Abstract
Background
Glioblastoma (GBM) is a malignant tumor with a short survival and poor prognosis and a lack of clinically validated biomarkers for diagnosis and prognosis.
Methods
We collected cerebrospinal fluid (CSF) samples and normal CSF sample from recurrent GBM patients and paired tissue samples. Methylation profiles of CSF circulating tumor DNA (ctDNA) and transcriptional profiles of tumor tissues were analyzed. The China Glioma Genome Atlas (CGGA) database and Gene Expression Omnibus (GEO) was used for data analysis.
Results
Lasso analysis and multiplex Cox analysis were performed using intersecting genes of differentially methylated regions and differentially expressed genes. 8 hub genes were screened to construct diagnostic and prognostic models. Based on these 8 hub genes, the diagnostic (AUC = 0.944) and prognostic (3-years, AUC = 0.876) models were accurate.
Conclusions
In this study, 8 hub genes were identified for the diagnosis and prognosis of recurrent GBM, providing new biomarkers for the clinical study of recurrent GBM.
Keywords: Recurrent glioblastoma, Liquid biopsy, ctDNA, Methylation, CSF
1. Introduction
Glioblastoma (GBM) is a common central nervous system (CNS) malignancy with a low survival rate and a lack of effective clinical treatments [1,2]. Maximum surgical resection was followed by chemoradiotherapy, is the current standard treatment for GBM [3]. However, the prognosis of aggressively treated GBM patients remains poor, with a median survival time of less than 15 months and a high risk of recurrence [4] due to rapidly growing, aggressive GBM cells in tumor mass that infiltrate into the normal brain parenchyma around the lesion [5]. Moreover, some GBM patients are diagnosed only when they develop symptoms and often miss the optimal time for treatment [6]. Therefore, the development of effective biomarkers to monitor primary or recurrent GBM is critical.
Biopsy is used as one of the strategies for the diagnosis and prognosis of patients with GBM, but its limitations are significant. Tumor tissue is usually obtained through surgery, a highly invasive procedure that may be difficult to obtain because of the anatomical location of the tumor and the inability to monitor the tumor for early diagnosis, residual status, and recurrence [7]. In addition, tumor histopathology biopsies do not fully represent tumor heterogeneity, limiting the accuracy of predictive results [8]. Therefore, there is an urgent clinical need for a method to diagnose tumors and monitor their progression and recurrence in real-time. Recently, liquid biopsies (LB), particularly the analysis of circulating tumor DNA (ctDNA), have demonstrated significant advantages and promising applications in the field of cancer diagnosis and surveillance [9]. LB identifies and monitors tumor biomarkers at the molecular level, such as circulating tumor cells (CTCs), ctDNA, exosomes, and circulating tumor RNA (ctRNA), using human blood, urine, saliva, and other body fluids. Among the LB analytes, ctDNA is a biomarker that contains tumor-specific genetic information. Therefore, it can be used as an alternative to tumor DNA for diagnosis and prognostic monitoring [10]. However, due to the blood-brain barrier (BBB), the amount of ctDNA in plasma in CNS tumors is much lower than that in solid tumors, making it more challenging to extract ctDNA from plasma. Fortunately, although the detection of ctDNA in plasma of brain tumor patients is challenging, the detection of ctDNA in CSF may provide a new low-cost approach to the clinic [11,12].
CSF, as one of the important components of LB, can provide nutrients, transmit signaling molecules and clear disease-causing substances in the brain [[13], [14], [15]]. CSF is an important diagnostic method for many central nervous system diseases, and the correct selection of CSF test is helpful for the clinical treatment and diagnosis of brain tumors [16]. Alexandra M. Miller et al. detected tumor-derived DNA in cerebrospinal fluid and was associated with disease burden and adverse outcomes [17]. Mutations in CSF ctDNA are consistent with the genetic changes seen in brain tumors and can be used as a reliable source for determining CNS tumor status [12,17,18]. Tumor cells release about 200bp of DNA into the surrounding environment and further spread to plasma and CSF [19]. Thus, CSF tends to diagnose ctDNA earlier than plasma under the influence of the BBB [20].
Therefore, the present study aimed to find biological markers capable of diagnosing and prognosticating recurrent GBM using CSF ctDNA genome-wide methylation profiling.
2. Materials and methods
2.1. Samples
CSF samples and paired tumor tissue samples were obtained from four recurrent GBM patients who underwent surgical procedures at the Department of Neurosurgery, Sanbo Brain Hospital, Capital Medical University (Beijing, China). T1 samples were taken from the left basal ganglia region of the thalamus, T2 and T3 samples from the left temporal lobe, and T4 samples from the right frontal lobe. In addition, one paracancerous tissue sample and one normal CSF sample were collected as controls. All samples were pathologically diagnosed, and tumor tissues were rapidly frozen in liquid nitrogen and stored in a −80 °C refrigerator. Patient information is presented in Supplementary Table 1. All patients signed an informed consent form according to the study protocol approved by the local institutional review board (SBNK-2018-011-01).
2.2. CSF ctDNA whole-genome methylation profile sequencing
The cell-free DNA (cfDNA) was extracted according to the recommendations of the manufacturer of QIAamp Circulating Acid Kit (QIAGEN). In brief, highly purified methylated DNA is obtained by purification from cerebrospinal fluid, and the purified DNA is maintained in methylation state, which can be subjected to bisulfite conversion assay for methylation status analysis.
High throughput sequencing was provided by CloudSeq Biotech Inc. (Shanghai, China). The phenol-chloroform method was used to extract genomic DNA, which was then purified and recovered by ethanol precipitation. Genomic DNA was broken into 100–500 bp DNA fragments by a Bioruptor ultrasonicator. NEBNext® Ultra™ DNA Library Prep Kit from NEB was used to repair, tailed, and add splice sequences to fragmented DNA. Methylated DNA immunoprecipitation (MeDIP) was performed on DNA spiked with a methylated DNA antibody from Active Motif. The DNA library after MeDIP was accurately quantified by Quant-iT PicoGreen dsDNA Kits (Life Technologies). The DNA libraries were then sequenced for 150 bp double-end sequencing on Illumina's NovaSeq sequencing platform.
Raw reads (Raw Data) were generated after sequencing on an Illumina NovaSeq 6000 sequencer, image analysis, base identification, and quality control (QC) (Supplementary Table 2, Suppplementary Table 3 and Supplementary Fig. 1). First, Q30 was used to perform QC. Cutadapt software (v1.9.3) was used to adaptor-trimming, remove low-quality reads, and produce high-quality clean reads [21]. The clean reads were then aligned to the human reference genome (HG19) using bowtie2 software (v2.2.4) with default parameters [22]. Methylation enrichment peak identification was performed through MACS software (v2.0), and diffReps software (v1.55.4) was used to identify differentially methylated regions that were then annotated using the UCSC RefSeq database, peaks, and gene information [[23], [24], [25]].
2.3. Total mRNA expression profile sequencing
RNA high throughput sequencing was performed by Cloud-Seq Biotech (Shanghai, China). Ribosomal RNA (rRNA) was removed from the samples using the NEBNext rRNA Depletion Kit (New England Biolabs, Inc., Massachusetts, USA) under the supplier's instructions. NEBNext® Ultra™ II Directional RNA Library Prep Kit (New England Biolabs, Inc., Massachusetts, USA) was used to construct sequencing libraries. The libraries were quality-controlled and quantified using a BioAnalyzer 2100 system (Agilent Technologies, USA), and 150 bp double-end sequencing was performed using an Illumina NovaSeq 6000 instrument. The raw data was sequenced using the Illumina NovaSeq 6000 sequencer. Raw data QC was first performed using Q30 (Supplementary Table 4, Supplementary Table 5 and Supplementary Fig. 2). To remove low-quality reads and obtain high-quality clean reads, adaptor-trimming was performed using cutadapt software (v1.9.3) [21]. The clean reads were compared to the human reference genome (HG19) using hisat2 software (v2.0.4) (http://ccb.jhu.edu/software/hisat2/index.shtml). Then, using HTSeq software (v0.9.1), raw count numbers were obtained and normalized using edgeR. Ploidy changes and p-values were calculated between the samples to screen for differentially expressed genes [26,27].
2.4. Identification of methylation markers
To screen biomarkers for recurrent glioblastoma, we first compared 5-hydroxymethylcytomidine (5hmC) in plasma cfDNA from the GEO database, which included 111 glioma patients and 111 age - and sex-matched healthy individuals (GSE132118). 9199 differential genes in GSE132118 were analyzed by DESeq2 package, and these genes will be included in hub gene screening [28]. From CGGA database (Home | CGGA - Chinese Glioma Genome Atlas) of 109 cases of recurrent glioma samples (including secondary recurrence) mRNA expression and 20 cases of normal samples, whitch will be used for prognostic model building.
2.5. Statistical analysis and image construction
The present study used R (version 3.6.3), SPSS software (version 24, IBM Company), and Graphpad Prism (v. 8.0.2) for statistical analysis and graph plotting. The “survival” and “survminer” R packages (https://CRAN.R-project.org/package=survival
The “survival” R package was used to calculate the risk scores for the hub gene, divided into high and low-risk groups based on the median risk score. Diagnostic recipient operating characteristics (ROC) for multi-gene associations were analyzed using SPSS software (version 24), predictors of genes were calculated using binary logistic regression analysis, and then their ROC was analyzed based on the predictors. In addition, visualization of some of the data in the present study was provided by an online analysis website (https://www.xiantao.love/).
3. Result
3.1. Characterization of CSF ctDNA methylation regions and distribution on chromosomes
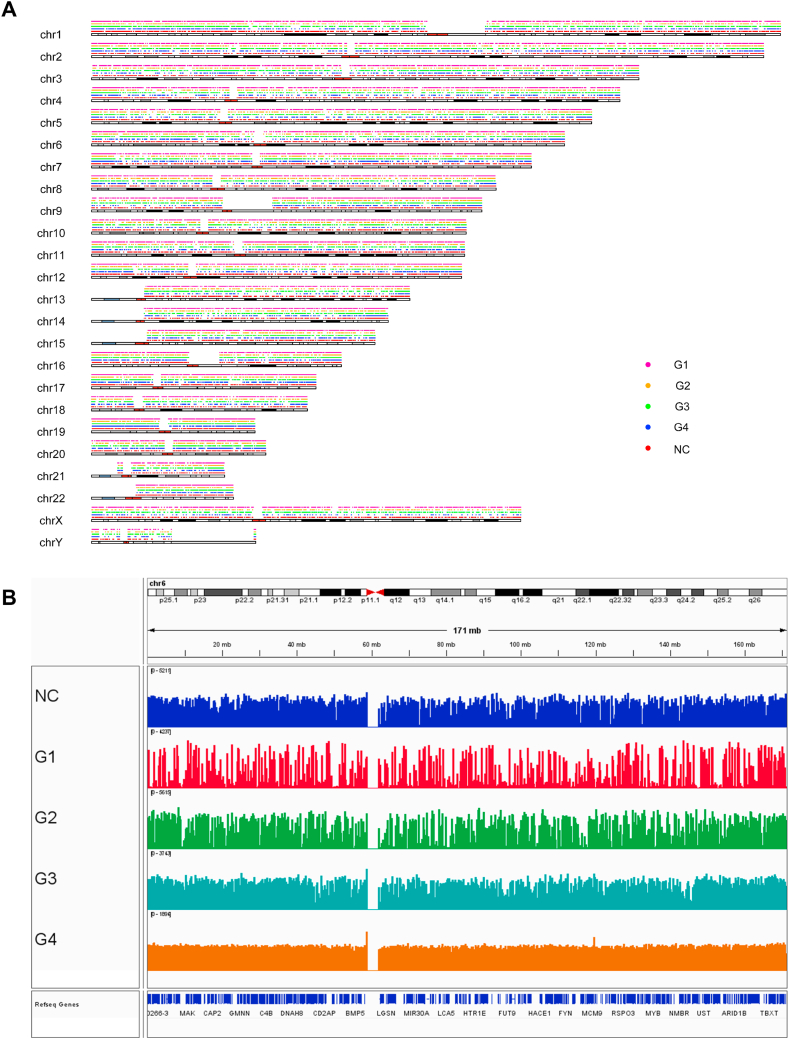
For each group of samples, we visualized the distribution of CSF ctDNA methylation regions on chromosomes, including tumor CSF samples (G1, G2, G3, and G4) and normal CSF samples (NC) (Fig. 1A). To determine the distribution and size of ctDNA methylated regions in CSF from GBM patients, IGV software (version 2.11.8) was used to visualize the sequencing results. (Fig. 1B). The results showed that the distribution and peak of CSF ctDNA methylation regions on chromosomes did not show regularity.
Fig. 1.
Distribution and visualization of methylation regions on chromosomes. (A) Distribution of CSF ctDNA methylation regions on chromosomes. (B) The methylation level of each CSF sample group. The higher the methylation level, the more significant the peak.
3.2. Characteristics of methylation regions of CSF ctDNA samples in recurrent GBM
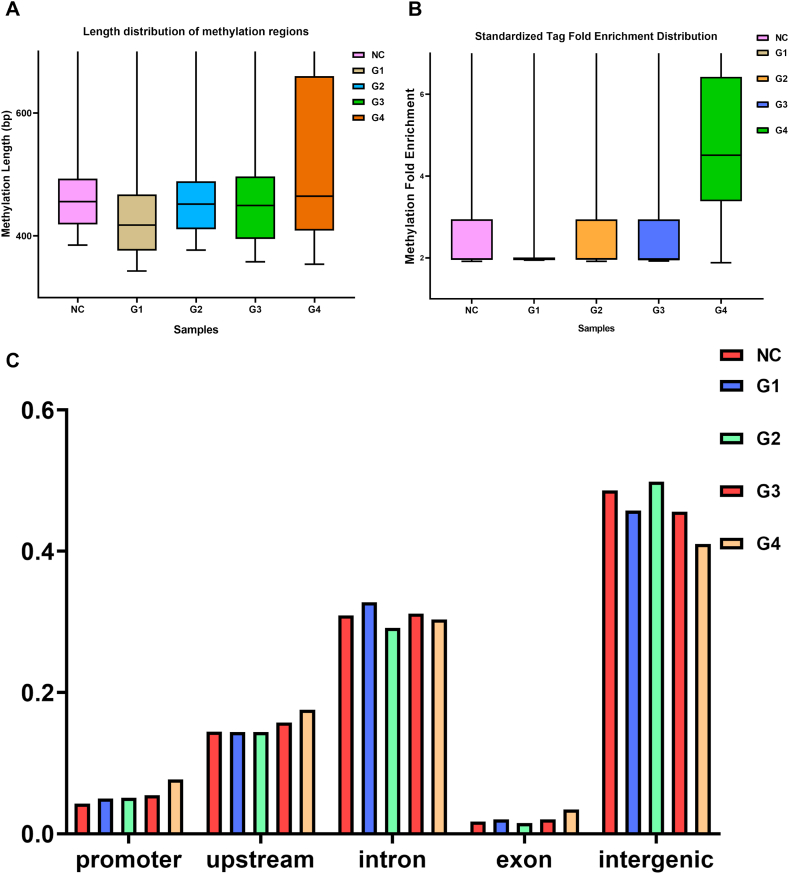
The length, tag fold enrichment distribution and ratio on genomic elements of CSF ctDNA methylation regions are shown and used to analyze the variability between tumor samples and normal samples in a holistic manner. The length of enrichment peak was higher in the G4 group (354–98298 bp) than in other tumor samples (Fig. 2A and B). Such results may be related to the individual variability among tumor samples. It also had a higher standardized tag fold enrichment than different samples. Furthermore, G1 demonstrated the lowest while G4 depicted the highest percentage of fold enrichment distribution. However, the majority of the methylation fold enrichment of tumor samples was concentrated between 2 and 3, with only a few methylation regions showing a high level of methylation. The analysis of the distribution ratio of genomic elements in each group of samples revealed that the distribution of recurrent GBM methylation regions on genomic elements did not differ significantly (Fig. 2C).
Fig. 2.
Characterization of the CSF ctDNA methylation region (A) The length distribution of enrichment peak in each group of CSF samples. (B) Fold enrichment distribution of methylation regions in each group of CSF samples. (C) Distribution of methylated regions on genomic elements.
3.3. Methylation levels of CSF ctDNA and screening of differential genes
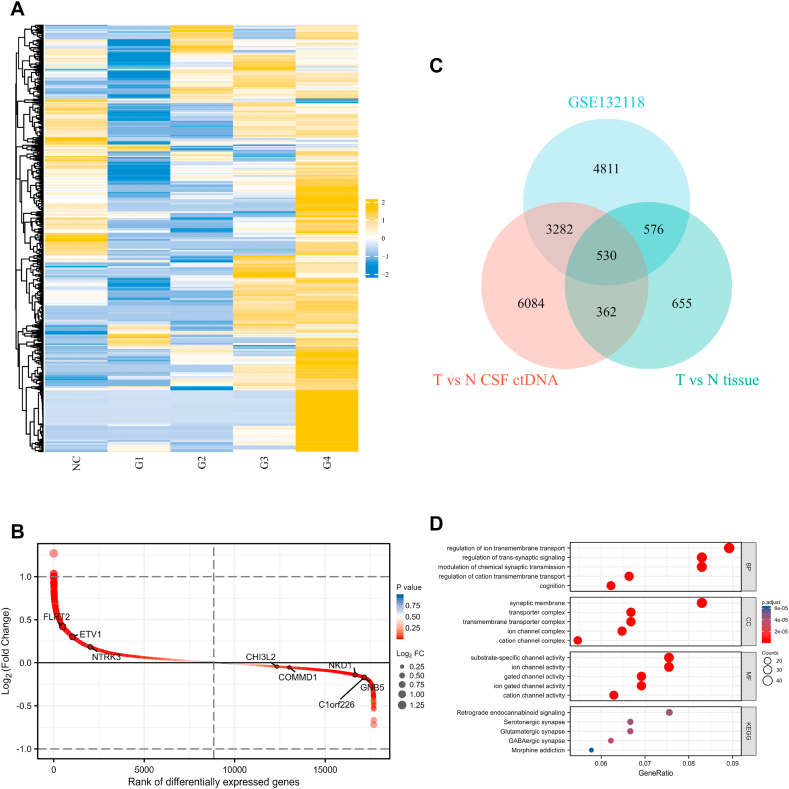
To screen genes related to prognosis and diagnosis of recurrent GBM, we analyzed methylation profiles of GSE132118, ctDNA differential methylation regions of CSF, and differential genes in tumor tissue. The methylation regions of CSF samples in the tumor group (G1–G4) and in the normal group (NC) were identified, duplicate identified regions were removed, and the methylation levels of 1348 methylation regions (Log FC) were then plotted in a heatmap (Fig. 3A). Due to the specificity between tumor samples, the methylation level of G4 was higher than that of other tumor samples, and G1 had the lowest methylation level among all tumor samples. It is important to note that one gene may correspond to multiple methylated regions. On the other hand, mRNA expression profiles of tumor tissue samples were analyzed, and a total of 2123 genes were different between the tumor group and the normal group (p < 0.05) (Supplementary Fig. 3). GSE132118 consisted of 111 glioma cfDNA samples and 111 paired normal cfDNA samples. 9199 differential methylated genes were identified in the analysis (Fig. 3B). After combined analysis of three sets of data (CSF ctDNA differential methylation region, tumor tissue differential gene, and GSE132118), 530 genes were screened for subsequent analysis (Fig. 3C). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were analyzed for enrichment of 530 genes. The identification contents included biological process (BP), molecular function (MF), and cell component (CC) (Fig. 3D). The results showed that the top 5 items in BP ranking are regulation of ion transmembrane transport, modulation of chemical synaptic transmission, and regulation of trans-synaptic signaling, cognition and regulation of cation transmembrane transport. In addition, KEGG top 5 pathways for GABAergic synapse, Glutamatergic synapse and Serotonergic synapse, Retrograde endocannabinoid signaling and Morphine addiction.
Fig. 3.
CSF ctDNA methylation levels and screening of hub genes. (A) Heatmap of ctDNA methylation levels in CSF. (B) Analysis of plasma cfDNA difference between tumor and normal samples in GSE132118. In the figure, the horizontal coordinate is the position of differentially expressed genes (from largest to smallest) in order of differentially expressed genes, and the vertical coordinate is differentially expressed genes. The closer the points are to the left and right sides, the greater the absolute value of the difference multiples. (C) Venn diagram of the intersection of CSF differential methylation domains (T vs N CSF ctDNA), tumor tissue differential genes (T vs N Tissue), and plasma cfDNA differential methylation genes (GSE132118). (D) GO and KEGG analysis of 530 intersection genes.
3.4. Screening hub genes from 530 genes
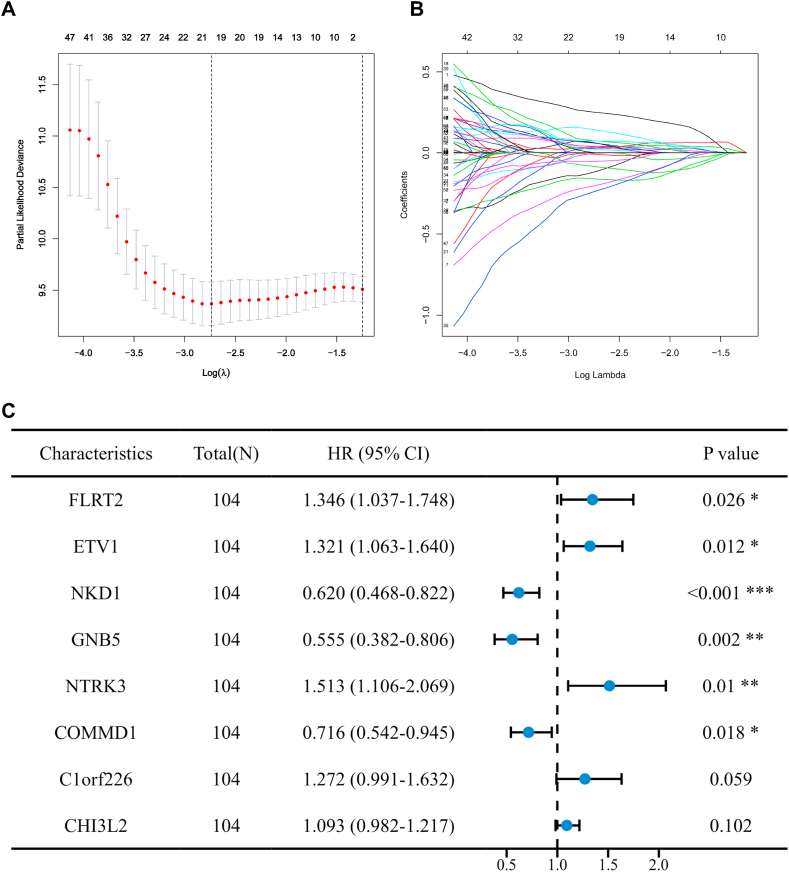
In order to screen out hub genes associated with prognosis and diagnosis of recurrent GBM from 530 genes, transcriptome data and clinical information of 109 recurrent GBM and 20 normal brain tissues from the CGGA database were collected as a training cohort and included in the following analysis. Based on univariate cox regression model, a total of 68 hub genes were identified to be significantly correlated with overall survival (Supplementary Fig. 4). The 20 genes selected in Lasso penalized cox analysis were analyzed by multivariate cox analysis (Fig. 4A and 4B), and 8 genes were finally selected to construct the prognostic model. They include FLRT2, ETV1, NKD1, GNB5, NTRK3, COMMD1, C1orf226, and CHI3L2 (Fig. 3, Fig. 4C).
Fig. 4.
Screening of hub genes based on CGGA database mRNA expression profiles. (A–B) 20 genes were selected from the Lasso regression analysis. (C) 8 hub genes were screened by multivariate cox analysis.
3.5. Construction of a prognosis model based on 8 hub genes
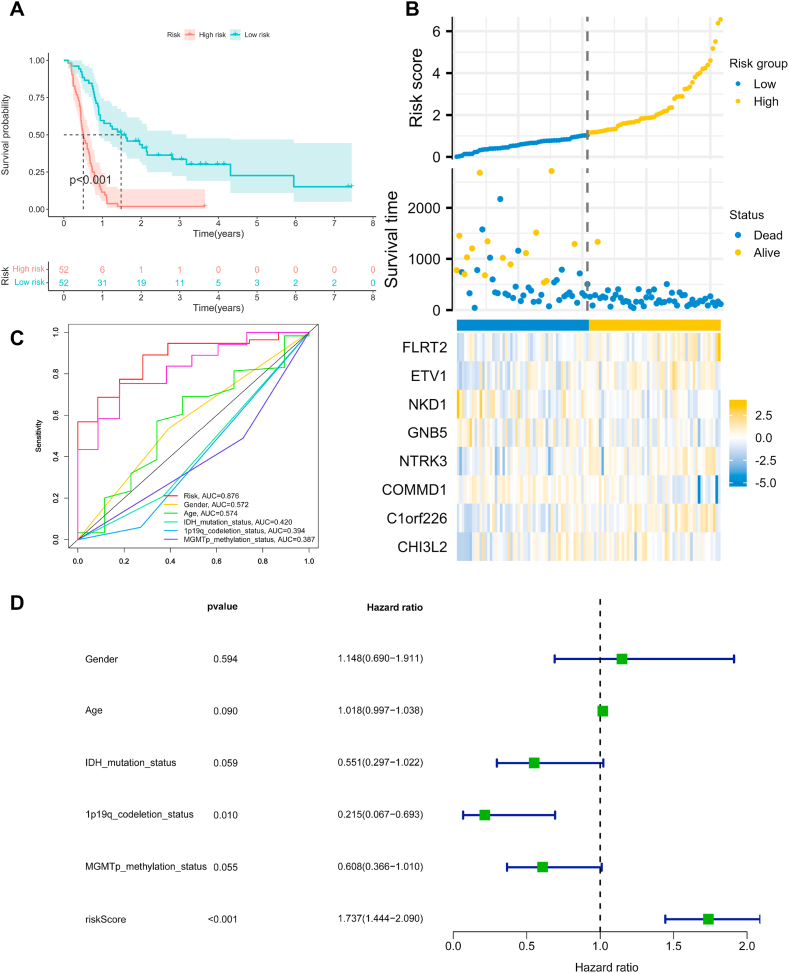
The risk score of the genes in each sample was calculated based on the expression of the 8 hub genes in the CGGA dataset, and the samples were classified into high-risk and low-risk groups based on the median of the risk scores. Results showed that patients in the high-risk group had significantly lower OS than those in the low-risk group (p < 0.001) (Fig. 5A). Moreover, with the increase of survival time, patients' risk score and mortality also increased, which meant that patients with higher hub gene expression levels generally had worse prognosis (Fig. 5B). The heatmap of hub gene expression showed that FLRT2, ETV1, NTRK3, and C1orf226 were directly proportional to the risk score, and NKD1, GNB5, COMMD1, and CHI3L2 were inversely proportional to the risk score. To assess the prognostic ability of hub genes, time-dependent receiver operational characteristic (ROC) curves (3 years) were used to calculate risk score and other clinical information including gender, age, IDH mutation status, and 1p19q codeletion satatus, MGMTp methyaltion status) area under curve (AUC) (Fig. 5C). Interestingly, these genes had higher sensitivity and specificity in predicting OS than other clinical information (AUC = 0.876). In addition, risk scores were used as independent prognostic factors to predict patient outcomes (p < 0.001) (Fig. 5D). Overall, we established a clinical prognostic model based on the expression levels of these hub genes in recurrent GBM, which can be used as an independent post-parameter to assess patients' clinical risk.
Fig. 5.
Construction of a prognostic model for recurrent GBM based on 8 hub genes. (A) KM survival curves for the high-risk and low-risk groups. The low-risk group is represented by blue, while the high-risk group is represented by red. (B) The top picture shows that the risk score increases with survival time. The picture in the middle shows the distribution of patients' survival over time. The bottom image shows the expression of hub genes in high-risk (yellow) and low-risk (blue) patients. (C) Time-dependent ROC for prognostic models and other clinical information. (D) Independent prognostic analysis of clinical information including risk score. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Constructing diagnostic model base on the 8 hub genes
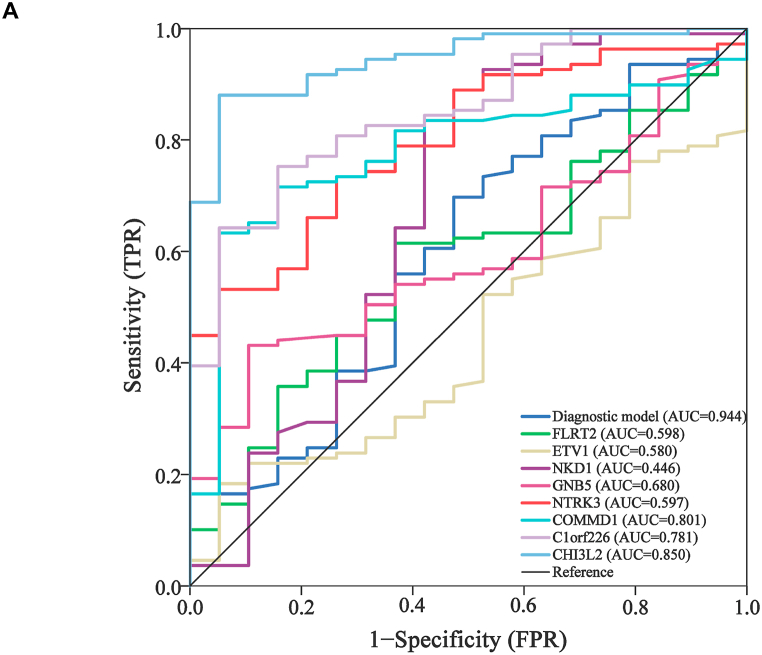
Based on the methylation levels of 111 gliomas and 111 normal plasma cfDNA in GSE132118 samples, binary logistic regression analysis was performed for the above 8 hub genes. The combined diagnostic ROC of eight genes was calculated using the diagnostic predictors of these genes (Fig. 6). The results showed that the AUC values of COMMD1 (AUC = 0.801), C1orf226(AUC = O.781) and CH3L2(AUC = 0.850) were relatively high. The AUC values of FLRT2 (AUC = 0.598), ETV1 (AUC = 0.580), NKD1 (AUC = 0.446), GNB5 (AUC = 0.680) and NTRK3 (AUC = 0.597) were low. It should be noted that the combined diagnostic ROC of hub gene (AUC = 0.944) was greater than that of single gene. This means that the diagnostic model constructed based on 8 hub genes has high accuracy in predicting the occurrence of recurrent glioblastoma, but it still needs to be further verified.
Fig. 6.
A diagnostic model based on the 8 hub genes. (A) Univariate diagnostic ROC of 8 hub genes and multifactor combined diagnostic ROC.
4. Discussion
As one of the first epigenetic regulatory patterns identified, DNA methylation has shown widespread application in the diagnosis and prognosis of GBM [29,30]. The epigenetic characteristics of the primary tumor can be expressed in CSF ctDNA. Detecting the methylation level of CSF ctDNA is a sensitive and accurate method to monitor tumorigenesis, progression, and recurrence and to predict outcomes [31]. Thus, we tried to find biomarkers for the diagnosis and prognosis of recurrent GBM by analyzing the genome-wide methylation profile of CSF ctDNA, which provide a better clinical approach to monitoring tumor recurrence.
In this study, the distribution of chromosome methylation regions in CSF samples was visualized. The results showed that the distribution of methylated regions did not differ significantly between tumor and normal samples. However, when we used IGV software to visualize the tumor samples, the normalized (log scale) G4 demonstrated smooth methylation levels, whereas the methylation levels of different regions in the G1 was quite variable. When the length distribution and standardized tag fold enrichment distribution of methylation regions in tumor samples were analyzed, the G4 showed a distinct group different from other samples. The G4 did not show significant differences from other tumor samples when analyzing the distribution of sample methylation regions on genomic elements. Such results may be due to the specificity between tumors or the insufficient number of samples in GBM patients. We will further expand the number of sample to verify these results.
Subsequently, we plotted a heatmap to compare the methylation levels of tumor samples (G1–G4) and normal samples (NC). The results depicted the methylation levels of the G4 was significantly higher than the other tumor samples, consistent with previous findings. However, in the mRNA expression profiles, there were significant expression differences between normal tissue samples (T1) and tumor tissue samples (T1–T4), and the T4 group did not show higher intergroup differences than before. Therefore, we concluded that the methylation variability in the G4 was acceptable and included in the analysis of the subsequent genetic screen. In addition, DNA methylation, an epigenetic regulator of cell differentiation and development, is achieved by manipulating gene expression without altering the genome sequence [32]. So we will CSF ctDNA methylation differences between area and mRNA expression of tumor tissue, the combination of joint GSE132118 data at the same time used to improve the accuracy of genetic screening.
Based on the transcriptional profiles of 109 recurrent GBM samples and 20 normal samples from the CGGA database, 530 genes were analyzed by univariate Cox analysis, Lasso regression analysis and multivariate Cox analysis. Through statistical analysis, 8 hub genes out of 530 genes were screened to construct prognostic and diagnostic models. The prognostic model built on 8 hub genes could accurately predict the survival of patients (AUC = 0.876), and the samples with high-risk scores had lower survival than those with low-risk scores. Furthermore, as expected, the risk score of patients increased with increasing survival time. The diagnostic model based on the methylation levels of the 8 hub genes showed high accuracy. The results suggest that the hub genes could not only predict a patient's prognosis, but also be capable of diagnosing recurrent GBM. However, on the basis of the available information, it is difficult to obtain clinical follow-up data over a long period of time, and longer follow-up and clinical monitoring are needed for further study and use to fully assess the accuracy of this risk score model in terms of clinical prognosis. We identified 8 hub genes associated with the diagnosis and prognosis of recurrent GBM using CSF ctDNA methylation profiling as biomarkers. These hub genes provide a new strategy for the diagnosis and prognosis of recurrent GBM. Based on the results of this study, we will further explore the mechanism of action of these genes on recurrent GBM.
Declarations
Author contribution statement
Lin Dai: Analyzed and interpreted the data; Wrote the paper.
Zhihui Liu: Contributed reagents, materials, analysis tools or data.
Yi Zhu: Performed the experiments.
Lixin Ma: Conceived and designed the experiments.
Funding statement
Lixin Ma was supported by the National Key R&D Program [2019YFC1316104]; National Science Foundation of China [22077120]; the China Postdoctoral Science Foundation [2019M660715].
Data availability statement
Data associated with this study has been deposited at GENE EXPRESSION OMNIBUS (GEO) under the accession number ercvisqszlybjgr.
Declaration of competing interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at.https://doi.org/10.1016/j.heliyon.2023.e14339.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Urbanska K., Sokolowska J., Szmidt M., Sysa P. Glioblastoma multiforme - an overview. Contemp. Oncol. 2014;18(5):307–312. doi: 10.5114/wo.2014.40559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birko Z., Nagy B., Klekner A., Virga J. Novel molecular markers in glioblastoma-benefits of liquid biopsy. Int. J. Mol. Sci. 2020;21(20) doi: 10.3390/ijms21207522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weller M., van den Bent M., Preusser M., Le Rhun E., Tonn J.C., Minniti G., et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021;18(3):170–186. doi: 10.1038/s41571-020-00447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Ma Y., Wu Z., Xie R., Zeng F., Cai H., et al. Advanced imaging techniques for differentiating pseudoprogression and tumor recurrence after immunotherapy for glioblastoma. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.790674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vollmann-Zwerenz A., Leidgens V., Feliciello G., Klein C.A., Hau P. Tumor cell invasion in glioblastoma. Int. J. Mol. Sci. 2020;21(6) doi: 10.3390/ijms21061932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao M., van Straten D., Broekman M.L.D., Preat V., Schiffelers R.M. Nanocarrier-based drug combination therapy for glioblastoma. Theranostics. 2020;10(3):1355–1372. doi: 10.7150/thno.38147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulet G., Massias J., Taly V. Liquid biopsy: general concepts. Acta Cytol. 2019;63(6):449–455. doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 8.Constancio V., Barros-Silva D., Jeronimo C., Henrique R. Known epigenetic biomarkers for prostate cancer detection and management: exploring the potential of blood-based liquid biopsies. Expert Rev. Mol. Diagn. 2019;19(5):367–375. doi: 10.1080/14737159.2019.1604224. [DOI] [PubMed] [Google Scholar]
- 9.Luo H., Wei W., Ye Z., Zheng J., Xu R.H. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol. Med. 2021;27(5):482–500. doi: 10.1016/j.molmed.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Diaz L.A., Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Springer S., Zhang M., McMahon K.W., Kinde I., Dobbyn L., et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl. Acad. Sci. USA. 2015;112(31):9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pentsova E.I., Shah R.H., Tang J., Boire A., You D., Briggs S., et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J. Clin. Oncol. 2016;34(20):2404–2415. doi: 10.1200/JCO.2016.66.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehtinen M.K., Walsh C.A. Neurogenesis at the brain-cerebrospinal fluid interface. Annu. Rev. Cell Dev. Biol. 2011;27:653–679. doi: 10.1146/annurev-cellbio-092910-154026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva-Vargas V., Maldonado-Soto A.R., Mizrak D., Codega P., Doetsch F. Age-dependent niche signals from the choroid plexus regulate adult neural stem cells. Cell Stem Cell. 2016;19(5):643–652. doi: 10.1016/j.stem.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Lun M.P., Johnson M.B., Broadbelt K.G., Watanabe M., Kang Y.J., Chau K.F., et al. Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J. Neurosci. 2015;35(12):4903–4916. doi: 10.1523/JNEUROSCI.3081-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leen W.G., Willemsen M.A., Wevers R.A., Verbeek M.M. Cerebrospinal fluid glucose and lactate: age-specific reference values and implications for clinical practice. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller A.M., Shah R.H., Pentsova E.I., Pourmaleki M., Briggs S., Distefano N., et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Mattos-Arruda L., Mayor R., Ng C.K.Y., Weigelt B., Martinez-Ricarte F., Torrejon D., et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z., Zhang C., Li M., Shen Y., Feng S., Liu J., et al. Applications of cerebrospinal fluid circulating tumor DNA in the diagnosis of gliomas. Jpn. J. Clin. Oncol. 2020;50(3):325–332. doi: 10.1093/jjco/hyz156. [DOI] [PubMed] [Google Scholar]
- 21.Kechin A., Boyarskikh U., Kel A., Filipenko M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 2017;24(11):1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puchalski R.B., Shah N., Miller J., Dalley R., Nomura S.R., Yoon J.G., et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360(6389):660–663. doi: 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X., Zhang Y., Hu T., He X., Zou Y., Deng Q., et al. A novel cell-free DNA methylation-based model improves the early detection of colorectal cancer. Mol Oncol. 2021;15(10):2702–2714. doi: 10.1002/1878-0261.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maire C.L., Fuh M.M., Kaulich K., Fita K.D., Stevic I., Heiland D.H., et al. Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol. 2021;23(7):1087–1099. doi: 10.1093/neuonc/noab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H.Y., Li J., Tang Y., Huang Y.X., Chen Y.G., Xie Y.Y., et al. MethHC 2.0: information repository of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2021;49(D1):D1268–D1275. doi: 10.1093/nar/gkaa1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at GENE EXPRESSION OMNIBUS (GEO) under the accession number ercvisqszlybjgr.