Abstract
We report here a novel method to simultaneously detect CpG methylation and single nucleotide polymorphisms (SNPs) using denaturing high performance liquid chromatography (DHPLC). PCR products of bisulfite-modified CpG islands were separated using DHPLC. BstUI digestion and DNA sequencing were used in confirmation studies. Consistent with the BstUI digestion assay, the 294 bp PCR product of the modified hMLH1 promoter showed different retention times between the methylated cell lines (RKO and Cla, 6.7 min) and the unmethylated cell lines (PACM82 and MGC803, 6.2 min). No hMLH1 methylation was observed in 13 primary gastric carcinomas and their matched normal tissues. One hMLH1 SNP was detected in gastric cancer patients, in both cancer and normal tissues. DNA sequencing revealed that the SNP is a G→A variation at –93 nt of the hMLH1 promoter. A two-peak chromatogram was also obtained in the 605 bp PCR product of the Cox-2 promoter of the AGS, HEK293 and MKN45 cell lines by DHPLC. Another peak corresponding to methylated CpG islands was observed on the chromatogram of the Cox-2-methylated AGS cell line after bisulfite treatment. In conclusion, methylation in homoallelic and heteroallelic CpG islands could be detected rapidly and reliably by bisulfite–DHPLC. A SNP in the target sequence could also be detected at the same time.
INTRODUCTION
Most housekeeping genes and 40% of tissue-specific genes contain CpG islands in their promoter regions (1). Methylation of such CpG islands is associated with transcription silencing and gene imprinting (2). Many tumor suppressor genes are down-regulated by promoter methylation during the development and progression of cancer (3), thus detection of CpG methylation is important to our understanding of cancer gene regulation.
Most current approaches to methylation detection use sequence-specific DNA methylation assays, including Southern blotting–restriction fragment length polymorphism coupled with methylation-sensitive enzymes, the combined bisulfite restriction assay (COBRA), methylation-specific PCR (MSP) and the MethyLight assay (4–7). These approaches depend on methylation of specific nucleotides located at cleavage sites or primer/probe mating regions. Their efficiency and specificity are influenced by the extent of methylation of CpGs in these regions. Methylation of CpGs in other regions of target sequences is undetectable using such methods. Although sequencing combined with cloning detects CpG methylation accurately, it is costly and not suitable for analysis of a number of samples. Thus, an analysis which overcomes these pitfalls of current methods for analysis of CpG methylation is currently a pressing issue.
Here we report in detail a bisulfite–denaturing high performance liquid chromatography (DHPLC) method for simultaneous detection of methylation of CpG islands and single nucleotide polymorphisms (SNPs) by DHPLC (Fig. 1), based on our previous pilot methylation study in 2000 (8). This assay detected all CpG methylation in a target sequence and is not influenced by the extent of CpG methylation.
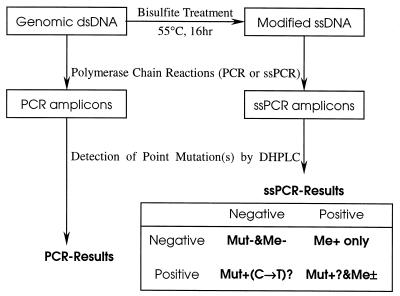
Figure 1.
Scheme of the detection strategy for CpG methylation and point mutations by DHPLC. The heteroduplexed ssPCR products of the bisulfite-treated templates were analyzed by DHPLC at the PDT of the unmethylated templates. The mutation chromatogram of ssPCR products may result from CpG methylation or genomic point mutation or both. Therefore, the presence or absence of a point mutation(s) in the PCR products of the untreated templates should be analyzed. Mut, point mutation; Me, CpG methylation; –, negative; +, positive; ±, positive or negative.
MATERIALS AND METHODS
Cell lines, DNA preparation and bisulfite modification
Human colon carcinoma cell lines RKO and Cla (from the University of California, San Francisco, CA), human gastric carcinoma cell lines PACM82, MGC803, MKN45 and AGS and the human embryonic kidney cell line HEK293 were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Thirteen pairs of primary gastric carcinoma and corresponding normal tissue samples were collected surgically at Beijing Cancer Hospital. Genomic DNA from these cells in log phase and tissue samples was isolated using phenol/chloroform as previously described (9). One microgram of genomic DNA was treated with sodium bisulfite as described (6) and the Wizard DNA Clean-Up System Kit (A7280; Promega), subsequently used prior to PCR amplification. During this modification unmethylated C residues are converted to U (T in PCR products), while methylated C residues are not. Thus the sense and antisense sequences no longer match each other.
Design of primers and PCR conditions
Primers were designed according to the CpG islands of the sense strand of the hMLH1 (GenBank accession no. U83845, gi:2511457) and Cox-2 (GenBank accession no. AF276953, gi:9230774) genes. Exactly the same sequence areas were amplified from the templates with and without bisulfite treatment. The strand-specific primers for the treated CpG islands were hMLH1-mF (5′-gtatttttgtttttattggttggata-3′), hMLH1-mR (5′-aataccttcaaccaatcacctcaata-3′), Cox2-mF (5′-tttggagaggaagttaagtgttt-3′) and Cox2-mR (5′-gccaaatactcacctatataacta-3′). These primers for the modified templates amplify both methylated and unmethylated ones, because no CpG sites exist in the primer sequences. Primers for the templates without bisulfite treatment were hMLH1-wF (5′-gcatctctgctcctattggctggata-3′), hMLH1-wR (5′-agtgccttcagccaatcacctcagtg-3′), Cox2-wF (5′-cctggagaggaagccaagtgtcc-3′) and Cox2-wR (5′-gccaggtactcacctgtatggctg-3′).
A PTC-200 DNA Engine (MJ Research) was used. Regular PCR (35 cycles of 95°C for 40 s, 65°C for 75 s and 72°C for 75 s) was used to amplify Cox-2 without bisulfite treatment. Hot-start touchdown PCR (35 cycles, 72 to 58°C, –1.0°C per cycle) was used to amplify hMLH1 without bisulfite treatment and the sense strand templates with bisulfite treatment [strand-specific PCR (ssPCR) 65 to 45°C for hMLH1, 57 to 47°C for Cox-2]. The other conditions in the touchdown PCR (including ssPCR) were the same as for regular PCR. The PCR reaction mixture comprised 2.5 µl of Davis 10× buffer, 1.0 µl of 25 mM dNTPs, 1.3 µl of DMSO, 2.0 µl of 50 mM MgCl2, 16 µl of H2O, 0.3 µl of DNA polymerase 5 (U/µl) and 0.5 µl of forward and reversed primers (350 ng/µl). Taq DNA polymerase (Biostar, Canada) (added to the PCR reaction held at 95°C) and platinum-Taq polymerase (Gibco) were used to amplify hMLH1 and Cox-2, respectively. The hMLH1 and Cox-2 PCR products were separated in 1.5 and 2.5% agarose gels, respectively.
BstUI COBRA analysis for methylation
The bisulfite-modified and unmodified hMLH1 templates were amplified by ssPCR and PCR as described above. One microgram of PCR products was digested with 5 U BstUI (New England Biolabs) in 30 µl total volume at 60°C for 3 h. A 2.5% agarose gel was used to separate the digested fragments.
Analysis for methylation and SNP by DHPLC
Detection of point mutations in PCR products and methylation in ssPCR products was performed by DHPLC on a WAVE DNA Fragment Analysis System (Transgenomic). PCR and ssPCR products were introduced into the mobile phase at an injection volume of 5 µl by the autosampler. A DNASep analytical column (Transgenomic) was used as the solid phase and the products were eluted from the column with a binary gradient of 0.1 M triethylammonium acetate (TEAA) and 0.1 M TEAA/25% acetonitrile as the mobile phase at a flow rate of 0.9 ml/min. Elution gradients and analysis temperatures were automatically predicted from the target sequences by WAVEMaker 4.0 software (Transgenomic) and eluted products detected by UV analysis at 260 nm. Non-denaturing analysis was conducted at 48°C and partially denaturing analyses were conducted at the temperatures predicted by WAVEMaker.
Sequencing of the hMLH1 and Cox-2 promoters
The PCR and ssPCR products obtained using the above primers were purified by electrophoresis on a 1.5% agarose gel and eluted with a QIAquick gel extraction kit (Qiagen). The eluted DNA was mixed with primer and sequenced on a Perkin-Elmer ABI PRISM 3700 DNA Analyzer with Big Dye terminators.
RESULTS AND DISCUSSIONS
Efficiency and specificity of PCR were not influenced by the extent of methylation of CpG
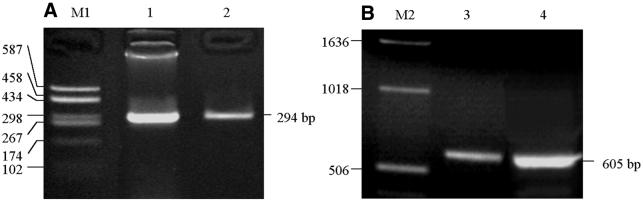
In order to optimize the PCR and DHPLC conditions for analysis of CpG islands, a single 294 bp fragment of the hMLH1 promoter was obtained after PCR amplification in each of the cell lines, with and without sodium bisulfite treatment (Fig. 2A). A single 605 bp fragment of the Cox2 promoter was also obtained (Fig. 2B). We observed that both the PCR and ssPCR products of these genes resulted in a single peak chromatogram on DHPLC DNA sizing analysis at 48°C (Fig. 3A), indicating that the quantity and the quality of the two kinds of PCR products met the requirements for mutation detection by DHPLC. Because of the high A/T content in amplicons of bisulfite-treated templates, 48 instead of 50°C should be used in double-stranded DNA fragment sizing by DHPLC.
Figure 2.
PCR amplification of CpG islands. (A) 294 bp human hMLH1 promoter from colon cancer cell line RKO; (B) 605 bp human Cox-2 promoter from gastric cancer cell line MKN45. M1, puC18 DNA/HeaIII marker; M2, 1 kb DNA ladder (Gibco no. 15615-016); lanes 1 and 4, untreated DNA; lanes 2 and 3, bisulfite-modified template.
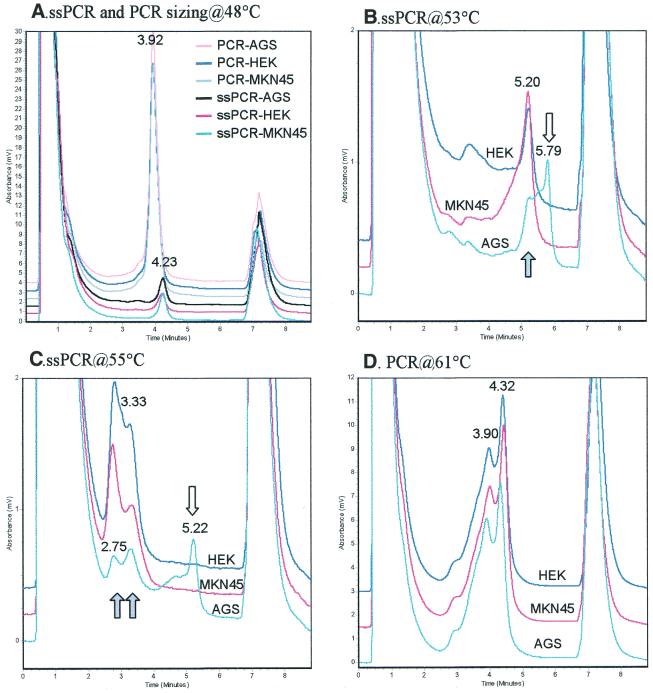
Figure 3.
DHPLC chromatograms of ssPCR and PCR products of CpG islands in the Cox-2 promoter. The PCR products were heteroduplexed, then sized at 48°C and detected at the PDT. Open arrow, methylated fragments/peaks; closed arrow, ummethylated fragments/peaks. (A) Sizing of Cox2 PCR and ssPCR products from the AGS, HEK and MKN45 cell lines at 48°C. (B and C) Cox-2 ssPCR products detected at 53 and 55°C, the PDT for the unmethylated and methylated templates, respectively. (D) Cox-2 PCR products detected at 61°C.
Because the extent of methylation at various CpG sites of most genes is unknown, it is hard to design good MSP primers or MethyLight probes for methylated templates, which require full methylation at all CpG sites in their mating regions. Unlike MSP and MethyLight, the efficiency and specificity of ssPCR for bisulfite-modified templates are not influenced by the extent of methylation of CpGs, because there are no CpG sites in the primer sequences. Furthermore, these kinds of primers can amplify both methylated and unmethylated templates. This advantage is useful for detection of methylation of heterozygous templates from tissue samples.
Detection of ssPCR products of methylated and unmethylated CpG islands by DHPLC
It has been reported that hMLH1 is silenced by CpG methylation in RKO and Cla colon cancer cell lines (10). We found that the ssPCR products of the bisulfite-modified promoter of the homozygous hMLH1 gene with 15 CpG sites from RKO and Cla colon cancer cell lines were completely separated from those of gastric cancer cell lines PACM82 and MGC803 at 53 and 55°C, the partial denaturing temperature (PDT) of the unmethylated and methylated templates predicted by the WAVEMaker4.0 software. The retention time was 6.2 min for the gastric cancer cell lines and 6.7 min for the colon cancer cell lines. These results suggest that the hMLH1 promoter in the two colon cancer cell lines is methylated, while in PACM82 and MGC803 it is not, since methylation can protect against the conversion of C to U and then to T in ssPCR and maintains a higher C/G content after bisulfite treatment (11), leading to a higher denaturing temperature and a delayed retention time. This conclusion was consistent with the results of the BstUI COBRA analysis below.
Both heteroduplex and homoduplex peaks could be observed in a typical DHPLC chromatogram of the mutant PCR products at the PDT after mixing and hybridization with wild-type amplicons (12). As on the chromatogram of the hybridized mixture of ssPCR products, only two peaks were observed on the chromatogram of the unhybridized ssPCR mixture of bisulfite-modified hMLH1 of the RKO and PACM82 cell lines. This phenomenon suggests no obvious formation of heteroduplexes from the mixed ssPCR products of these two cell lines through hybridization. A total of 15 CpG sites in the target sequence might account for the predominant formation of homoduplexes, because of the existence of 15 ‘mutations’ in the 294 bp sequence: conversion of C to T in the unmethylated CpG sites and no conversion in the methylated ones following bisulfite modification. The widespread multi-mutations might completely inhibit the formation of heteroduplexes between the methylated and unmethylated amplicons. Therefore, the detection of methylated and unmethylated amplicons by DHPLC would not interfere with each other. From this point of view, the mechanism of detection of CpG methylation (multi-mutations) is different from that of point mutations. The retention times were the same (5.5 min at 62°C) for the hybridized PCR products of the templates without bisulfite treatment from the RKO and PACM82 cell lines and their mixture. These indicate that no point mutations exist in the target sequence of these cell lines. Therefore, methylation must be the only reason accounting for the delayed retention time of the PCR product of the bisulfite-modified templates from RKO cells (Fig. 1, Me+ only).
Similar results were obtained in the more complex case of the heterozygous Cox-2 gene by DHPLC analysis. The one-peak chromatograms (3.9 min for PCR products or 4.2 min for ssPCR products) of the AGS, MKN45 and HEK293 cell lines were the same at 48°C, the non-denaturing temperature (Fig. 3A). Two peaks (2.8 and 3.3 min) were detected in the hybridized ssPCR products of the three cell lines at 55°C, the PDT of the methylated templates. However, an extra peak (5.2 min) was observed on the chromatogram of the heteroduplexed ssPCR products of the AGS cell line (Fig. 3C). This result suggests that the Cox-2 ssPCR product from the AGS cell line is different from those of MKN45 and HEK293. Because identical two-peak chromatograms (3.9 and 4.3 min) were obtained from all three of the tested cell lines (Fig. 3D), these results suggest that the genomic sequences of the PCR products of Cox-2 in all three of the cell lines are the same and that the extra peak on the chromatogram of the ssPCR product of the AGS cell line is the result of CpG methylation in Cox-2, while Cox-2 in MKN45 and HEK293 is not methylated. This conclusion is supported by the results of DNA sequencing; a number of CpG sites were included in the Cox-2 ssPCR products of the AGS cell line, but not in the MKN45 cell line. This is also consistent with the report that Cox-2 is unmethylated in the MKN45 cell line and hemimethylated in the AGS cell line (13). Moreover, a two-peak mutation chromatogram was not detectable in the MKN45 and HEK293 cell lines at 53°C, the PDT of the unmethylated templates, but the extra peak was still detectable in the AGS cell line (Fig. 3C). These results indicate that CpG methylation is detectable at both PDTs.
There are many CpG sites in the ssPCR amplicons of CpG islands. Methylation of multiple CpGs will lead to a delay of the ssPCR product in DHPLC chromatograms. Thus, DHPLC can analyze methylation at multiple CpG sites between primers, an advantage over existing methods for methylation analysis, such as methylation-sensitive enzyme digestion, COBRA, MSP, MethyLight, etc., which detect only methylation at a specific site(s). In conclusion, the methylated and unmethylated ssPCR products of both homoallele (hMLH1) and heteroallele (Cox-2) CpG islands could be unambiguously separated by DHPLC, making it amenable for high throughput, routine use.
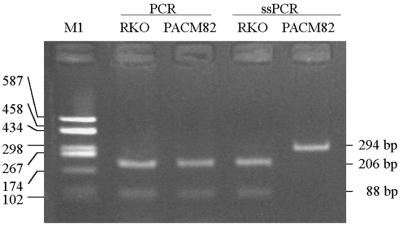
Comparison with BstUI COBRA analysis
Two fragments (88 and 206 bp) were obtained from the 294 bp PCR products of each sample in the BstUI COBRA analysis, since the hMLH1 promoter has a BstUI cleavage site (CGCG) that is fully methylated in the hMLH1-silenced cell lines and unmethylated in the hMLH1-expressing cell lines (10). Therefore, the ssPCR product of the methylated hMLH1 promoter must be sensitive to BstUI cleavage and that of the unmethylated one resistant, because of inhibition of the conversion of C to T by methylation. We observed that the ssPCR product from cell line RKO was sensitive to BstUI cleavage, but that from cell line PACM82 was resistant (Fig. 4). These results indicate that the hMLH1 promoter in RKO cells is methylated, as previously reported (10), and that in PACM82 cells is unmethylated. These results are consistent with those of the DHPLC assay.
Figure 4.
Detection of methylation in the hMLH1 promoter by BstUI COBRA assay in the RKO and PACM82 cell lines. DNA without or with bisulfite treatment was amplified by PCR or ssPCR. The amplicon mixture was digested with BstUI at 60°C for 3 h. M1, PCR and ssPCR as in Figure 1.
Application of DHPLC to simultaneously detect methylation and SNP in human gastric tissues
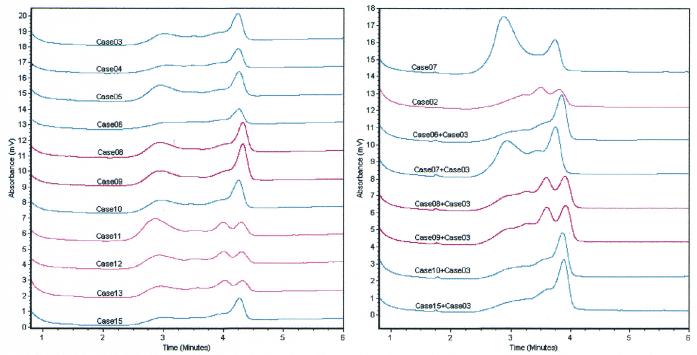
Methylation of CpG islands in the hMLH1 promoter was analyzed in 13 gastric carcinomas and corresponding normal gastric mucosal samples using DHPLC and COBRA analysis. No methylation was observed among the 13 pairs of specimens. Two-peak chromatograms were detected at 53°C in both PCR and ssPCR products of 4 of the 13 (30.8%) pairs of specimens (cases 02 and 11–13) after the products were self-hybridized (Fig. 5). Only one peak could be observed in these four samples at the non-denaturing temperature of 48°C. This indicates that the double-stranded DNA fragment size in the tested samples was the same. The CpG islands in hMLH1 in these four cases must be in heteroalleles (Fig. 1, Mut+&Me–). DNA sequencing was also used to identify the SNP in three samples with two chromatographic peaks (cases 11–13) and three samples with only one peak (cases 03–05). The results showed that all three sequenced samples with two peaks contained a G/A polymorphism at –93 nt of hMLH1 and those with one peak were A/A homozygous.
Figure 5.
Chromatograms of PCR products of the hMLH1 promoter in 13 normal gastric mucosa samples detected by DHPLC at the partial denaturing temperature, 62°C. Each PCR product or the mixture was hybridized (held at 95°C for 5 min and then decreasing gradually by –1°C/min to room temperature).
Then, the PCR products of case 03 (A/A homozygous) were hybridized with the PCR products of the remaining six cases with one peak to look for possible homozygous G/G samples by DHPLC assay. A two-peak chromatogram was observed in two samples (cases 08 and 09) after hybridization with the PCR products of case 03 (Fig. 5). Because the G/A variant in gastric carcinoma samples also exists in the corresponding normal gastric mucosa samples, allelic loss of the hMLH1 gene appears unlikely. Therefore, cases 08 and 09 (15.4%) should be G/G homozygous, and this was confirmed by the results of sequencing (data not shown). These results are also consistent with the report that there is a SNP at –93 nt of hMLH1 (14).
Baumer et al. used the same approach to detect gene imprinting (15). However, they did not exclude possible interference of a genomic point mutation or SNP on detection of CpG methylation by DHPLC. In the case of homoallelic CpG islands, only a single methylation analysis of the ssPCR products by DHPLC is required to detect CpG methylation for each sample. In the case of heteroalleles, however, a genomic point mutation in the templates without bisulfite treatment would be detected at the same time (see Fig. 1).
Conclusions
This bisulfite–DHPLC assay can be used to detect methylation and SNP in homoallelic and heteroallelic CpG islands in homogeneous and heterogeneous samples simultaneously and conveniently. That methylation at almost any CpG site in the PCR-amplified sequences could be detected shows the advantage of this novel method over others, which detect CpG methylation only at specific sites.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr W. Davis Parker (Department of Neurology, University of Virginia Hospital) for letting us use the DHPLC set to detect methylation of Cox-2 in his laboratory. This work was supported by grants from Peking University Center for Human Disease Genomics and Peking University Cancer Research Center, a grant from Beijing Laboratory of Molecular Oncology and a grant from the National Natural Science Foundation of China.
REFERENCES
- 1.Lewin B. (1997) Genes, 6th Edn. Oxford University Press, Oxford, UK.
- 2.Bird A. (1999) DNA methylation de novo. Science, 286, 2287–2288. [DOI] [PubMed] [Google Scholar]
- 3.Jones P.A. and Laird,P.W. (1999) Cancer epigenetics comes of age. Nature Genet., 21, 163–167. [DOI] [PubMed] [Google Scholar]
- 4.McClelland M. (1981) The effect of sequence specific DNA methylation restriction endonuclease cleavage. Nucleic Acids Res., 9, 5859–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Z. and Laird,P.W. (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res., 25, 2532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman J.G., Graff,J.R., Myöhänen,S., Nelkin,B.D. and Baylin,S.B. (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA, 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eads C.A., Danenberg,K.D., Kawakami,K., Saltz,L.B., Blake,C., Shibata,D., Danenberg,P.V. and Laird,P.W. (2000) MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res., 28, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng D.J., Deng,G.R., Zhou,J. and Xin,H.J. (2000) Detection of CpG methylations in human mismatch repair gene hMLH1 promoter by denaturing high-performance liquid chromatography (DHPLC). Chin. J. Cancer Res., 12, 171 and 191. [Google Scholar]
- 9.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Deng G., Chen,A., Hong,J., Chae,H.S. and Kim,Y. (1999) Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res., 59, 2029–2033. [PubMed] [Google Scholar]
- 11.Wang R.Y.H., Gehrke,C.W. and Ehrlich,M. (1980) Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res., 8, 4777–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao W. and Oefner,P.J. (2001) Denaturing high-performance liquid chromatography: a review. Hum. Mutat., 17, 439–474. [DOI] [PubMed] [Google Scholar]
- 13.Akhtar M., Cheng,Y., Magno,R.M., Ashktorab,H., Smoot,D.T., Meltzer,S.J. and Wilson,K.T. (2001) Promoter methylation regulates Helicobacter pylori-stimulated cyclooxygenase-2 expression in gastric epithelial cells. Cancer Res., 61, 2399–2403. [PubMed] [Google Scholar]
- 14.Ito E., Yanagisawa,Y., Iwahashi,Y., Suzuki,Y., Nagasaki,H., Akiyama,Y., Sugano,S., Yuasa,Y. and Maruyama,K. (1999) A core promoter and a frequent single-nucleotide polymorphism of the mismatch repair gene hMLH1. Biochem. Biophys. Res. Commun., 256, 488–494. [DOI] [PubMed] [Google Scholar]
- 15.Baumer A., Wiedemann,U., Hergersberg,M. and Schinzel,A. (2001) A novel MSP/DHPLC method for the investigation of the methylation status of imprinted genes enables the molecular detection of low cell mosaicisms. Hum. Mutat., 17, 423–430. [DOI] [PubMed] [Google Scholar]