., Novakova Z., Topinka J. Genotoxicant exposure, activation of the aryl hydrocarbon receptor, and lipid peroxidation in cultured human alveolar type II A549 cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2020;853 doi: 10.1016/j.mrgentox.2020.503173. [DOI] [PubMed] [Google Scholar]
- 51.Sztolsztener K., Chabowski A., Harasim-Symbor E., Bielawiec P., Konstantynowicz-Nowicka K. Arachidonic acid as an early indicator of inflammation during non-alcoholic fatty liver disease development. Biomolecules. 2020;10 doi: 10.3390/biom10081133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atarashi K., Suda W., Luo C., Kawaguchi T., Motoo I., Narushima S., Kiguchi Y., Yasuma K., Watanabe E., Tanoue T., et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science. 2017;358:359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 54.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng J.S.Y., Ryan U., Trengove R.D., Maker G.L. Development of an untargeted metabolomics method for the analysis of human faecal samples using Cryptosporidium-infected samples. Mol. Biochem. Parasitol. 2012;185:145–150. doi: 10.1016/j.molbiopara.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Want E.J., Masson P., Michopoulos F., Wilson I.D., Theodoridis G., Plumb R.S., Shockcor J., Loftus N., Holmes E., Nicholson J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 59.Sangster T., Major H., Plumb R., Wilson A.J., Wilson I.D. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst. 2006;131:1075–1078. doi: 10.1039/b604498k. [DOI] [PubMed] [Google Scholar]
- 60.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D.S., Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets supporting the conclusions of this article have been deposited at the Sequence Read Archive (SRA) repository (https://www.ncbi.nlm.nih.gov/) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
No new code was generated in this study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.