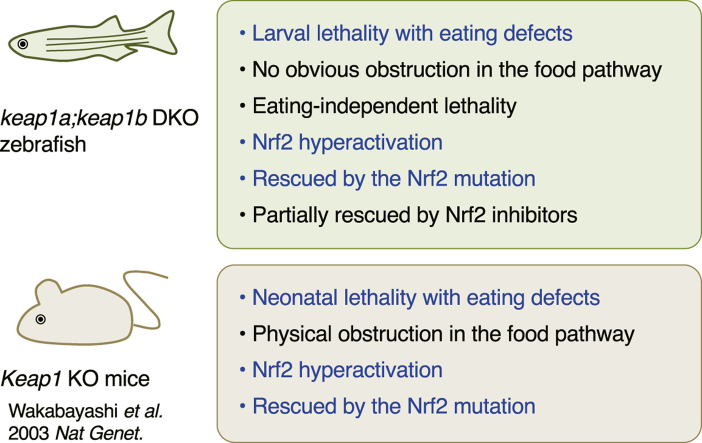
Abstract
The Keap1–Nrf2 pathway is an evolutionarily conserved mechanism that protects cells from oxidative stress and electrophiles. Keap1 is a repressor of Nrf2 in normal cellular conditions but also a stress sensor for Nrf2 activation. Interestingly, fish and amphibians have two Keap1s (Keap1a and Keap1b), of which Keap1b is the ortholog of mammalian Keap1. Keap1a, on the other hand, is a gene found only in fish and amphibians, having been lost during the evolution to amniotes. We have previously shown that keap1b-knockout zebrafish have increased Nrf2 activity and reduced response to certain Nrf2-activating compounds but that they grow normally to adulthood. This may be because the remaining keap1a suppresses the hyperactivation of Nrf2, which is responsible for the post-natal lethality of Keap1-knockout mice. In this study, we analyzed keap1a;keap1b-double-knockout zebrafish to test this hypothesis. We found that keap1a;keap1b-double-knockout zebrafish, like Keap1-knockout mice, showed eating defects and were lethal within a week of hatching. Genetic introduction of the Nrf2 mutation rescued both the eating defects and the larval lethality, indicating that Nrf2 hyperactivation is the cause. However, unlike Keap1-knockout mice, keap1a;keap1b-double-knockout zebrafish showed no physical blockage of the food pathway; moreover, the cause of death was not directly related to eating defects. RNA-sequencing analysis revealed that keap1a;keap1b-double-knockout larvae showed extraordinarily high expression of known Nrf2-target genes as well as decreased expression of visual cycle genes. Finally, trigonelline or brusatol partially rescued the lethality of keap1a;keap1b-double-knockout larvae, suggesting that they can serve as an in vivo evaluation system for Nrf2-inhibiting compounds.
Keywords: Eating defects, Keap1–Nrf2 pathway, keap1a;keap1b-double knockout zebrafish, Nrf2-inhibiting compounds, Visual cycle genes
Graphical abstract
Highlights
-
•
keap1a;keap1b-double-knockout in zebrafish caused larval lethality and eating defects.
-
•
Double-knockout larvae showed lethality even under non-feeding conditions.
-
•
Nrf2 mutation rescued all the phenotypes in keap1a;keap1b-double-knockout zebrafish.
-
•
Nrf2 inhibitors partially rescued the lethality of keap1a;keap1b-double-knockout larvae.
1. Introduction
The Keap1–Nrf2 pathway is a cellular defense system against oxidative and electrophilic stresses and is widely conserved among vertebrates [1,2]. Nrf2 is a transcriptional activator that regulates antioxidant and drug-metabolizing genes and is normally degraded by ubiquitin-proteasome-dependent proteolysis but is stabilized when oxidative stress or electrophiles accumulate in the cell. Keap1 suppresses Nrf2 as a mediator of Nrf2-specific ubiquitination and functions as a sensor molecule to sense Nrf2-activating compounds [[3], [4], [5]]. Interestingly, mammals, birds, and reptiles have only one Keap1 gene, whilst fish and amphibians have two isoforms, Keap1a and Keap1b [[6], [7], [8]]. Phylogenetic analysis revealed that Keap1b corresponds to Keap1 in mammals/birds/reptiles, whilst Keap1a disappeared during the evolution to amniotes [2,8].
Keap1-knockout (KO) mice were lethal within 3 weeks of birth, and this lethality was rescued by disruption of the Nrf2 gene, indicating that Nrf2 hyperactivation caused the lethality [9]. In contrast, keap1b-KO zebrafish were viable to adulthood, although they showed some activation of Nrf2 [8]. We hypothesized that the keap1b-KO zebrafish did not accumulate Nrf2 as much as did the Keap1-KO mice because of the presence of keap1a, which can also suppress Nrf2. In this study, we generated keap1a;keap1b-double-KO (DKO) zebrafish and analyzed their phenotypes to test this hypothesis.
2. Materials and methods
2.1. Zebrafish
keap1a-KO (keap1ait302) [8], keap1b-KO (keap1bit308) [8], Nrf2 mutant (nfe2l2afh318) [10], and wild-type (AB strain) zebrafish were used. Genotyping was performed as previously described [8,11]. keap1ait302 and keap1bit308 are available from the National BioResource Project (https://shigen.nig.ac.jp/zebra), and nfe2l2afh318, from the Zebrafish International Resource Center (http://zebrafish.org).
2.2. Other methods
See the Supplemental Materials and Methods for chemicals, details of the survival assay, eating analysis, histological analysis, Alcian blue staining, gene expression analysis, statistical analysis, regulation for animal experiments, and primers used in quantitative reverse transcription PCR (qRT-PCR).
3. Results
3.1. keap1a;keap1b-DKO larvae were lethal and showed eating defects
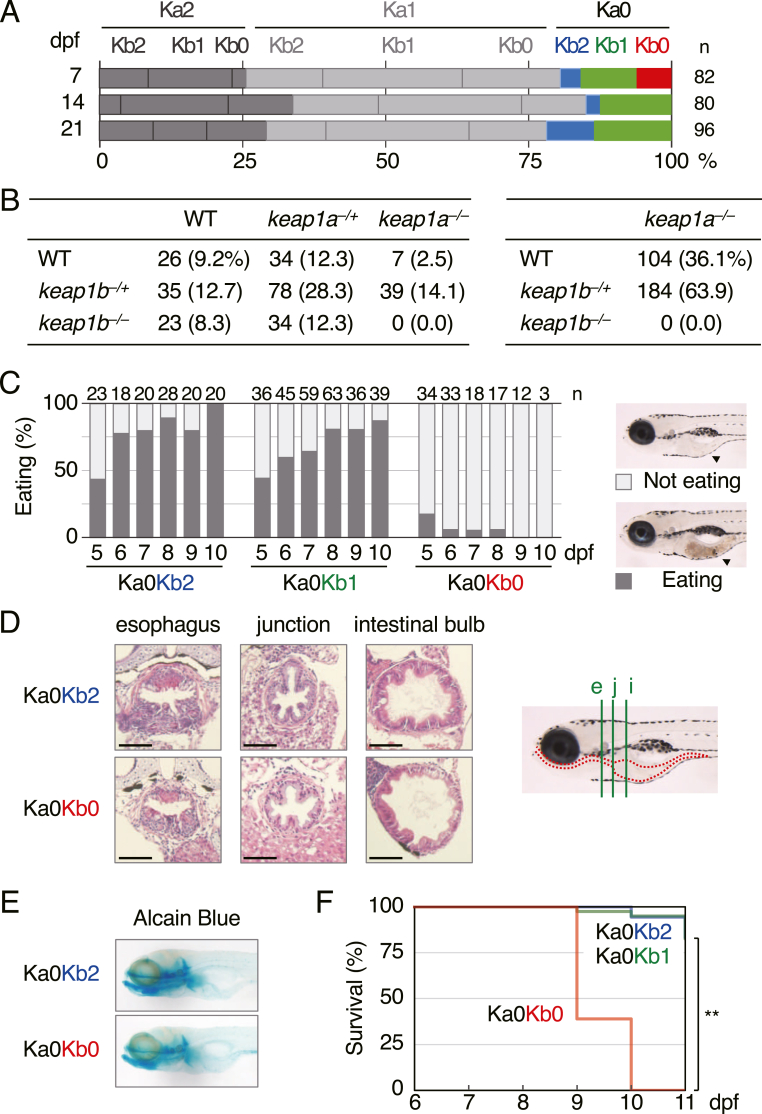
Heterozygous keap1ait302;keap1bit308 DKO (Ka1Kb1) parents were mated to generate homozygous DKO (Ka0Kb0) offspring together with siblings. At 7, 14, and 21 days-post-fertilization (dpf), about 100 of each offspring were collected and genotyped (Fig. 1A). All the genotypes appeared at 7 dpf, whilst there were no Ka0Kb0 larvae but others at 14 or 21 days, suggesting their lethality at the larval stage. We next raised 276 offspring from Ka1Kb1 parents (Fig. 1B, left) and 288 offspring from Ka0Kb1 parents (Fig. 1B, right) to 3 months-post-fertilization (mpf) adult fish and genotyped them and found no Ka0Kb0 adults. All these results indicate that zebrafish without Keap1a and Keap1b are lethal during larval development.
Fig. 1.
Phenotypes of keap1a;keap1b-DKO zebrafish. (A) Survival rates of offspring from heterozygous keap1a;keap1b-DKO (Ka1Kb1) parents at 7, 14, and 21 dpf. The red bar represents homozygous keap1a;keap1b-DKO (Ka0Kb0) larvae. (B) Survival rates of 3-mpf offspring from Ka1Kb1 parents (left) and Ka0Kb1 parents (right). WT, −/+, and −/− indicate wild-type, heterozygous, and homozygous fish, respectively. Data from 3 independent experiments were combined. (C) Eating analysis of 5- to 10-dpf offspring from Ka0Kb1 parents. The numbers at the top are the number of larvae tested. It was noted that eating in Ka0Kb0 larvae was reduced. The black arrowheads in the right panels indicate the food inside the gut of the larvae. (D) Hematoxylin and eosin staining of paraffin sections of 7-dpf larvae. In the right panel, the larval digestive tract is indicated with red dotted lines, and vertical sections at the locations of the esophagus (e, green line), the esophagus-gut junction (j), and the intestinal bulb (i) are shown on the left panel. Scale bars, 50 μm. (E) Alcian blue staining of head cartilages of 7-dpf larvae. (F) Survival rates of offspring from Ka0Kb1 parents without feeding. Survival was observed every 24 h from 6 to 11 dpf. The tested numbers were as follows: Ka0Kb2 n = 18, Ka0Kb1 n = 40, Ka0Kb0 n = 18. The asterisks indicate significant differences (**p < 0.01). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Keap1-KO post-natal mice die probably because of eating defects caused by a narrowed esophagus and forestomach [9]. Zebrafish larvae hatch at 2.5 dpf and start to eat after 5 dpf. To elucidate the reason for the lethality of keap1a;keap1b-DKO larvae, the eating defects of the larvae obtained from Ka0Kb1 parents were analyzed (Fig. 1C). The results showed that the majority of the Ka0Kb0 larvae were not eating even at 8 dpf, whilst their siblings started to eat from 5 dpf. To investigate the cause of the eating defect, the thicknesses of the esophagus and gut of Ka0Kb0 and Ka0Kb2 larvae were histologically analyzed (Fig. 1D). Unexpectedly, no significant difference was found between the Ka0Kb0 and the Ka0Kb2 larvae, suggesting that the gastrointestinal tract was normal. The structure of the mouth was also analyzed by means of cartilage staining, and again no obvious difference between the Ka0Kb0 and the Ka0Kb2 larvae was found (Fig. 1E). These results suggest that the eating defect in keap1a;keap1b-DKO larvae was not due to physical disturbances in the food pathway. We next performed a survival assay of larvae without feeding (Fig. 1F). The results showed that Ka0Kb2 and Ka0Kb1 larvae could survive without feeding at 11 dpf, whereas all the Ka0Kb0 larvae died by 10 dpf. This result suggests that the eating defect was not the only cause of the lethality of keap1a;keap1b-DKO larvae.
3.2. Lethality of keap1a;keap1b-DKO larvae was rescued by Nrf2 mutation
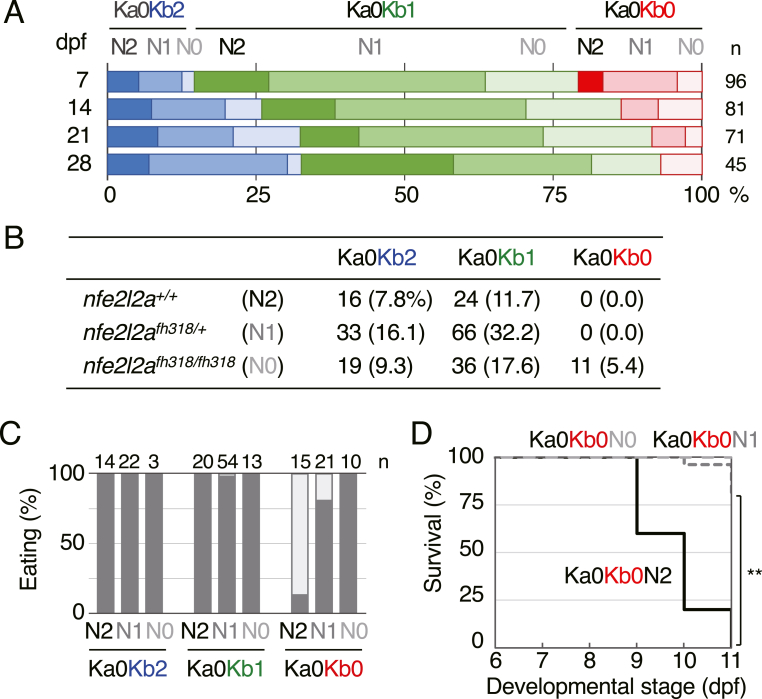
The lethality of Keap1-KO mice was rescued by Nrf2 disruption, indicating that the lethality is due to Nrf2 hyperactivation [9]. To determine if the lethality of the keap1a;keap1b-DKO larvae was also due to Nrf2 hyperactivation, the Nrf2 mutation (nfe2l2afh318) was introduced into Ka0Kb0 zebrafish. We first generated keap1ait302 homozygous, keap1bit308 heterozygous, and nfe2l2afh318 heterozygous adults (Ka0Kb1N1) and then performed a survival assay using their offspring (Fig. 2A). Ka0Kb0N2 larvae, like the Ka0Kb0 larvae shown in Fig. 1A, were not found at later than 7 dpf, whilst Ka0Kb0N1 and Ka0Kb0N0 larvae were present at up to 21 dpf and 28 dpf, respectively. We also raised 205 offspring from Ka0Kb1N1 parents until 3- to 6-mpf adults and found Ka0Kb0N0, but not Ka0Kb0N2 and Ka0Kb0N1, adults among them (Fig. 2B). These results suggest that the lethality of the keap1a;keap1b-DKO larvae was due to Nrf2 hyperactivation, as in Keap1-KO mice, although the Nrf2-independent effects cannot be ruled out.
Fig. 2.
Effects of the Nrf2 mutation on the phenotypes of keap1a;keap1b-DKO zebrafish. (A) Survival rates of offspring from Ka0Kb1N1 parents at 7, 14, 21, and 28 dpf. The red, pink, and lavenderblush bars represent Ka0Kb0N2, Ka0Kb0N1, and Ka0Kb0N0 larvae, respectively. N0, N1, and N2 designate homozygous, heterozygous, and wild-type of the Nrf2 mutant (nfe2l2afh318), respectively. (B) Survival rates of 3- to 6-mpf offspring from Ka0Kb1N1 parents. Data from 3 independent experiments were combined. (C) Eating analysis of offspring from Ka0Kb1N1 parents at 8 dpf. Data from 3 independent experiments were combined. (D) Survival rates of offspring from Ka1Kb0N1 parents without feeding. The survival was observed every 24 h from 6 to 11 dpf. The tested numbers were as follows: Ka0Kb0N2 n = 5, Ka0Kb0N1 n = 25, Ka0Kb0N0 n = 11. The asterisks indicate significant differences (**p < 0.01). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To investigate the effect of Nrf2 mutation on eating defects in keap1a;keap1b-DKO larvae, we next performed eating analysis using 8-dpf larvae obtained from Ka0Kb1N1 parents (Fig. 2C). The eating defects found in Ka0Kb0N2 larvae were partially rescued in Ka0Kb0N1 and fully rescued in Ka0Kb0N0 larvae. Similarly, survival assays without feeding were performed showing that the lethality was also rescued in Ka0Kb0N1 and Ka0Kb0N0 larvae (Fig. 2D). All these results indicate that Nrf2 hyperactivation is the cause of both eating defects and lethality in non-feeding conditions of keap1a;keap1b-DKO larvae.
3.3. Nrf2 hyperactivation in keap1a;keap1b-DKO larvae
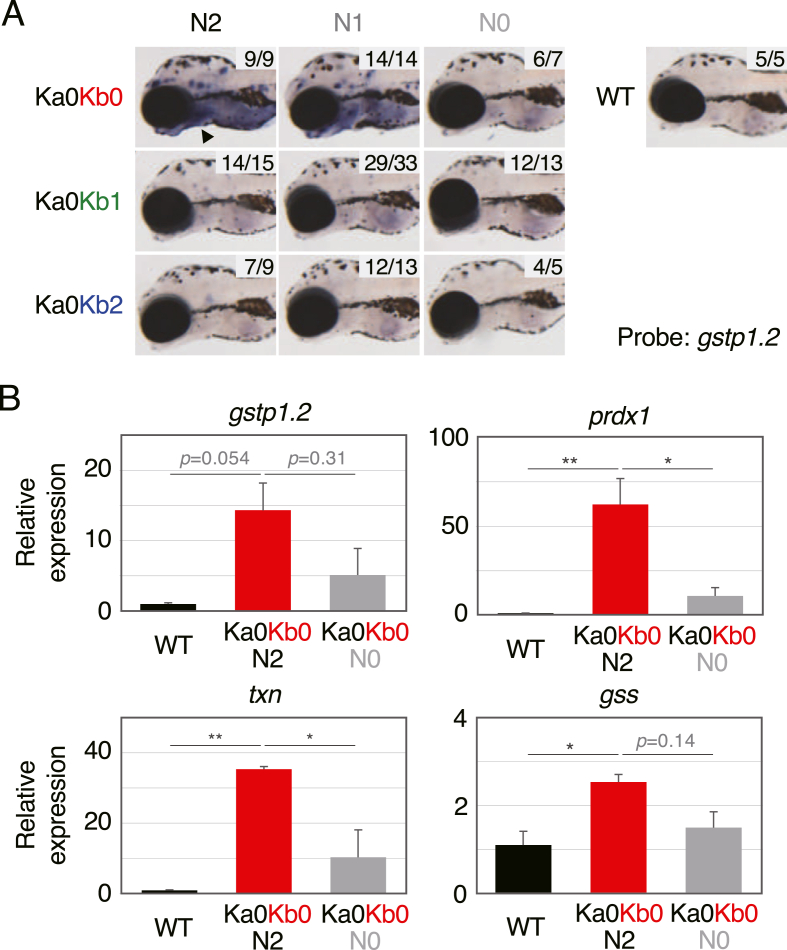
We next examined the expression of Nrf2 target genes in keap1a;keap1b-DKO larvae. First, gstp1.2 expression in 5-dpf larvae was analyzed by whole-mount in situ hybridization (WISH) (Fig. 3A). The results showed that gstp1.2 was expressed drastically higher in Ka0Kb0N2 larvae than in their siblings or wild-type controls. This expression was reduced in Ka0Kb0N0 larvae and partially so in Ka0Kb0N1 larvae, indicating that it is Nrf2-dependent. Next, we quantified the expression of gstp1.2 by means of qRT-PCR (Fig. 3B). The results showed that gstp1.2 expression in Ka0Kb0N2 larvae was 14.35-fold higher than in wild-type larvae, whereas in Ka0Kb0N0 larvae, it was only 5.13-fold higher. Because the upregulated expression of gstp1.2 was not statistically significant, we examined the expression of other Nrf2 target genes (prdx1, txn, and gss) and found a similar Nrf2-dependent upregulation that was statistically significant in Ka0Kb0N2 larvae (Fig. 3B). These results suggest that Nrf2 is abnormally activated in keap1a;keap1b-DKO larvae.
Fig. 3.
Upregulation of Nrf2-target genes in keap1a;keap1b-DKO larvae. (A) WISH analysis of gstp1.2 expression in 5-dpf offspring from Ka0Kb1N1 parents. The arrowhead indicates the gill. The numbers in each picture indicate the numbers of larvae with similar staining profiles and tested larvae. (B) qRT-PCR analysis of the relative expressions of gstp1.2, prdx1, txn, and gss in wild-type, Ka0Kb0N2, and Ka0Kb0N0 larvae at 5 dpf. The expression of the wild-type specimens was normalized to 1. Asterisks indicate significant differences (*p < 0.05, **p < 0.01).
3.4. Genes abnormally expressed in keap1a;keap1b-DKO larvae
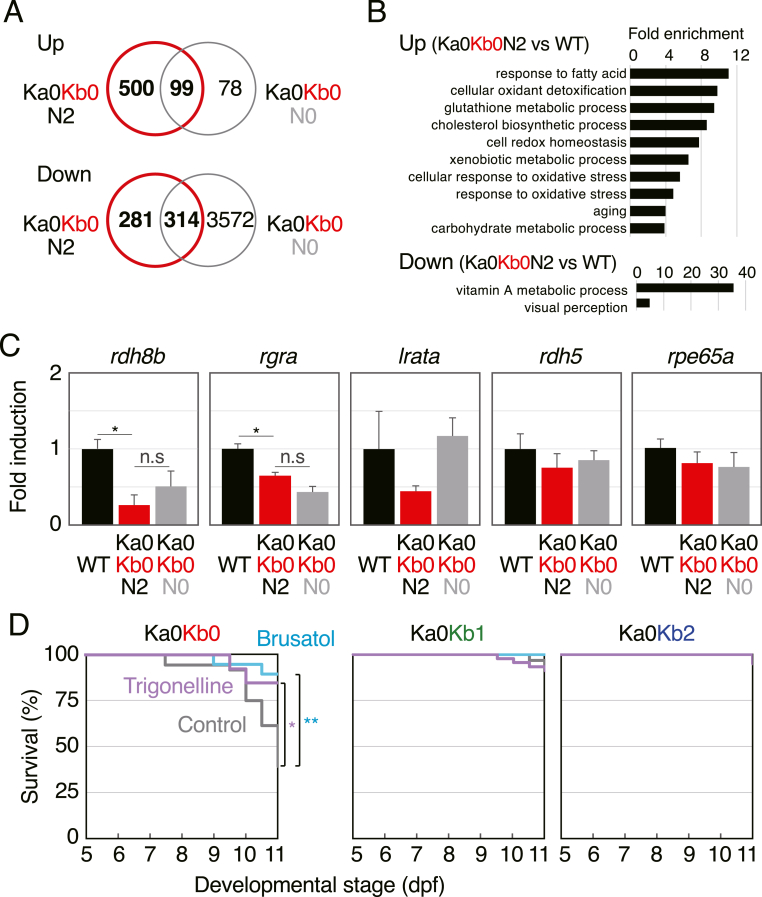
We next performed RNA-sequencing analysis (RNA-seq) using Ka0Kb0N2, Ka0Kb0N0, Ka2Kb2N0, and wild-type larvae to comprehensively identify genes that are aberrantly expressed in keap1a;keap1b-DKO larvae (Fig. 4A, Tables S1 and S2). As a result, we identified 599 genes as those expressed more than 2-fold higher and 595 genes as those expressed less than half in Ka0Kb0N2 larvae than in wild-type larvae. Using these genes, gene ontology (GO) analysis was performed to identify biological processes modulated by keap1a;keap1b-DKO (Fig. 4B, Table S3). We did not exclude 99 and 314 genes that were expected to be Nrf2-independent (see Fig. 4A) because known target genes of zebrafish Nrf2, such as prdx1, txn, fthl31, sqor, gsr, histh1l1, gpx1a, tcnbb, and fthl27 [[12], [13], [14]], were included in these categories (see also Table S1). Most of the biological processes that emerged in the GO analysis were known processes that are regulated by the Keap1–Nrf2 pathway (Fig. 4B). On the other hand, “vitamin A metabolic process” and “visual perception” identified as biological processes downregulated in keap1a;keap1b-DKO larvae (Fig. 4B, lower panel) have not been previously reported. We therefore examined the expression of visual cycle genes by qRT-PCR. Although Nrf2 dependence was unclear, there was an overall trend indicating that these visual cycle genes were downregulated in keap1a;keap1b-DKO larvae (Fig. 4C). Finally, the effects of known Nrf2 inhibitors on the lethality of keap1a;keap1b-DKO larvae were examined, as a result of which the lethality was shown to be partially rescued by treatment with trigonelline or brusatol [15,16].
Fig. 4.
RNA-seq and the effects of Nrf2 inhibitors. (A) Venn diagrams showing upregulated and downregulated genes in 5-dpf Ka0Kb0N2 and Ka0Kb0N0 larvae identified by means of RNA-seq in comparison with wild-type larvae. (B) GO enrichment analysis of the biological processes in keap1a;keap1b-DKO larvae. (C) qRT-PCR analysis of the relative expression of visual cycle genes (rdh8b, rgra, lrata, rdh5, and rpe65a) in wild-type, Ka0Kb0N2, and Ka0Kb0N0 larvae at 5 dpf. Asterisk and n.s. denote significant difference (*p < 0.05) and not significant, respectively. For rpe65a, lrata, and rdh5, one-way analysis of variance did not show significant differences. (D) Survival rates of offspring from Ka0Kb1 parents without feeding. Larvae were treated without (control, gray) or with 0.3 μM trigonelline (purple) or 0.3 μM brusatol (turquoise blue) from 5 to 11 dpf (replaced every 2 days). The tested numbers were as follows: brusatol n = 19, trigonelline n = 13, control n = 36 (Ka0Kb0); brusatol n = 39, trigonelline n = 45, control n = 63 (Ka0Kb1); brusatol n = 19, trigonelline n = 20, control n = 26 (Ka0Kb2). Asterisks indicate significant differences (*p < 0.05, **p < 0.01). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In the present study, we found that keap1a;keap1b-DKO zebrafish are lethal at the larval stage and have eating and other defects and that their larvae showed extraordinarily high expression of Nrf2 target genes. Both the larval lethality and the high expression of Nrf2 targets were rescued by the Nrf2 mutation, resulting in viable and fertile adults. The lethality of keap1a;keap1b-DKO zebrafish, which die around a week after hatching (10 dpf), is similar to that of Keap1-KO mice, which die within 3 weeks of birth [9]. Furthermore, the lethality of keap1a;keap1b-DKO zebrafish was rescued by the Nrf2 mutation, which is also similar to that of Keap1-KO mice [9]. These facts indicate that Nrf2 hyperactivation is fatal to post-natal mammals and post-hatched fish larvae. In both zebrafish and mice, one of the critical targets for hyperactivated Nrf2 was eating. In Keap1-KO mice, hyperkeratosis of the esophagus and forestomach inhibited food intake into the gastrointestinal tract [9], and its lethality was rescued by Keratin 5-Cre-specific KO of Nrf2 [17], suggesting that gastric obstruction of food may be the cause of the eating defects. In zebrafish, however, no such physical disturbance was found in the gastrointestinal tract, therefore suggesting another reason. Moreover, the lethality of keap1a;keap1b-DKO larvae has a cause different from eating defects. Whether these phenotypes are also present in mice remains to be clarified.
All these defects in keap1a;keap1b-DKO zebrafish and Keap1-KO mice were rescued by reducing Nrf2 activity, so Nrf2 hyperactivation is definitely the cause of the effects. Intriguingly, lethality did not appear at the embryonic stage, but after hatching/birth. Similar regulation may exist in invertebrates, because the disruption of Keap1 in Drosophila was also lethal in the larval stage [18,19]. A recently study of Keap1 in a group of birds, Neoaves, is also interesting [20]. The Neoaves Keap1 protein has a deletion in the Nrf2-binding domain and is unable to inhibit Nrf2, but Neoave chicks are viable and have no eating problems. It will be interesting to elucidate how Neoaves overcome the detrimental effects of Nrf2 hyperactivation. In any case, the lethality of keap1a;keap1b-DKO larvae fully rescued by Nrf2 mutation and partially so by chemical Nrf2 inhibitors could be a useful animal assay system to evaluate the efficacy/toxicity of Nrf2-inhibiting compounds currently under development to inhibit cancer growth.
We previously performed microarray analysis using Nrf2-overexpressing embryos and found that proteasome subunit genes were targets of zebrafish Nrf2 [14]. In contrast, none of the proteasome subunit genes were upregulated in keap1a;keap1b-DKO larvae. Because it has recently been shown that the expression of proteasome subunit genes is regulated by Nrf1 in mammalian cells [21], it is likely that these are not the original Nrf2 targets, but only artificially induced in embryos overexpressing Nrf2 by mRNA injection. On the other hand, a group of cancer-promoting genes observed in Nrf2-hyperactivated cancer cells [22,23], including growth-related factors, serine/glycine metabolic enzymes, and oncogenic proteins, were not upregulated in keap1a;keap1b-DKO larvae. Hyperactivation of Nrf2 alone may not be sufficient to upregulate these cancer-promoting genes.
The only downregulated pathways in keap1a;keap1b-DKO larvae that emerged in the current GO analysis were “vitamin A metabolic process” and “visual perception” pathways, to which the same genes belong, with no reduction in proinflammatory cytokine genes observed in Keap1-knockdown mice [24]. Some genes in these pathways are known to be induced by retinoic acid [25], and it is possible that abnormally increased Nrf2 interacted with retinoic acid receptor α [26] and/or retinoid X receptor α [27] to inhibit transcriptional activation of their target genes. Interestingly, the expression of visual cycle genes such as lrata and rdh5, which are downregulated in keap1a;keap1b-DKO larvae, is known to increase with age in mice [28]. Considering that Nrf2 activity attenuates with age [29], it is possible that Nrf2 suppression is released with age, resulting in increased expression of visual cycle genes.
Authors contributions
Investigation, L.B., V.T.N.; materials, Y.E., G.D., A.S.; data analysis, J.T.; writing, L.B., M.K.
Declaration of competing interest
The authors have no conflict of interests to declare.
Acknowledgements
We thank the Tsukuba i-Laboratory LLP for performing RNA-seq and F. Miyamasu (Medical English Communications Center, University of Tsukuba) for excellent English proofreading. This work was supported by JSPS/MEXT KAKENHI Grant Numbers 26520101 and 22H05556 (to M.K.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102673.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
No data was used for the research described in the article.
References
- 1.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuse Y., Kobayashi M. Conservation of the Keap1-Nrf2 system: an evolutionary journey through stressful space and time. Molecules. 2017;22:436. doi: 10.3390/molecules22030436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/mcb.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi M., Itoh K., Suzuki T., Osanai H., Nishikawa K., Katoh Y., Takagi Y., Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Gene Cell. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 7.Li L., Kobayashi M., Kaneko H., Nakajima-Takagi Y., Nakayama Y., Yamamoto M. Molecular evolution of Keap1: two Keap1 molecules with distinctive intervening region structures are conserved among fish. J. Biol. Chem. 2008;283:3248–3255. doi: 10.1074/jbc.m708702200. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen V.T., Bian L., Tamaoki J., Otsubo S., Muratani M., Kawahara A., Kobayashi M. Generation and characterization of keap1a- and keap1b-knockout zebrafish. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D.R., Harada T., Engel J.D., Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 10.Mukaigasa K., Nguyen L.T., Li L., Nakajima H., Yamamoto M., Kobayashi M. Genetic evidence of an evolutionarily conserved role for Nrf2 in the protection against oxidative stress. Mol. Cell Biol. 2012;32:4455–4461. doi: 10.1128/mcb.00481-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuse Y., Nguyen V.T., Kobayashi M. Nrf2-dependent protection against acute sodium arsenite toxicity in zebrafish. Toxicol. Appl. Pharmacol. 2016;305:136–142. doi: 10.1016/j.taap.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima H., Nakajima-Takagi Y., Tsujita T., Akiyama S., Wakasa T., Mukaigasa K., Kaneko H., Tamaru Y., Yamamoto M., Kobayashi M. Tissue-restricted expression of Nrf2 and its target genes in zebrafish with gene-specific variations in the induction profiles. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn M.E., McArthur A.G., Karchner S.I., Franks D.G., Jenny M.J., Timme-Laragy A.R., Stegeman J.J., Woodin B.R., Cipriano M.J., Linney E. The transcriptional response to oxidative stress during vertebrate development: effects of tert-butylhydroquinone and 2,3,7,8-tetrachlorodibenzo-p-dioxin. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen V.T., Fuse Y., Tamaoki J., Akiyama S., Muratani M., Tamaru Y., Kobayashi M. Conservation of the Nrf2-mediated gene regulation of proteasome subunits and glucose metabolism in zebrafish. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/5720574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boettler U., Sommerfeld K., Volz N., Pahlke G., Teller N., Somoza V., Lang R., Hofmann T., Marko D. Coffee constituents as modulators of Nrf2 nuclear translocation and ARE (EpRE)-dependent gene expression. J. Nutr. Biochem. 2011;22:426–440. doi: 10.1016/j.jnutbio.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T., Seki S., Hiramoto K., Naganuma E., Kobayashi E.H., Yamaoka A., Baird L., Takahashi N., Sato H., Yamamoto M. Hyperactivation of Nrf2 in early tubular development induces nephrogenic diabetes insipidus. Nat. Commun. 2017;8 doi: 10.1038/ncomms14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sykiotis G.P., Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castillo-Quan J.I., Li L., Kinghorn K.J., Ivanov D.K., Tain L.S., Slack C., Kerr F., Nespital T., Thornton J., Hardy J., Bjedov I., Partridge L. Lithium promotes longevity through GSK3/NRF2-dependent hormesis. Cell Rep. 2016;15:638–650. doi: 10.1016/j.celrep.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castiglione G.M., Xu Z., Zhou L., Duh E.J. Adaptation of the master antioxidant response connects metabolism, lifespan and feather development pathways in birds. Nat. Commun. 2020;11:2476. doi: 10.1038/s41467-020-16129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine H., Motohashi H. Roles of CNC transcription factors NRF1 and NRF2 in cancer. Cancers. 2021;13:541. doi: 10.3390/cancers13030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami S., Motohashi H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic. Biol. Med. 2015;88:168–178. doi: 10.1016/j.freeradbiomed.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7 doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connor C., Varshosaz P., Moise A.R. Mechanisms of feedback regulation of vitamin A metabolism. Nutrients. 2022;14:1312. doi: 10.3390/nu14061312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X.J., Hayes J.D., Henderson C.J., Wolf C.R. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc. Natl. Acad. Sci. USA. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Liu K., Geng M., Gao P., Wu X., Hai Y., Li Y., Li Y., Luo L., Hayes J.D., Wang X.J., Tang X. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.can-12-3386. [DOI] [PubMed] [Google Scholar]
- 28.Butler J.M., Supharattanasitthi W., Yang Y.C., Paraoan L. RNA-seq analysis of ageing human retinal pigment epithelium: unexpected up-regulation of visual cycle gene transcription. J. Cell Mol. Med. 2021;25:5572–5585. doi: 10.1111/jcmm.16569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumaru D., Motohashi H. The KEAP1-NRF2 system in healthy aging and longevity. Antioxidants. 2021;10:1929. doi: 10.3390/antiox10121929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.