Abstract
Lipid-formulated RNA vaccines have been widely used for disease prevention and treatment, yet their mechanism of action and individual components contributing to such actions remain to be delineated. Here, we show that a therapeutic cancer vaccine composed of a protamine/mRNA core and a lipid shell is highly potent in promoting cytotoxic CD8+ T cell responses and mediating anti-tumor immunity. Mechanistically, both the mRNA core and lipid shell are needed to fully stimulate the expression of type I interferons and inflammatory cytokines in dendritic cells. Stimulation of interferon-β expression is exclusively dependent on STING, and antitumor activity from the mRNA vaccine is significantly compromised in mice with a defective Sting gene. Thus, the mRNA vaccine elicits STING-dependent antitumor immunity.
Key words: Cancer, Immunotherapy, mRNA, Vaccine, Dendritic cells, EDOPC, STING, MAVS
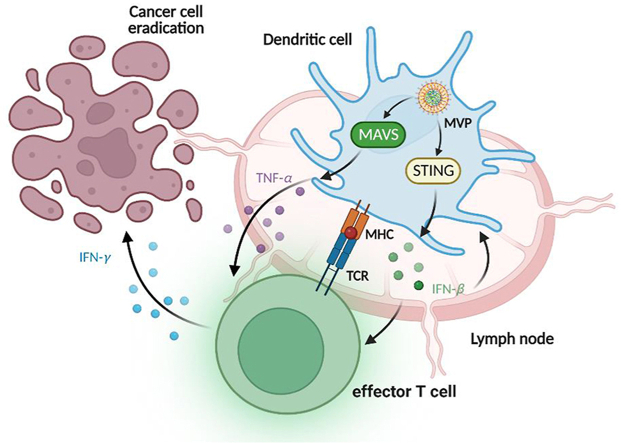
Graphical abstract
The mRNA vaccine particle (MVP) uptake by DC triggers expression of IFN-β and TNF-α through STING and MAVS dependent pathways, respectively. T cells are subsequently primed, leading to antigen-specific cancer cell killing and tumor eradication.
1. Introduction
Rapid development and worldwide application of mRNA vaccines for the prevention of SARS-CoV-2 infection have demonstrated the power of mRNA-based drugs in healthcare1,2. Due to their large molecular weight and negative charge, mRNA molecules need to be packaged into delivery vehicles in order to effectively enter mammalian cells3. Packaging into the nanometer-size delivery vehicles also has the benefit of protecting mRNA molecules from enzymatic degradation. Multiple delivery platforms have been developed to suit the purpose, such as lipid nanoparticle4, lipopolyplex (LPP)5, liposome-protamine-RNA (LPR)6, RNA-lipoplex (RNA-LPX)7, and virus-like vaccine particle (VLVP)8. While each platform has its own unique structure and composition, most vehicles contain an ionic lipid molecule that facilitates mRNA packaging and the escape of mRNA molecules from the endosomes.
With the success of the prophylactic vaccines, there is a general realization that mRNA therapeutics can be used to treat perhaps most, if not all, disease types9, 10, 11, 12. Indeed, mRNA-based therapeutic cancer vaccines have been studied for many years13,14. Recent advances in clinical trials have also demonstrated their application potential in selected cancer patients15, 16, 17. Unlike peptide cancer vaccines that are prepared with adjuvant molecules18, 19, 20, mRNA vaccine particles can also serve as self-adjuvants21. For example, a two-component mRNA-based cancer vaccine containing free and protamine-complexed mRNA can also activate the toll-like receptor 7 (TLR7) signaling22. However, with the increasing concern on acute innate immune toxicity from naked mRNA, most investigators and companies are using modified RNA to avoid innate recognition by the TLRs23. Consequently, the lipid components are playing an important role in enhancing adjuvant activity in the mRNA vaccine particle, preferentially by activating non-TLR signaling. A recent study on the lipid-formulated, negatively charged RNA-LPX constituted with 1,2-di-octadecenyl-3-trimethylammonium (DOTMA, a cationic lipid)/dioleoylphosphatidylethanolamine (DOPE, a helper lipid) liposome revealed activation of the interleukin 1 (IL1)-interleukin 1 receptor antagonist (IL-1ra) axis in regulating secretion of proinflammatory cytokines, and the essential role of activating the two-step inflammasome pathway in monocytes24. Interestingly, another recent investigation on the LNP-based BNT162b2 prepared with ALC-0315 (an ionized lipid), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC, a helper lipid), polyethylene glycol-2000-N,N-ditetradecylacetamide (PEG2000-DTA), and cholesterol showed the key role of activating type I interferon-dependent MDA5 signaling, but not TLRs or inflammasome, in stimulating both innate and adaptive immunity of the COVID-19 vaccine25. These studies point to the possibility that delivery platforms comprised of variable lipid molecules may rely on different signal transduction pathways for vaccine activity. Thus, it is important to fully investigate the function of key molecules and their combinations in order to further improve mRNA therapeutics.
In the current study, we set up experiments to dissect the functional role of individual components in a therapeutic cancer vaccine. The mRNA vaccine particle (MVP) is composed of a protamine/mRNA core that is encapsulated in a lipid shell consisting of a cationic lipid, a helper lipid, a pegylated lipid, and cholesterol (Fig. 1A). It has been demonstrated that inclusion of charged lipid can facilitate targeted RNA delivery26, and dioleoylethylphosphatidylcholine (EDOPC) and dioleoyl-3-trimethylammonium propane (DOTAP) are two of the cationic lipids that have been tested for this purpose5,27. We examined stimulation of expression of interferon-β (IFN-β), IL-1β, and tumor necrosis factor-α (TNF-α) by the mRNA core, mRNA-free vehicle, and the whole MVP, and correlated such activities to the TLR7, mitochondrial antiviral signaling (MAVS, also known as IPS-1), stimulator of IFN genes (STING), and TIR-domain-containing adapter-inducing IFN-β (TRIF) signaling. Subsequently, we investigated the role of protamine in the core and cationic lipid in the shell in stimulating IFN-β and TNF-α expression. Finally, we compared anti-tumor immune responses from the MVP in wild-type and gene knockout mice.
Figure 1.
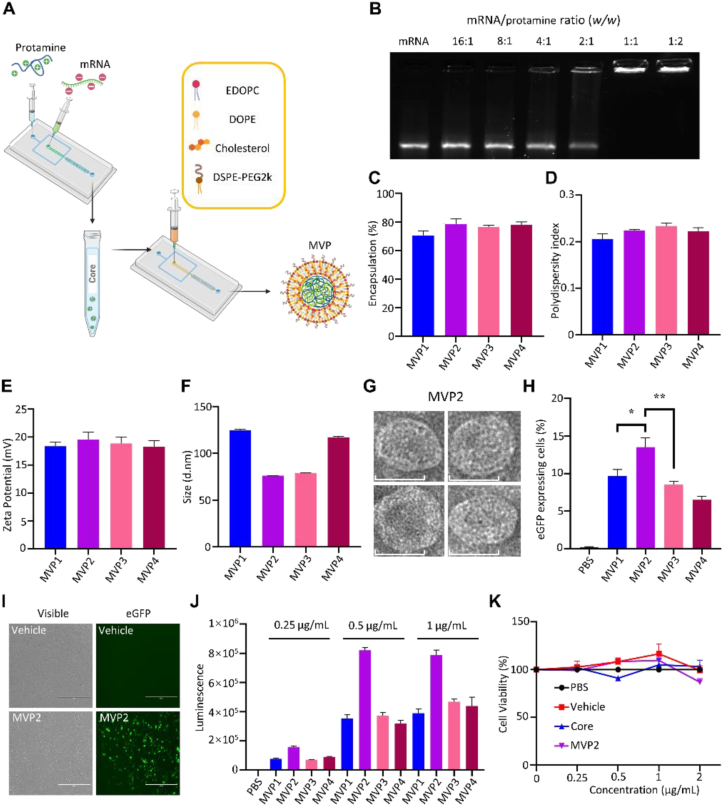
Preparation and characterization of mRNA particles. (A) Schematic view of vaccine particle preparation. (B) Agarose gel electrophoresis shows that mRNA molecules were retained in the sample-loading well in samples prepared with mRNA/protamine at 1:1 to 1:2 ratio. (C–F) Characterization of mRNA vaccine particles (MVPs) based on percentage of encapsulation, polydispersity index, zeta potential, and size. (G) Representative TEM images of MVP2 particles, scale bar = 50 nm. (H) Percentage of eGFP expression after DC2.4 cells were treated with eGFP-MVPs for 16 h. (I) Fluorescent imaging of eGFP-expressing DC2.4 cells, scale bar = 400 μm. (J) Quantitative analysis on bioluminescence in BMDCs treated with MVPs encapsulated with luciferase-encoding mRNA for 16 h. (K) Changes in the viability of BMDCs treated with PBS, mRNA-free vehicle, mRNA/protamine core, or mRNA-encapsulated MVP2. Treatment concentrations were mRNA-equivalent. Data are presented as mean ± SEM (n = 3). ∗P < 0.05; ∗∗P < 0.01.
2. Materials and methods
2.1. Materials
1,2-Dioleoyl-sn-glycero-3-ethylphosphocholine (EDOPC) (890704), 1,2-dioleoyl-sn-glycero-3-phosphatidyl-ethanolamine (DOPE) (850725), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000 (DSPE-PEG2k) (880128), 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) (890890), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (850375) were purchased from Avanti Polar Lipids, Inc. (Birmingham, AL, USA). Cholesterol (C8667) was obtained from Sigma–Aldrich (Saint Louis, MO, USA). The reagents were dissolved in ethanol at a concentration of 2 mg/mL for DSPE-PEG2k, 10 mg/mL for cholesterol and DOPC, and 20 mg/mL for EDOPC, DOPE, DOTAP, respectively. Protamine sulfate (P4020) was obtained from Sigma–Aldrich. mRNA molecules encoding ovalbumin (OVA-mRNA) (L-7210), eGFP (eGFP-mRNA) (L-7201), and luciferase (Luc-mRNA) (L-7204) were purchased from TriLink Biotechnologies (San Diego, CA, USA). RNase free water (W0805-010) was obtained from GeneDEPOT (Baker, TX, USA). The TLR7 agonist imiquimod (tlr-imq) and Sting agonist 2′3′-cGAMP (tlrl-nacga23) were product from Invivogen (San Diego, CA, USA). Lipofectamine 2000 (11668019) and Quant-iT™ RiboGreen™ RNA assay kit (R11490) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Recombinant mouse GM-CSF (554586) was purchased from BD Biosciences (San Diego, CA, USA). 500 × Protein transporter inhibitor cocktail (00-4980-93) was a product from Invitrogen. Cytofix/Cytoperm kit (AB_2869010) was purchased from BD Biosciences. EasySep™ Mouse T Cell Isolation Kit (19851) was obtained from STEMCELL Technologies, Inc. (Vancouver, BC, CAN). The following antibodies were acquired from BioLegend (San Diego, CA, USA): anti-CD80-PE-Cy7 (104734), anti-CD86-FITC (105006), anti-CD11b-APC-Cy7 (101226), anti-CD8-BV510 (100752), anti-CD44-APC (103012), anti-CD69-APC (104514) and anti-H-2Kb (SIINFEKL)-PE (141604), anti-CD64-PE-Cy7 (139314), anti-B220-APC (103212), anti-MHCII-BV711 (107643), and anti-CD103-PE (121406). Anti-CD40-FITC (553723), anti-LY6C-AF700 (557979), and anti-IFN-γ-PE (554412) were from BD Biosciences. OVA257–264-MHCI dextramer-PE (JD2163) was purchased from ImmuDex (Fairfax, VA, USA). ELISA kits for IL-1β (EM2IL1B) and TNF-α (BMS607-3) were purchased from Invitrogen, and ELISA kits for CCL5 (DY478) and IFN-β (DY8234) were obtained from R&D Systems, Inc. (Minneapolis, MN, USA). Antibodies for Western blot analysis including anti-MAVS (4983), anti-STING (13647), anti-TBK1 (3504), anti-phospho-TBK1 (5483), anti-β-actin (4970) and anti-GAPDH (5174) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA).
2.2. Cell lines and cell culture
The DC2.4 murine dendritic cell line was obtained from ATCC (Manassas, VA, USA), and was cultured in RPMI-1640 containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The murine melanoma cell line B16-OVA was acquired from Dr. Kenneth Rock, Dana-Farber Cancer Institute (Boston, MA, USA). B16-OVA cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. The murine colon adenocarcinoma cell line MC38 was purchased from ATCC. Cells were engineered with OVA expression, and were cultured in DMEM with 10% FBS and 1% penicillin-streptomycin. Cell culture was maintained at 37 °C with 5% CO2.
2.3. mRNA vaccine particle preparation
All mRNA vaccine particles were prepared using the NanoAssemblr Benchtop microfluidic instrument from Precision Nanosystems, Inc., (Vancouver, BC, CAN) by mixing the organic phase and aqueous phase. To prepare the organic phase, EDOPC (20 mg/mL), DOPE (20 mg/mL), cholesterol (10 mg/mL) and bis-DSPE-PEG2k (2 mg/mL) were dissolved in ethanol and mixed at 34:30:35:1 M ratio with a flow rate of 9 mL/min. To prepare the aqueous phase mRNA core, mRNA was mixed with protamine sulfate at 1:1 (w/w) in the microfluidic instrument at a flow rate of 9 mL/min. After 20 min incubation at 20 °C, the aqueous phase mRNA core was mixed with the organic phase to generate mRNA vaccine particles. After 20 min, the vaccine particle suspension was transferred into an Amicon® ultra centrifugal filter (MWCO 30 kDa), and 9 × volume molecular grade water was added to dilute ethanol concentration from 25% to 2.5%. The particle suspension was then concentrated by centrifugation at 2000×g (Multifuge X4, Thermo Fisher Scientific, Waltham, MA, USA) at 4 °C. An equal volume of 2 × PBS was added to the concentrated product to adjust the osmotic pressure.
2.4. Gel retardation assay
The mRNA/protamine core was first analyzed by gel retardation. Briefly, mRNA/protamine core containing 0.5 μg of mRNA was loaded into the well of a 1% agarose gel in 1 × TBE solution and electrophoresis was performed at 120 V for 30 min (PowerPac™, BioRad Co., Ltd., Hercules, CA). RNA bands were stained with Gelred (Biotium, Hayward, CA) and detected with a GelDoc system (Gel Doc XR+, BioRad Co., Ltd.).
2.5. Characterization of mRNA vaccine particles
Size distribution and zeta potential were measured with a dynamic light scattering Zetasizer (Zetasizer Nano, Malvern Pananalytical, Inc., Westborough, MA, USA). Encapsulation rate was determined using a Quant-iT™ RiboGreen™ RNA assay kit. Briefly, a vaccine particle sample containing 0.5 μg of mRNA was diluted with a Tris–EDTA buffer (10 mmol/L Tris–HCl, 1 mmol/L EDTA, pH 7.5) with or without 2% Triton-X100 in a 96-well plate. After 10 min of incubation at 37 °C, RiboGreen was added into each well and fluorescent intensity was measured. Encapsulation efficiency was calculated as shown in Eq. (1):
| (1) |
Transmission electron microscopy (TEM) of vaccine particles was performed following a previously described procedure9.
2.6. Animals
All the animal operation were approved by the Animal Care and Use Committee of Houston Methodist Hospital (protocol number AUP06200042). Mice were housed under the environment which completely fulfilling the regulatory standards of the National Institute of Health and American Association of Laboratory Animal Care standards. Wild-type C57BL/6J mice and genetically engineered mice including Tlr7−/−, Sting−/−, Mavs−/−, Trif−/− mice were purchased from Jackson Laboratory.
2.7. Generation of murine bone marrow-derived dendritic cells (BMDCs)
Bone marrow cells were flushed out from femur and tibia with complete RPMI 1640. Red blood cells were lysed with an ACK lysis buffer, and immature bone marrow cells were cultured in RPMI 1640 supplemented with 20 ng/mL recombinant murine GM-CSF for 10 days at 37 °C with 5% CO2. Cell culture medium was refreshed on Days 3, 6 and 8, non-adherent dendritic cells were collected on Day 10.
2.8. Measurement of cytokines and chemokines
BMDCs were seeded in a 24-well plate at a density of 5 × 105 cells/well. After 1 h of settlement, cells were treated with 1 mg/mL imiquimod, 20 mg/mL 2′3′-cGAMP, (1 mg/mL mRNA-equivalent) vaccine particles or controls. Cell culture media were collected 24 h later, and levels of TNF-α, CCL5, IFN-β, IL-1β were measured using enzyme-linked immunosorbent assay (ELISA) kits by following manufacturer suggested procedures.
2.9. Analyses on DC transfection, DC maturation and stimulation
To analyze DC transfection efficiency in vitro, DC2.4 cells were seeded at 2.5 × 105 cells/well in a 24-well plate. Cells were treated with eGFP- or luciferase-encoding mRNA-encapsulated particles at a final concentration of 1 mg mRNA/mL. Cells were harvested 24 h later, washed with 2% FBS in PBS solution, and then resuspended in the same solution before they were applied for flow cytometry analysis with a BD LSR II flow cytometer (LSR II, BD Biosciences, San Jose, CA). To measure DC maturation and antigen presentation in vitro, BMDCs were seeded at 2.5 × 105 cells/well in a 24-well plate and treated with 1 μg/mL OVA-MVP for 24 h. BMDCs were stained for 30 min with antibodies specific for CD11c, MHC I, MHC II, CD40, CD80 and CD86 for DC maturation, and anti-H-2Kb (SIINFEKL) for antigen presentation. To determine stimulating potency in vivo, C57BL/6J mice were vaccinated once with 10 μg of OVA-MVP per mouse in the footpad, and draining popliteal LNs were harvested 24, 48 and 72 h later. Tissues were processed to generate single-cell suspensions, and were stained with antibodies specific for the following cell surface markers: CD11c, MHC I, CD11b, B220, LY6C, CD64, CD8, CD103, CD86.
2.10. Flow cytometry analyses on T cell activation and proliferation
To measure T cell activation in vivo, mice were vaccinated once with 10 μg of OVA-MVP, and popliteal LNs were isolated at 24 or 48 h. T cells were stained with the following antibodies: CD45, CD3, CD4, CD8, and CD69. To detect antigen-specific T cells, tumor, spleen and popliteal lymph nodes were harvested 5 days after the second vaccination. Tissues were processed to generate single cell suspensions, and cells were stained with T cell-specific antibodies and OVA257–264-MHCI dextramer following manufacturer's instructions. To measure intracellular IFN-γ level, 2 × 106 splenocytes or cells isolated from popliteal LNs and tumors were stimulated with 10 μg/mL OVA257–264 peptide in complete RPMI 1640 medium supplemented with 55 μmol/L β-mercaptoethanol and 1 × protein transporter inhibitor cocktail for 18 h. Cells were harvested and stained with T cell-specific antibodies. After fixation and permeabilization with Cytofix/Cytoperm kit, cells were stained with anti-IFN-γ antibody and applied for analysis on BD LSR II flow cytometer (BD Biosciences). To measure T cell proliferation, T cells were isolated from the spleens of OT-I transgenic mice using a mouse T cell isolation kit. They were stained with 1 μmol/L CFSE in RPMI 1640 containing 200 μg/mL BSA for 10 min at 37 °C. CFSE labeled T cells were washed twice with 5 × volumes of cold complete RPMI 1640. In the meantime, mature BMDCs were treated with 2 μg/mL OVA mRNA-encapsulated MVP for 24 h before T cell isolation. Once T cells were ready, BMDCs and T cells were co-cultured at a ratio of 1:5 (DC:T) for 72 h. Cells were collected and stained with surface antibodies for T cell, the proliferation was tested using BD LSR II flow cytometer (BD Biosciences) and analyzed. All flow cytometry results were analyzed with the FlowJo v10 software (Ashland, OH, USA).
2.11. Tracking protein expression in live mice
BALB/c mice were treated by intra-footpad injection with 10 μg of Luc mRNA-encapsulated MVP. They were injected intraperitoneally with 30 μg of RediJect D-luciferin per mouse 6, 12, 24 or 48 h later, and bioluminescence was measured with the Xenogen IVIS-200 imaging system (Caliper Life Sciences, Inc., Hopkinton, MA, USA).
2.12. ELISpot assay
IP filter plates were pre-rinsed with 50 μL of 35% ethanol, and washed thoroughly 3 times with PBS before the ethanol evaporates. The plates were subsequently coated with anti-IFN-γ capture antibodies at 4 °C overnight. The plates were blocked with RPMI 1640 complete medium containing 55 μmol/L β-mercaptoethanol for 2 h at 37 °C. Cells (1 × 105 splenocytes, 1 × 105 cells from popliteal LN) were seeded into each well, and stimulated for 36 h with 10 μg/mL OVA257–264 peptide. Media were discarded, and cells were washed 4 times with PBST (PBS containing 0.05% Tween 20) and twice with PBS. After the final wash, anti-IFN-γ detection antibody was added and incubated for 2 h at room temperature in dark. Plates were washed 4 times with PBST and twice with PBS, and avidin-HRP was added and incubated for another 45 min at room temperature in dark. Finally, the plates were washed with PBST and PBS again as described above, and 50 μL of AEC substrate was applied and watched for spots reaction. When spots became visible, the reaction was stopped by discarding AEC substrate and flushing with double distilled water 5 times. After air-dried overnight, the plates were scanned and analyzed using the CTL ImmunoSpot SeriesS5 Versa ELISpot Analyzer (S5Versa-02-9038, CTL Inc., Cleveland, OH, USA).
2.13. Western blot analysis
BMDCs derived from wild-type, Mavs KO or Sting KO mice were collected, and cells were lysed in RIPA cell lysis buffer supplemented with protease and phosphatase inhibitors. Protein concentration in cell lysate was measured with BCA protein assay kit. Protein samples were separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a polyvinylidene difluoride (PVDF) membrane. MAVS and STING expression was detected after the membrane was incubated with an anti-MAVS or anti-STING antibody at 1:2000 dilution followed by immunodetection with SuperSignal West Pico and Femto Chemiluminescent Substrate. To measure activation of the STING pathway, BMDCs derived from wild-type mice were treated with vehicle or OVA mRNA-encapsulated MVP at indicated concentration for 2 h. Cells were lysed, and proceeded for Western blot analysis with anti-TBK1 and anti-phospho-TBK1 antibodies at 1:2000 dilution.
2.14. Antitumor assay
Murine tumor models were generated by inoculating 2 × 105 cells B16-OVA melanoma cells or 5 × 105 MC38-OVA cells per mouse subcutaneously into the left flank of 6 to 8-week-old female C57BL/6J mice. Mice were randomly allocated into treatment groups and treated twice separated by 7 days with 10 μg of OVA-MVP or controls in the footpad. Tumor growth was monitored every 2 days. Tumor volume was calculated according to Eq. (2):
| Tumor volume = 0.5 × Length × Width2 | (2) |
Mice were euthanized 5 days after the second vaccination, and weight of tumor tissues was recorded.
2.15. Statistical analysis
All results are presented as mean ± SEM. Statistics were assessed with one-way ANOVA test using Tukey's correction for multiple group comparison and unpaired two-tailed t-test for two group comparation. Data were analyzed with the GraphPad Prism v8.0.2 software. P < 0.05 was considered statistically significant (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.005; ∗∗∗∗P < 0.001).
3. Results
3.1. MVP mediates robust mRNA expression in dendritic cell (DC)
We prepared core–shell vaccine particles in a two-step microfluidic approach (Fig. 1A), and systematically evaluated individual components in the nanoparticle in order to assemble a vaccine particle with high stability, high cellular uptake and high expression potential in DCs. Gel electrophoresis revealed that a stable mRNA core could be formed once mRNA-to-protamine ratio was at 1 or below (Fig. 1B). Using eGFP-encoding mRNA as a surrogate marker, we found that a lipid composition at a molar ratio of 34% EDOPC/30% DOPE/35% cholesterol/1% DSPE-PEG2k provided an ideal encapsulation rate and the best expression efficiency (Table 1, Supporting Information Fig. S1A–S1D). Altering charge ratio did not significantly change encapsulation rate, polydispensity index, or surface charge of the particles (Table 2, Fig. 1C–E); however, size of the resulting particles was between 70 and 80 nm in diameter when the ratio was between 12 and 16, compared to a diameter of 120–130 nm once the ratio fell out of the range (Fig. 1F and G). The best expression was detected in DC2.4 cells treated with MVP2 that had a charge ratio of 12 (Fig. 1H and I). A similar pattern was observed when particles were prepared with mRNA encoding luciferase, and dose-dependent expression was detected (Fig. 1J). The mRNA-encapsulated particles demonstrated desirable stability, since they did not loss activity after lyophilization or freeze-thaw (Fig. S1E and S1F). In addition, there was no cytotoxicity after bone marrow-derived dendritic cells (BMDCs) were treated with the mRNA core alone, a vehicle particle prepared without mRNA molecules, or mRNA-containing MVP2 (Fig. 1K). Thus, MVP2 displayed the best expression potential. It was used for all further experiments in the study, and labeled as MVP in the rest of the text. The MVP particles could effectively deliver mRNA molecules in vivo, and there was no detectable toxicity from the mRNA-encapsulated MVP based on body weight changes (Fig. S1G–S1I).
Table 1.
Composition of nanoparticles encapsulated with eGFP-encoding mRNA.
| Sample | Composition (mol%) |
|||
|---|---|---|---|---|
| EDOPC | DOPE | Cholesterol | DSPE-PEG2k | |
| eGFP1 | 16 | 48 | 35 | 1 |
| eGFP2 | 32 | 32 | 35 | 1 |
| eGFP3 | 34 | 30 | 35 | 1 |
| eGFP4 | 36 | 28 | 35 | 1 |
| eGFP5 | 40 | 24 | 35 | 1 |
| eGFP6 | 42 | 22 | 35 | 1 |
| eGFP7 | 44 | 20 | 35 | 1 |
| eGFP8 | 48 | 16 | 35 | 1 |
Table 2.
Lipid composition and charge ratio of MVP particles.
| Sample | Composition (mol%) |
Charge ratio | |||
|---|---|---|---|---|---|
| EDOPC | DOPE | Cholesterol | DSPE-PEG2k | ||
| MVP1 | 34 | 30 | 35 | 1 | 8 |
| MVP2 | 34 | 30 | 35 | 1 | 12 |
| MVP3 | 34 | 30 | 35 | 1 | 16 |
| MVP4 | 34 | 30 | 35 | 1 | 20 |
3.2. MVP effectively induces antigen presentation and T cell activation in vitro and in vivo
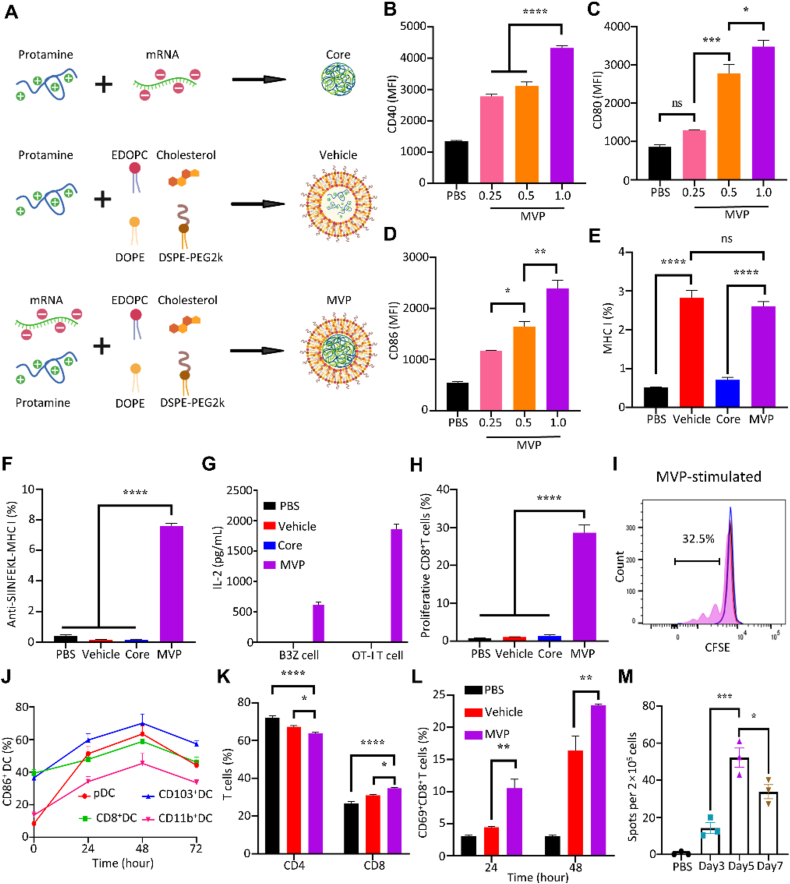
We prepared mRNA core, mRNA-free vehicle, and OVA mRNA-encapsulated MVP, and applied them to test DC maturation and antigen presentation (Fig. 2A). BMDCs treated with MVP displayed dose-dependent overexpression of DC maturation markers including CD40, CD80 and CD86 (Fig. 2B–D). Treatment with either the mRNA-free vehicle or mRNA-encapsulated MVP triggered MHC I overexpression (Fig. 2E), indicating that the effect was associated with the vehicle rather than the mRNA complex. In addition, MVP treatment resulted in cell surface display of the OVA257–264 antigen epitope-MHC I complex (SIINFEKL-MHC I) which was detected with an anti-SIINFEKL-MHCI antibody (Fig. 2F), demonstrating effective antigen processing and presentation by BMDCs. Furthermore, co-incubation of MVP-treated BMDCs with B3Z, a CD8+ T cell line that specifically recognized the OVA257–264 epitope, or T cells isolated from the spleen of an OT-I mouse that expressed an OVA257–264-specific T cell receptor, triggered IL-2 secretion, a result not observed in T cells co-incubated with DCs treated with vehicle alone or the mRNA/protamine core alone (Fig. 2G). Flow cytometry analysis detected 32.5% proliferative CD8+ T cells after co-incubation with MVP (Fig. 2H and I).
Figure 2.
Stimulation of immune responses by MVP. (A) Schematic view of mRNA/protamine core, mRNA-free vehicle, and complete MVP. (B–D) Maturation of BMDCs after treated with OVA-MVP for 24 h. (E and F) MHC I expression and OVA257–264 antigen epitope-MHC I complex in BMDCs after treated with OVA-MVP for 24 h. (G) Level of IL-2 from treated BMDCs co-incubated with B3Z or OT-I T cells. (H and I) Flow cytometry analysis of proliferative T cells. Representative chromatograms of T cells are shown. (J) DC maturation after MVP treatment in vivo. C57BL/6J mice were treated with OVA-MVP, and DCs in popliteal lymph nodes were analyzed. (K and L) T cell population analysis. C57BL/6J mice were treated with vehicle or OVA-MVP, and T cells in popliteal lymph nodes were analyzed with flow cytometry. (M) ELISpot assay. C57BL/6J mice were treated with OVA-MVP, and lymph nodes were collected at the indicated time points. Isolated cells were applied for measurement of IFN-γ-expressing cells. Data are presented as mean ± SEM (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.005; ∗∗∗∗P < 0.001; ns, not significant.
OVA mRNA-encapsulated MVP particles were also applied to treat mice by intradermal injection. Examination of cells in the draining lymph nodes revealed steady increase in CD86 expression among both classical DCs (CD8+ DC, CD11b+ DC, CD103+ DC) and plasmacytoid DCs (pDC) during the first 48 h, indicating stimulation of DC maturation (Fig. 2J). Treatment also promoted enrichment of CD8+ T cells in the lymph nodes, with a significant increase in the population of CD69+CD8+ T cells (Fig. 2K and L), indicating T cell activation by the treatment. To determine T cell activation, we collected cells in the draining lymph nodes 3, 5 or 7 days after MVP treatment, and performed ELISpot analyses after cells were challenged with the OVA257–264 antigen peptide (Supporting Information Fig. S2). MVP treatment triggered a big jump in IFN-γ-secreting cells during the time, with a peak activity on Day 5 (Fig. 2M). These results demonstrated robust DC stimulation, antigen presentation and T cell activation after treatment with MVP.
3.3. MVP elicits potent anti-tumor immunity in murine models of colorectal tumor and melanoma
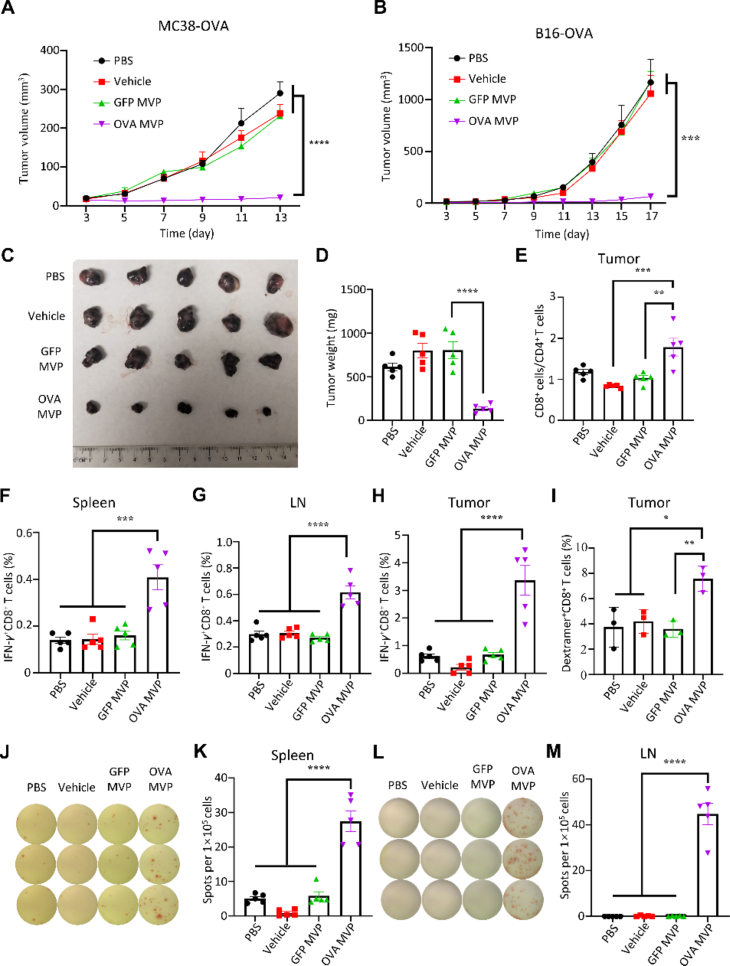
MVP was applied to treat mice with MC38 colon cancer and B16 melanoma. In both models, treatment with OVA mRNA-encapsulated MVP twice completely blocked tumor growth subcutaneously; in comparison, treatment with eGFP mRNA-encapsulated MVP or vehicle alone did not have any detectable inhibitory effect on tumor growth (Fig. 3A–D, Supporting Information Fig. S3). In line with the pattern of a CD4 to CD8 shift in lymph nodes (Fig. 2), we observed an increase in CD8+/CD4+ T cell ratio in tumors from mice treated with OVA mRNA-encapsulated MVP (Fig. 3E). In addition, we detected elevated levels in IFN-γ+CD8+ T cells in the spleens, lymph nodes, and tumors in mice treated with OVA mRNA-encapsulated MVP, but not with vehicle alone or GFP mRNA-encapsulated MVP (Fig. 3F–H, Supporting Information Fig. S4). Furthermore, there was a significant increase in OVA257–264-MHCI dextramer-positive CD8+ T cells in tumors from the OVA mRNA-encapsulated MVP treatment group indicating proliferation of antigen-specific CD8+ T cells (Fig. 3I). ELISpot assay with cells isolated from the spleens and lymph nodes provided further support on T cell activation (Fig. 3J–M).
Figure 3.
Anti-tumor activity from MVP. (A) Inhibition of MC38-OVA tumor growth. Female C57BL/6J mice were inoculated subcutaneously with 5 × 105 MC38-OVA tumor cells/mouse on Day 0 and treated with PBS control, vehicle control, GFP-MVP, or OVA-MVP on Days 3 and 10. (B) Inhibition of B16-OVA tumor growth. Female C57BL/6J mice were inoculated subcutaneously with 2 × 105 B16-OVA tumor cells/mouse on Day 0, and treated with PBS control, vehicle control, GFP-MVP, or OVA-MVP on Days 3 and 10. (C and D) Image and weight of B16-OVA tumors on Day 17. (E) T cell population in tumors of post-treatment mice. (F–H) IFN-γ+CD8+ T cells in spleens, lymph nodes (LN), and tumors of post-treatment mice. (I) OVA-specific CD8+ T cells in tumors of post-treatment mice. (J–M) Images and quantitative analysis of ELISpot assay of splenic and LN cells from post-treatment mice. Data are presented as mean ± SEM (n = 5). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.005; ∗∗∗∗P < 0.001.
3.4. MVP stimulates innate immune responses by activating STING-dependent signaling
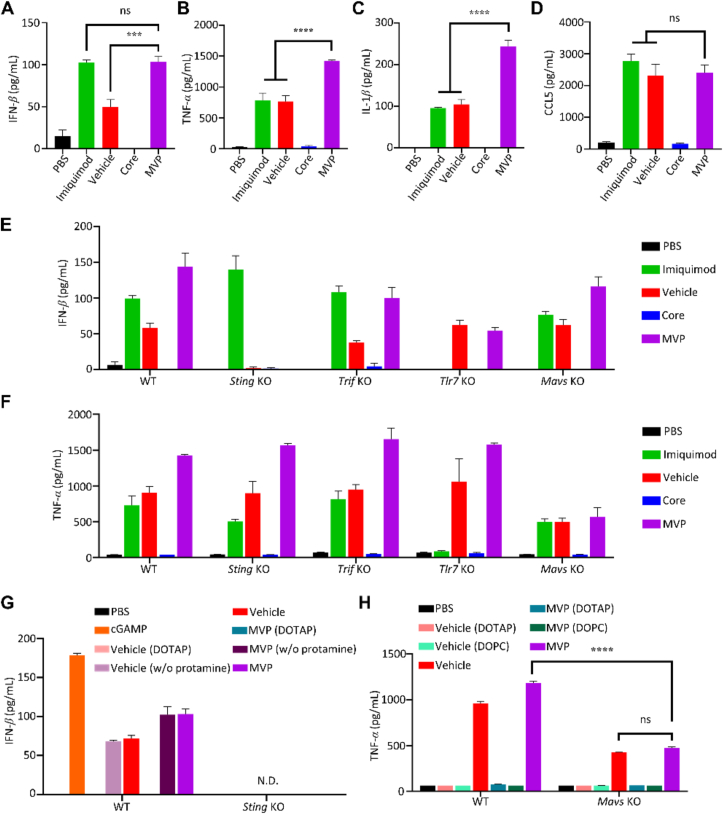
We applied mRNA core, mRNA-free vehicle, and mRNA-encapsulated MVP to treat BMDCs in order to identify key factors and pathways responsible for stimulation of type I IFN and inflammatory cytokine expression. Interestingly, MVP was as potent as the Tlr7 agonist imiquimod in triggering IFN-β secretion, and vehicle alone also showed activity in promoting IFN-β secretion, although its potency was not as high as MVP; however, the mRNA core alone was ineffective in stimulating IFN-β secretion (Fig. 4A). The result implied that both mRNA and components in the vehicle were needed to maximize IFN-β expression. In the meantime, imiquimod and vehicle showed similar activity in promoting TNF-α and IL-1β expression, while MVP was much more potent than either one of them (Fig. 4B and C). Expression of CCL5, a cytokine commonly associated with NF-κB and IFN regulatory factors, was equally stimulated in cells treated with imiquimod, vehicle only, or MVP (Fig. 4D).
Figure 4.
Promotion of innate immune responses by MVP. (A–D) BMDCs treated with 1 mg/mL imiquimod, 1 mg/mL mRNA-equivalent core, vehicle, or MVP for 24 h. IFN-β, TNF-α, IL-1β, and CCL5 levels were measured with ELISA. (E and F) IFN-β and TNF-α expression of BMDCs derived from wild-type and gene knockout (KO) mice. BMDCs were treated with the indicated reagents for 24 h. IFN-β and TNF-α were measured with ELISA. (G) IFN-β in BMDCs derived from wild-type and Sting knockout mice. BMDCs were treated with 20 mg/mL cGAMP, 1 mg/mL mRNA-equivalent core, vehicle, or MVP for 24 h. Vehicle (DOTAP): prepared by replacing EDOPC with DOTAP; vehicle (w/o protamine): prepared by omitting protamine. N.D.: not detectable. (H) TNF-α in BMDCs derived from wild-type and Mavs knockout mice. BMDCs treated with 1 mg/mL mRNA-equivalent core, vehicle, or MVP for 24 h. Vehicle (DOTAP): prepared by replacing EDOPC with DOTAP; vehicle (DOPC): prepared by replacing EDOPC with DOPC. Data are presented as mean ± SEM (n = 3). ∗∗∗P < 0.005; ∗∗∗∗P < 0.001; ns, not significant.
TLR7, MAVS, STING, and TRIF are key factors in major signal transduction pathways that mediate cellular responses to viral RNA and damaged DNA28, 29, 30. We generated BMDCs from mice with Tlr7, Mavs, Sting or Trif gene knockout (KO), and treated cells with mRNA core, mRNA-free vehicle, or mRNA-encapsulated MVP to examine IFN-β and TNF-α expression. KO status of Mavs and Sting in BMDCs were confirmed with Western blot analysis (Supporting Information Fig. S5A). Surprisingly, Sting KO wiped out stimulatory activity on IFN-β secretion from both vehicle alone and MVP (Fig. 4E), indicating that MVP activated type I IFN expression through STING. To support the notion, we detected phosphorylation of tank-binding kinase 1 (TBK1) (Fig. S5B), a kinase that is commonly associated with STING activity31. In addition, this pathway was independent of TLR7 signaling, since IFN-β expression was not compromised in cells treated with imiquimod (Fig. 4E). In comparison, Trif or Mavs KO had a minimal impact on IFN-β expression promoted by MVP. As expected, imiquimod was ineffective in stimulating IFN-β secretion in BMDCs derived from Tlr7 KO; however, Tlr7 KO had a negative impact on stimulatory effect from MVP (Fig. 4E). Interestingly, a different profile was observed on TNF-α secretion from the same treatments. Knockout of Sting, Trif, or Tlr7 had no to minimal impact on MVP-stimulated cytokine expression. Knockout of Mavs, on the other hand, dramatically reduced TNF-α levels in cells treated with either vehicle alone or MVP (Fig. 4F). The result indicates that MAVS is a key regulator of TNF-α expression in DCs upon MVP treatment.
3.5. MVP stimulates STING and MAVS pathways to promote secretion of IFN-I and inflammatory cytokines
To identify components in the vehicle responsible for stimulation of STING and MAVS signaling, we prepared vehicles and MVPs by replacing or omitting key components and applied them to treat BMDCs. The Sting agonist cyclic GMP-AMP (cGAMP) served as a positive control in stimulating IFN-β secretion. Replacing the cationic lipid EDOPC in the vehicle with another positively charged lipid, DOTAP, completely wiped out IFN-β secretion. In comparison, stimulatory effect from the vehicle and MVP was retained with or without protamine, and such stimulatory activity was dependent on an intact Sting gene (Fig. 4G). Interestingly, BMDCs treated with individual components of the lipid shell including EDOPC did not promote strong IFN-β secretion (Supporting Information Fig. S6). Thus, EDOPC was essential for STING activation, and its activity is dependent on the formation of a lipid nanoparticle (i.e., vehicle or MVP). Vehicle and MVP prepared with EDOPC or DOTAP were also applied to treat BMDCS derived from wild-type and Mavs KO mice, and TNF-α levels in cell growth media were determined. As expected, replacing EDOPC with DOTAP or DOPC eliminated stimulatory effort from the vehicle and MVP. In addition, TNF-α level was significantly reduced in Mavs KO cells compared to wild-type cells, but not diminished (Fig. 4H), indicating additional factors might be involved in mediating MVP-stimulated TNF-α expression.
3.6. STING pathway is essential for MVP-mediated anti-tumor immune responses
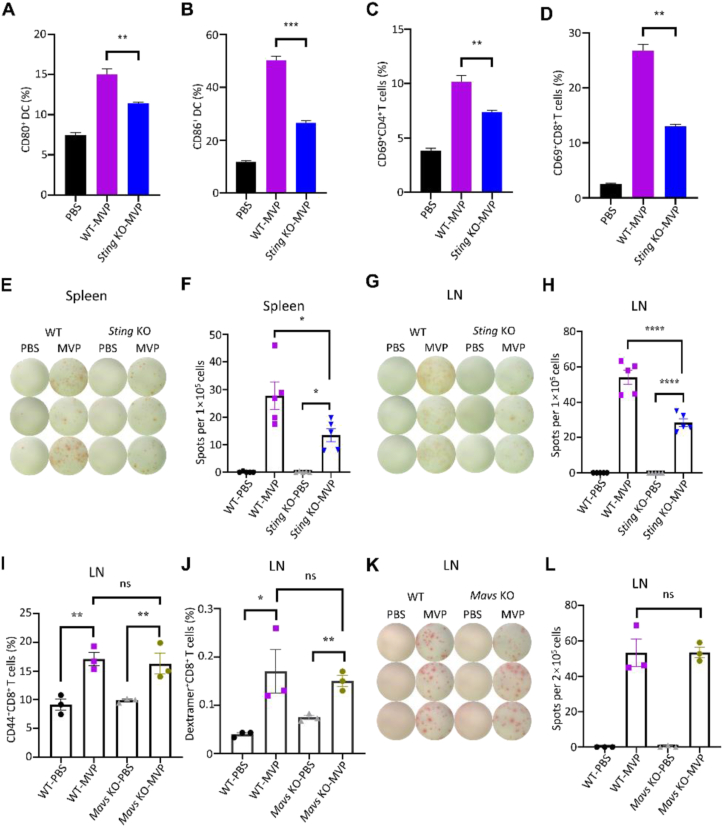
Both wild-type and knockout mice were treated with OVA mRNA-encapsulated MVP, and DC maturation and T cell proliferation were examined. Sting KO significantly reduced percentage of CD80+ and CD86+ DC cells and CD69+ T cells (Fig. 5A–D). ELISpot assay revealed significantly reduced IFN-γ-producing cells in the spleens and lymph nodes from Sting KO mice compared to wild-type mice after they were treated with the same dose of MVP (Fig. 5E–H). The impact from Mavs KO was minimal, as no difference was monitored on CD44+CD8+ memory T cells, OVA257–264-MHCI dextramer-positive CD8+ T cells, or number of IFN-γ-producing cells in the lymph nodes compared to wild-type mice (Fig. 5I–L). The result reinforces the notion that additional factors besides MAVS may be involved in MVP-stimulated inflammatory cytokine expression.
Figure 5.
Antitumor immune responses in Sting and Mavs knockout mice. (A–D) Diminished DC and T cell activation in Sting knockout mice. Both wild-type and Sting knockout mice were treated with OVA mRNA-encapsulated MVP, and lymph nodes were collected 48 h later for DC and T cell measurement. (E–H) Reduced IFN-γ-expressing T cells in spleen and lymph nodes from Sting knockout mice. Both images of spots and quantitative analyses are shown. (I–L) No impact on T cell activity in Mavs knockout mice. Both wild-type and Mavs knockout mice were treated with PBS control or OVA mRNA-encapsulated MVP, and lymph nodes were collected 48 h later for measurement of CD44+CD8+ memory T cells (I), OVA257–264-MHCI dextramer-positive CD8+ T cells (J), and number of IFN-γ-producing cells (K and L). Data are presented as mean ± SEM (n = 3 or 5). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.005; ∗∗∗∗P < 0.001; ns, not significant.
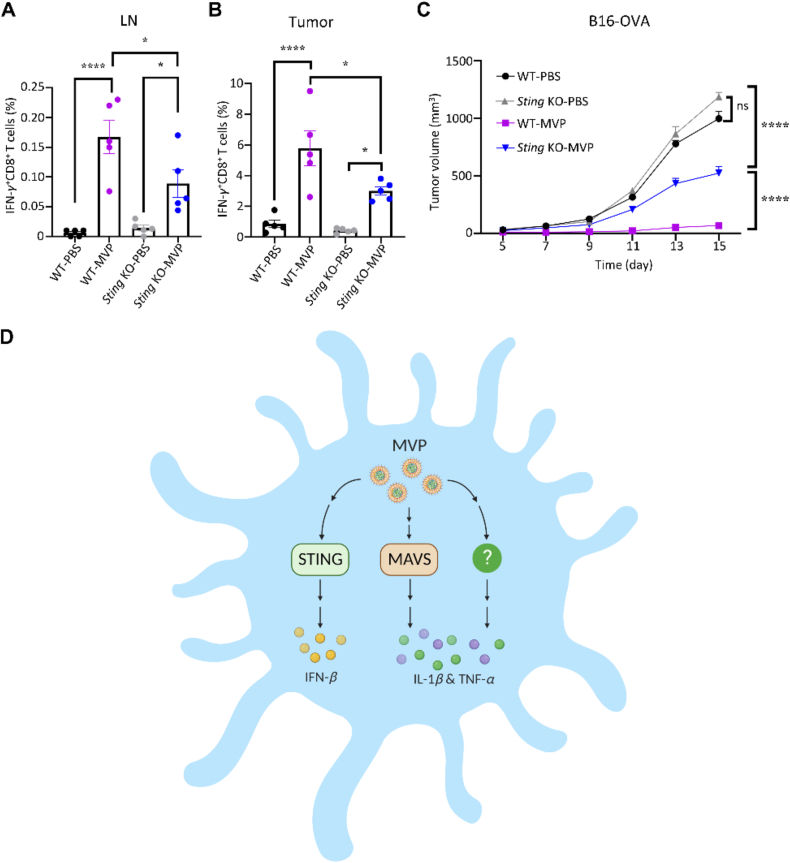
Both wild-type and Sting KO mice were applied to inoculate B16-OVA tumors, and mice were treated with PBS control or OVA mRNA-encapsulated MVP. Flow cytometry analysis on lymph node and tumor samples collected from vaccinated mice revealed significantly reduced number of IFN-γ-producing CD8+ T cells in the Sting KO mice (Fig. 6A and B). As a result, tumor growth was completely inhibited in wild-type mice treated with MVP, compared to only partial inhibition in the Sting KO mice (Fig. 6C).
Figure 6.
Sting-dependent anti-tumor immunity. (A and B) Analysis of IFN-γ-expressing T cells in lymph nodes and tumor tissues after wild-type and Sting knockout mice bearing B16-OVA tumors were treated with OVA mRNA-encapsulated MVP. Mice were treated on Days 3 and 10, and lymph nodes and tumors were collected on Day 15. Single cells were analyzed with flow cytometry. (C) Inhibition of tumor growth in wild-type and Sting knockout mice. Mice were treated with PBS control or MVP on Days 3 and 10. Data are presented as mean ± SEM (n = 5). ∗P < 0.05; ∗∗∗∗P < 0.001; ns, not significant. (D) Schematic view on MVP stimulation of signal transduction pathways leading to cytokine production in DCs. While stimulation of IFN-β expression is exclusively mediated by STING, there are multiple factors including MAVS that mediate TNF-α and IL-1β expression.
4. Discussion
In the current study, we have systematically examined functional roles of individual components in the MVP on stimulation of anti-tumor immune responses. Our cell-based study has clearly demonstrated that both mRNA molecules and the RNA-free vehicle in MVP are needed for its full activity in promoting expression of IFN-β and a number of inflammatory cytokines including IL-1β and TNF-α in dendritic cells. Such cytokines have been shown to play essential roles on dendritic cell activation and vaccine-mediated anti-tumor immunity20. Our study has also revealed that stimulation of cytokine production is dependent on a number of key signal transduction pathways involved in innate immune sensing including those mediated by STING and MAVS. While STING is essential for IFN-β expression, MAVS is mainly involved in regulating TNF-α expression (Fig. 6D). The significance of STING signaling on MVP-mediated anti-tumor immunity is further demonstrated by reduced T cell activity and consequently compromised tumor growth inhibition in Sting KO mice. This result is in line with other reports that STING activation determines cytotoxic T cell-mediated antitumor immunity32.
MAVS is a key intermediator in the RIG-I-receptor (RLR) signaling in response to viral infection. Activation of this pathway leads to a cascade of inflammatory responses33. It is intriguing that T cell activity is not compromised in Mavs KO mice, given that the MAVS signaling is clearly playing a big role in regulating expression of inflammatory cytokines including TNF-α. A possible reason is that stimulation of such cytokines by MVP is mediated by multiple pathways, and disruption of the MAVS pathway does not reduce cytokine expression to a low enough level to cause a significant impact. Alternatively, loss of MAVS signaling is compensated by the other pathways. Further studies are needed to identify these pathways to further understand the mechanism of action of MVP vaccine. Although TLR7 has traditionally been associated with mRNA therapeutics21, the TLR7 signaling does not seem to play a major role in mediating MVP activity. Application of fully methylated mRNA molecules in the current study may have rendered the TLR7 signaling less relevant. It has been well demonstrated that RNA methylation suppresses recognition by the TLRs23. TRIF is a key adaptor protein for TLR3/7/8 signaling28. Knockout of Trif does not affect MVP activity either, reenforcing the notion that the above TLRs including TLR7 do not play a major role in mediating cellular responses to MVP.
Multiple Sting agonists have been applied in cancer vaccine development including cyclic dinucleotides and small and large molecular compounds34, 35, 36, 37. Such Sting agonists need to be added as external adjuvants during vaccine preparation which adds another layer of complexity on the vaccine composition. In addition, the externally added Sting agonist may cause undesirable adverse effects once it separates from the mRNA complex. In comparison, our MVP serves as a self-adjuvant and works in context of a cancer vaccine, since none of the individual components of the vaccine including EDOPC can promote cytokine expression. Interestingly, the cationic phospholipid EDOPC cannot be replaced with the non-phospholipid DOTAP which is also positively charged. It is intriguing to speculate the potential mechanism for the difference between these two cationic lipids. However, it has been documented that other properties of a lipid molecule component such as hydrophobicity may have a profound impact on the overall function of a nanoparticle and its delivery efficacy for nucleic acids38. Thus, care must be taken in selecting the proper molecules to prepare a fully functional mRNA vaccine.
In summary, we have developed a potent mRNA-based therapeutic cancer vaccine and assigned individual components in the vaccine with their functionality. Its mechanism of action is quite different from other mRNA platforms used for prophylactic and therapeutic interventions. The knowledge derived from the current study will definitely guide future development of additional mRNA therapeutics.
Acknowledgments
This work was partially supported with a sponsored research grant from Stemirna Therapeutics, China, and from internal funding sources in Houston Methodist Research Institute, USA.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.11.013.
Author contributions
Haifa Shen designed and supervised this research. Zhe Chen carried out experiments and performed data analysis. Chaoyang Meng, Junhua Mai and Yongbin Liu participated in part of the study. Zhe Chen, Haifa Shen and Hangwen Li wrote the manuscript. All authors have read and approved the final manuscript.
Conflicts of interest
Haifa Shen is an inventor of a patent that is related to MVP.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.El Sahly H.M., Baden L.R., Essink B., Doblecki-Lewis S., Martin J.M., Anderson E.J., et al. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N Engl J Med. 2021;385:1774–1785. doi: 10.1056/NEJMoa2113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreira E.D., Jr., Kitchin N., Xu X., Dychter S.S., Lockhart S., Gurtman A., et al. Safety and efficacy of a third dose of BNT162b2 Covid-19 vaccine. N Engl J Med. 2022;386:1910–1921. doi: 10.1056/NEJMoa2200674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 4.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persano S., Guevara M.L., Li Z., Mai J., Ferrari M., Pompa P.P., et al. Lipopolyplex potentiates anti-tumor immunity of mRNA-based vaccination. Biomaterials. 2017;125:81–89. doi: 10.1016/j.biomaterials.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Su H.H., Yang Y., Hu Y., Zhang L., Blancafort P., et al. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 8.Meng C., Chen Z., Mai J., Shi Q., Tian S., Hinkle L., et al. Virus-mimic mRNA vaccine for cancer treatment. Adv Ther. 2021;4 doi: 10.1002/adtp.202100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rurik J.G., Tombacz I., Yadegari A., Mendez Fernandez P.O., Shewale S.V., Li L., et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375:91–96. doi: 10.1126/science.abm0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteban I., Pastor-Quinones C., Usero L., Plana M., Garcia F., Leal L. In the era of mRNA vaccines, Is there any hope for HIV functional cure?. Viruses. 2021;13:501. doi: 10.3390/v13030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krienke C., Kolb L., Diken E., Streuber M., Kirchhoff S., Bukur T., et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 13.Miao L., Zhang Y., Huang L. mRNA vaccine for cancer immunotherapy. Mol Cancer. 2021;20:41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Hoecke L., Verbeke R., Dewitte H., Lentacker I., Vermaelen K., Breckpot K., et al. mRNA in cancer immunotherapy: beyond a source of antigen. Mol Cancer. 2021;20:48. doi: 10.1186/s12943-021-01329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Lower M., et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 16.Sahin U., Oehm P., Derhovanessian E., Jabulowsky R.A., Vormehr M., Gold M., et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585:107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 17.Hilf N., Kuttruff-Coqui S., Frenzel K., Bukur V., Stevanovic S., Gouttefangeas C., et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 18.Carreno B.M., Magrini V., Becker-Hapak M., Kaabinejadian S., Hundal J., Petti A.A., et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai J., Li Z., Xia X., Zhang J., Li J., Liu H., et al. Synergistic activation of antitumor immunity by a particulate therapeutic vaccine. Adv Sci. 2021;8 doi: 10.1002/advs.202100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobiyama K., Ishii K.J. Making innate sense of mRNA vaccine adjuvanticity. Nat Immunol. 2022;23:474–476. doi: 10.1038/s41590-022-01168-4. [DOI] [PubMed] [Google Scholar]
- 22.Fotin-Mleczek M., Duchardt K.M., Lorenz C., Pfeiffer R., Ojkic-Zrna S., Probst J., et al. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 23.Kariko K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Tahtinen S., Tong A.J., Himmels P., Oh J., Paler-Martinez A., Kim L., et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat Immunol. 2022;23:532–542. doi: 10.1038/s41590-022-01160-y. [DOI] [PubMed] [Google Scholar]
- 25.Li C., Lee A., Grigoryan L., Arunachalam P.S., Scott M.K.D., Trisal M., et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat Immunol. 2022;23:543–555. doi: 10.1038/s41590-022-01163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LoPresti S.T., Arral M.L., Chaudhary N., Whitehead K.A. The replacement of helper lipids with charged alternatives in lipid nanoparticles facilitates targeted mRNA delivery to the spleen and lungs. J Control Release. 2022;345:819–831. doi: 10.1016/j.jconrel.2022.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charbe N.B., Amnerkar N.D., Ramesh B., Tambuwala M.M., Bakshi H.A., Aljabali A.A.A., et al. Small interfering RNA for cancer treatment: overcoming hurdles in delivery. Acta Pharm Sin B. 2020;10:2075–2109. doi: 10.1016/j.apsb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuertes M.B., Woo S.R., Burnett B., Fu Y.X., Gajewski T.F. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34:67–73. doi: 10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., Greenberg R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivick K.E., Desbien A.L., Glickman L.H., Reiner G.L., Corrales L., Surh N.H., et al. Magnitude of therapeutic STING activation determines CD8+ T cell-mediated anti-tumor immunity. Cell Rep. 2018;25:3074–3085 e5. doi: 10.1016/j.celrep.2018.11.047. [DOI] [PubMed] [Google Scholar]
- 33.Reikine S., Nguyen J.B., Modis Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu J., Kanne D.B., Leong M., Glickman L.H., McWhirter S.M., Lemmens E., et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E., et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 37.Luo M., Wang H., Wang Z., Cai H., Lu Z., Li Y., et al. A STING-activating nanovaccine for cancer immunotherapy. Nat Nanotechnol. 2017;12:648–654. doi: 10.1038/nnano.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Koynova R., Parikh H., MacDonald R.C. Transfection activity of binary mixtures of cationic o-substituted phosphatidylcholine derivatives: the hydrophobic core strongly modulates physical properties and DNA delivery efficacy. Biophys J. 2006;91:3692–3706. doi: 10.1529/biophysj.106.092700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.