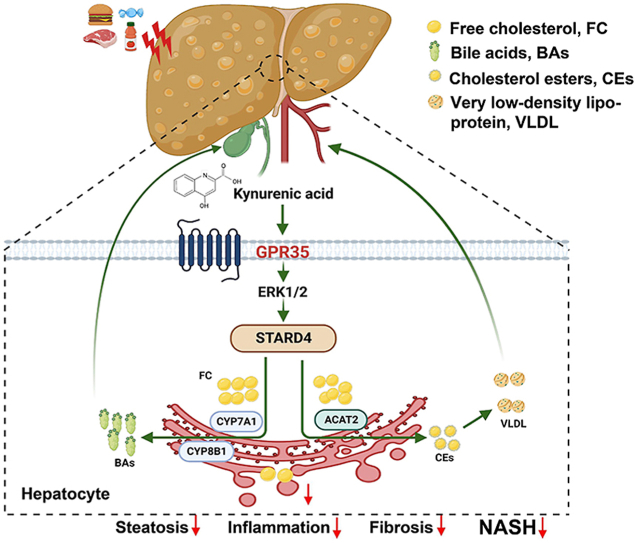
Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease worldwide. Fat accumulation “sensitizes” the liver to insult and leads to nonalcoholic steatohepatitis (NASH). G protein-coupled receptor 35 (GPR35) is involved in metabolic stresses, but its role in NAFLD is unknown. We report that hepatocyte GPR35 mitigates NASH by regulating hepatic cholesterol homeostasis. Specifically, we found that GPR35 overexpression in hepatocytes protected against high-fat/cholesterol/fructose (HFCF) diet-induced steatohepatitis, whereas loss of GPR35 had the opposite effect. Administration of the GPR35 agonist kynurenic acid (Kyna) suppressed HFCF diet-induced steatohepatitis in mice. Kyna/GPR35 induced expression of StAR-related lipid transfer protein 4 (STARD4) through the ERK1/2 signaling pathway, ultimately resulting in hepatic cholesterol esterification and bile acid synthesis (BAS). The overexpression of STARD4 increased the expression of the BAS rate-limiting enzymes cytochrome P450 family 7 subfamily A member 1 (CYP7A1) and CYP8B1, promoting the conversion of cholesterol to bile acid. The protective effect induced by GPR35 overexpression in hepatocytes disappeared in hepatocyte STARD4-knockdown mice. STARD4 overexpression in hepatocytes reversed the aggravation of HFCF diet-induced steatohepatitis caused by the loss of GPR35 expression in hepatocytes in mice. Our findings indicate that the GPR35–STARD4 axis is a promising therapeutic target for NAFLD.
Key words: G protein-coupled receptor 35, Kynurenic acid, Steatohepatitis, Cholesterol, Bile acid, STARD4, ACAT2, CYP7A1
Graphical abstract
kynurenic acid and G protein-coupled receptor 35 (GPR35) play critical protective roles in nonalcoholic steatohepatitis (NASH) through star-associated lipid transfer protein 4 (STARD4)-mediated hepatocyte cholesterol homeostasis.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is a series of liver diseases ranging from asymptomatic steatosis (nonalcoholic fatty liver, NAFL) to nonalcoholic steatohepatitis (NASH). NASH is characterized by varying degrees of inflammation and fibrosis in the liver1,2. Central obesity, diabetes mellitus (DM), and insulin resistance (IR) are important risk factors for the development of hepatic steatosis3. The underlying mechanisms responsible for the development of inflammation and fibrosis that characterize progressive NASH are unclear, but emerging evidence suggests that hepatic lipotoxicity is critical for understanding NASH pathogenesis4, 5, 6. The concept of hepatic lipotoxicity implies that exposure to, or accumulation of, certain lipid species in hepatocytes may cause cytotoxicity directly or act in a proinflammatory or profibrotic manner4. Some studies have suggested that triglycerides (TGs, which make up most hepatic lipids in NASH and steatosis) are safe storage lipids with little lipotoxic potential7,8. Relatively small quantities of lipotoxic lipid species can exert disproportionately large negative effects on NASH development. Potentially lipotoxic molecules include free cholesterol (FC) and free fatty acid (FFA) and their derivatives9. These molecules may act in one or more hepatic cell types: hepatocytes (the cells that store most hepatic lipids), Kupffer cells, hepatic stellate cells, and sinusoidal endothelial cells. These lipids also affect the function of subcellular organelles (e.g., mitochondria and the endoplasmic reticulum [ER]) and can inhibit insulin signaling and induce apoptosis.
Dysregulation of cholesterol homeostasis has been documented in NAFLD10,11. This phenomenon can cause increased synthesis and uptake of cholesterol as well as relatively reduced efflux and excretion of cholesterol and lead to an increased cholesterol level in the liver. The main pathways by which hepatocytes acquire cholesterol are endogenous synthesis of cholesterol (the rate-limiting enzyme being 3-hydroxy-3-methylglutaryl coenzyme A reductase [HMGCR])12, uptake of low-density lipoprotein particles and chylomicron remnants via low-density lipoprotein receptor (LDLR)-mediated endocytosis11, and uptake of circulating high-density lipoprotein cholesterol esters (CEs) directly via scavenger receptor class B type I (SR-BI)13. The main pathways by which cholesterol can be removed from hepatocytes are conversion to bile acids (the rate-limiting enzymes being cholesterol 7 alpha-hydroxylase [CYP7A1] and sterol-27-hydroxylase [CYP27A1]) and excretion of the bile acids into bile by the bile salt export pump (BSEP)14,15, excretion of cholesterol into bile by ATP-binding cassette subfamily G member 5 or G member 8 (ABCG5/G8)16, and efflux of cholesterol onto circulating apolipoprotein-AI and nascent high-density lipoprotein particles by ATP-binding cassette transporter family A protein 1 (ABCA1)-transported free cholestenone16. In addition, hepatic FC can be esterified to CEs by acyl-coenzyme A:cholesterol acyltransferase 2 (ACAT2)17. ACAT2 also promotes the incorporation of CEs into very-low-density lipoprotein particles that are then secreted into the bloodstream. Thus, reducing the accumulation of hepatic FC (lipotoxic molecules) by intervening in these pathways may be a feasible strategy to ameliorate NAFLD. For instance, mice lacking hepatic zinc finger protein 36 like 1 (ZFP36L1) are protected from diet-induced obesity and steatosis through increased bile acid synthesis (BAS) mediated by CYP7A118. Overexpression of nuclear factor-4α in hepatocytes protects against high-fat/cholesterol/fructose (HFCF) diet-induced steatohepatitis by coordinating the regulation of CYP7A1- and CYP8B1-mediated bile-acid signaling pathways19.
G protein-coupled receptors (GPCRs) detect a wide array of extracellular molecules, activate intracellular signaling, and lead to multiple cellular responses6. GPCRs have been shown to have essential roles in NAFLD20, such as G protein-coupled receptor (GPR)12021, GPR13222, GPR5523, and GPR9124, through their function as receptors for bile acids and FFA. A bile acid-activated membrane-bound GPCR called Takeda G protein-coupled receptor 5 (TGR5) appears to have a role in stimulating energy metabolism, protecting the liver and intestine from inflammation and steatosis, and improving insulin sensitivity25,26. Another recently identified bile acid-activated GPCR called sphingosine-1-phosphate receptor 2 may also play an important part in regulation of hepatic metabolism of glucose, bile acids, and lipids via coordinated activation of extracellular signal-regulated protein kinase-1/2, protein kinase B, and farnesoid X receptor (FXR)27. However, the GPCRs responsible for signaling the conversion of cholesterol into bile acids have not been identified.
GPR35 (also known as CXCR8) is a poorly characterized “orphan” GPCR28. GPR35 has been shown to have a role in obesity and DM29,30. Increased circulating levels of kynurenic acid (Kyna, the endogenous agonist of GPR35) have been observed in experimental and physiological conditions of inflammation and metabolic stress29,30. In obesity and type-2 diabetes mellitus (T2DM), GPR35 in adipose tissue mediates several of the effects elicited by Kyna, including reduced bodyweight gain and increased energy expenditure in adipose tissue31. Interestingly, genetic abnormalities in the coding and intergenic regions of GPR35 have been linked to diseases of metabolic character, such as hypertension, atherosclerotic plaque formation32, and T2DM33. Evidence strongly supports a role for GPR35 signaling in inflammation and metabolic stress. However, the convergent molecular mechanisms and target toxic lipids have not been clarified. In particular, the role of GPR35 in hepatocytes in obesity-related NAFLD is not known.
In the current study, we aimed to evaluate the specific role of hepatocyte GPR35 expression in metabolism. We generated mouse models with overexpression or knockout of Gpr35 via adeno-associated virus 8 (AAV8) and then investigated the impact of the overexpression or knockout under physiological (normal control diet [NCD]) and pathological (HFCF diet) conditions.
2. Materials and methods
2.1. Mice and diets
The protocol for animal experiments were approved by the Ethics Committee of Anhui Medical University (No. LLSC20200725). C57BL/6J and global GPR35-knockout (Gpr35-KO) mice were purchased from GemPharmatech (Nanjing, China). C57BL/6J mice were administered AAV8-TBG-SaCas9-2A-EGFP-U6-sgRNA (negative control, NC) and AAV8-TBG-SaCas9-2A-EGFP-U6-sgRNA (Gpr35) (purchased from Genechem Biotech Inc., Shanghai, China) to generate NC (Gpr35hep-NC1) mice and hepatocyte GPR35 knockout (Gpr35hep−/−) mice, respectively. Briefly, the vector genome was packaged into capsids from AAV8 by triple transfection in 293 cells. The vector genome contains a thyroxine-binding globulin (TBG) promoter (a liver-specific promoter), Staphylococcus aureus Cas9 nuclease (SaCas9) and its sgRNA. Cas9 is able to target indel formation at a genomic site complementary to the sgRNA. An indel can result in a frameshift, causing early termination and either production of nonfunctional protein or nonsense-mediated decay of the mRNA transcript. C57BL/6J mice were injected with AAV8-TBG-MCS-EGFP-3Flag-SV40 PolyA (NC) or AAV8-TBG-MCS-Gpr35-EGFP-3Flag-SV40 PolyA (purchased from Genechem Biotech Inc., Shanghai, China) to generate NC (Gpr35hep-NC2) mice and hepatocyte GPR35-overexpressing (Gpr35hep-OE) mice. AAV8-TBG-m-NC-3xFlag-mCherry or AAV8-TBG-m-Stard4 (StAR-related lipid transfer protein 4)-3xFlag-mCherry (purchased from Hanheng Biotechnology, Shanghai, China) was injected into Gpr35hep-NC1 and Gpr35hep−/− mice to generate NC (Stard4hep-NC1-Gpr35hep-NC1 and Stard4hep-NC1-Gpr35hep−/−) mice and hepatocyte Stard4-overexpressing (Stard4hep-OE-Gpr35hep-NC1 and Stard4hep-OE-Gpr35hep−/−) mice, respectively. AAV8-TBG-Mir30-m-shRNA(NC)-mCherry or AAV8-TBG-Mir30-m-shRNA(Stard4)-mCherry (purchased from Hanheng Biotechnology, Shanghai, China) was injected into Gpr35hep-NC2 and Gpr35hep-OE mice to generate NC (Stard4hep-NC2-Gpr35hep-NC2 and Stard4hep-NC2-Gpr35hep-OE) mice and hepatocyte STARD4-knockdown (Stard4hep−KD-Gpr35hep-NC2 and Stard4hep−KD-Gpr35hep-OE) mice, respectively. The AAV miR30-based shRNA knockdown vector system is a highly efficient viral tool for knocking down the expression of target gene(s) in vivo. The shRNAmiR transcript is processed by endogenous cellular micro-RNA pathways to produce mature shRNAs, which facilitate degradation of target gene mRNAs. All vectors were injected via the tail vein (1 × 1011 vg/mouse). One week after intravenous (i.v.) injection, the knockout, overexpression or knockdown efficiency of the target gene was verified at the mRNA level and total protein level.
The HFCF diet contained 40% fat/0.2% cholesterol (TP26304; Trophic Diets, Nantong, China) and 4.2% fructose (in drinking water). Mice were fed the HFCF diet for 16 weeks. A normal chow diet (D12450J; Research Diets, New Brunswick, NJ, USA) was used as the NCD. Male eight-week-old genetically obese mice (ob/ob, B6/JGpt-Lepem1Cd25/Gpt, T001461; GemPharmatech, Nanjing, China) were fed the NCD and served as another model of fatty liver disease.
2.2. Culture and treatment of primary hepatocytes
Primary hepatocytes were isolated from male C57BL/6J mice. In situ liver perfusion with Hank's balanced salt solution was followed by collagenase I (MilliporeSigma, Burlington, MA, USA) digestion. After digestion, liver tissue was dissected, placed in a sterile Petri dish containing cold Dulbecco's modified Eagle's medium/high glucose (DMEM/high glucose; Gibco, Grand Island, NY, USA) and passed through a 75-μm cell strainer. Then, the cell suspension was centrifuged twice at 50 × g for 5 min at 4 °C. The hepatocyte pellet was collected and confirmed by trypan blue staining. The hepatocytes were cultured in standard medium comprising DMEM/high glucose, 10% fetal bovine serum and 1% penicillin–streptomycin, and maintained in a humidified atmosphere of 5% CO2 in a cell incubator at 37 °C.
Palmitic acid (PA) powder (catalog number, P0500; MilliporeSigma) and oleic acid (OA; O1008, MilliporeSigma) were dissolved in 25% bovine serum albumin (BSA; BAH66-0050; Equitech-Bio, Kerrville, TX, USA) solution. Then, primary hepatocytes were treated with the final concentrations of PA/OA (0.5 mmol/L/1 mmol/L) and 1.25% BSA for 24 h.
2.3. Treatment of AML-12 cells
The pEX-3 (pGCMV/MCS/Neo)-control or pEX-3-Mus Gpr35 plasmid (GenePharma, Shanghai, China) was transfected into AML-12 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendation, and the cells were then incubated at 37 °C for 48 h. To further investigate the involvement of ERK1/2 and CREB signaling in the expression of STARD4, AML-12 cells were preincubated with or without the ERK1/2 inhibitor PD98059 (20 μmol/L; MCE) or the CREB inhibitor 666-15 (1 μmol/L; MCE) for 2 h before PA/OA (0.5 mmol/L/1 mmol/L) treatment for 24 h. Finally, the cells were collected and prepared for the following experiments.
2.4. Mouse experiments
Mouse body weight and fasting blood glucose levels were determined at different time points during the experiments. Fasting blood glucose was assessed using a glucometer after the mice were fasted for 6 h. For glucose tolerance tests (GTTs), mice were injected intraperitoneally with 1 g/kg glucose after a 6-h fast, whereas for insulin tolerance tests (ITTs), mice were injected intraperitoneally with 0.75 U/kg insulin after a 6-h fast. Blood glucose concentrations in tail blood samples were detected using a glucometer at baseline and at 15, 30, 45, 60, and 120 min after injection.
2.5. Histopathology
Liver tissues were embedded in paraffin and stained with hematoxylin & eosin (H&E) or Sirius Red. Liver tissues were embedded in optimum cutting temperature (OCT) medium and frozen. Then, frozen sections were prepared and stained with Oil Red O. Immunohistochemical staining for F4/80 (70076; Cell Signaling Technology, Danvers, MA, USA), and alpha-smooth muscle actin (α-SMA; 19245; Cell Signaling Technology) was performed on the liver sections. Images were acquired with a light microscope (Olympus, Tokyo, Japan).
2.6. Biochemical assays
i) Serum levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured by an automatic biochemistry analyzer (Mindray Biomedical Electronics, Shenzhen, China) according to the instructions provided in commercial assay kits (Mindray Biomedical Electronics). ii) Liver TG extractant was prepared by mixing n-heptane and isopropyl alcohol at a volume ratio of 1:1. Every 100 mg of liver tissue was homogenized with 1 mL of extract in an ice bath and then centrifuged at 8000 × g at 4 °C for 10 min. The supernatant was taken for testing. Hepatic TG content was measured using a TG content detection kit (BC0625; Beijing Solarbio Science & Technology). iii) After the liver tissue was washed with normal saline, the surface water was absorbed with absorbent paper. Each 100 mg of liver tissue was homogenized with 1 mL of FFA extract, which was obtained from a Micro Free Fatty Acid Content Assay Kit (BC0595; Beijing Solarbio Science & Technology, Beijing, China). The tissue homogenates were centrifuged at 8000 × g at 4 °C for 10 min, and the supernatant was taken for testing. Hepatic FFA content was measured using the Micro Free Fatty Acid Content Assay Kit. iv) Isopropanol was used as liver total cholesterol (TC) or FC extractant. Every 100 mg of liver tissue was homogenized with 1 mL of extract in ice bath. The hepatic levels of TC and FC were measured using a Micro Total Cholestenone Content Assay Kit (BC1985, Beijing Solarbio Science & Technology) and a Micro Free Cholestenone Content Assay Kit (BCBC1895; Beijing Solarbio Science & Technology), respectively. The hepatic level of CEs was determined as the TC content minus the FC content. v) Every 100 mg of liver tissue was homogenized with 1 mL of 0.9% normal saline in ice bath and then centrifuged at 3500 rpm (Centrifuge 5424R, Eppendorf, Germany) at 4 °C for 10 min. The supernatant was taken for testing of bile-acid content. The hepatic bile-acid content was measured using a total bile acid kit (E003-2-1; Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.7. Real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells or tissues with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). The total RNA was reverse-transcribed into complimentary DNA using a PrimeScript RT Reagent Kit (Takara Biotechnology, Shiga, Japan) in a thermal cycler (T100; Bio-Rad Laboratories, Hercules, CA, USA) with the following conditions: 37 °C for 15 min, 85 °C for 5 s, and a hold at 4 °C. PCR was performed using SYBR Premix Ex Taq II (Takara Biotechnology) in a CFX Connect Real-Time System (Bio-Rad Laboratories) under the following conditions: predenaturation at 95 °C for 30 s and thermal cycling at 95 °C for 5 s and 60 °C for 30 s for 40 cycles. The primer sequences used in our study are described in Supporting Information Table S1. The relative mRNA expression of each gene was normalized to β-actin expression and is reported as the fold change from the basal level.
2.8. Western blotting
Liver tissues or cells were homogenized in RIPA buffer (P0013B, Beyotime, China) containing a mixture of protease and phosphatase inhibitors (P1045, Beyotime, China). Protein extracts were loaded onto 10% acrylamide gels and transferred onto PVDF membranes. The membranes were blotted with primary antibodies and secondary antibodies. The primary antibodies are described in Supporting Information Table S2. Protein bands were visualized with an enhanced chemiluminescence (ECL) substrate kit (A38554, Thermo, USA).
2.9. Statistical analyses
The data are presented as the mean ± standard deviation (SD). Prior to performing specific statistical tests, we performed tests for normality and homogeneity of variance. To compare significance between 2 groups, the two-tailed Student t test was performed. To compare significance among 3 or more than 3 groups, one-way ANOVA followed by Tukey–Kramer multiple comparisons test was performed. Two-way ANOVA with a Bonferroni post hoc test was used for multiparameter analyses or repeated-measures data analysis (body weight, fasting blood glucose, and GTT and ITT results). Differences were considered significant at P < 0.05. The statistical analyses were carried out using Prism 8 (GraphPad, La Jolla, CA, USA). Differences were considered significant at P < 0.05. Statistical analyses were carried out using Prism 8 (GraphPad, La Jolla, CA, USA).
3. Results
3.1. Global KO of GPR35 exacerbates HFCF diet-induced steatohepatitis
To determine the relevance of GPR35 in hepatic metabolism, we measured GPR35 expression in the livers of mice with NAFLD. Eight-week-old mice were fed a HFCF diet or a NCD for 16 weeks. The protein and mRNA expression of GPR35 in the liver was upregulated in HFCF mice compared with control mice (Supporting Information Fig. S1A and S1B). As expected, GPR35 expression was also increased in the liver of ob/ob mice compared with those of lean mice (Fig. S1C and S1D). To ascertain whether this increase occurred in hepatocytes, primary hepatocytes were isolated from wild-type (WT) mice and stimulated with vehicle or PA&OA (inducer of lipid accumulation and IR in hepatocytes) in vitro14. Compared with vehicle treatment, PA&OA treatment triggered a marked increase in GPR35 expression in hepatocytes (Fig. S1E and S1F).
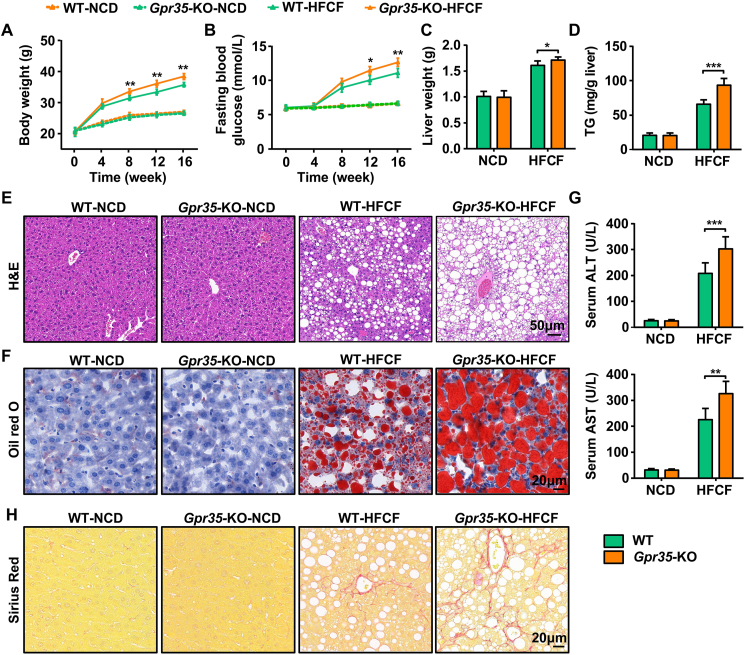
Next, we investigated the role of GPR35 in NAFLD. Global Gpr35-KO and C57BL/6J WT mice were fed a NCD or a HFCF diet for 16 weeks. Among the HFCF diet-fed mice, Gpr35-KO mice had significantly larger increases in body weight and fasting blood glucose levels than WT mice (Fig. 1A and B). At the end of HFCF diet feeding for 16 weeks, the liver weight and TG content were higher in Gpr35-KO mice than in WT mice (Fig. 1C and D). Oil Red O and H&E staining showed more severe steatosis and lipid accumulation in Gpr35-KO mice (Fig. 1E and F). In line with the changes in steatosis, loss of global GPR35 also increased plasma ALT and AST levels (Fig. 1G) and induced severe fibrosis (Fig. 1H). Thus, loss of GPR35 aggravated the development of diet-induced NASH.
Figure 1.
Global knockout of GPR35 exacerbates HFCF diet-induced steatohepatitis (n = 6). WT and global GPR35-knockout (Gpr35-KO) C57BL/6J mice were fed a NCD or HFCF diet for 16 weeks. (A) Body weight was measured every 4 weeks from 0 to 16 weeks. (B) Fasting blood glucose levels were measured every 4 weeks from 0 to 16 weeks. (C) Liver weight. (D) TG level in the liver. (E) H&E staining of liver tissue sections. Scale bar, 50 μm. (F) Oil Red O staining of liver tissue sections. Scale bar, 20 μm. (G) Serum levels of ALT and AST. (H) Representative images of Sirius Red staining of liver tissue sections. Scale bar, 20 μm. Data are the mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.2. Deficiency of GPR35 expression in hepatocytes exacerbates HFCF diet-induced steatohepatitis and IR
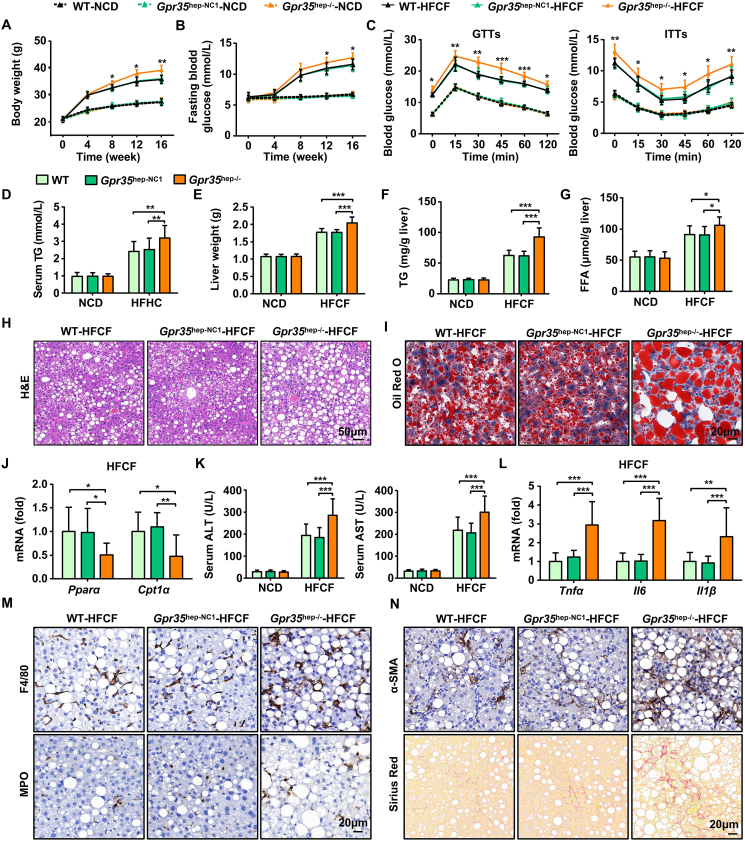
To clarify the effect of GPR35 expression in hepatocytes on steatohepatitis and IR in vivo, we generated hepatocyte GPR35 KO (Gpr35hep−/−) mice via injection (i.v.) with AAV8-TBG-SaCas9-2A-EGFP-sgGpr35 in WT mice (Supporting Information Fig. S2A and S2B). Subsequently, Gpr35hep−/−, control (Gpr35hep-NC1), and WT mice were fed the HFCF diet or NCD separately for 16 weeks. Deficiency of GPR35 expression in hepatocytes had no effect on bodyweight, IR, or liver steatosis in mice fed with the NCD (Fig. S2C and S2D, Fig. 2). Consumption of the HFCF diet led to significant enhancement of body weight and fasting glucose levels in WT mice. Gpr35hep−/− mice fed the HFCF diet had consistently increased body weight and fasting glucose levels compared with those in Gpr35hep-NC1 and WT mice (Fig. 2A and B). Glucose tolerance was reduced and insulin sensitivity was compromised in Gpr35hep−/− mice compared with control mice, as shown by the GTTs and ITTs, respectively (Fig. 2C). Serum TG levels were higher in Gpr35hep−/− mice than in Gpr35hep-NC1 and WT mice at the end of 16 weeks of HFCF diet feeding (Fig. 2D). These data reveal that deficiency of GPR35 expression in hepatocytes promoted obesity and IR after HFCF diet feeding.
Figure 2.
GPR35 deficiency in hepatocytes exacerbates HFCF diet-induced steatohepatitis and insulin resistance (n = 10). C57BL/6J mice were injected (i.v.) with AAV8-TBG-SaCas9-2A-EGFP-U6-sgRNA (NC) or AAV8-TBG-SaCas9-2A-EGFP-U6-sgRNA (Gpr35) to generate control (Gpr35hep-NC1) mice and hepatocyte GPR35-knockout (Gpr35hep−/−) mice, which were then fed a NCD or HFCF for 16 weeks. (A) Body weight was measured every 4 weeks from 0 to 16 weeks. (B) Fasting blood glucose levels were measured every 4 weeks from 0 to 16 weeks. (C) GTTs and ITTs were carried out on WT, Gpr35hep-NC1, and Gpr35hep−/− mice after NCD or HFCF diet feeding for 16 weeks. (D–N) WT, Gpr35hep-NC1, and Gpr35hep−/− mice at the end of 16 weeks of HFCF-diet feeding. (D) Serum TG levels. (E) Liver weight. (F) TG and (G) FFA levels in the liver. (H) H&E staining of liver tissue sections. Scale bar, 50 μm. (I) Oil Red O staining of liver tissue sections. Scale bar, 20 μm. (J) Relative mRNA expression of Pparα and Cpt1α in the liver. (K) Serum levels of ALT and AST. (L) Relative mRNA expression of cytokines (Tnfα, Il6, Il1β) in the liver. (M) Representative images of F4/80 and MPO immunostaining of liver tissue sections. Scale bar, 20 μm. (N) Representative images of Sirius Red staining and α-SMA immunostaining of liver tissue sections. Scale bar, 20 μm. The mRNA expression of the genes was normalized to that of β-actin. Data are the mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Compared with Gpr35hep-NC1 and WT mice, Gpr35hep−/− mice had higher liver weights (Fig. 2E). Gpr35hep−/− mice exhibited remarkable increases in hepatic TG and FFA levels (Fig. 2F and G). Sections stained with H&E and Oil Red O (Fig. 2H and I) revealed steatosis to be aggravated by deficiency of GPR35 expression in hepatocytes. Although there were no changes in the mRNA expression of the genes involved in de novo lipogenesis (DNL) (Srebp1c, Acc) or the protein expression of fas (Supporting Information Fig. S3), the mRNA expression of genes involved in fatty acid oxidation (FAO) (Pparα, Cpt1α) was inhibited by the loss of GPR35 expression in hepatocytes (Fig. 2J).
Next, we examined whether deficiency of GPR35 expression in hepatocytes could exacerbate HFCF diet-induced damage, inflammation, and fibrosis of the liver. Serum levels of ALT and AST were higher in HFCF diet-fed Gpr35hep−/− mice than in Gpr35hep-NC1 and WT mice (Fig. 2K). Moreover, the mRNA expression of proinflammatory cytokines (tumor necrosis factor-alpha [Tnfα], interleukin-6 [Il6], Il1β) was significantly elevated in the livers of Gpr35hep−/− mice (Fig. 2L). Consistent with the increased expression of proinflammatory cytokines, Gpr35hep−/− mice had increased hepatic infiltration of inflammatory cells, which was confirmed by immunohistochemical staining of F4/80 (a macrophage marker) and myeloperoxidase (a neutrophil marker) (Fig. 2M). HFCF diet-fed Gpr35hep−/− mice had increased liver fibrosis, as indicated by the α-SMA level upon Sirius Red staining (Fig. 2N). Collectively, these results suggest that deficiency of GPR35 expression in hepatocytes exacerbated HFCF diet-induced steatohepatitis and IR.
3.3. Mice with GPR35 overexpression in hepatocytes are resistant to HFCF diet-induced steatohepatitis and IR
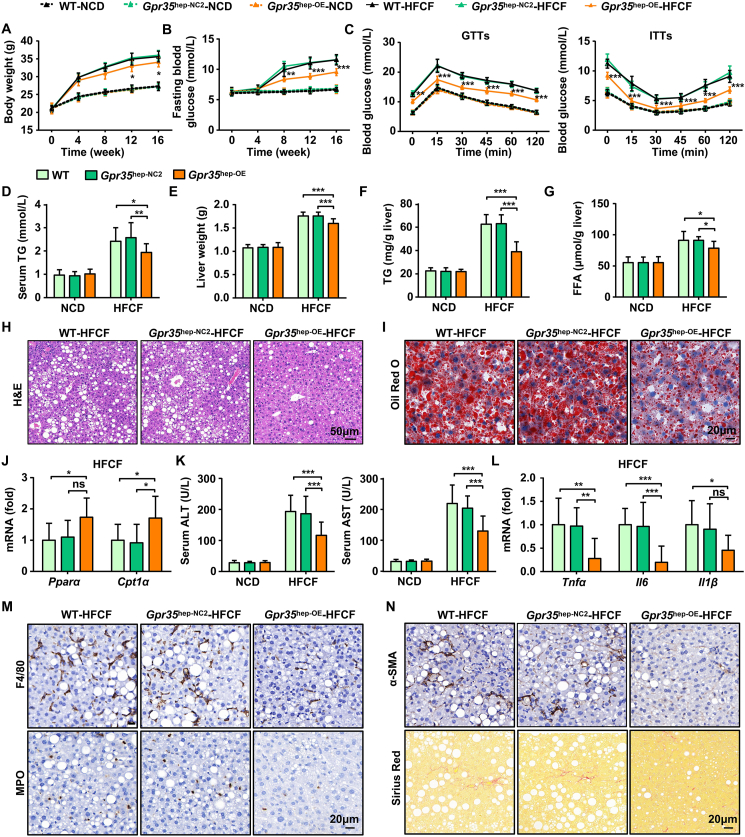
In addition to implementing the loss-of-function approach, we established mice with GPR35 overexpression in hepatocytes (Gpr35hep-OE mice) by injecting AAV8-TBGp-MCS-EGFP-3Flag-SV40 PolyA-Gpr35 into WT mice (Supporting Information Fig. S4A and S4B) to determine whether GPR35 overexpression in hepatocytes could attenuate HFCF diet-induced NAFLD. Gpr35hep-oe, control (Gpr35hep-NC2), and WT mice were fed the HFCF diet or the NCD for 16 weeks. GPR35 overexpression in hepatocytes had no effect on body weight, IR, or liver steatosis in mice fed the NCD (Fig. S4C and S4D, Fig. 3). Consumption of the HFCF diet induced smaller increases in body weight and fasting glucose levels in Gpr35hep-OE mice than in Gpr35hep-NC2 or WT mice (Fig. 3A and B). GTTs and ITTs revealed that glucose tolerance and insulin sensitivity were enhanced significantly in HFCF diet-fed Gpr35hep-oe mice (Fig. 3C). Similarly, HFCF diet-fed Gpr35hep-OE mice had lower serum levels of TGs (Fig. 3D). These results indicate that GPR35 overexpression in hepatocytes attenuated HFCF diet-induced obesity and IR.
Figure 3.
Mice with overexpressed GPR35 in hepatocytes are resistant to HFCF diet-induced steatohepatitis and IR (n = 10). C57BL/6J mice were injected (i.v.) with AAV8-TBGp-MCS-EGFP-3Flag-SV40 PolyA or AAV8-TBG-MCS-Gpr35-EGFP-3Flag-SV40 PolyA to generate control (Gpr35hep-NC2) mice and hepatocyte GPR35-overexpressing (Gpr35hep-OE) mice, which were then fed a NCD or HFCF diet for 16 weeks. (A) Body weight was measured every 4 weeks from 0 to 16 weeks. (B) Fasting blood glucose levels were measured every 4 weeks from 0 to 16 weeks. (C) GTTs and ITTs were undertaken on WT, Gpr35hep-NC2, and Gpr35hep-OE mice after NCD or HFCF-diet feeding for 16 weeks. (D–N) WT, Gpr35hep-NC2, and Gpr35hep-OE mice at the end of 16 weeks of HFCF-diet feeding. (D) Serum TG levels. (E) Liver weight. (F) TG and (G) FFA levels in the liver. (H) H&E staining of liver tissue sections. Scale bar, 50 μm. (I) Oil Red O staining of liver tissue sections. Scale bar, 20 μm. (J) Relative mRNA expression of Pparα and Cpt1α in the liver. (K) Serum levels of ALT and AST. (L) Relative mRNA expression of cytokines (Tnfα, Il6, Il1β) in the liver. (M) Representative images of F4/80 and MPO immunostaining of liver tissue sections. Scale bar, 20 μm. (N) Representative images of Sirius Red staining and α-SMA immunostaining of liver tissue sections. Scale bar, 50 μm. mRNA expression of genes was normalized to that of β-actin. The data are the mean ± SD. ns, not significant; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Liver weight, TG content, and FFA content decreased following consumption of the HFCF diet in Gpr35hep-oe mice compared with Gpr35hep-NC2 and WT mice on the same diet (Fig. 3E–G). Consistent with the results shown above, gain of GPR35 expression in hepatocytes increased the mRNA expression of genes involved in FAO (Fig. 3J) but not DNL (Supporting Information Fig. S5). Lipid accumulation and steatosis of the liver were decreased significantly in HFCF diet-fed Gpr35hep-OE mice, as indicated by stained liver sections (H&E and Oil Red O) (Fig. 3H and I).
HFCF diet-induced liver damage (indicated by the serum levels of ALT and AST) was reduced in Gpr35hep-OE mice (Fig. 3K). GPR35 overexpression in hepatocytes inhibited the expression of proinflammatory cytokines and the infiltration of inflammatory cells in the liver induced by consumption of the HFCF diet (Fig. 3L and M). Furthermore, HFCF diet-induced liver fibrosis was relieved by GPR35 overexpression in hepatocytes (Fig. 3N). Taken together, these results suggest a protective effect of GPR35 overexpression in hepatocytes in mice with HFCF diet-induced NAFLD.
3.4. GPR35 regulates cholesterol homeostasis via cholesterol esterification and BAS in the liver
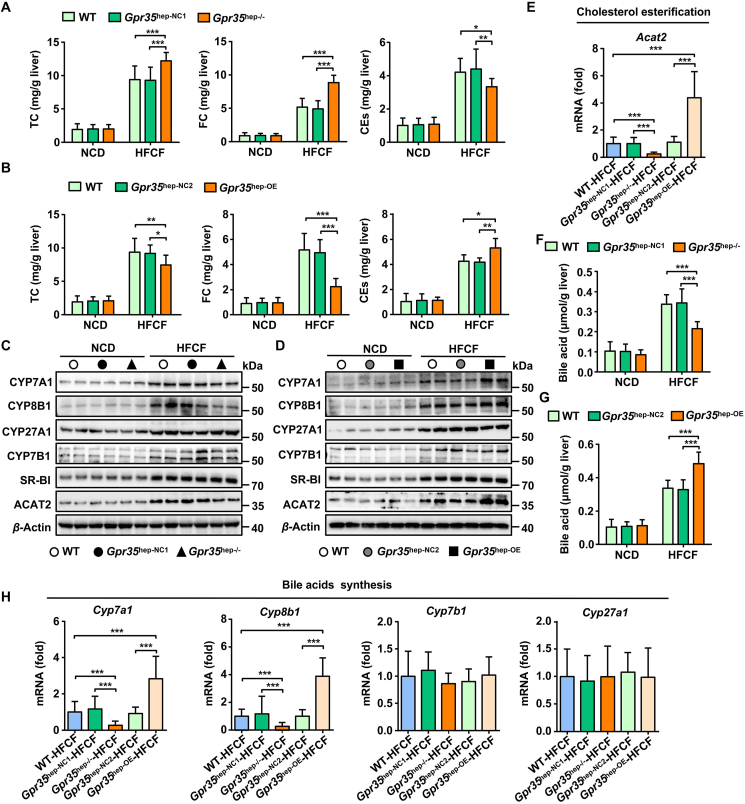
An increasing amount of evidence is connecting altered cholesterol homeostasis and hepatic FC accumulation with NASH pathogenesis10,11. In NAFLD patients, the development of NASH and fibrosis parallels hepatic FC accumulation34. Experimental induction of hepatic FC accumulation promotes steatohepatitis and fibrosis, whereas correction of hepatic FC overload improves liver disease severity in NASH35. Notably, the levels of TC and FC were markedly increased in the livers of Gpr35hep−/− mice, whereas the levels of CEs were decreased (Fig. 4A). Similarly, TC and FC levels were reduced in the livers of Gpr35hep-OE mice, whereas CEs levels were increased (Fig. 4B). Esterification of cholesterol is an important way of excreting FC in the liver. Formation of CEs in the liver by ACAT2 in hepatocytes increases secretion of hepatic cholesterol directly into the bloodstream36. Consistent with changes in CEs levels, GPR35 expression in hepatocytes induced increased protein and mRNA expression of Acat2 in the liver of HFCF diet-fed mice (Fig. 4C–E), which suggested that GPR35 promoted cholesterol esterification. The liver is a major site for the biosynthesis, uptake, and excretion of cholesterol as well as the metabolism of cholesterol to bile acids. We found that the gain or loss of GPR35 expression in hepatocytes did not regulate the excretion (Abcg5, Abcg8, Abca1), synthesis (Hmgcr), or uptake (Ldlr, Srb1) of cholesterol in the liver (Supporting Information Fig. S6A–S6C). In addition to the liver, the intestine has important roles in regulating cholesterol levels36. The intestine also maintains the cholesterol level in the whole body (i) by mediating intestinal absorption of dietary and biliary cholesterol and (ii) via direct transintestinal cholesterol excretion36. These processes are regulated by interactions between the liver and intestine. To test whether the absorption and excretion of cholesterol in the intestine was regulated by GPR35 expression in hepatocytes, we measured the expression of the genes involved in the absorption (Niemann-Pick C1-like 1 [Npc1l1]) and excretion (Abcg5 and Abcg8) of cholesterol in the intestine. GPR35 expression in hepatocytes had no effect on the absorption and excretion of cholesterol in the intestine (Supporting Information Fig. S7).
Figure 4.
GPR35 regulates hepatic bile acid synthesis and cholesterol esterification (n = 10). (A–D) WT, Gpr35hep-NC1, Gpr35hep−/−, Gpr35hep-NC2, and Gpr35hep-OE mice were fed a NCD or HFCF diet for 16 weeks. (A) and (B) Levels of TC, FC and CEs in the liver. (C) Representative Western blot of CYP7A1, CYP8B1, CYP27A1, CYP7B1, SR-BI, and ACAT2 proteins in the livers of WT, Gpr35hep-NC1, and Gpr35hep−/− mice. (D) Representative Western blot of CYP7A1, CYP8B1, CYP27A1, CYP7B1, SR-BI, and ACAT2 proteins in the livers of WT, Gpr35hep-NC2, and Gpr35hep-OE mice. (E) Relative mRNA expression of a cholesterol esterification gene (Acat2) in the livers of WT, Gpr35hep-NC1, Gpr35hep−/−, Gpr35hep-NC2, and Gpr35hep-OE mice fed the HFCF diet for 16 weeks. (F) Levels of bile acids in the livers of WT, Gpr35hep-NC1, and Gpr35hep−/− mice fed the NCD or HFCF diet for 16 weeks. (G) Levels of bile acids in the livers of WT, Gpr35hep-NC2, and Gpr35hep-OE mice fed the NCD or HFCF diet for 16 weeks. (H) Relative mRNA expression of bile acids synthesis genes (Cyp7a1, Cyp8b1, Cpy27a1 and Cyp7b1) in the livers of WT, Gpr35hep-NC1, Gpr35hep−/−, Gpr35hep-NC2, and Gpr35hep-OE mice fed the HFCF diet for 16 weeks. The mRNA expression of genes was normalized to that of β-actin. Data are the mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
In humans, conversion of cholesterol to bile acids is essential for maintaining cholesterol homeostasis and preventing the liver damage induced by FC accumulation37. In accordance with decreased FC levels, compared with those in HFCF diet-fed control mice, the levels of bile acids in the liver were reduced in Gpr35hep−/− mice and increased in Gpr35hep-OE mice (Fig. 4F and G). By participating in BAS pathways, different cytochrome P450 (P450 or CYP) enzymes have important roles in the maintenance of cholesterol elimination38. Among the cytochrome enzymes affected by deletion of hepatic GPR35 expression, CYP7A1 and CYP8B1 (key cytochrome enzymes involved in the classic pathway of BAS in the ER) exhibited reduced expression in Gpr35hep−/− mice (Fig. 4C and H). The expression of CYP27A1 and CYP7B1, which are involved mostly in the alternative pathway of BAS in mitochondria, was not affected. Consistent with these data, GPR35 overexpression in hepatocytes resulted in increased expression of CYP7A1 and CYP8B1 (Fig. 4D and H). These results indicate that GPR35 promoted the conversion of cholesterol to bile acids through the neutral pathway (initiated by CYP7A1) rather than the acidic pathway (initiated by CYP27A1). Moreover, we quantified the factors involved in the reabsorption of bile acids (Ntcp) (Supporting Information Fig. S8A), bile-acid conjugation (Bal, Bat, Hnf4a1) (Fig. S8B), and export (Abcb11, Abcb4, Ostb) of bile acids from the liver (Fig. S8C). We measured markers of bile acid reabsorption in the intestine (Osta, Ostb, Ibabp, Asbt) (Supporting Information Fig. S9). The expression of most of these markers was not affected by hepatic GPR35 expression. Importantly, we also obtained consistent results in Gpr35-KO mice. Global deletion of GPR35 prevented ACAT2-mediated cholesterol esterification and CYP7A1-initiated BAS in the livers of HFCF diet-fed mice, which aggravated hepatic FC accumulation (Supporting Information Fig. S10). Collectively, these data suggest that GPR35 expression in hepatocytes might protect against HFCF diet-induced NASH by a mechanism involving cholesterol homeostasis.
3.5. GPR35 promotes hepatic expression of STARD4
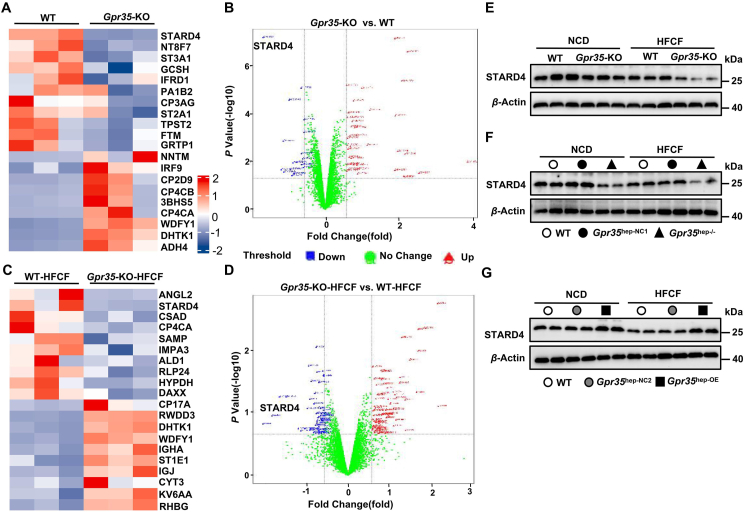
We wished to further explore the mechanism by which hepatic GPR35 regulates hepatic cholesterol homeostasis. We undertook in-depth quantitative proteomic analysis of liver tissues in Gpr35-KO and WT mice fed a HFCF diet or a NCD through tandem mass tag (TMT) labeling, which led to the identification of 7049 proteins. A multiple-sample test controlled by a false-detection rate (FDR) threshold of 0.05 was applied to identify significant differences in protein abundance (≥1.5-fold change and P < 0.05). In NCD-fed mice, 38 proteins had upregulated expression and 18 proteins had downregulated expression in Gpr35-KO mice compared with WT mice (Fig. 5A and B). Among these, STARD4 (Q99JV5; www.uniprot.org/uniprot/Q99JV5), a protein involved in the intracellular transport of cholesterol, exhibited the most significant expression downregulation. In total, 77 proteins had upregulated expression and 55 proteins had downregulated expression in HFCF diet-fed Gpr35-KO mice compared with WT mice (Fig. 5C and D), and STARD4 expression was downregulated significantly. Moreover, we verified the difference in STARD4 expression in the liver between Gpr35-KO and WT mice via Western blotting (Fig. 5E). STARD4 is a member of the evolutionarily conserved StAR-related lipid transfer gene family and is expressed in most tissues, with the highest levels found in the liver and kidneys39,40. STARD4 has been implicated as an important high-efficiency soluble sterol transport protein involved in maintaining cholesterol homeostasis39, 40, 41, 42. Notably, STARD4 has been shown to promote ACAT2-dependent cholesterol esterification by transporting cholesterol to the ER40. STARD4 expression in primary mouse hepatocytes leads to marked increase in the intracellular cholesteryl ester concentration and the BAS rate40,43. Interestingly, STARD4 plays an important role in the delivery of cholesterol to the ER44, which is rich in the key enzyme CYP7A1 for the neutral pathway of BAS. Experimental studies have demonstrated that GPR35 promotes the cholesterol to bile acid conversion and cholesterol esterification in the liver. Furthermore, compared with that in the livers of control mice, STARD4 expression was decreased in the livers of HFCF diet-fed Gpr35hep−/− mice but increased in the livers of HFCF diet-fed Gpr35hep-OE mice according to Western blotting (Fig. 5F and G), which suggested that GPR35 positively regulated STARD4 expression. Therefore, we hypothesized that STARD4 might be responsible for the protective effect of GPR35 in hepatocytes against steatohepatitis via maintenance of cholesterol homeostasis.
Figure 5.
GPR35 promotes hepatic expression of STARD4. (A–D) In-depth quantitative proteomic analysis of the liver. A multiple sample test controlled by an FDR threshold of 0.05 was applied to identify significant differences in protein abundance (≥1.5-fold change). (A) Hierarchical clustering map of total proteins in the livers from WT and Gpr35-KO mice, where the top 10 proteins with the most significant upregulation (red) or downregulation (blue) are shown. (B) Volcano plot of total proteins in the livers from WT and Gpr35-KO mice. (C) Hierarchical clustering map of total proteins in the livers from WT and Gpr35-KO mice fed a HFCF diet for 16 weeks, where the top 10 proteins with the most significant upregulation (red) or downregulation (blue) are shown. (D) Volcano plot of total proteins in the livers from WT and Gpr35-KO mice fed the HFCF diet for 16 weeks. (E) Representative Western blot of STARD4 proteins in the livers of WT and Gpr35-KO mice fed the NCD or HFCF diet for 16 weeks. (F) Representative Western blot of STARD4 proteins in the livers of WT, Gpr35hep-NC1, and Gpr35hep−/− mice fed the NCD or HFCF diet for 16 weeks. (G) Representative Western blot of STARD4 proteins in the livers of WT, Gpr35hep-NC2, and Gpr35hep-OE mice fed the NCD or HFCF for 16 weeks.
3.6. STARD4 is essential for GPR35-mediated protection against HFCF-induced steatohepatitis
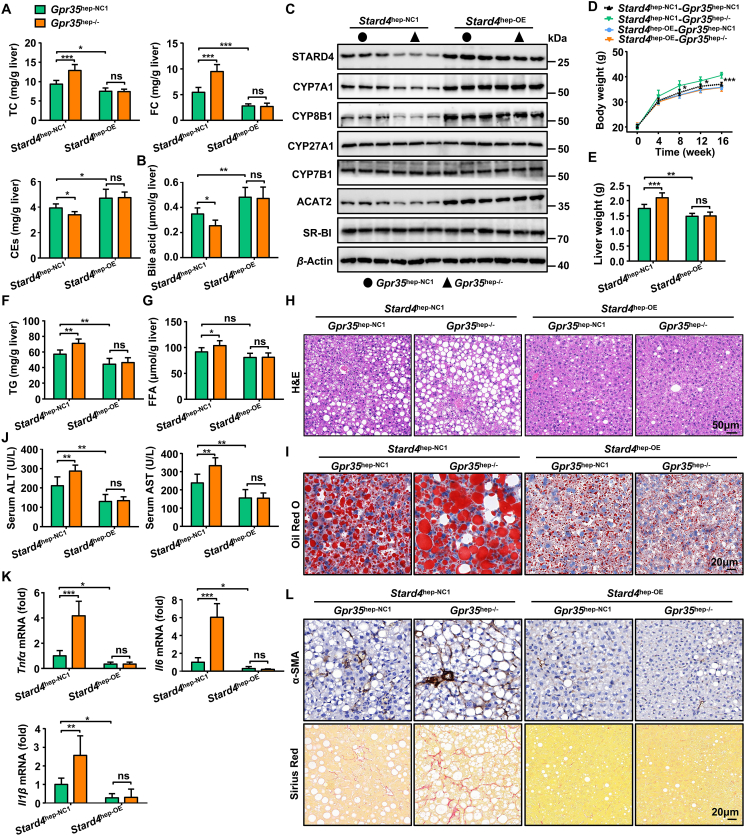
To test the hypothesis stated above, we injected (i.v.) AAV8-TBG-m-Stard4-3xFlag-mCherry into Gpr35hep-NC1 or Gpr35hep−/− mice to restore STARD4 expression in hepatocytes and then fed the mice a HFCF diet for 16 weeks. Knockout of GPR35 expression in hepatocytes increased the hepatic levels of TC and FC and reduced the levels of CEs and bile acids in Stard4hep-NC1 mice but not in Stard4hep-OE mice (Fig. 6A and B). Reduced expression of CYP7A1, CYP8B1, and ACAT2 induced by knockout of GPR35 expression in hepatocytes was restored in Stard4hep-OE mice (Fig. 6C). Consistent with these data, loss of GPR35 expression in hepatocytes exacerbated steatohepatitis (according to the body weight, liver weight, TG and FFA content in the liver, hepatic steatosis, liver injury, and fibrosis) in Stard4hep-NC1 mice but not in Stard4hep-OE mice (Fig. 6D–L).
Figure 6.
A gain in function of STARD4 can reverse the exacerbation of steatohepatitis caused by GPR35 deletion (n = 6). Gpr35hep-NC1 and Gpr35hep−/− mice were injected (i.v.) with AAV8-TBG-m-NC-3xFlag-mCherry or AAV8-TBG-m-Stard4-3xFlag-mCherry to generate control (Stard4hep-NC1-Gpr35hep-NC1 and Stard4hep-NC1-Gpr35hep−/−) mice and hepatocyte STARD4-overexpressing (Stard4hep-OE-Gpr35hep-NC1 and Stard4hep-OE-Gpr35hep−/−) mice, which were then fed a HFCF diet for 16 weeks. (A) TC, FC, CE and (B) bile acid levels in the liver. (C) Representative Western blot of CYP7A1, CYP8B1, CYP27A1, CYP7B1, SR-BI, and ACAT2 proteins in the liver. (D) Body weight was measured every 4 weeks from 0 to 16 weeks. (E) Liver weight. (F) TG and (G) FFA levels in the liver. (H) H&E staining of liver tissue sections. Scale bar, 50 μm. (I) Oil Red O staining of liver tissue sections. Scale bar, 20 μm. (J) Serum levels of ALT and AST. (K) Relative mRNA expression of cytokines in the liver. (L) Representative images of Sirius Red staining and α-SMA immunostaining of liver-tissue sections. Scale bar, 20 μm. The mRNA expression of genes was normalized to that of β-actin. Data are the mean ± SD; ns, not significant; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
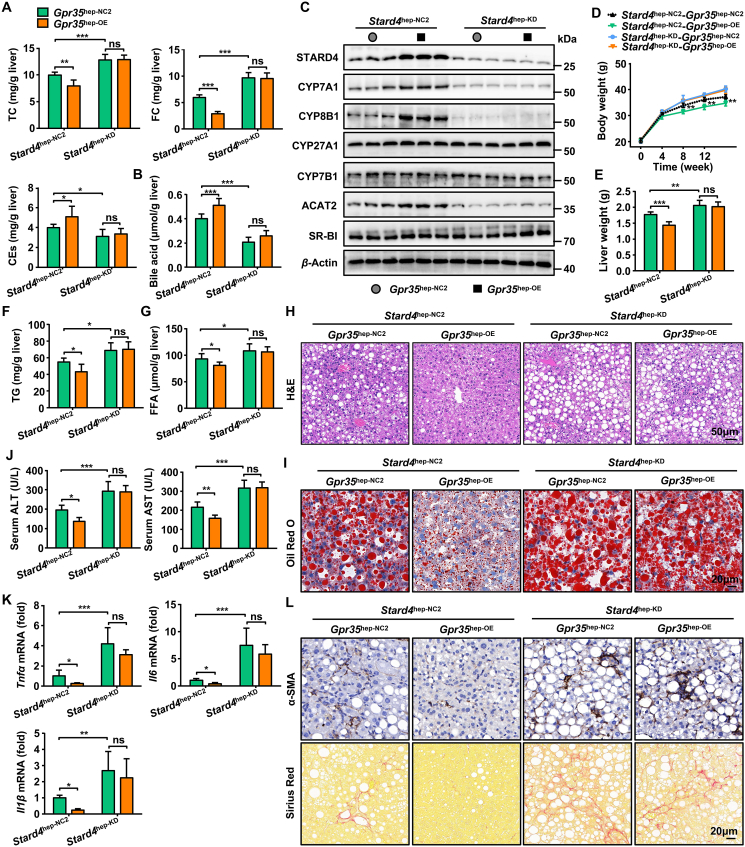
Furthermore, we injected (i.v.) Gpr35hep-NC2 or Gpr35hep-OE mice with AAV8-TBG-Mir30-m-shRNA(Stard4)-mCherry to downregulate STARD4 expression. Downregulation of STARD4 expression completely eliminated the protective effect of GPR35 against steatohepatitis by reducing bile acid production and cholesterol esterification (Fig. 7A–L). However, neither gain nor loss of STARD4 expression regulated the excretion (Abcg5, Abcg8 and Abca1), synthesis (Hmgcr), or uptake (Ldlr and Srb1) of cholesterol as well as DNL (Acc and Fas) in the liver (Supporting Information Figs. S11 and S12). Taken together, these data demonstrate that gain of hepatic GPR35 function protected against HFCF diet-induced steatohepatitis by positively regulating STARD4 expression to maintain cholesterol homeostasis.
Figure 7.
Loss of STARD4 can eliminate the protective effect of GPR35 against steatohepatitis (n = 6). Gpr35hep-NC2 and Gpr35hep-OE mice were injected (i.v.) with AAV8-TBG-Mir30-m-shRNA(NC)-mCherry or AAV8-TBG-Mir30-m-shRNA(Stard4)-mCherry to generate control (Stard4hep-NC2-Gpr35hep-NC2 and Stard4hep-NC2-Gpr35hep-OE) mice and hepatocyte STARD4-knockout (Stard4hep-KD-Gpr35hep-NC2 and Stard4hep-KD-Gpr35hep-OE) mice, which were then fed a HFCF diet for 16 weeks. (A) TC, FC, CE and (B) bile acid levels in the liver. (C) Representative Western blot of CYP7A1, CYP8B1, CYP27A1, CYP7B1, SR-BI, and ACAT2 proteins in the liver. (D) Body weight was measured every 4 weeks from 0 to 16 weeks. (E) Liver weight. (F) TG and (G) FFA levels in the liver. (H) H&E staining of liver tissue sections. Scale bar, 50 μm. (I) Oil Red O staining of liver tissue sections. Scale bar, 20 μm. (J) Serum levels of ALT and AST. (K) Relative mRNA expression of cytokines in the liver. (L) Representative images of Sirius Red staining and α-SMA immunostaining of liver tissue sections. Scale bar, 20 μm. The mRNA expression of genes was normalized to that of β-actin. Data are the mean ± SD; ns, not significant; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.7. Hepatocyte GPR35 induces STARD4 expression via the ERK1/2-CREB signaling pathway
In response to ligand binding, GPCRs initiate diverse downstream signaling pathways through four groups of G proteins and other interacting proteins. Key components in GPCR-induced intracellular signaling are four groups of mitogen-activated protein kinase (MAPK) cascades: the extracellular signal-related kinase (ERK), Jun N-terminal kinase (JNK), p38MAPK and big MAPK (BMK; ERK5)45. The hallmark of MAPK signaling is the stimulation-dependent nuclear translocation of the involved kinases, which regulate transcription, cellular metabolism and the cytoplasmic acute response to stress-related apoptosis46. Our results show that the levels of p-ERK1/2 were increased in the livers of GPR35-overexpressing mice fed a HFCF diet compared with those of WT and control mice, while the levels of members of other MAPK pathways (p-p38, p-JNK and p-ERK5) and p-AKT were not significantly changed (Supporting Information Fig. S13A). This finding is consistent with previous studies of Kyna/GPR35-induced ERK1/2 phosphorylation in intestinal epithelial cells and adipocytes31,47. Moreover, an ERK1/2 inhibitor (PD98059) significantly reversed GPR35-induced STARD4 protein expression in AML-12 cells treated with PA&OA (Fig. S13B). These results indicate that the ERK1/2 pathway participated in the induction of STARD4 by GPR35.
In adipocytes, Kyna/GPR35 signals activate ERK1/2-CREB signaling and induce downstream gene expression31. Furthermore, ERK1/2 and p38-mediated CREB signaling can upregulate steroidogenic acute regulatory protein (StAR) transcription48. However, whether CREB regulates STARD4 expression remains unclear. We then examined whether CREB is involved in the induction of STARD4 by GPR35-ERK1/2. The results show that CREB phosphorylation was induced by GPR35 in the livers of HFCF diet-fed mice (Fig. S13C) and was inhibited by ERK1/2 inhibitors in AML-12 cells treated with PA&OA (Fig. S13B). Similarly, the CREB inhibitor (666-15) significantly reversed GPR35-induced STARD4 protein expression in AML-12 cells treated with PA&OA (Fig. S13D). Taken together, these results suggest that hepatocyte GPR35 increased the expression of STARD4 via the ERK1/2-CREB signaling pathway in the NAFLD model.
3.8. A pharmacological agonist of GPR35 prevents HFCF diet-induced steatohepatitis in mice
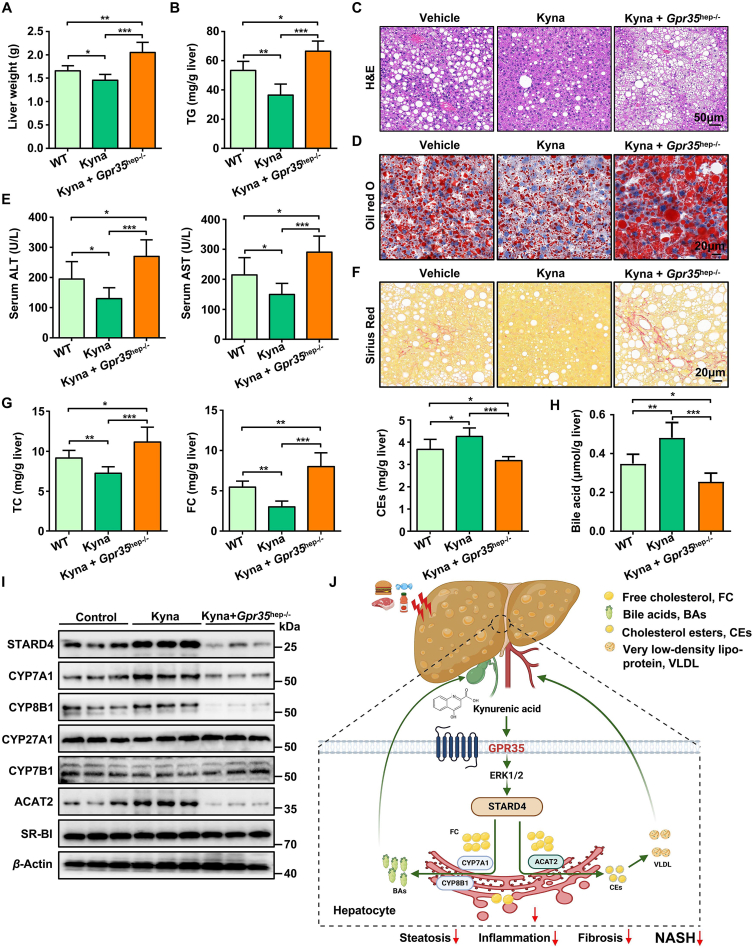
Kyna is an endogenous agonist of the orphan receptor GPR3549. Kyna has been reported to increase energy expenditure by activating GPR35 in adipose tissue31. We sought to investigate whether Kyna can be used to inhibit the development of HFCF diet-induced steatohepatitis in the liver. C57BL/6J WT and Gpr35hep−/− mice were fed a HFCF diet for 8 weeks until they showed significant body weight gain and signs of impaired glucose tolerance. At that point, we started daily administration of Kyna (5 mg/kg body weight, i.p.) to the C57BL/6J mice and continued feeding them the HFCF diet for 8 weeks. After Kyna administration, WT mice, but not Gpr35hep−/− mice, exhibited decreases in liver weight and TG content compared with the levels in vehicle-injected mice (Fig. 8A and B). Chronic Kyna administration ameliorated the steatosis, injury, and fibrosis of the liver induced by HFCF-diet feeding for 16 weeks, whereas loss of GPR35 expression in hepatocytes prevented the protective effect of Kyna (Fig. 8C–F). Kyna administration resulted in remarkable reductions in the hepatic levels of TC and FC and increases in the hepatic levels of CEs and bile acids (Fig. 8G and H). This effect was accompanied by Kyna-mediated upregulation of the expression of proteins involved in cholesterol metabolism (CYP7A1, CYP8B1, and ACAT2) (Fig. 8I). Deletion of GPR35 (the receptor of Kyna) eliminated the promoting effect of Kyna on bile acid production and cholesterol esterification (Fig. 8G–I). In addition, consistent with the results obtained in GPR35-overexpressing mice, Kyna treatment activated the ERK1/2–CREB–STARD4 signaling pathway in the liver (Supporting Information Figs. S14 and 8I). Taken together, these results show that Kyna prevented HFCF diet-induced steatohepatitis by inducing GPR35 activation in the liver.
Figure 8.
The GPR35 agonist Kyna prevents HFCF diet-induced steatohepatitis in mice (n = 6). After HFCF-diet feeding for 8 weeks, WT and Gpr35hep−/− mice were injected with Kyna (5 mg/kg body weight, i.p.) daily and continued to be fed the HFCF diet for 8 weeks. (A) Liver weight. (B) TG level in the liver. (C) H&E staining of liver tissue sections. Scale bar, 50 μm. (D) Oil Red O staining of liver tissue sections. Scale bar, 20 μm. (E) Serum levels of ALT and AST. (F) Sirius Red staining of liver-tissue sections. Scale bar, 20 μm. (G) TC, FC, CEs, and (H) bile acid levels in the liver. (I) Representative Western blot of STARD4, CYP7A1, CYP8B1, CYP27A1, CYP7B1, SR-BI, and ACAT2 proteins in the liver. (J) Model depicting the critical role GPR35 signaling plays in controlling NASH by reprogramming cholesterol homeostasis in hepatocytes, thereby attenuating NASH. The picture was created by BioRender.com. Data are the mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
We found that GPR35 expression was increased in the hepatocytes of mice with NASH or ob/ob mice. Upon observing increased GPR35 expression in hepatocytes in the NASH model, we investigated the role of hepatocyte GPR35 expression in steatohepatitis development. GPR35 overexpression in hepatocytes protected against HFCF diet-induced steatohepatitis, inflammation, and fibrosis. In contrast, GPR35 deletion in hepatocytes aggravated steatohepatitis upon HFCF diet feeding. We showed that the cholesterol transporter-associated protein STARD4 was responsible for the protective function of hepatic GPR35 against steatohepatitis. STARD4 also promoted bile acid production by inducing CYP7A1 and CYP8B1 expression, and mediated cholesterol esterification by inducing ACAT2 expression. Ultimately, these actions led to reduced FC in the liver. Furthermore, in vivo administration of Kyna (an endogenous agonist of GPR35) increased hepatic cholesterol output and BAS and ameliorated the symptoms of steatohepatitis in mice (Fig. 8J).
Lipotoxicity drives the development of progressive hepatic inflammation and fibrosis in a subgroup of patients with NASH. The underlying molecular mechanisms responsible for the development of inflammation and fibrosis that characterize progressive NASH are unclear. However, emerging evidence from experimental and clinical studies has linked altered hepatic cholesterol homeostasis and FC accumulation to NASH pathogenesis9,10. Several in vitro studies have indicated that GPR35 activation inhibits lipid accumulation in hepatocytes50,51. Nevertheless, the identity of the class of toxic lipids regulated by GPR35 and the mechanism are not known.
The role of hepatocyte GPR35 in regulating hepatic cholesterol homeostasis has not been reported. Using approaches based on gain of function and loss of function, we showed that GPR35 significantly reduced liver TC and FC levels and increased CEs levels by inducing STARD4 expression. STARD4 is a soluble sterol transport protein implicated in “sensing” of cholesterol and maintenance of cellular homeostasis. STARD4 is expressed widely and has been shown to transfer sterol between liposomes as well as cell organelles. Overexpression of STARD4 has also been shown to increase CEs formation43. STARD4 overexpression improves sterol delivery to the endocytic recycling compartment and leads to ACAT2-dependent accumulation of sterol esters in lipid droplets, whereas STARD4 silencing increases cellular FC levels dramatically40. We showed that GPR35 increased STARD4 expression through the ERK1/2 signaling pathway, thereby promoting the conversion of FC to CEs in the liver. GPR35 did not affect hepatic genes involved in the biosynthesis, uptake, or excretion of cholesterol.
Conversion of cholesterol to bile acids is a critical pathway by which cholesterol can be removed from hepatocytes, which prevents the accumulation of cholesterol and toxic metabolites44,52. GPR35 promoted BAS in the livers of HFCF diet-fed mice through STARD4 regulation. Furthermore, GPR35 did not affect the reabsorption, conjugation, or export of bile acids in the liver or the export of bile acids in the ileum. The conversion of cholesterol to bile acids occurs via two main pathways. The neutral (classic) pathway is initiated by CYP7A1 in the ER of the liver, which synthesizes two primary bile acids: cholic acid (CA) and chenodeoxycholic acid (CDCA). CYP8B1 is required for CA synthesis (without CYP8B1, the product is CDCA). A fraction (3%–18%) of the bile acid “pool” is synthesized via the acidic (or alternative) pathway, which is initiated by CYP27A137,52,53. The latter is located in the inner mitochondrial membrane, where the cholesterol content is very low. CYP7B1 participates in a series of alternative pathway reactions. Overexpression of the steroidogenic acute regulatory protein has been reported to lead to a marked increase in the rate of BAS initiated by delivery of cholesterol to mitochondria containing CYP27A154,55. However, the pathway and mechanism by which STARD4 regulates BAS are not known. We found that STARD4 promoted the neutral pathway of BAS but not the acidic pathway, mainly by regulating the expression of CYP7A1 and CYP8B1. STARD4 mainly delivers cholesterol to the ER44, which is rich in CYP7A1, which may explain why STARD4 regulates the neutral BAS pathway. GPR35 promoted the expression of the genes involved in FAO and decreased the content of FFA in the livers of HFCF diet-fed mice but did not affect DNL in hepatocytes. Bile acids produced by CYP7A1 and CYP8B1 are endogenous ligands for FXR19, whose activation induces FAO. The selective regulation of FAO (but not DNL) may be attributable to FXR activation by bile acids.
SREBPs are a class of transcription factors that sense intracellular cholesterol homeostasis. There are 3 SREBP proteins: SREBP-2 primarily activates genes involved in cholesterol synthesis, whereas SREBP-1a and SREBP-1c have greater effects on genes involved in fatty acid synthesis. SREBPs are synthesized as transcriptionally inactive ER transmembrane proteins56. When the cholesterol content of ER membranes drops below 5% (mol/mol), SREBPs is escorted from the ER to the Golgi by the cholesterol-sensing protein SREBP cleavage-activating protein (SCAP)56. SREBPs is then proteolytically processed to generate a cytoplasmic form that translocates into the nucleus and activates transcription of target genes implicated in sterol biosynthesis, uptake, and metabolism. Above the 5% (mol/mol) threshold, cholesterol-bound SCAP and SREBPs are retained in the ER, preventing SREBPs processing. Although overexpression of STARD4 promoted ER cholesterol import, our results showed that there were no significant differences in the expression of the SREBPs target genes Hmgcr, Ldlr and Srb1 in the livers of STARD4-overexpressing mice compared with those of control mice (Fig. S11). In addition to STARD4's ability to mobilize cholesterol into the ER, our results and those of previous studies40,43 also confirm that STARD4 has a strong ability to convert cholesterol into bile acids and CEs in the ER, which may eventually lead to a lack of a significant increase or even a decrease in the ER cholesterol level. STARD4, a strong candidate participant in intracellular cholesterol homeostasis, is an SREBP2 target gene. Interestingly, a previous study40 showed that STARD4 is necessary for SREBP2 to sense changes in cellular cholesterol. In STARD4-silenced cells, cholesterol-mediated regulation of SREBP-2 is markedly prevented despite the higher FC levels in these cells40. These findings may explain why knockdown of STARD4 had no effect on the expression of SREBP2 target genes in the livers of HFCF diet-fed mice. These results suggest that the SREBPs-mediated cholesterol feedback system is a complex and fine process, which is worthy of further investigation in future work.
Pharmacological agonists of GPR35 have been shown to be involved in thermogenesis and anti-inflammatory responses in adipose tissue31,57. However, the effect of GPR35 agonists in the treatment of hepatic inflammatory diseases (including NASH) are not known. Knowing how hepatocytes respond to GPR35 agonists is necessary to better understand how these targeted therapies might be applied in patients with NASH. In the current study, an endogenous agonist of GPR35, Kyna, promoted cholesterol esterification and bile acid production and reduced liver FC content, thereby resulting in significant amelioration of experimental steatohepatitis. Moreover, treatment with Kyna increased the expression of STARD4 in the liver tissues of HFCF diet-fed mice, thereby promoting the expression of CYP7A1 and ACAT2. Notably, the effect of Kyna on hepatic cholesterol homeostasis and the STARD4–CYP7A1/ACAT2 axis was dependent upon GPR35. This result provides a potential rationale for clinical trials to investigate the use of targeted GPR35 agonists for NASH treatment.
5. Conclusions
We reveal GPR35 expression in hepatocytes to be a key regulator of cholesterol homeostasis in NASH. GPR35 expression in hepatocytes promotes (i) the classic pathway of BAS in the liver and (ii) cholesterol secretion by cholesterol esterification. Reducing FC levels could reduce lipotoxicity. Targeting GPR35 expression in hepatocytes may be a promising approach for NASH treatment.
Acknowledgments
We want to thank the Inflammation and Immune Mediated Diseases Laboratory of Anhui Province and the Laboratory Animal Center, University of Science and Technology of China. This work was supported by the National Science Fund for Distinguished Young Scholars (#82225008, China), the National Natural Science Foundation of China (#82070608), the Anhui Provincial Natural Science Foundation (#2108085Y28, China) and the Research Improvement Program of Anhui Medical University (#2019xkjT007, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.10.011.
Contributor Information
Hua Wang, Email: wanghua@ahmu.edu.cn.
Xuefu Wang, Email: wangxuefu@ahmu.edu.cn.
Author contributions
Xuefu Wang and Hua Wang designed the research. Xiaoli Wei, Fan Yin and Miaomiao Wu carried out the experiments and performed data analysis. Qianqian Xie, Xueqin Zhao, Ruiqian Xie, Cheng Zhu, Chongqing Chen, Menghua Liu, Xueying Wang, Ruixue Ren, Guijie Kang, Chenwen Zhu and Jingjing Cong participated in part of the experiments. Xuefu Wang and Hua Wang provided experimental drugs and quality control. Xiaoli Wei, Fan Yin and Miaomiao Wu wrote the manuscript. Xuefu Wang, Hua Wang, Xiaoli Wei, Fan Yin and Miaomiao Wu revised the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Younossi Z. Non-alcoholic fatty liver disease—a global public health perspective. J Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 2.Oduro P., Zheng X., Wei J., Yang Y., Wang Y., Zhang H., et al. The cGAS–STING signaling in cardiovascular and metabolic diseases: future novel target option for pharmacotherapy. Acta Pharm Sin B. 2022;12:50–75. doi: 10.1016/j.apsb.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilg H., Moschen A., Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol. 2017;14:32–42. doi: 10.1038/nrgastro.2016.147. [DOI] [PubMed] [Google Scholar]
- 4.Neuschwander-Tetri B. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–788. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 5.Dong J., Viswanathan S., Adami E., Singh B., Chothani S., Ng B., et al. Hepatocyte-specific IL11 cis-signaling drives lipotoxicity and underlies the transition from NAFLD to NASH. Nat Commun. 2021;12:66. doi: 10.1038/s41467-020-20303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., Pan Q., Cao H., Xin F., Zhao Z., Yang R., et al. Lipotoxic hepatocyte-derived exosomal microRNA 192-5p activates macrophages through rictor/akt/forkhead box transcription factor O1 signaling in nonalcoholic fatty liver disease. Hepatology. 2020;72:454–469. doi: 10.1002/hep.31050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi K., Yang L., McCall S., Huang J., Yu X., Pandey S., et al. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology. 2007;45:1366–1374. doi: 10.1002/hep.21655. [DOI] [PubMed] [Google Scholar]
- 8.Wouters K., van Bilsen M., van Gorp P., Bieghs V., Lütjohann D., Kerksiek A., et al. Intrahepatic cholesterol influences progression, inhibition and reversal of non-alcoholic steatohepatitis in hyperlipidemic mice. FEBS Lett. 2010;584:1001–1005. doi: 10.1016/j.febslet.2010.01.046. [DOI] [PubMed] [Google Scholar]
- 9.Ioannou G. The role of cholesterol in the pathogenesis of NASH. Trends Endocrinol Metab. 2016;27:84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Musso G., Gambino R., Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog Lipid Res. 2013;52:175–191. doi: 10.1016/j.plipres.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Yu X., Ou X., Ouyang X., Tang C. Hepatic cholesterol transport and its role in non-alcoholic fatty liver disease and atherosclerosis. Prog Lipid Res. 2021;83 doi: 10.1016/j.plipres.2021.101109. [DOI] [PubMed] [Google Scholar]
- 12.Min H., Kapoor A., Fuchs M., Mirshahi F., Zhou H., Maher J., et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15:665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linton M., Tao H., Linton E., Yancey P. SR-BI: a multifunctional receptor in cholesterol homeostasis and atherosclerosis. Trends Endocrinol Metab. 2017;28:461–472. doi: 10.1016/j.tem.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T., Matozel M., Boehme S., Kong B., Nilsson L., Guo G., et al. Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzolo D., Kong B., Taylor R.E., Brinker A., Goedken M., Buckley B., et al. Bile acid homeostasis in female mice deficient in Cyp7a1 and Cyp27a1. Acta Pharm Sin B. 2021;11:3847–3856. doi: 10.1016/j.apsb.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plummer A., Culbertson A., Liao M. The ABCs of sterol transport. Annu Rev Physiol. 2021;83:153–181. doi: 10.1146/annurev-physiol-031620-094944. [DOI] [PubMed] [Google Scholar]
- 17.Parini P., Davis M., Lada A., Erickson S., Wright T., Gustafsson U., et al. ACAT2 is localized to hepatocytes and is the major cholesterol-esterifying enzyme in human liver. Circulation. 2004;110:2017–2023. doi: 10.1161/01.CIR.0000143163.76212.0B. [DOI] [PubMed] [Google Scholar]
- 18.Tarling E., Clifford B., Cheng J., Morand P., Cheng A., Lester E., et al. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Investig. 2017;127:3741–3754. doi: 10.1172/JCI94029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y., Zhu Y., Hu S., Xu Y., Stroup D., Pan X., et al. Hepatocyte nuclear factor 4α prevents the steatosis-to-NASH progression by regulating p53 and bile acid signaling (in mice) Hepatology. 2021;73:2251–2265. doi: 10.1002/hep.31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura T., Pydi S., Pham J., Tanaka N. Metabolic functions of G protein-coupled receptors in hepatocytes-potential applications for diabetes and NAFLD. Biomolecules. 2020;10:1445. doi: 10.3390/biom10101445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh D., Walenta E., Akiyama T., Lagakos W., Lackey D., Pessentheiner A., et al. A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nat Med. 2014;20:942–947. doi: 10.1038/nm.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson L., Elias M., Bolick D., Skaflen M., Green R., Hedrick C. The G protein-coupled receptor G2A: involvement in hepatic lipid metabolism and gallstone formation in mice. Hepatology. 2008;48:1138–1148. doi: 10.1002/hep.22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipina C., Walsh S., Mitchell S., Speakman J., Wainwright C., Hundal H. GPR55 deficiency is associated with increased adiposity and impaired insulin signaling in peripheral metabolic tissues. FASEB J. 2019;33:1299–1312. doi: 10.1096/fj.201800171R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Xie L., Du K., Liu C., Zhang N., Gu C., et al. Succinate-GPR-91 receptor signalling is responsible for nonalcoholic steatohepatitis-associated fibrosis: effects of DHA supplementation. Liver Int. 2020;40:830–843. doi: 10.1111/liv.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 26.Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studer E., Zhou X., Zhao R., Wang Y., Takabe K., Nagahashi M., et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Dowd B., Nguyen T., Marchese A., Cheng R., Lynch K., Heng H., et al. Discovery of three novel G-protein-coupled receptor genes. Genomics. 1998;47:310–313. doi: 10.1006/geno.1998.5095. [DOI] [PubMed] [Google Scholar]
- 29.Oxenkrug G. Increased plasma levels of xanthurenic and kynurenic acids in type 2 diabetes. Mol Neurobiol. 2015;52:805–810. doi: 10.1007/s12035-015-9232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Favennec M., Hennart B., Caiazzo R., Leloire A., Yengo L., Verbanck M., et al. The kynurenine pathway is activated in human obesity and shifted toward kynurenine monooxygenase activation. Obesity. 2015;23:2066–2074. doi: 10.1002/oby.21199. [DOI] [PubMed] [Google Scholar]
- 31.Agudelo L., Ferreira D., Cervenka I., Bryzgalova G., Dadvar S., Jannig P., et al. Kynurenic acid and Gpr35 regulate adipose tissue energy homeostasis and inflammation. Cell Metab. 2018;27:378–392.e5. doi: 10.1016/j.cmet.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y., Bielak L., Peyser P., Turner S., Sheedy P., Boerwinkle E., et al. Application of machine learning algorithms to predict coronary artery calcification with a sibship-based design. Genet Epidemiol. 2008;32:350–360. doi: 10.1002/gepi.20309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horikawa Y., Oda N., Cox N., Li X., Orho-Melander M., Hara M., et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 34.Caballero F., Fernández A., De Lacy A., Fernández-Checa J., Caballería J., García-Ruiz C. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. J Hepatol. 2009;50:789–796. doi: 10.1016/j.jhep.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Van Rooyen D., Gan L., Yeh M., Haigh W., Larter C., Ioannou G., et al. Pharmacological cholesterol lowering reverses fibrotic NASH in obese, diabetic mice with metabolic syndrome. J Hepatol. 2013;59:144–152. doi: 10.1016/j.jhep.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 36.Luo J., Yang H., Song B. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 37.Vlahcevic Z., Heuman D., Hylemon P. Regulation of bile acid synthesis. Hepatology. 1991;13:590–600. [PubMed] [Google Scholar]
- 38.Pikuleva I. Cytochrome P450s and cholesterol homeostasis. Pharmacol Ther. 2006;112:761–773. doi: 10.1016/j.pharmthera.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Soccio R., Adams R., Romanowski M., Sehayek E., Burley S., Breslow J. The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc Natl Acad Sci U S A. 2002;99:6943–6948. doi: 10.1073/pnas.052143799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesmin B., Pipalia N., Lund F., Ramlall T., Sokolov A., Eliezer D., et al. STARD4 abundance regulates sterol transport and sensing. Mol Biol Cell. 2011;22:4004–4015. doi: 10.1091/mbc.E11-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garbarino J., Pan M., Chin H., Lund F., Maxfield F., Breslow J. STARD4 knockdown in HepG2 cells disrupts cholesterol trafficking associated with the plasma membrane, ER, and ERC. J Lipid Res. 2012;53:2716–2725. doi: 10.1194/jlr.M032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iaea D., Dikiy I., Kiburu I., Eliezer D., Maxfield F. STARD4 membrane interactions and sterol binding. Biochemistry. 2015;54:4623–4636. doi: 10.1021/acs.biochem.5b00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Agudo D., Ren S., Wong E., Marques D., Redford K., Gil G., et al. Intracellular cholesterol transporter StarD4 binds free cholesterol and increases cholesteryl ester formation. J Lipid Res. 2008;49:1409–1419. doi: 10.1194/jlr.M700537-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iaea D., Maxfield F. Cholesterol trafficking and distribution. Essays Biochem. 2015;57:43–55. doi: 10.1042/bse0570043. [DOI] [PubMed] [Google Scholar]
- 45.Cobb M. MAP kinase pathways. Prog Biophys Mol Biol. 1999;71:479–500. doi: 10.1016/s0079-6107(98)00056-x. [DOI] [PubMed] [Google Scholar]
- 46.Pearson G., Robinson F., Beers Gibson T., Xu B., Karandikar M., Berman K., et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 47.Wang D., Li D., Zhang Y., Chen J., Zhang Y., Liao C., et al. Functional metabolomics reveal the role of AHR/GPR35 mediated kynurenic acid gradient sensing in chemotherapy-induced intestinal damage. Acta Pharm Sin B. 2021;11:763–780. doi: 10.1016/j.apsb.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dang X., Zhu Q., He Y., Wang Y., Lu Y., Li X., et al. IL-1β upregulates StAR and progesterone production through the ERK1/2- and p38-mediated CREB signaling pathways in human granulosa-lutein cells. Endocrinology. 2017;158:3281–3291. doi: 10.1210/en.2017-00029. [DOI] [PubMed] [Google Scholar]
- 49.Divorty N., Mackenzie A., Nicklin S., Milligan G. G protein-coupled receptor 35: an emerging target in inflammatory and cardiovascular disease. Front Pharmacol. 2015;6:41. doi: 10.3389/fphar.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nam S., Park S., Im D. Protective effect of lodoxamide on hepatic steatosis through GPR35. Cell Signal. 2019;53:190–200. doi: 10.1016/j.cellsig.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Lin L., Quon T., Engberg S., Mackenzie A., Tobin A., Milligan G. G protein-coupled receptor GPR35 suppresses lipid accumulation in hepatocytes. ACS Pharmacol Transl Sci. 2021;4:1835–1848. doi: 10.1021/acsptsci.1c00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chávez-Talavera O., Tailleux A., Lefebvre P., Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017;152:1679–1694.e3. doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 53.Chiang J. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren S., Hylemon P., Marques D., Gurley E., Bodhan P., Hall E., et al. Overexpression of cholesterol transporter StAR increases in vivo rates of bile acid synthesis in the rat and mouse. Hepatology. 2004;40:910–917. doi: 10.1002/hep.20382. [DOI] [PubMed] [Google Scholar]
- 55.Ren S., Hylemon P., Marques D., Hall E., Redford K., Gil G., et al. Effect of increasing the expression of cholesterol transporters (StAR, MLN64, and SCP-2) on bile acid synthesis. J Lipid Res. 2004;45:2123–2131. doi: 10.1194/jlr.M400233-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Radhakrishnan A., Goldstein J., McDonald J., Brown M. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dadvar S., Ferreira D., Cervenka I., Ruas J. The weight of nutrients: kynurenine metabolites in obesity and exercise. J Intern Med. 2018;284:519–533. doi: 10.1111/joim.12830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.