Abstract
Nonalcoholic fatty liver disease (NAFLD) is a chronic liver disease affecting multiple cell types of the human liver. The high prevalence of NAFLD and the lack of approved therapies increase the demand for reliable models for the preclinical discovery of drug targets. In the last decade, multiple proof-of-principle studies have demonstrated human-specific NAFLD modeling in the dish. These systems have included technologies based on human induced pluripotent stem cell derivatives, liver tissue section cultures, intrahepatic cholangiocyte organoids, and liver-on-a-chip. These platforms differ in functional maturity, multicellularity, scalability, and spatial organization. Identifying an appropriate model for a specific NAFLD-related research question is challenging. Therefore, we review different platforms for their strengths and limitations in modeling NAFLD. To define the fidelity of the current human in vitro NAFLD models in depth, we define disease hallmarks within the NAFLD spectrum that range from steatosis to severe fibroinflammatory tissue injury. We discuss how the most common methods are efficacious in modeling genetic contributions and aspects of the early NAFLD-related tissue response. We also highlight the shortcoming of current models to recapitulate the complexity of inter-organ crosstalk and the chronic process of liver fibrosis-to-cirrhosis that usually takes decades in patients. Importantly, we provide methodological overviews and discuss implementation hurdles (eg, reproducibility or costs) to help choose the most appropriate NAFLD model for the individual research focus: hepatocyte injury, ductular reaction, cellular crosstalk, or other applications. In sum, we highlight current strategies and deficiencies to model NAFLD in the dish and propose a framework for the next generation of human-specific investigations.
Keywords: Induced Pluripotent Stem Cells, Liver-on-Chip, Precision-Cut Liver Slices, Liver Organoids
Abbreviations used in this paper: 3D, 3-dimensional; hiPSC, human induced pluripotent stem cell; HLO, human liver organoid; HSC, hepatic stellate cell; ICO, intrahepatic cholangiocyte organoid; KC, Kupffer cell; LSEC, liver sinusoidal endothelial cell; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; OA, oleic acid; PCLS, precision-cut liver slice; PNPLA3, patatin like phospholipase domain containing 3; PPAR, peroxisome proliferator activated receptor
Summary.
This review article presents the latest research on nonalcoholic fatty liver disease (NAFLD) in vitro modeling. Benefits and limitations of using human induced pluripotent stem cell-derived approaches, primary organoids, liver-on-a-chip, and precision-cut liver sections are discussed to help researchers choose the most appropriate model.
Up to one-fourth of the global population presents with some degree of nonalcoholic fatty liver disease (NAFLD). Furthermore, the incidence is expected to rise in the coming years primarily because of Western life habits.1 NAFLD can range from hepatocellular steatosis to chronic fibroinflammatory liver injury and cirrhosis with liver failure at end stages. Although some pathomechanisms, especially in early NAFLD development, are well-understood, there is currently no approved treatment for patients. For this reason, there is an urgent need to implement methods that reliably model human NAFLD for therapeutic target discovery or candidate drug testing. In recent years, a debate has emerged on the translational utility of NAFLD animal models. Despite the rationale for preclinical studies in vivo, significant inter-species differences have led to a vast collection of lost-in-translation cases such that therapeutic candidates validated in animals did not effectively ameliorate NAFLD in clinical trials. Here, we review the state of advanced in vitro models that allow a better understanding of human NAFLD. We focus on different human-specific modeling strategies, their promises, and progress, and finally emphasize their limitations. In our assessment, we analyze the different human platforms through a lens of how effectively they model the clinical hallmarks of the NAFLD spectrum.
Defining the Hallmarks of the Human NAFLD Spectrum for Effective in Vitro Modeling
Chronic substrate excess induces NAFLD, characterized by subcellular lipid accumulation in hepatocytes (steatosis). NAFLD may progress to nonalcoholic steatohepatitis (NASH) and a fibroinflammatory tissue response. Persistent steatosis triggers a lipotoxic cell response, progressively leading to a surge in oxidative stress, endoplasmic reticulum stress, and compromised mitochondrial function. Consequently, hepatocytes may undergo cell death and release damage-associated patterns and alarmins found in the liver and systemic circulation.2 This cascade of events activates liver resident macrophages (ie, Kupffer cells [KCs]) and liver sinusoidal endothelial cells (LSECs) to secrete a wide range of chemokines to recruit circulating myeloid and lymphoid immune cells.3, 4, 5, 6 The complex inflammatory response triggers the differentiation of hepatic stellate cells (HSCs) into myofibroblasts responsible for extracellular matrix deposition unless the injury subsides. As relevant for patient-specific models, genetic polymorphisms can determine the risk for NAFLD and its progression. Importantly, NAFLD progression usually builds up over several decades in patients. On the basis of the available literature, we focus on in vitro models recapitulating NAFLD-associated mechanisms in the liver. Notably, NAFLD is a medical condition affecting multiple organs, eg, in the setting of insulin resistance. However, we acknowledge the need for future systems to model inter-organ crosstalk. Other literature reviews described relevant in vitro models for the study of NAFLD in other organs.7
Human Induced Pluripotent Stem Cell-Based Platforms
Human induced pluripotent stem cells (hiPSCs) are generated by directly reprogramming somatic cells. The method most commonly involves the overexpression of transcription factors Oct-4, SOX2, KLF4, and C-Myc, which were initially identified by Takahashi et al8 in 2007. However, many transcription factors and methods have now been used, with “chemical” reprogramming being the latest addition in this panoply.9 Another critical aspect concerning hiPSCs is their state of pluripotency. Indeed, somatic cells can be reprogrammed into pre-implantation pluripotent stem cells (ground state hiPSCs)10,11 or post-implantation pluripotent stem cells (primed or conventional hiPSCs).12,13
Ground state hiPSCs can differentiate into a wide range of cell types, including both intra- and extra-embryonic tissues. Primed hiPSCs have been having a more limited ability to differentiate into other types of cells but can still differentiate directly into the 3 primary germ layers by being exposed to specific growth cocktails promoting the differentiation into a specific cell type or lineage. The adult human liver primarily comprises endoderm and mesoderm derivatives (hepatobiliary and stromal/immune lineages). Thus, this review will focus on primed hiPSC lines, which remain the primary choice for liver disease modeling applications. However, the full implications of ground states for liver injury modeling require further studies.
The property that makes hiPSC lines uniquely interesting is their capacity to proliferate while maintaining the capability to differentiate into many cell types. Thus, hiPSCs can be used to produce a large number of liver cells for clinical or research applications. In addition, hiPSCs can be derived from almost any patient to study the genetic impact on disease onset.14 However, modeling the complex interplays of genetic predisposition and chronic environmental factors, as in NAFLD/NASH, remains challenging. Here, we will review these advances in the context of hiPSC-based two-dimensional and three-dimensional (3D) platforms to model NAFLD/NASH and describe the advantages and limitations.
Generating the Different Hepatic Cells From hiPSCs
NAFLD/NASH is a multicellular disease that involves virtually all cell types composing the human liver.4 Thus, modeling the tissue response in NAFLD/NASH in vitro would at least require culture systems combining the following liver cells: hepatocytes, cholangiocytes, LSECs, KCs/macrophages, and HSCs. Robust protocols are now available to generate these cell types from hiPSCs, which could be reconstituted as co-cultures. In a separate section, we will also discuss how different liver cell types could be seeded on interconnected biochip compartments to model the interaction across cell types.
Hepatocytes are the main target of the disease because steatosis and then lipotoxicity mark the initial steps of NAFLD/NASH. Thus, robust production of functional hepatocytes from hiPSCs is essential to model NAFLD/NASH. There is a broad diversity of protocols for differentiating hiPSCs into hepatocytes.15,16 They can be divided into 2 main categories. First, directed differentiation is the most common approach and follows a natural path of development in vitro by first inducing differentiation of hiPSCs into endoderm, which is then patterned into foregut, hepatic endoderm, hepatoblast, and then hepatocyte-like cells. This process can take up to 20–35 days (Figure 1). In the end, the resulting hepatocyte-like cells express specific markers such as AAT and HNF4-alpha while displaying mature hepatocyte functional traits such as urea cycle activity, albumin secretion, and lipid metabolism.16 However, the cells also express fetal markers, such as alpha-fetoprotein, while expressing low levels of adult cytochrome P450, such as CyP3A4. Thus, hepatocytes generated by directed differentiation are generally considered “fetal-like" cells. However, a recent study comparing hepatocytes differentiating in vivo and in vitro showed that hiPSC-derived cells follow a specific developmental trajectory.17 Thus, hiPSC-derived hepatocyte-like cells are likely to represent an artificial state.
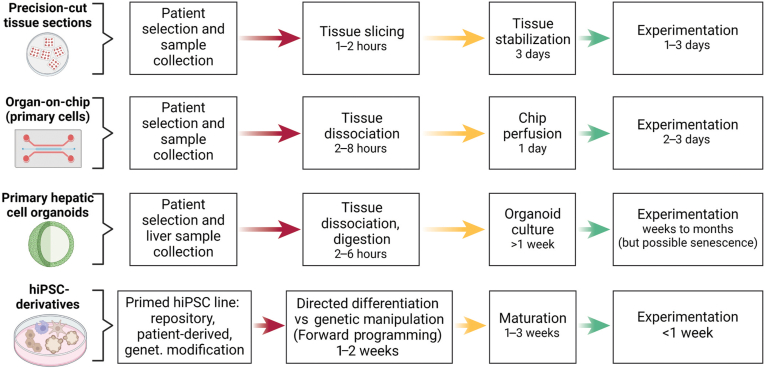
Figure 1.
Experimental milestones and time considerations in NAFLD models. Each model includes sequential steps of sample preparation before experimentation. hiPSC, human induced pluripotent stem cell. Created with biorender.com.
Nonetheless, this observation does not disqualify the use of hepatocyte-like cells for disease modeling. It only confirms that mimicking human liver development in vitro is extremely difficult. Accordingly, hepatocyte-like cells have been successfully used to model a diversity of liver diseases and to provide proof-of-principle for cell-based therapy applications.15,18
Forward programming is the second and more recent method available to generate hepatocytes from hiPSCs.19 This approach involves overexpressing transcription factors directly in hiPSCs to re-create the core transcriptional network characterizing a specific cell type. This approach has been successfully applied for hepatocytes,20,21 and the resulting protocols are less time-consuming and more robust and require less complex culture conditions. Nonetheless, forward programming still requires specialized culture media to maintain the identity of the cell type generated. This aspect is particularly challenging in the context of hepatocytes because there are no culture conditions allowing the expansion of these cells in vitro. Thus, the functionality of hepatocyte-like cells generated by forward programming could remain a challenge. Further validations are required to confirm that these cells display the full range of activities associated with primary cells.
Cholangiocytes are often ignored in the context of NAFLD/NASH, because the role of ductular reaction in disease progression is not fully understood.22 Nonetheless, their interaction with hepatocytes and their role in inflammation suggest that these cells could also drive disease progression. Several protocols are available to differentiate hiPSCs into cholangiocytes expressing KRT19 and SOX9 while displaying the capacity to transport bile acid, secrete gamma-GT, and react to hormonal stimuli.23,24 These cells have been used for disease modeling and drug screening in cystic fibrosis.23 Importantly, cholangiocytes represent one of the rare hepatic cell types that can be grown in vitro as epithelial organoids while maintaining their key characteristics25, 26, 27 (see separate section on primary tissue organoids) and may be used for regenerative cytotherapy applications.28
LSECs play a less direct, albeit essential, role in NAFLD/NASH. They interact with numerous cells, including stellate and immune cells, but more importantly, they are particularly affected by fibrosis. Specifically, the fibrotic liver undergoes significant alterations in its micro-circulation that lead to hypoxia and affect the physiological hepatic zonation.3 There is a diversity of protocols for generating generic endothelial cells from hiPSCs using directed differentiation.29 The production of liver-specific endothelial cells remains more challenging, but recent reports describe the production of LSECs with the characteristic fenestrae and the production of clotting factor VIII.30 The resulting cells have been used to correct hemophilia induced by factor VIII deficiency in animal models.31 Forward programming combined with directed differentiation has also been applied to generate endothelial cells.32 For this cell type, the simple expression of ETV2 seems sufficient to induce endothelial programming. The resulting cells have been combined with other hepatic cell types to model NAFLD/NASH. However, the functionality of these cells remains to be shown, especially in the context of NAFLD/NASH.
KCs are the tissue resident macrophages of the liver, and together with bone marrow-derived myeloid cells, they play a crucial role in the proinflammatory feedback loop driving fibrosis in NAFLD/NASH.33 Most protocols currently available to generate macrophages from hiPSCs follow the directed differentiation method first described by van Wilgenburg et al.34 This method relies on embryoid body formation to produce monocytes, which are further differentiated into macrophages. The embryoid body step renders it difficult to define the exact embryonic origin of these macrophages. This matters because tissue-specific macrophages are generally considered to derive from early hematopoietic progenitors localized in the yolk sac, whereas circulating macrophages are generated by adult hematopoietic stem cells.33
Nonetheless, this method has been recently modified to generate KCs by differentiating monocytes in medium conditioned by hiPSC-derived hepatocytes.35 The resulting cells express markers such as CLEC-4F, ID1, and ID3 but not MARCO, which has been identified as one of the rare Kupffer-specific markers by single cell transcriptomic analyses.36,37 There is currently no functional test to distinguish KCs from circulating macrophages. Thus, further investigation is needed to define better the nature of macrophages produced in vitro. Of note, a recent report has shown that microglia can also be generated using forward programming by simply overexpressing PU.1 and C/EBPβ in hiPSCs.38 It would be interesting to check whether this protocol could be adapted to produce KCs by co-culture with hiPSC-derived hepatocytes.
HSCs have been the central focus of many studies in the context of liver disease. Indeed, during liver disease, HSCs activate toward tissue-specific myofibroblasts and play a direct role in fibrosis by secreting extracellular matrix proteins such as collagens.4 Significantly, primary HSCs are challenging to grow in vitro because they quickly adopt an “activated” phenotype. A diversity of protocols is now available to generate HSCs from hiPSCs.39,40 They all rely on the production of stromal progenitor cells and display a relatively limited capacity to be fibrogenic by transforming growth factor-β treatment. The cells have already been used to model disease or to screen therapeutic agents against liver fibrosis. Finally, forward programming can be used to generate fibroblasts from hiPSCs by overexpression of Nkx3.1.41 Nonetheless, the hepatic-specific nature of these in vitro-generated myofibroblasts remains to be fully demonstrated, because these protocols are generic without a dedicated liver-specific step. This highlights a general challenge for HSCs, KCs, and LSECs. Non-parenchymal cells generated in vitro lack the microenvironment necessary for liver specification. This could potentially be addressed by protocols allowing the co-differentiation of interdependent cell types in multicellular, hiPSCs-organoids. The functional impact of the differences between in vitro-generated cells and primary cells remains to be fully evaluated in the context of disease modeling.
Overall, protocols exist to differentiate hiPSCs into different types necessary to model NAFLD/NASH. Despite some limitations around their tissue specificity and functionality, these cells display key activities necessary for modeling NASH/NAFLD. The next section will give a few examples of such applications.
HiPSC-based Platforms for Modeling NAFLD/NASH
Using the protocols mentioned above, several studies have shown that hiPSC-derived cells can be used to model NAFLD/NASH. Tilson et al42 have used hepatocyte-like cells generated from hiPSCs to study the functions of patatin like phospholipase domain containing 3 (PNPLA3) in lipid accumulation and lipid toxicity. The genetic variant in PNPLA3 has been linked to NAFLD progression by genome-wide association studies. To clarify the functional role of PNPLA3 and its variant, the authors generated isogenic hiPSC lines carrying the I148M variant or a full knockout of PNPLA3. The resulting cells were then differentiated into hepatocyte-like cells, which were then grown in the presence of oleic acid (OA) to induce lipid accumulation and palmitic acid to induce lipotoxicity. The mutant genotypes increased lipid accumulation while protecting against lipotoxicity. Furthermore, the same genetic changes increased the sensitivity of hepatocytes against other toxins such as alcohol. These results suggest that PNPLA3 variants decrease the activity of the triglyceride lipase that, in turn, seems to protect against fatty acid-induced lipotoxicity. Furthermore, the PNPLA3 variant may amplify other types of injuries. This indicates that genetic variants may determine NAFLD progression independent of lipid-mediated injuries. However, one major limitation of this study remains an absence of other PNPLA3-expressing liver cells, ie, HSCs and KCs, that could have a major impact on disease phenotypes.
This limitation is addressed in part by the 3D co-culture system developed by Kumar et al.43 This system combines hepatoblast-like cells, HSCs, macrophages, and endothelial cells. These different cells were generated from hiPSCs by either directed differentiation or forward programming and co-cultured together in a tailor-made polyethylene glycol hydrogel. The co-cultured cells maintain their functional characteristics, indicating that the culture conditions developed by the authors can maintain the identity of each cell type. Of note, hepatocyte-like cells generated under these co-culture conditions did not undergo functional maturation, especially when compared with primary hepatocytes. Thus, 3D co-culture might not suffice to address the limitations of current protocols for generating mature hepatocyte-like cells. Nonetheless, the authors elegantly demonstrate how adding OA results in lipid accumulation in hepatocyte-like cells while activating HSCs as evidenced by increased collagen and alpha-SMA expression. A surge in proinflammatory cytokines, such as interleukin-6 or tumor necrosis factor-alpha, was also detected, suggesting that different phases of NAFLD/NASH progression could be modeled: steatosis, proinflammatory response, and fibrogenesis. Finally, the authors show that this platform could be used for drug screening applications by demonstrating the positive effect of peroxisome proliferator activated receptor (PPAR) α/δ activator, elafibranor, on fibrosis in vitro. This proof-of-concept paves the way for future drug screening applications. However, this approach involves the differentiation of 4 different cell types in a relatively complex hydrogel and thus could be challenging to scale up for high throughput screening solutions currently used by the pharmaceutical industry.
For this reason, the group of Takanori Takebe has adapted a protocol originally developed for generating intestinal organoids to produce multicellular human liver organoids (HLOs).44 HLOs are generated by hiPSC-co-differentiation into endodermal and mesodermal progenitors.44,45 The co-developed hepatic endoderm and mesoderm derivatives include hepatocyte-like cells, biliary-like cells, stromal cells, and macrophages. Although the actual HLOs primarily contain hepatobiliary cell types, the overall 3D culture harbors a very rich stromal cell population. The in-depth assessment of functional maturation for each HLO cell type is challenging, but existing data indicate that HLO hepatocytes are fetal-like as evidenced by high alpha-fetoprotein and minimal CyP3A4 expression. Nonetheless, HLOs can model steatosis and proinflammatory response after OA treatment.44,45 Furthermore, they can be easily created from a broad number of different, patient-specific hiPSC lines, thereby allowing genetic studies otherwise impossible with alternative systems. Accordingly, HLOs have been recently tested under OA and insulin–high growth conditions to link the risk allele associated with the GCKR gene to identify potential drugs that could modulate the functional impact of this variant on mitochondria activity.46 In this study, pooling HLOs from different donors into a single 3D hydrogel condition decreased batch effects and allowed organoid phenotyping that could be traced back to the donor genotype after individual HLO isolation. Of note, variants affecting GCKR are commonly found in genome-wide associated studies because of its general function in cellular metabolic activity.47 Liver-specific risk alleles such as TM6SF2 rs58542926 were not picked up by this study. Thus, further studies will be necessary to demonstrate that such approach can indeed identify new liver-specific variants associated with NAFLD/NASH penetrance. Nonetheless, HLOs provide a new platform to study NAFLD/NASH allowing high throughput analyses, which are essential to study complex genetic mechanisms.
These 3 studies exemplify how hiPSC-derived hepatic cells can be used not only to model NAFLD/NASH but also to establish new knowledge about the disease and to validate drug efficacy. This represents a major step forward and provides a new assay for therapeutic development. Nonetheless, several aspects need to be considered when using hiPSC-based models, especially the functional state of their derivatives produced in the dish. The development of new methods of differentiation such as forward programming could address this drawback. In addition, hiPSC-based models are time- and resource-consuming and require major technical streamlining to be compatible with industrial standards. Only multidisciplinary teams will be able to achieve such objective.
Precision-Cut Liver Slices
Precision-cut liver slices (PCLSs) may be regarded as a mirror of methods using non-hepatic stromal cells and driving them toward a liver-like phenotype in terms of advantages and pitfalls. Indeed, PCLSs consist of slicing fresh liver tissue from either explants or biopsies, thus maintaining tissue architecture (eg, native extracellular matrix components) as well as cellular organization. However, precision-cut liver slicing presents major limitations for the study of circulating agent roles in liver disease such as blood immune cells, and in most setups PCLSs may only be used up to 5 days, thereby limiting the study of slowly progressing pathologic mechanisms (eg, fibrosis). Moreover, tissue slicing techniques are not trivial and require specific equipment to ensure consistent slice thickness and cell viability.48 Liver slices may be cut at a thickness as low as 0.1 mm for a diameter of generally 5–10 mm. The most challenging aspect of PCLSs resides in obtaining viable slices that will generate close to physiological data during the experimentation. It is generally accepted that liver slice viability may be maintained up to 6 hours in simple culture conditions or up to a few days in controlled conditions according to most studies. Nonetheless, PCLSs offer great opportunities for drug testing or molecular investigations in primary liver tissue from selected healthy or diseased donors. Indeed, the preservation of (patho)physiological cellular and extracellular context is valuable for functional and toxicity assays.48,49 This was evidenced per the use of the PPARα agonist Wy14643 on liver tissue slices, which demonstrated broad effects of the PPARα signaling on metabolism-related gene signatures driven by multiple cell types, when compared with the findings derived from a similar experiment performed on isolated primary hepatocytes.50 Similarly, the effects of obeticholic acid were investigated on PCLSs and led to the identification of novel farnesoid X receptor target genes, which opened new research avenues for understanding the effects of obeticholic acids on high-density lipoprotein levels in NASH patients.51 Importantly, PCLSs allow for circumventing most issues encountered in models that consist of the artificial reconstitution of cellular organization and, most significantly, preserves the extracellular matrix components and their location. This was particularly relevant in a study by Bansal et al,52 which demonstrated the anti-fibrogenic consequences of ITGA11 expression reduction through the inhibition of the hedgehog signaling pathway. An improved method for PCLS has recently been described and aimed at preserving the hepatocytes from cell death for PCLS use in toxicity assays.49 By using this protocol that relies on tissue immobilization and tissue preserving solutions, the authors claimed that almost no cell death was induced from the slicing procedure, and notably the mitochondrial respiration was similar to that of hepatocyte cultures, and caspase 3 activity was minimal. This setup led the authors to demonstrate the detrimental effects of tumor necrosis factor on hepatocyte survival from virus-infected livers.
However, it must be considered that precision-cut liver slicing requires specific equipment and expertise. Furthermore, tissue slicing inevitably leads to liver cell exposure to damage-associated molecular patterns of varying types. As such, it was evidenced tissue slicing activates inflammatory responses during the first 4 days of culture and suggested that experiments performed on liver slices must be conducted after 7 days, when the system is stabilized.53 Hence, the authors of this study claimed they could maintain liver slices in culture for more than 2 weeks, although this would need further validation by other groups to evaluate cell phenotypes and functionality after such a period. Moreover, liver slicing induces the activation of fibrogenic mechanisms, a response that may hamper the assessment of anti-fibrotic treatments. A study showed that HSC activation occurring after liver slicing could be prevented by the addition of valproic acid sodium salt. However, in such setup PCLSs may only be maintained up to 5 days.54 Overall, liver tissue slice culture protocols may require further optimization but provide opportunities for patient-specific investigations of NAFLD pathobiology and drug development while preserving most characteristics of the native tissues.
Primary Tissue-Derived Organoids for NAFLD Studies
Examples of disease modeling per tissue-derived organoids, as shown for stem cell-rich organs like the intestine,55 have also raised hope for liver disease modeling. However, a large body of literature indicates that replicating preexisting hepatocytes, and not stem cells, regenerate the mammalian liver.56,57 The robust organoid culture of proliferative primary human hepatocytes is mainly restricted to fetal hepatocytes.58 Nevertheless, human tissue organoids can model other cell states that emerge in persistent injury. In chronic liver diseases such as NASH or cholestatic diseases, the liver mounts a “ductular reaction” characterized by the expansion of cholangiocyte-like cells into the hepatic parenchyma. Cholangiocytes give rise to the ductular reaction, but also hepatocytes contribute when chronically injured.59,60 The hepatocyte contribution has been described as a metaplasia-like response into bipotential biliary-like progenitors that can re-differentiate back into hepatocytes upon injury withdrawal.60 The cholangiocyte-biased, bipotential progenitor cell state can be modeled in the dish using human intrahepatic cholangiocyte organoids (ICOs) that model a default biliary epithelial state but can differentiate into hepatocytes to model lipid metabolism and lipotoxicity.25 Because the ductular reaction usually occurs at more advanced disease stages, ICOs may aid in closing knowledge gaps relevant to the late-chronic epithelial injury response in NAFLD/NASH. Because NAFLD displays significant inter-patient variation in disease progression, patient-specific ICOs may also elucidate individual molecular drivers. Both liver biopsy material (>50 mg) and wedges from liver explants can give rise to ICOs. Within 2 weeks, these starting materials allow the growth of ICOs equipped with patient-specific genetic and epigenetic risk factors.61 For example, the I148M PNPLA3 variant is also expressed in cholangiocytes and has even been linked to the progression of biliary diseases.62 Although not demonstrated yet, ICOs carrying the risk variant may help decipher PNPLA3’s role in NAFLD/NASH’s ductular reaction. Although ICOs can be expanded for more than 15 passages25 and cryo-biobanked, cohort-wide phenotyping of gene mutations, as shown with hiPSC-derived organoids,46 remains an untapped potential.
In the few studies of their kind, McCarron et al61 observed differences in the lipid metabolism of primary liver tissue organoids from a small cohort (n = 6) of NASH-related cirrhosis as compared with controls (n = 3). ICOs were re-differentiated into hepatocyte-like cells using a defined medium. The authors assessed hepatically differentiated ICOs and found a resemblance to hiPSC-derived hepatocytes, specifically an immature hepatocyte-like cell state with a hybrid biliary signature. NASH organoids displayed 4.5 times higher OA-induced lipid uptake compared with non-NASH controls. A counterintuitive finding in the same NASH organoids has been that low-density lipoprotein accumulation is about 2-fold reduced.61 Unfortunately, the authors did not link these intriguing findings to clinical data, eg, serum low-density lipoprotein levels or statin therapy.
As part of the natural history of NAFLD-to-NASH progression, the liver undergoes a complex fibroinflammatory response. In persistent lipotoxic liver injury, liver fibrosis may progress into end-stage liver disease related to cirrhosis. Accordingly, the study by McCarron et al61 focused on end-stage NASH and generated ICOs from NASH patients at the time of transplantation. Noteworthy, hepatically differentiated ICOs lacked the multicellular complexity of the native liver. Nonetheless, NASH organoid transcriptomes showed up-regulation of pathogenic pathways usually attributed to non-parenchymal cells. Among the most up-regulated programs was the attraction of lymphocytes. This epithelial inflammatory profile is reminiscent of human ICOs generated from epithelial cells found in bile of primary sclerosing cholangitis patients. Here, the authors demonstrated an immune-reactive phenotype when ICOs were exposed to interleukin 17A or tumor necrosis factor.63 As a step toward intercellular crosstalk, a recent study showed how mouse ICOs could be reconstituted into droplets, allowing cell-cell contact with portal fibroblasts using droplet microfluidics.64 In a different report, scientists aggregated primary mouse hepatocytes, HSCs, LSECs, and KCs in U-bottom plates. They generated syngeneic multicellular organoids at a large scale and could perform proof-of-principle studies for anti-fibrotic medication candidates.65 These data suggest that ICO assembly with stromal or immune cells may be a promising avenue to decipher the human fibroinflammatory crosstalk. Notably, epithelial intestinal or pulmonary cells were shown to mature co-cultured innate lymphoid cells into tissue-specific phenotypes without requiring subset-specific cytokine supplementation.66 Complex organoid assembly may be cumbersome because of limitations related to the isolation of sufficient syngeneic cell types from human liver donor tissue or biopsy material. Using HLA-matched peripheral blood mononuclear cells from commercial or academic repositories or allogenic cell types for HLA-independent hypotheses may overcome the limitation.
ICOs from NASH cirrhosis revealed further findings that may serve as a model for common phenomena in liver regeneration. NASH cirrhosis organoids replicated faster but eventually underwent growth arrest earlier, limiting their expandability.61 In addition, oncogenic pathways were up-regulated in NASH cirrhosis organoids, which supports them as a model for genomic stress with possible implications to the “regenerative nodule” in cirrhosis. Furthermore, NASH cirrhosis-derived organoids dedifferentiated less efficiently from the hepatocyte state back to the ductal state. This loss of cellular plasticity in organoids may help elucidate molecular mechanisms in the loss of effective liver regeneration in vivo. Replicative senescence and loss of the native transcription factor machinery are possible explanations.
Intriguingly, NASH cirrhosis–derived organoids were also more susceptible to infection with severe acute respiratory syndrome–associated coronavirus 2 pseudovirus, reminiscent of poor coronavirus disease 2019 outcomes in patients with NAFLD and fibrosis.67 The authors demonstrate how viral permissiveness levels correlate better with ubiquitin D mRNA levels, a fibrosis marker and inhibitor of interferon response, than with the severe acute respiratory syndrome–associated coronavirus 2 entry factor ACE2 and TMPRSS2.61 Thus, human ICO technology can serve to study viral infection in the setting of NAFLD/NASH with high clinical implications. In turn, virally introduced CRISPR libraries into human expandable NAFLD/NASH-derived organoids may allow future genetic screens for determinants of viral permissiveness, regeneration, and lipid metabolism.
The limitations of functional phenotyping of ICOs from end-stage diseases, as reported for NASH61 or biliary atresia,68 are related to the spontaneous mutations and epigenetic remodeling that may occur in such severe chronic tissue injury. Thus, ICOs from end-stage liver diseases may not be ideal for early NAFLD pathogenesis studies. Furthermore, the bi-cellular contribution to the ductular reaction in chronic liver injury likely creates ICOs of mixed cholangiocyte and hepatocyte origin; thus, the unclear human cell source limits comparisons to healthy control organoids derived from livers without native ductular reaction. Limitations related to the immaturity and hybrid cell phenotypes of hepatically differentiated ICOs will hopefully be overcome in the future with protocols describing robust postnatal hepatocyte organoid culture. Furthermore, current go-to protocols rely on mouse tumor-derived hydrogels for culturing human liver ICOs, which raises ethical concerns and may create a growth advantage for proliferative subsets of ICO cells.
Overall, the potential of primary human organoids for the studies of NAFLD is broad and may include the following foci in the near future: patient- or gene mutation-specific lipid metabolism studies, biobanking and personalized drug screening of assembled multicellular organoids, innate immunity in the setting of NAFLD, CRISPR screens in lipotoxic settings, NAFLD-related genotoxicity, and organoid-on-a-chip technology.
Perfused Liver-on-Chip Models
Organ-on-chip defines a type of in vitro model that allows seeding multiple cell types on a device mimicking the typical organ architecture or fluidic dynamics. Major advances of these approaches rely on a much higher flexibility in term of experimental design, from the choice of the nature and relative densities of the cells being seeded to the modulation of the culture milieu. Several solutions have been developed in recent years. Options include those biochips with dynamic perfusion of the cell culture milieu, allowing for the modeling of blood or other fluid content (eg, nutrients, cytokines, therapeutic agents, cellular waste disposal) as well as the modulation of the perfusion flow (eg, differential shear stress). This is exemplified by a growing number of solutions for the culture of primary or immortalized hepatocytes under fluidic conditions.69, 70, 71, 72, 73 As a multifactorial, multicellular, and interorgan-affecting disease (eg, insulin resistance), NAFLD models should be geared toward such complexity. Raasch et al74 recently developed a perfusable biochip and demonstrated the strength of their model in assessing endothelial cell functions when compared with static culture conditions. This group further developed this setup by seeding 2 chambers, each containing hepatocytes and HSCs on one side and macrophages and endothelial cells on the other. The compartments were separated by a porous membrane, allowing for the diffusion of macromolecules and intercellular communication.75 The authors further demonstrated the value of dynamic perfusion in establishing or maintaining hepatocyte metabolic functions. Likewise, others reported on a microfluidic NASH-on-a-chip model of the concurrent culture of primary human hepatocytes, KCs, HSCs, and LSECs in a collagen-based hydrogel.76 Here, the authors could demonstrate that their model mimics some aspects of NASH such that inflammatory cytokine secretion was increased upon lipotoxic conditions, which was reduced by introducing the drug candidate Elafibranor. In the same fashion, it was shown that free fatty acids could effectively induce inflammation and fibrosis-related gene expression as well as pro-collagen 1 secretion in a microfluidic system using primary liver cells.77 In this study, the authors demonstrated by using multiple cell donors that chips seeded with PNPLA3 mutant cells had increased NASH phenotypes (ie, inflammation, lipotoxicity, and fibrogenesis). Finally, the authors claimed their system was viable and relevant for a period of at least 2 weeks. In a follow-up study, the same group demonstrated that their system showed a similar fibroinflammatory transcriptomic signature to NASH patients and response to NASH therapeutic candidates.78
These studies reveal the difficulties of generating high throughput read-outs in multicellular culture biochip systems with limited material for further analysis. On the basis of the literature, culturing cells for up to 2 weeks in a liver-on-chip system is possible but challenging. Nonetheless, similar to all other approaches described in this review, such a culture period cannot appropriately recapitulate NAFLD progression that occurs over decades in patients. Future incremental developments are expected to establish liver-on-chip models that further optimize their fidelity in modeling native pathophysiology. Optimizations will likely include improving primary cell isolation and culture protocols or further advancing hiPSC-derived liver cell applications. Intriguingly, organ-on-chip systems have been developed to mimic the consequences of overweight in organs such as adipose tissue or the potentials of multiorgan chips as discussed elsewhere.7,79, 80, 81 As such, the possibility of connecting several organ-on-a-chip systems represents a largely untapped potential to investigate NAFLD under the influence of metabolic and inflammatory crosstalk between the liver, blood, adipose tissue, and other organs.
Conclusions
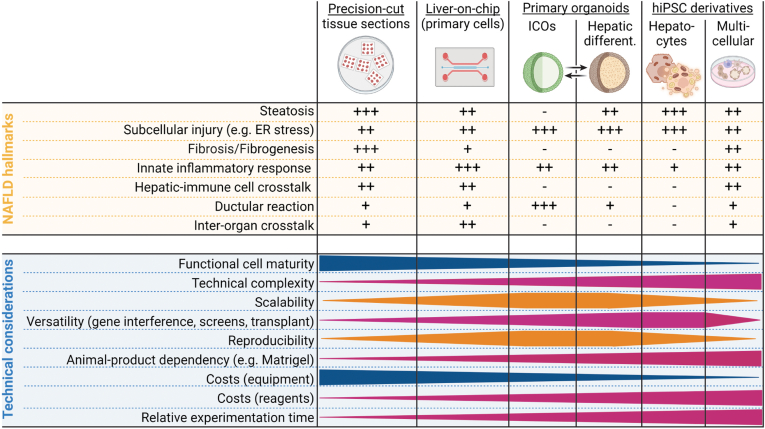
NAFLD/NASH is a comparably well-understood, multifactorial disease that involves multiple liver cell types and evolves in crosstalk with other organs over many years. The public health burden and the need for effective therapies increase the demand for faithful and reproducible human disease models. Despite significant progress in the last decade in proof-of-principle liver disease modeling in the dish, no single platform recapitulated all nuances of NAFLD/NASH pathobiology, let alone discovered approved drug targets. The specific research hypothesis or question indicates which model is best suited (Table 1, Figure 2). HiPSC-based platforms allow scalable, parallel differentiation of the different cell types comprising the tissue in NAFLD/NASH, but at the expense of cellular maturity and liver tissue-specific context. Human PCLSs preserve the spatial tissue architecture for a limited period but are not scalable. Primary tissue-derived ICOs may help better understand epithelial injury and hepatocyte-biliary cell plasticity but lack multicellular context. This shortcoming can be addressed by liver-on-chip models with dedicated cell compartments, which may be challenged by deriving primary human liver cells but could be overcome per hiPSC derivation. The metabolic and immune crosstalk between the liver, muscle, fat, and other tissues is only rudimentarily captured by current models and represents a largely untapped potential for future efforts. Ultimately, neither the hiPSC-derived platform nor any other model system can be used in isolation. Results generated in vitro need to be validated using animal models, patient data, and ultimately in clinical trials.
Table 1.
Take Home Messages on Current NAFLD Models in the Dish
| NAFLD models | Main advantages | Limitations |
|---|---|---|
| Human precision-cut liver slices |
|
|
| Liver-on-chip |
|
|
| Primary ICOs |
|
|
| hiPSC-derivatives |
|
|
DAMP, damage-associated molecular pattern.
Figure 2.
Technical considerations and NAFLD hallmark fidelity of NAFLD models. This figure depicts the NAFLD models discussed in this review in relation to either their relevance for the study of typical NAFLD hallmarks or material needs. ER, endoplasmic reticulum; hiPSC, human induced pluripotent stem cell; ICO, intrahepatic cholangiocyte organoid; NAFLD, nonalcoholic fatty liver disease. Created with biorender.com.
Footnotes
Conflicts of interest This author discloses the following: L.V. is a shareholder in DefiniGen, Billitech, and bit.bio. The remaining authors disclose no conflicts.
Funding Supported by Deutsche Forschungsgemeinschaft-Emmy Noether Program (M.R.) and Einstein foundation strategic professorship and a core grant from the Berlin Institute of Health (L.V.). This work was funded by the German Research Foundation (DFG SFB/TRR 296 and CRC1382, Project-ID 403224013) and the German Ministry of Education and Research (BMBF ImmunAvatar consortium).
References
- 1.Karlsen T.H., Sheron N., Zelber-Sagi S., et al. The EASL-Lancet Liver Commission: protecting the next generation of Europeans against liver disease complications and premature mortality. Lancet. 2022;399:61–116. doi: 10.1016/S0140-6736(21)01701-3. [DOI] [PubMed] [Google Scholar]
- 2.Lambrecht J., Tacke F. Controversies and opportunities in the use of inflammatory markers for diagnosis or risk prediction in fatty liver disease. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.634409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasiri-Ansari N., Androutsakos T., Flessa C.M., et al. Endothelial cell dysfunction and nonalcoholic fatty liver disease (NAFLD): a concise review. Cells. 2022:11. doi: 10.3390/cells11162511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace S.J., Tacke F., Schwabe R.F., et al. Understanding the cellular interactome of non-alcoholic fatty liver disease. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiseler M., Schwabe R., Hampe J., et al. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease: novel insights into cellular communication circuits. J Hepatol. 2022;77:1136–1160. doi: 10.1016/j.jhep.2022.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Bruneau A., Hundertmark J., Guillot A., et al. Molecular and cellular mediators of the gut-liver axis in the progression of liver diseases. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.725390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shroff T., Aina K., Maass C., et al. Studying metabolism with multi-organ chips: new tools for disease modelling, pharmacokinetics and pharmacodynamics. Open Biol. 2022;12 doi: 10.1098/rsob.210333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K., Tanabe K., Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Guan J., Wang G., Wang J., et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature. 2022;605:325–331. doi: 10.1038/s41586-022-04593-5. [DOI] [PubMed] [Google Scholar]
- 10.Ying Q.L., Wray J., Nichols J., et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gafni O., Weinberger L., Mansour A.A., et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 12.Brons I.G., Smithers L.E., Trotter M.W., et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 13.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 14.Brooks I.R., Garrone C.M., Kerins C., et al. Functional genomics and the future of iPSCs in disease modeling. Stem Cell Reports. 2022;17:1033–1047. doi: 10.1016/j.stemcr.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yiangou L., Ross A.D.B., Goh K.J., et al. Human pluripotent stem cell-derived endoderm for modeling development and clinical applications. Cell Stem Cell. 2018;22:485–499. doi: 10.1016/j.stem.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Sampaziotis F., Segeritz C.P., Vallier L. Potential of human induced pluripotent stem cells in studies of liver disease. Hepatology. 2015;62:303–311. doi: 10.1002/hep.27651. [DOI] [PubMed] [Google Scholar]
- 17.Wesley B.T., Ross A.D.B., Muraro D., et al. Single-cell atlas of human liver development reveals pathways directing hepatic cell fates. Nat Cell Biol. 2022;24:1487–1498. doi: 10.1038/s41556-022-00989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett J.L., Duncan S.A. iPSC-derived hepatocytes as a platform for disease modeling and drug discovery. Front Med (Lausanne) 2019;6:265. doi: 10.3389/fmed.2019.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pawlowski M., Ortmann D., Bertero A., et al. Inducible and deterministic forward programming of human pluripotent stem cells into neurons, skeletal myocytes, and oligodendrocytes. Stem Cell Reports. 2017;8:803–812. doi: 10.1016/j.stemcr.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boon R., Kumar M., Tricot T., et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat Commun. 2020;11:1393. doi: 10.1038/s41467-020-15058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomaz R.A., Zacharis E.D., Bachinger F., et al. Generation of functional hepatocytes by forward programming with nuclear receptors. Elife. 2022:11. doi: 10.7554/eLife.71591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou T., Kundu D., Robles-Linares J., et al. Feedback signaling between cholangiopathies, ductular reaction, and non-alcoholic fatty liver disease. Cells. 2021:10. doi: 10.3390/cells10082072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampaziotis F., Cardoso de Brito M., Madrigal P., et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa M., Ogawa S., Bear C.E., et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 25.Huch M., Gehart H., van Boxtel R., et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimland C.A., Tilson S.G., Morell C.M., et al. Regional differences in human biliary tissues and corresponding in vitro-derived organoids. Hepatology. 2021;73:247–267. doi: 10.1002/hep.31252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roos F.J.M., van Tienderen G.S., Wu H., et al. Human branching cholangiocyte organoids recapitulate functional bile duct formation. Cell Stem Cell. 2022;29:776–794 e713. doi: 10.1016/j.stem.2022.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Sampaziotis F., Muraro D., Tysoe O.C., et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 2021;371:839–846. doi: 10.1126/science.aaz6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams I.M., Wu J.C. Generation of endothelial cells from human pluripotent stem cells. Arterioscler Thromb Vasc Biol. 2019;39:1317–1329. doi: 10.1161/ATVBAHA.119.312265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gage B.K., Liu J.C., Innes B.T., et al. Generation of functional liver sinusoidal endothelial cells from human pluripotent stem-cell-derived venous angioblasts. Cell Stem Cell. 2020;27:254–269 e259. doi: 10.1016/j.stem.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Gage B.K., Merlin S., Olgasi C., et al. Therapeutic correction of hemophilia A by transplantation of hPSC-derived liver sinusoidal endothelial cell progenitors. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110759. [DOI] [PubMed] [Google Scholar]
- 32.Elcheva I., Brok-Volchanskaya V., Kumar A., et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krenkel O., Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 34.van Wilgenburg B., Browne C., Vowles J., et al. Efficient, long term production of monocyte-derived macrophages from human pluripotent stem cells under partly-defined and fully-defined conditions. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tasnim F., Xing J., Huang X., et al. Generation of mature kupffer cells from human induced pluripotent stem cells. Biomaterials. 2019;192:377–391. doi: 10.1016/j.biomaterials.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 36.Andrews T.S., Atif J., Liu J.C., et al. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol Commun. 2021 doi: 10.1002/hep4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aizarani N., Saviano A., Sagar, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speicher A.M., Korn L., Csatari J., et al. Deterministic programming of human pluripotent stem cells into microglia facilitates studying their role in health and disease. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2123476119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallverdu J., Martinez Garcia de la Torre R.A., Mannaerts I., et al. Directed differentiation of human induced pluripotent stem cells to hepatic stellate cells. Nat Protoc. 2021;16:2542–2563. doi: 10.1038/s41596-021-00509-1. [DOI] [PubMed] [Google Scholar]
- 40.Koui Y., Himeno M., Mori Y., et al. Development of human iPSC-derived quiescent hepatic stellate cell-like cells for drug discovery and in vitro disease modeling. Stem Cell Reports. 2021;16:3050–3063. doi: 10.1016/j.stemcr.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng A.H.M., Khoshakhlagh P., Rojo Arias J.E., et al. A comprehensive library of human transcription factors for cell fate engineering. Nat Biotechnol. 2021;39:510–519. doi: 10.1038/s41587-020-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilson S.G., Morell C.M., Lenaerts A.S., et al. Modelling PNPLA3-associated non-alcoholic fatty liver disease using human induced pluripotent stem cells. Hepatology. 2021;74:2998–3017. doi: 10.1002/hep.32063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar M., Toprakhisar B., Van Haele M., et al. A fully defined matrix to support a pluripotent stem cell derived multi-cell-liver steatohepatitis and fibrosis model. Biomaterials. 2021;276 doi: 10.1016/j.biomaterials.2021.121006. [DOI] [PubMed] [Google Scholar]
- 44.Ouchi R., Togo S., Kimura M., et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384 e376. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinozawa T., Kimura M., Cai Y., et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology. 2021;160:831–846 e810. doi: 10.1053/j.gastro.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura M., Iguchi T., Iwasawa K., et al. En masse organoid phenotyping informs metabolic-associated genetic susceptibility to NASH. Cell. 2022;185:4216–4232 e4216. doi: 10.1016/j.cell.2022.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun B.B., Kurki M.I., Foley C.N., et al. Genetic associations of protein-coding variants in human disease. Nature. 2022;603:95–102. doi: 10.1038/s41586-022-04394-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Graaf I.A., Olinga P., de Jager M.H., et al. Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat Protoc. 2010;5:1540–1551. doi: 10.1038/nprot.2010.111. [DOI] [PubMed] [Google Scholar]
- 49.Brugger M., Laschinger M., Lampl S., et al. High precision-cut liver slice model to study cell-autonomous antiviral defense of hepatocytes within their microenvironment. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janssen A.W., Betzel B., Stoopen G., et al. The impact of PPARalpha activation on whole genome gene expression in human precision cut liver slices. BMC Genomics. 2015;16:760. doi: 10.1186/s12864-015-1969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ijssennagger N., Janssen A.W.F., Milona A., et al. Gene expression profiling in human precision cut liver slices in response to the FXR agonist obeticholic acid. J Hepatol. 2016;64:1158–1166. doi: 10.1016/j.jhep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 52.Bansal R., Nakagawa S., Yazdani S., et al. Integrin alpha 11 in the regulation of the myofibroblast phenotype: implications for fibrotic diseases. Exp Mol Med. 2017;49:e396. doi: 10.1038/emm.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X., Roberto J.B., Knupp A., et al. Precision-cut human liver slice cultures as an immunological platform. J Immunol Methods. 2018;455:71–79. doi: 10.1016/j.jim.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dewyse L., De Smet V., Verhulst S., et al. Improved precision-cut liver slice cultures for testing drug-induced liver fibrosis. Front Med (Lausanne) 2022;9 doi: 10.3389/fmed.2022.862185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 56.Yanger K., Knigin D., Zong Y., et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 2014;15:340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaub J.R., Malato Y., Gormond C., et al. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu H., Gehart H., Artegiani B., et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606 e1519. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Rodrigo-Torres D., Affo S., Coll M., et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology. 2014;60:1367–1377. doi: 10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tarlow B.D., Pelz C., Naugler W.E., et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarron S., Bathon B., Conlon D.M., et al. Functional characterization of organoids derived from irreversibly damaged liver of patients with NASH. Hepatology. 2021;74:1825–1844. doi: 10.1002/hep.31857. [DOI] [PubMed] [Google Scholar]
- 62.Friedrich K., Rupp C., Hov J.R., et al. A frequent PNPLA3 variant is a sex specific disease modifier in PSC patients with bile duct stenosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soroka C.J., Assis D.N., Alrabadi L.S., et al. Bile-derived organoids from patients with primary sclerosing cholangitis recapitulate their inflammatory immune profile. Hepatology. 2019;70:871–882. doi: 10.1002/hep.30470. [DOI] [PubMed] [Google Scholar]
- 64.Cordero-Espinoza L., Dowbaj A.M., Kohler T.N., et al. Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell Stem Cell. 2021;28:1907–1921 e1908. doi: 10.1016/j.stem.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Os E.A., Cools L., Eysackers N., et al. Modelling fatty liver disease with mouse liver-derived multicellular spheroids. Biomaterials. 2022;290 doi: 10.1016/j.biomaterials.2022.121817. [DOI] [PubMed] [Google Scholar]
- 66.Jowett G.M., Read E., Roberts L.B., et al. Organoids capture tissue-specific innate lymphoid cell development in mice and humans. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Targher G., Mantovani A., Byrne C.D., et al. Risk of severe illness from COVID-19 in patients with metabolic dysfunction-associated fatty liver disease and increased fibrosis scores. Gut. 2020;69:1545–1547. doi: 10.1136/gutjnl-2020-321611. [DOI] [PubMed] [Google Scholar]
- 68.Amarachintha S.P., Mourya R., Ayabe H., et al. Biliary organoids uncover delayed epithelial development and barrier function in biliary atresia. Hepatology. 2022;75:89–103. doi: 10.1002/hep.32107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kostrzewski T., Cornforth T., Snow S.A., et al. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J Gastroenterol. 2017;23:204–215. doi: 10.3748/wjg.v23.i2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gori M., Simonelli M.C., Giannitelli S.M., et al. Investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kidambi S., Yarmush R.S., Novik E., et al. Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc Natl Acad Sci U S A. 2009;106:15714–15719. doi: 10.1073/pnas.0906820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang Y.B.A., Eo J., Mert S., et al. Metabolic patterning on a chip: towards in vitro liver zonation of primary rat and human hepatocytes. Sci Rep. 2018;8:8951. doi: 10.1038/s41598-018-27179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ehrlich A., Duche D., Ouedraogo G., et al. Challenges and opportunities in the design of liver-on-chip microdevices. Annu Rev Biomed Eng. 2019;21:219–239. doi: 10.1146/annurev-bioeng-060418-052305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raasch M., Rennert K., Jahn T., et al. Microfluidically supported biochip design for culture of endothelial cell layers with improved perfusion conditions. Biofabrication. 2015;7 doi: 10.1088/1758-5090/7/1/015013. [DOI] [PubMed] [Google Scholar]
- 75.Rennert K., Steinborn S., Groger M., et al. A microfluidically perfused three dimensional human liver model. Biomaterials. 2015;71:119–131. doi: 10.1016/j.biomaterials.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 76.Freag M.S., Namgung B., Reyna Fernandez M.E., et al. Human nonalcoholic steatohepatitis on a chip. Hepatol Commun. 2021;5:217–233. doi: 10.1002/hep4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kostrzewski T., Maraver P., Ouro-Gnao L., et al. A microphysiological system for studying nonalcoholic steatohepatitis. Hepatol Commun. 2020;4:77–91. doi: 10.1002/hep4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kostrzewski T., Snow S., Battle A.L., et al. Modelling human liver fibrosis in the context of non-alcoholic steatohepatitis using a microphysiological system. Commun Biol. 2021;4:1080. doi: 10.1038/s42003-021-02616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCarthy M., Brown T., Alarcon A., et al. Fat-on-a-chip models for research and discovery in obesity and its metabolic comorbidities. Tissue Eng Part B Rev. 2020;26:586–595. doi: 10.1089/ten.teb.2019.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Picollet-D’hahan N., Zuchowska A., Lemeunier I., et al. Multiorgan-on-a-chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021;39:788–810. doi: 10.1016/j.tibtech.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 81.Slaughter V.L., Rumsey J.W., Boone R., et al. Validation of an adipose-liver human-on-a-chip model of NAFLD for preclinical therapeutic efficacy evaluation. Sci Rep. 2021;11 doi: 10.1038/s41598-021-92264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]