Abstract
The term PFAS encompasses diverse per- and polyfluorinated alkyl (and increasingly aromatic) chemicals spanning industrial processes, commercial uses, environmental occurrence, and potential concerns. With increased chemical curation, currently exceeding 14,000 structures in the PFASSTRUCTV5 inventory on EPA’s CompTox Chemicals Dashboard, has come increased motivation to profile, categorize, and analyze the PFAS structure space using modern cheminformatics approaches. Making use of the publicly available ToxPrint chemotypes and ChemoTyper application, we have developed a new PFAS-specific fingerprint set consisting of 129 TxP_PFAS chemotypes coded in CSRML, a chemical-based XML-query language. These are split into two groups, the first containing 56 mostly bond-type ToxPrints modified to incorporate attachment to either a CF group or F atom to enforce proximity to the fluorinated portion of the chemical. This focus resulted in a dramatic reduction in TxP_PFAS chemotype counts relative to the corresponding ToxPrint counts (averaging 54%). The remaining TxP_PFAS chemotypes consist of various lengths and types of fluorinated chains, rings, and bonding patterns covering indications of branching, alternate halogenation, and fluorotelomers. Both groups of chemotypes are well represented across the PFASSTRUCT inventory. Using the ChemoTyper application, we show how the TxP_PFAS chemotypes can be visualized, filtered, and used to profile the PFASSTRUCT inventory, as well as to construct chemically intuitive, structure-based PFAS categories. Lastly, we used a selection of expert-based PFAS categories from the OECD Global PFAS list to evaluate a small set of analogous structure-based TxP_PFAS categories. TxP_PFAS chemotypes were able to recapitulate the expert-based PFAS category concepts based on clearly defined structure rules that can be computationally implemented and reproducibly applied to process large PFAS inventories without need to consult an expert. The TxP_PFAS chemotypes have the potential to support computational modeling, harmonize PFAS structure-based categories, facilitate communication, and allow for more efficient and chemically informed exploration of PFAS chemicals moving forward.
1. Introduction
Per- and polyfluoroalkyl substances (PFAS) is a broadly encompassing term that has come to represent a wide range of structurally and functionally diverse chemicals. As public awareness and concerns for perfluorooctanoic acid (PFOA) and perfluorooctanesulfonic acid (PFOS), the two most environmentally widespread and well-studied PFAS, have increased,1−3 so have efforts in the regulatory and scientific communities to compile and categorize larger and more structurally diverse lists of PFAS-type chemicals for potential monitoring and study.4,5 In 2018, the Organisation for Economic Cooperation and Development (OECD) published a “Global PFAS list” of over 4700 unique PFAS substances, spanning the regulatory, usage, and occurrence landscapes.6 Accompanying this list, each of the PFAS substances was manually assigned to one of 106 expert-defined categories. Coordinating with the OECD Global PFAS list release, the U.S. Environmental Protection Agency’s (EPA) Distributed Structure-Searchable Toxicity (DSSTox) Database project7 published a companion curated PFASOECD list on the EPA CompTox Chemicals Dashboard (referred to henceforth as Dashboard)8,9 that included more than 3800 defined structures, the remainder consisting of polymers and mixtures, some linked to structure components or Markush representations.10 This DSSTox PFASOECD list was subsequently combined with several publicly available PFAS lists, designated as such by source authors, to create the first DSSTox PFASMASTER list, totaling just over 5000 substances (https://comptox.epa.gov/dashboard/chemical-lists/PFASMASTER, accessed August 25, 2022). Shortly thereafter, DSSTox published its first PFASSTRUCT inventory in which a small set of structure filters was used to objectively query the entire DSSTox inventory to identify a total of 4357 PFAS structures.
In the years since, the size of DSSTox’s PFASSTRUCT inventory (https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCTV5, accessed October 25, 2022), has grown considerably to more than 14,000 PFAS structures, out of 1.2 million total substances in the current DSSTox inventory, due to a combination of expanded curation of public sources and the refinement of the set of structure-based filters designed to be sufficiently encompassing of small and large PFAS-type structures of potential regulatory interest to EPA. A recent publication by Williams et al. detailed the evolution of EPA’s DSSTox PFAS curation efforts and list expansion through to PFASSTRUCTV4 (https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCTV4), comparing this list definition to several other proposed working definitions, including those serving varied needs of EPA regulatory programs.11,12 The most expansive PFAS definition to date has been put forth by the OECD PFAS workgroup as “containing an aliphatic and saturated −CF2–”.13 Although many would consider this an overly simplistic definition, yielding more than 40,000 PFAS in the current DSSTox database, it provides a practical and unambiguous filter to identify potential PFAS, with the understanding that the list is likely to be further filtered to serve research and regulatory needs across OECD Member Countries. Clearly, there is no one-size-fits-all definition of PFAS given the innumerable ways in which chemicals can be fluorinated and when the term takes on varied meanings related to industrial and commercial use, as well as regulatory and societal concerns. For example, a single −CF2– present in a small chemical with a high percentage contribution of fluorine to the whole is likely more indicative of PFAS’s unique properties and concerns than a large drug molecule containing a single trifluoromethyl group. Providing further evidence of an evolving and context-dependent definition of PFAS, the most recent DSSTox PFASSTRUCTV5 inventory additionally includes structures containing 30% or greater fluorine content by mass (not including hydrogen) to account for mostly smaller PFAS-like substances of potential regulatory interest falling outside of the PFASSTRUCT substructure definitions.12
Chemical categories have long played an important role in chemical hazard and risk assessment and represent attempts to group chemicals according to chemical structure features believed or predicted to be most influential in determining properties and activities. The use of structure-based chemical similarity metrics and categories for inferring properties is also closely aligned with the practice of read-across and application of quantitative structure–activity methods in both the U.S. and Europe.14,15 To date, the PFAS community has promoted standardized terminology and largely relied on the use of expert-defined categories, in which each chemical is assigned to a single PFAS category. In the past, this was reasonable due to the relatively simple PFAS structures under consideration, typically with a single functional group, as well as the small size of PFAS space for which occurrence, fate or bioactivity data were available. The practice persists to the present due to a natural desire within the research and regulatory communities to simplify complex chemistry. Such expert-based approaches typically require manual application of chemical nomenclature and category definitions lacking precise structural boundaries such that new chemicals, not already named or categorized by experts, are not easily assigned by nonexperts in a consistent manner, particularly for more complex structures that span multiple categorization features. Examples of expert-based approaches include the original PFAS nomenclature categories proposed by Buck et al.,16 the 106 manual expert-assigned categories associated with the OECD Global PFAS list, OECD terminology guideline recommendations,17 and the recently published OECD Fact Cards detailing properties and data references for 115 PFAS manually assigned to 15 categories.18 Only in the last case were 10 of the 15 categories defined by one or more Markush-type structures, which convey clear, structure-defined boundaries on the category.10
The sheer size and structural diversity of the current PFAS landscape, as represented by the most recent PFASSTRUCT list, necessitates moving away from manual, imprecise, expert-based PFAS category definitions to adopting more systematic structure-based approaches. The few published efforts that have attempted to algorithmically categorize within PFAS space have shown promise but have fallen short of general applicability. Sha and co-workers proposed a novel “splitPFAS” approach that attempted to separate a perfluoroalkyl moiety (CnF2n+1) from the remaining portion (X–R) of the chemical, where X was initially restricted to CO, SO2, CH2 and CH2CH2.19 After separating out this PFAS portion, they applied the open-ontology “ClassyFire” tool to categorize the non-PFAS portion of the chemical.20 In practice, however, the approach was limited to relatively few PFAS categories and fully fluorinated CnF2n+1 terminal chains, and the subsequent application of ClassyFire met with only limited success largely due to lack of PFAS-specific knowledge. A second published approach, PFAS-Map, took a hybrid, semiempirical approach using modern machine-learning methods.21 The authors assigned a subset of 7,866 PFAS structures contained in a 2020 version of the DSSTox PFASSTRUCT file (https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCTV3) to expert-based categories consistent with the Buck et al. or OECD approaches, where possible. Next, they generated approximately 2000 1D and 2D PaDEL descriptors22 for each of the PFASSTRUCT structures and reduced these to 74 principal components (PCs). The core modules were optimized to predict the preassigned PFAS membership in the expert-based categories, whereas additional modules used statistical methods and the structural descriptors to cluster the remaining PFASSTRUCT chemicals. A new chemical, not included in PFASSTRUCT, could then be associated with the nearest category-cluster using structure-similarity metrics. The PFAS-Map method achieved broader success than the splitPFAS method and included both primary and secondary classifications. However, its reliance on a relatively small number of predefined expert categories and lack of clear chemical articulation of the structural basis for the remaining clusters are limiting, as are the several “other” categories that simply capture when the method is unable to assign a chemical to a clearly articulated category.
Considering the increasingly large and diverse PFAS structure space of potential interest, and the inability of expert-based categorization approaches to keep pace or be consistently applied, the goals of the present study are 2-fold: (1) to create a customized set of PFAS substructural features that are well represented within and across the known PFAS universe, as represented here by the PFASSTRUCT inventory, and that can be computationally implemented as a fingerprinting method; and (2) to demonstrate how such features can be used alone or as building blocks to construct structure-based categories that align with PFAS expert categories and known chemical-reactivity concepts. An automated approach to support structure-based PFAS profiling and categorization could provide an objective basis and tool-kit for read-across assessments, harmonize classification efforts across different groups, facilitate communication within the PFAS research community, support computational modeling efforts to predict PFAS properties and activities, and provide for more efficient and meaningful data aggregation across the PFAS space moving forward. Additional requirements that are essential for uptake of the approach by the broader PFAS research and regulatory communities include: (1) the method should be publicly available, i.e., should not require proprietary software; (2) the method should be relatively easy for chemists and nonexperts to understand and apply to small and large lists of new and existing PFAS chemicals; (3) the approach should provide the broadest possible coverage of the full diversity of PFAS chemical space, i.e., avoiding the use of overly general or “other” categories; and (4) the method and results should be based on transparent and intuitive PFAS chemical concepts that can be clearly communicated and understood and, where possible, aligned with existing expert-based PFAS categories and chemical features of importance to the PFAS research and regulatory communities. The latter include, but are not limited to, perfluoroalkyl chains (branched or linear) of varying lengths attached to various functional groups (e.g., carboxylic acids, sulfonic acids, alcohols), fluorotelomers, polyfluoro chains with other halogens (Cl, Br, I) or partial hydrogenation, per-or polyfluoro chains or cyclics with ether and ester linkages..
In the remainder of this article, we describe a new PFAS fingerprint approach, labeled TxP_PFAS, that is an extension of the public ToxPrints (https://toxprint.org/) fingerprint set, uses a consistent substructure naming convention, and is derived from the same XML-based Chemical Subgraphs and Reactions Markup Language (CSRML).23 The TxP_PFAS fingerprint set consists of a set of chemical substructure features, also referred to as chemotypes, that are specifically tailored to support structure-based profiling and categorization of PFAS and promises to address the above-stated requirements for uptake, as well as overcome many limitations of the PFAS categorization efforts put forth to date. In the first section, we provide background and history of development of the TxP_PFAS fingerprint set, including how a subset of ToxPrint chemotypes was identified based on sufficient representation within PFASSTRUCT and then modified by the addition of a -F or −CF group and evaluated for utility against the most recent PFASSTRUCT landscapes. The TxP_PFAS fingerprint set was further expanded to provide coverage of PFAS-specific concepts not previously included in the public ToxPrints. These included perfluoroalkyl chains of various lengths, terminated by CF3 or not, fluorotelomer-type linkages, alternate halogenation, polyfluoro chain indicators, and branching elements within perfluoroalkyl chains. Subsequently, we assess how well the TxP_PFAS fingerprint set captures category-relevant PFAS features and present examples of how the TxP_PFAS fingerprint set can be used in conjunction with the publicly available ChemoTyper application (https://chemotyper.org/) to visualize, organize, and profile the diverse PFASSTRUCT space. In addition, we demonstrate use of the ChemoTyper’s hierarchical chemotype organization coupled with its Boolean filtering options to create PFAS categories and, for a set of examples, examine the degree to which selected categories align with prior expert-based categories. We conclude by describing limitations of this approach, shared by any discrete fingerprint-type of approach to PFAS categorization, and ways in which these limitations can be addressed moving forward. The TxP_PFAS CSRML file and all associated results are available in the Supporting Information. In addition, the TxP_PFAS CSRML file and associated files will be made available for download on the ToxPrint Web site (https://toxprint.org/).
2. History of Development of TxP_PFAS
2.1. Some Prehistory
In 2017, shortly after DSSTox published its first set of PFAS-related lists on the Dashboard, including a list spanning EPA PFAS research activities (https://comptox.epa.gov/dashboard/chemical-lists/EPAPFASRL), researchers in EPA’s Center for Computational Toxicology and Exposure used these lists as candidates to successfully procure a library of 480 PFAS chemicals from commercial sources. Filters on library chemical selection, aside from commercial availability and cost, included a minimum of 4 fluorines and excluded aromatics, inorganics, and low molecular weight compounds. Toward the goal of supporting future “read-across” assessments,24,25 PFAS library chemicals were manually assigned to expert-based categories using guidance from Buck et al.16 These category assignments, in turn, were used to inform selection of an initial set of 75 PFAS chemicals to undergo Tier 1 testing in medium and high-throughput ToxCast screening assays (https://comptox.epa.gov/dashboard/chemical-lists/EPAPFAS75S1).26,27 Not long after, a second set of 75 PFAS chemicals were selected for Tier 1 screening (https://comptox.epa.gov/dashboard/chemical-lists/EPAPFAS75S2) using an expanded set of expert-based category approaches that included the Buck et al. nomenclature guidance, as well as expert-category assignments from the OECD Global database publication and DSSTox PFAS Markush structures, where available. Due to limited coverage of the library by these existing categories and Markush, it was necessary to use in-house expertise to extend category assignments to the full PFAS 430 library. (Note that although Markush structures can provide definitive generalized representations of structure-based categories, these require manual curation and their use is limited to the algorithmic implementation in current cheminformatics tools (ChemAxon, LLC, Newark, DE)). During this exercise, it became apparent that not only were manual, expert-based PFAS category assignments difficult to consistently apply and reproduce, but also that as soon as one went beyond the simple perfluoro alkyl chain + functional group (e.g., alcohol, carboxylic acid, sulfonic acid) type of PFAS to more complex structures containing multiple functional groups, ether linkages, partial hydrogenation, etc., the challenge of assigning PFAS substances to consistent and unique categories became more problematic. Hence, the increasing structural diversity of the full, testable (i.e., soluble in DMSO) 430 PFAS library (https://comptox.epa.gov/dashboard/chemical-lists/EPAPFASINV) resulted in a large number of poorly populated (i.e., singlet) categories, ill-suited to read-across, as well as chemicals that were difficult to assign to a single category. It was at that time that we began to explore structure-based fingerprinting approaches to provide a more automated, transparent, and consistent means of grouping and categorizing PFAS chemicals.
Molecular fingerprinting methods based on substructure keys are commonly used as the basis for structure similarity searching, wherein binary bit vector representations of molecules are compared to quantify overall similarity.28 Each bit in the vector, in turn, typically represents a substructure or chemical feature determined to be either present (1) or absent (0) in the molecule. Several publicly available fingerprinting methods, each providing a somewhat different feature “lens” through which to view structures, are available for computational implementation. Examples include MACCS,29 PubChem,30 and Morgan31 (or ECFP4) fingerprints. Each of these methods was primarily developed for generalized structure-similarity searching and clustering across large publicly available chemical structure databases, such as DSSTox or PubChem, and most often with drug discovery and virtual screening objectives in mind. In contrast, the ToxPrints fingerprint set adapted for the present study was specifically developed to provide coverage of large, structurally diverse environmental and industrial chemical landscapes, with toxicity pathways in mind.23
ToxPrints consist of 729 discrete chemical features, also referred to as chemotypes, that include atoms, functional bonds, chains, rings, ligands, and scaffolds. For present purposes of characterizing the PFAS chemical domain, ToxPrints were attractive from several added perspectives: (1) ToxPrints are encoded in the open source, flexible, and extensible CSRML language;23 (2) a ToxPrint fingerprint file can be generated, and ToxPrint features can be named, visualized, and hierarchically filtered on a set of structures from within the publicly available ChemoTyper application; and (3) ToxPrints capture a wide array of functional groups, bonding patterns, and toxicity alerts, as well as a small set of PFAS-specific substructures–namely, linear perfluoroalkyl ethyl, butyl, hexyl and octyl chain features. Demonstrating their extensible nature, a next generation of ToxPrints customized for organic flame retardants (OFR), some of which are PFAS, was reported by the National Academy of Science.32 For these reasons, a small subset of 34 ToxPrints, alone and in combination, were used to reproduce the simpler expert-assigned categories and more efficiently classify most of the remainder of EPA’s 430 PFAS chemical library to inform the selection of additional Tier 1 test chemicals.33 This approach served as a proof-of-principle and worked reasonably well, in large part because EPA’s PFAS library at the time was small and consisted mostly of simple combinations of linear perfluoroalkyl chains either directly attached to a functional group (such as an alcohol, sulfonic acid, carboxylic acid, etc.), or with intervening C1–C3 alkyl groups in the case of fluorotelomers.
The challenge we faced in extending this approach to the much larger, more structurally diverse PFASSTRUCT universe was the same as that faced by prior PFAS categorization approaches, namely, how to define and focus in on, what we shall refer to as PFAS-proximate features (as opposed to PFAS-distant features) across the wide structural diversity of more than 10,000 PFAS-labeled structures. For purposes of illustration, see Figure 1.
Figure 1.
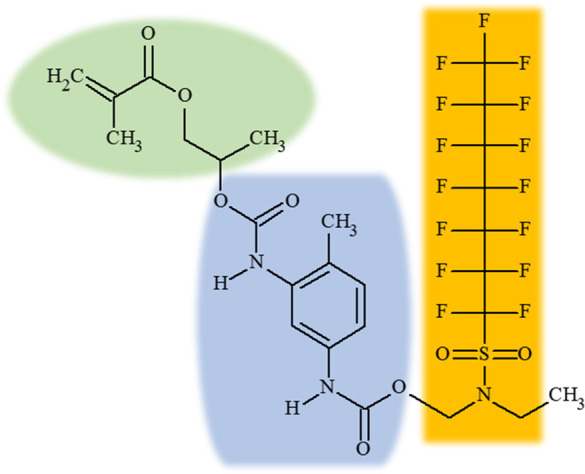
Three functionally important chemical features are highlighted for the above chemical, 2-((((5-(((2-(Ethyl((pentadecafluoroheptyl)sulphonyl)amino)ethoxy)carbonyl)amino)-2-methylphenyl)amino)carbonyl)oxy)propyl methacrylate, CASRN 68298–73–7, DTXSID40880601 (DSSTox Substance Identifier): a methacrylate (green oval), a carbamic acid derivative (blue bulge shape), and a perfluoroheptylsulfonyl amide (orange rectangle).
Based on functional group precedence, and reflected in its name, it would be reasonable to categorize this chemical as a methacrylate. In addition, the central carbamic acid derivative, will likely impact overall properties of this chemical. Viewed through the lens of PFAS categorization, however, we are most keenly interested in detecting the perfluoroheptyl sulfonyl aspect of the chemical and would attempt to first and foremost categorize the chemical based on this feature. The problem with a standard fingerprint method, such as ToxPrints, is that whereas several prominent features of this molecule would likely be recognized and assigned a bit value of 1, including the perfluorohexyl portion of the perfluoroheptyl chain, the proximate relationship of the separate features would not be captured. Hence, there would be no way of knowing that the perfluoroheptyl chain is attached to the sulfonyl group rather than the methacrylate. This independence of fingerprint bits is also a limitation of standard similarity search methods. The only solution within this fingerprint paradigm is to not only detect hundreds of possible distinct features (e.g., 729 ToxPrint features), but also features in close proximity to one another, which quickly leads to a combinatorial explosion in the number of feature combinations one could consider. However, we are not interested in every possible combination of features; rather, for present purposes, we are interested in focusing on features attached to the PFAS portion of the molecule that are also present in sufficient numbers to support categorical representation. These proximate features will largely influence and dictate how easily the PFAS portion of the chemical dissociates and what PFAS degradation products are ultimately formed. This is not to say that the PFAS-distant features of the molecule are unimportant, given that they influence the properties of the whole, but rather that these could be detected secondarily using generic (i.e., non-PFAS tailored) fingerprint methods, such as the full set of public ToxPrints. In the next section, we detail the process we undertook to modify and extend a portion of the public ToxPrints to create the PFAS-proximate TxP_PFAS fingerprint set.
2.2. Development of TxP_PFAS: Workflow
Figure 2 presents a flowchart summarizing the process used to create TxP_PFAS_v1.0, starting from the public set of ToxPrint features (abbreviated as TxPs in Figure 2) and based on iterative modifications, additions, and both objective and empirical assessment of chemotypes deemed to provide PFAS-relevant feature coverage of EPA’s 2021 PFASSTRUCTV4 list of 10,776 structures (https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCTV4). The upper half of the figure details the process of filtering out ToxPrints that were either underrepresented in PFASSTRUCTV4 (i.e., 463 TxPs, each present in fewer than 30chemicals), or that were less relevant to PFAS category considerations. The latter included 77 ToxPrints that referred to generic halogenation bonding patterns (i.e., whose names include the terms “X”, “halo”, or “halide”) and various types of nonfluorinated, generic “alkane” and “alkene” chains, which were to be replaced by fluorinated versions in TxP_PFAS. In addition, based on visual inspection and review within the ChemoTyper, we removed 96 TxPs that were most often found to be distant from the per- or polyfluoro portion of the molecule or that were redundant in coverage to a related TxP. Moving from the top half of Figure 2 into the bottom section, the remaining 93 ToxPrints were modified by direct attachment to either a CF group or an F atom at one or more sites (e.g., in the case of esters) depending on the nature of the chemotype and whether it already included an appropriate carbon attachment site for fluorine. The CSRML code modifications were manually debugged to achieve successful import and visual confirmation of the intended chemical feature concept in the ChemoTyper. Examining the frequency counts of the 93 modified chemotypes across the full PFASSTRUCTV4 inventory led to the further removal of 43 CF,F-TxPs having fewer than 30 incidences. The remaining 50 CF,F-TxPs were mostly “bond” type functional groups that were subject to further verification and, following manual inspection, possible modification, and confirmation of the chemotype concept using substructure-searching of the feature within the ACD/Lab’s Spectrus software (Advanced Chemistry Development, Inc., Toronto, Ontario, Canada). Finally, an additional set of expert-defined, PFAS-relevant chemotypes, mostly not derived from ToxPrints, was added to the TxP_PFAS CSRML file. These provided coverage of perfluoro linear alkyl chains of various lengths, terminating (capped) or not (uncapped), perfluoro branching and polyfluorination patterns, alternate halogenation (Cl, Br, I), n1-n3 fluorotelomers, and fluorinated cyclics, heterocyclics and aromatics.
Figure 2.
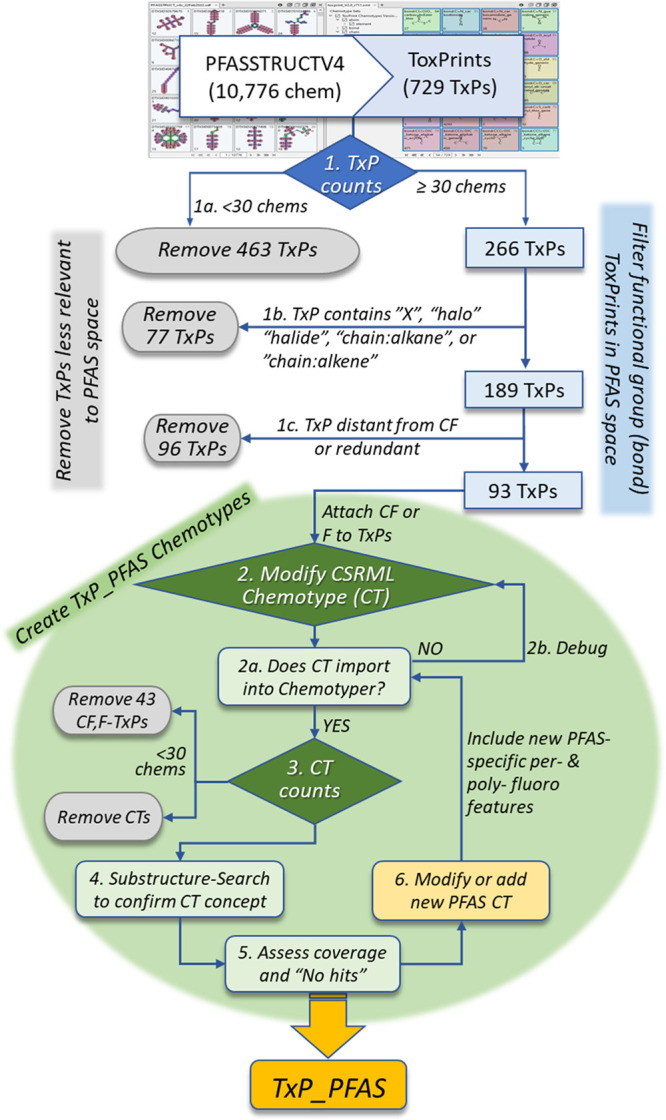
Workflow steps used to create the initial TxP_PFAS fingerprint set: Step 1. match 10,776 PFASSTRUCTV4 structures to 729 ToxPrints (TxPs) in the ChemoTyper, export the fingerprint file, generate total TxP counts; 1a. remove 463 TxPs, each in fewer than 30 chemicals; 1b. remove 77 TxPs whose name includes the terms “X”, “halogen”, “halide”, “chain:alkane” or “chain:alkene”; 1c. remove 96 TxPs found by manual inspection to be distant from the perfluoro portion of the structure, or redundant or closely related to another TxP; 2. for the remaining TxPs, either a CF or an F (depending on the feature) is added to the CSRML chemotype (CT) block; 2a. check to see that the CT imports into the ChemoTyper and the concept is properly visualized and conveyed; 2b. if not, manually inspect and debug the CT CSRML code and reimport; 3. import the CT into the ChemoTyper and generate total CT counts, removing 43 CTs in fewer than 30 chemicals from further consideration; 3a. assess how well the CT covers the feature space within PFASSTRUCTV4; 4. use substructure searching to independently validate the CT concept; 5. assess coverage of the individual CT and collection of TxP_PFAS CTs across the entire PFASSTRUCTV4 inventory and examine “NoHits” for possible missing features; 6. modify existing CT(s) or propose new CT(s) attached to a fluorine or fluoroalkyl moiety and introduce new PFAS-specific per- and polyfluoro features to capture capped (CF3) and noncapped (CF2) chains, partial hydrogenation, fluorotelomers, alternative halogenation (Cl, Br, I), branching, etc.
2.3. Assessing Overall Coverage
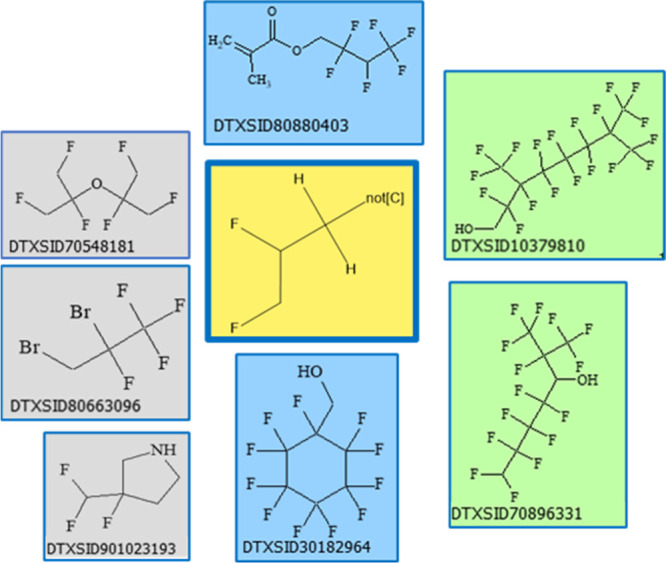
At each stage in the process of assessing the PFAS-relevance and coverage of individual PFAS-specific chemotypes, the goal was to capture sufficient detail with each chemotype so as to be useful for PFAS categorical specificity, while providing sufficient representation across the PFASSTRUCTV4 inventory to be useful for categorical generalization (i.e., present in >30 chemicals), i.e., balancing specificity with generalizability while maximizing coverage. An early version of the TxP_PFAS set consisting of 99 chemotypes was assessed for its overall coverage of PFASSTRUCTV4. An initial count of 1058 “NoHits” (i.e., chemicals without a single TxP_PFAS feature present) was visually surveyed for missing chemotype patterns and 27 chemotypes were modified or newly added to the TxP_PFAS set to reduce the final number of NoHits to 11 (free radicals, charged species, and small molecules). This TxP_PFAS_v1.0 version contained a total of 126 chemotypes. (Note that NoHits can be viewed in the ChemoTyper by selecting all TxP_PFAS chemotypes and using the option “Filter Structures> Not Containing Any Selected Chemotype (NOT OR)”.)
2.4. TxP_PFAS Chemotype Naming Convention
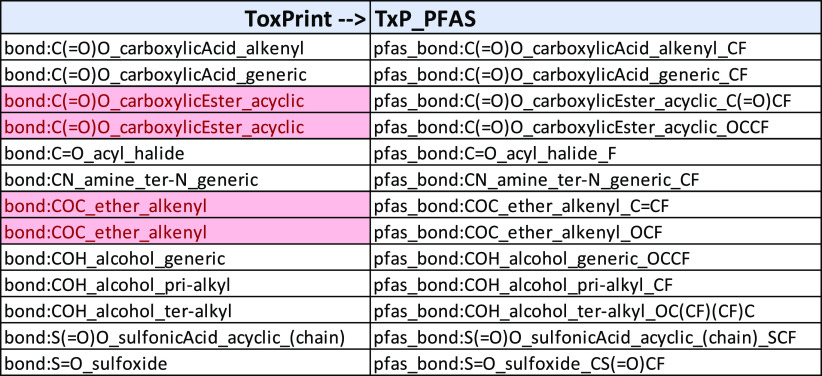
During the course of TxP_PFAS development, it became necessary to implement a naming convention for the modified and newly added chemotypes that would be PFAS-specific, chemically informative, and consistent with the naming convention used in the public ToxPrints, from which many were derived. The argument for the latter went beyond attribution. Using the original root ToxPrint chemotype name, where applicable, conveys appropriate chemical concepts such as the type of C moiety attached to functional groups, as in, e.g., bond:C(=O)O_carboxylicAcid_generic vs bond:C(=O)O_carboxylicAcid_alkenyl. In addition, maintaining such a name correspondence enables a direct comparison between the ToxPrint and corresponding TxP_PFAS fingerprint represented in the same chemical inventory.
Next was deciding how to modify the ToxPrint names so as to convey their PFAS domain-specific content, where applicable. The solution was to apply a common prefix of “pfas_” to each chemotype in the newly created TxP_PFAS set and append a suffix to the end of the name to specify the fluorinated modification. Examples of ToxPrint names and their corresponding modified version in the TxP_PFAS set are listed in Table 1. Notice that even though the added PFAS-related feature is typically restricted to either a CF or F, the appended SMILES-type suffix is often longer when additional contextual information is needed to specify the bonding location of the CF or F when multiple sites are possible. There is also potential ambiguity when an F (rather than a CF) is added to a C that is already part of the original ToxPrint. In addition, when multiple binding sites are possible, and each is well represented across the PFASSTRUCT inventory, a single ToxPrint can spawn more than one TxP_PFAS chemotype, as in the two highlighted pairs in Table 1. Lastly, pfas_bond:COH_alcohol_ter-alkyl_OC(CF)(CF)C illustrates a case where two F’s were added to two of the terminal C’s in the original ToxPrint due to this pattern being almost as well represented and more specific than the case of a single CF, i.e., 234 hits for one C(F) vs 204 hits for two C(F)s (note that if three C(F)s were added, there were only 9 hits).
Table 1. Sample of Names of Corresponding ToxPrint and Modified TxP_PFAS Chemotypes, Highlighting Two Cases Where a Single ToxPrint Gives Rise to More than One TxP_PFAS.
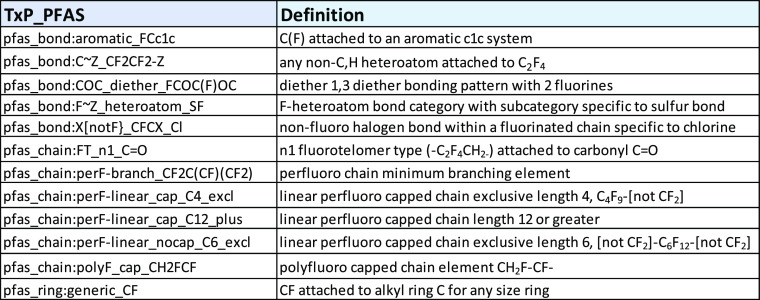
In addition to the ∼50 TxP_PFAS chemotypes directly derived from public ToxPrints, more than 70 newly created chemotypes were coded in CSRML to capture categories of fluorinated chains and bonding patterns particular to PFAS. A representative sample of these new chemotype names containing the “pfas_” prefix and followed by the same top-level “bond”, “chain”, and “ring” hierarchical organization of ToxPrints is provided in Table 2. As with ToxPrints, the names are intended to be concise while providing some chemically descriptive information.
Table 2. Sample TxP_PFAS Names and Expanded Definitions of Newly Created TxP_PFAS Chemotypes Specific to the PFAS Domain.
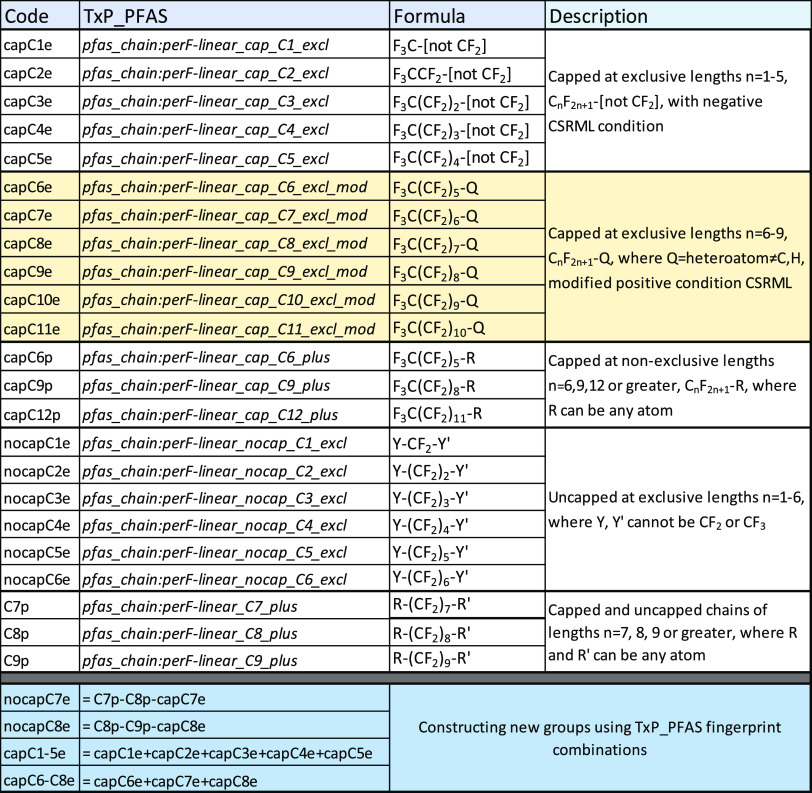
Listed in Table 2 are sample chemotypes for perfluoro linear chains of varying lengths (C#) within two groups: CF3 terminated, or capped (cap), and CF2 not terminated or not capped (nocap). The “excl” suffix refers to an exclusive length chain, e.g., pfas_chain:perF-linear_cap_C4_excl identifies only linear perfluoro chains of length 4 (i.e., C4F9), i.e., the chain CANNOT be longer than C4. There were no exclusive conditions of this sort in the original ToxPrints nor is it possible to code this type of negative condition using public fingerprint coding methods other than CSRML. The “plus” suffix, on the other hand, refers to the more typical, nonexclusive condition whereby pfas_chain:perF-linear_nocap_C12_plus indicates a linear perfluoro uncapped chain of carbon length 12 or greater, i.e., a C12F25 linear chain with a terminal CF3.
2.5. Practical Considerations
A problem that arose during the development of the TxP_PFAS CSRML file was the large increase in CSRML computational processing time within either the public ChemoTyper or the Corina Symphony software (MN-AM, Molecular Networks GmbH and Altamira LLC, Nuremburg, Germany) when implementing the negative exclusive perfluoro chain length condition beyond 4 or 5 carbon lengths. This problem was sufficiently serious that initial attempts to include the full set of 22 exclusive chain length chemotypes for C1–C11 capped and not capped led to application failure when attempting to process the large PFASSTRUCTV4 inventory, and several hours to process if the file were split into smaller inventories. As a result, we implemented a practical solution to reduce processing time. For the terminated “cap” set, we coded the exclusive negative conditions for C1–C5. For the C6–C11 cap chemotypes, we implemented a modified (mod) positive condition for the exclusives, i.e., the inverse of the negative condition that allowed termination of the end CF2 only from a specified list of atoms that could not include either H or C (the latter to maintain the original exclusive condition). This worked remarkably well, only missing the C6 or greater perfluoroalkyl chemicals terminating in CF3 on both ends, such as n-perfluorohexane, n-perfluoroheptane, etc.; these few cases were captured in the corresponding C6, C9 and C12 “plus” chemotypes. For the corresponding nocap cases, however, the logic of the mod condition could not be effectively implemented. Hence, we included C1–C6_excl for the nocaps and 3 generic plus conditions for C7, C8 and C9 to capture both nocap and cap cases, from which some higher nocap_excl counts could be inferred. With these changes and given their importance to the TxP_PFAS fingerprint set, we were able to provide complete coverage of the exclusive capped (through C11) and uncapped (through C6) perfluoro linear chains in PFASSTRUCTV4, as well as all longer perfluoro linear chains through the “plus” chemotypes. The full listing of the 23 perfluoro linear chain chemotypes and their definitions are provided in Table 3. Additional perfluoro-linear chain groups can be created from combinations of TxP_PFAS chemotype columns within the fingerprint file, with examples indicated at the bottom of Table 3.
Table 3. Complete List of New TxP_PFAS Chemotypes Representing Linear Perfluoro Chains of Various Lengths, Both Capped (CF3) or Uncapped (CF2); Highlighted (Yellow) “Mod” Subset Required CSRML Modifications, and Last 4 Rows (Blue) Indicate How New Perfluoro-Linear Chain Groups Can Be Constructed from Combinations of Chemotypes within the Fingerprint File.
2.6. TxP_PFAS ChemoTyper Hierarchy
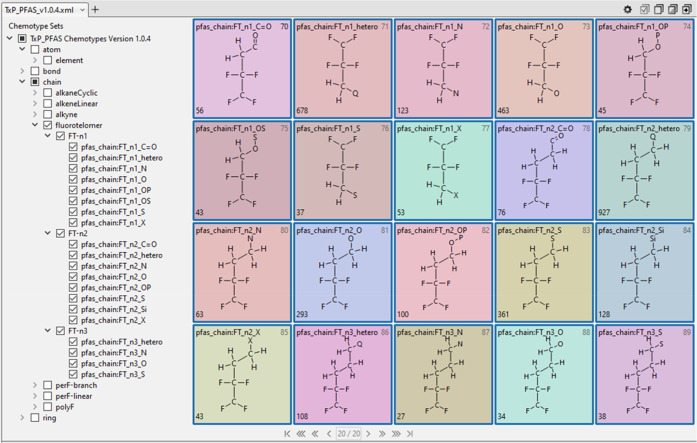
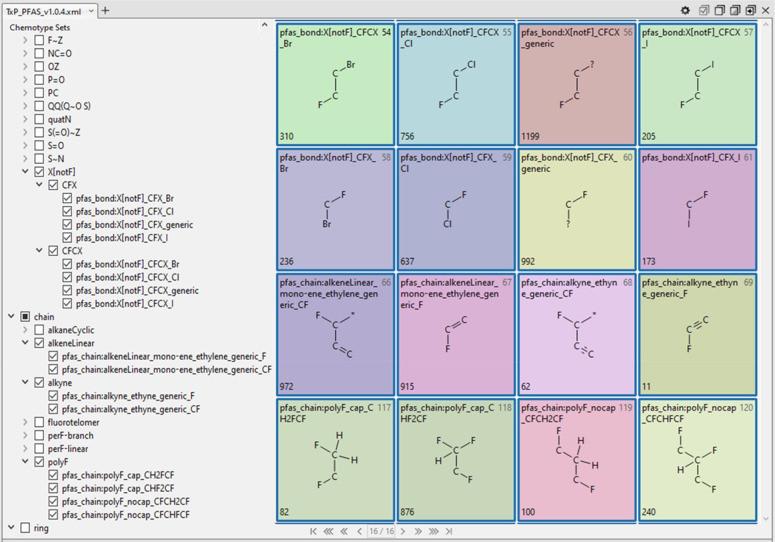
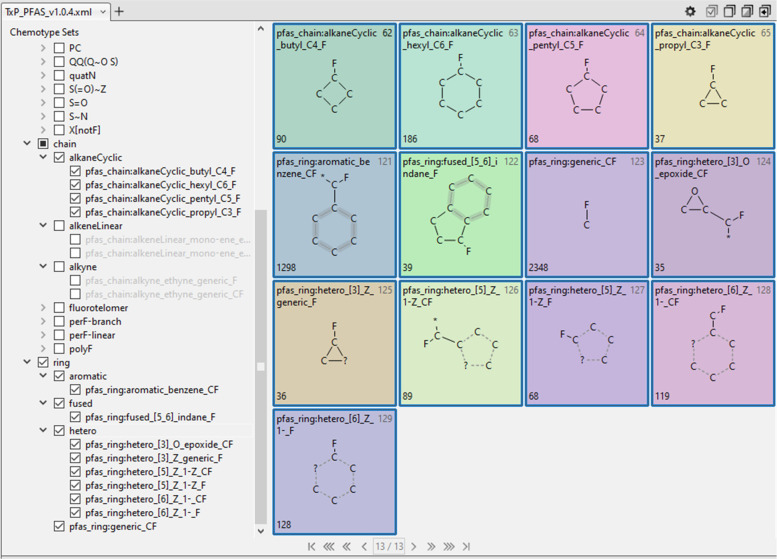
A valuable feature of CSRML when processed in the public ChemoTyper is the ability to create a hierarchical index of chemotypes that can be used to both convey relationships as well as to selectively filter chemotypes for viewing and export. The naming convention of the public ToxPrints was designed with this hierarchical organization ability in mind and we have adopted and extended this hierarchical structure in the TxP_PFAS CSRML. A sample view of the resulting TxP_PFAS hierarchy as it appears in the ChemoTyper, with the ability to collapse or expand from the topmost levels (atom, bond, chain, ring), is shown in Figure 3.
Figure 3.
Sample view of the hierarchical organization of TxP_PFAS_v1.0.4 chemotypes as displayed in the ChemoTyper showing representative samples of group and subclass headings.
2.7. Finalizing Public Version TxP_PFAS_v1.0.4 CSRML against PFASSTRUCTV5
After concluding the initial phase of TxP_PFAS development against PFASSTRUCTV4, TxP_PFAS_v1.0 was subjected to internal EPA testing and review. Based on review feedback, a small number of corrections to chemotype names were made (v1.0.1) and one additional chemotype was subsequently added (v1.0.2); pfas_bond:CC(=O)C_dione_(1_3-)_CF was associated with early reported activity in a ToxCast Tier 1 assay and, with over 80 counts, exceeded the minimum criteria for sufficient representation in PFASSTRUCTV4. This TxP_PFAS_v1.0.2 CSRML file contained a total of 127 chemotypes with the same small set of NoHits (11/10,776) as reported earlier.
A new version of PFAS structure file, PFASSTRUCTV5, was released to the public in September 2022. This version contained ∼4000 new DSSTox structures largely resulting from increased curation of public resources, but a minor fraction resulting from an added filtering criteria applied to the full DSSTox inventory, namely, a threshold count of 30% fluorine (excluding hydrogen) to allow for inclusion of complex, highly fluorinated structures missed by the PFASSTRUCTV4 substructure filters.12 This significantly larger PFASSTRUCTV5 inventory, totaling 14,735 structures, provided a valuable opportunity to test the applicability and coverage of the TxP_PFAS_v1.0.2 fingerprint set. Matching the PFASSTRUCTV5 file with the TxP_PFAS_v1.0.2 CSRML in the ChemoTyper yielded 353 NoHits. The majority of these could be eliminated by the modification of one existing F∼Z chemotype to include additional heteroatoms, and by the addition of two new chemotypes in which fluorine is directly attached to an alkene or alkyne carbon (TxP_PFAS_v1.0.3). Finally, during the writing of this manuscript, an error in the fluorotelomer hetero chemotypes (pfas_bond:FT_n#_hetero) was found and corrected. With these changes, the final TxP_PFAS_v1.0.4 CSRML achieved 99.6% coverage of PFASSTRUCTV5, with only 25 NoHits, 9 of which were determined to be mixtures on further curation review, with structures removed. Of the remaining 16: all contained fewer than 10 heavy atoms; 9 contained a CF2 double bonded to a heteroatom; 5 contained only CF-C groups, and 2 were small metal-containing, charged species. (Note that the DTXSIDs for these 25 NoHits can be identified within the Chemotyper or exported fingerprint file by their blank or all zero entries.) The final PFASSTRUCTV5 V2000 SDF file, along with the TxP_PFAS_v1.0.4 CSRML file and corresponding fingerprint file exported from the ChemoTyper, are provided in the Supporting Information In addition, along with this publication, the TxP_PFAS_v1.0.4 CSRML file is to be made publicly available for download on the ChemoTyper Web site, http://chemotyper.org.
3. Methods
All DSSTox structure files with chemical identifiers (DTXSID, CAS RN, name) used in the present study were exported in v2000 SDF format from the EPA CompTox Chemicals Dashboard. These included the following lists: PFASOECD, PFASSTRUCTV4, EPAPFASRL, EPAPFAS, EPAPFASINV, EPAPFAS75S1, EPAPFAS75S2 available for download at https://comptox.epa.gov/dashboard/chemical-lists via the Batch Search.34 The PFASSTRUCTV5 inventory is provided as a Supporting Information (SI) SDF file (V2000) file.
ACD/Lab Spectrus DB v2020.1.1 (Advanced Chemistry Development, Inc., Toronto, Ontario, Canada) was used to perform all structure and substructure search functions within PFASSTRUCT files.
A reduced version of the ToxPrint CSRML file containing 93 chemotypes after filters were applied (see Figure 2) was parsed and edited using the Python ElementTree XML API (https://docs.python.org/3/library/xml.etree.elementtree.html) to add CF or F features onto the first atom in each chemotype. The modified CSRML was checked for successful import into the ChemoTyper and the visual rendering of each chemotype within the ChemoTyper was checked to ensure that CF and F features were attached to the intended atom. The XML file was manually edited using the Sublime Text 3 shareware application (https://www.sublimetext.com/) to correct the CSRML code for chemotypes in which the CF or F was attached to the wrong atom. The remaining, newly added TxP_PFAS chemotypes, not directly corresponding to a ToxPrint, were either created de novo in CSRML or using the SMARTS to CSRML converter tool within the publicly available Chemotype Editor (https://chemotyper.org/). Additional minor edits to CSRML chemotypes were made using the Microsoft WordPad text editor.
The hierarchy code within the ToxPrint CSRML served as an initial template for creating the TxP_PFAS hierarchy. The code was checked with each iteration based on successful import and visual rendering in the ChemoTyper. Chemotype blocks within the CSRML file were arranged alphabetically so as to result in an alphabetically ordered set of columns in the resulting fingerprint file exported from the ChemoTyper.
The publicly available ChemoTyper application (v1.0) and toxprint_V2.0_r711 CSRML file were downloaded from the ChemoTyper Web site (https://chemotyper.org) and run under a Windows 10 Enterprise operating system. After importing a TxP_PFAS CSRML file into the ChemoTyper and mapping to an PFASSTRUCT SDF file, the fingerprint file was exported in csv format. The ToxPrint and TxP_PFAS_v1.0.4 fingerprint tables for PFASSTRUCTV5 are provided in csv format in SI Tables S1 and S2, respectively, indexed by DTXSID. The TxP_PFAS v1.0.4 CSRML file is provided as an SI xml format file and also will be made available for download from the ChemoTyper Web site. SI Table S3 compares total counts of TxP_PFAS_v1.0.4 chemotypes for both PFASSTRUCTV4 and PFASSTRUCTV5 inventories along with the corresponding ToxPrint chemotype name where available. SI Table S4 includes total counts for ToxPrints vs PFASSTRUCTV5, comparing counts for 51 closely corresponding TxP_PFAS_v1.0.4 chemotypes for which the ToxPrint count exceeded 45. SI Table S4 also includes the earlier ToxPrint counts for PFASSTRUCTV4 along with indicators for the various excluded subsets (1a–c) in Figure 2. SI Table S5 provides additional DTXSID chemical identifiers (name, CAS RN, Formula) for PFASSTRUCTV5 chemicals along with inventory indicators (PFASSTRUCTV4 and OECD Global PFAS list subsets), OECD PFAS category assignments for the OECD Global PFAS list subset, and indicators of overlapping TxP_PFAS chemotype-categories. OECD categories for the overlapping portion of the PFASSTRUCTV5 file, provided for all overlapping chemicals in the OECD Global chemical list with defined structures, were extracted from the downloadable OECD PFAS Global list.6 These counts, inventory indicators, and category assignments, along with the corresponding fingerprint tables in the SI Tables, were used to generate all profiling figures and tables in this manuscript. Tables and graphics were created in Microsoft Office 365 Enterprise versions of Word, Excel, and PowerPoint.
4. Results and Discussion
Results reported in this section are based on use of the latest TxP_PFAS_v1.0.4 CSRML file and the most recent PFASSTRUCTV5 structures file unless otherwise indicated; these latest versions will be henceforth referred to simply as TxP_PFAS and PFASSTRUCT. We consider below a number of applications of the TxP_PFAS fingerprint set to examining assumptions made in the course of this work. In addition, we attempt to validate assumptions regarding the ability of TxP_PFAS chemotypes to capture important PFAS concepts, as well as offer examples of how the TxP_PFAS chemotypes, alone or in combinations, can provide objective, structure-based building blocks for constructing PFAS categories.
4.1. PFASSTRUCTV4 vs PFASSTRUCTV5
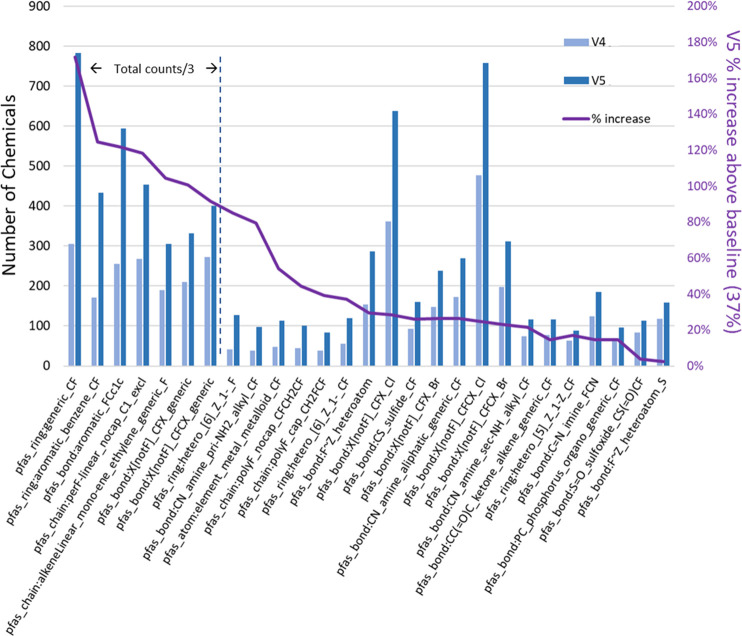
The iterative, and substantially manual process used to develop the TxP_PFAS fingerprint set heavily relied upon and was influenced by the contents of the older PFASSTRUCTV4 inventory, containing 10,776 PFAS structures. Approximately 4000 new PFAS structures were added to the latest PFASSTRUCTV5 inventory containing 14,735 structures, representing a 37% increase, which led to the question: is the newly added PFAS content sufficiently similar in profile to the old or are some ToxPrints or TxP_PFAS chemotypes more or less represented in the newly added structures, possibly due to a shift in focus within PFAS publications or data availability? And if there were a substantial shift in the PFAS profile, would it warrant revisiting the original ToxPrints to identify missing important features? To examine these questions, we applied both the original ToxPrint and the latest TxP_PFAS CSRML files to profile and compare the PFASSTRUCTV4 and V5 inventories (SI Table S3).
Figure 4 plots the frequency of hits, or structure counts, for a representative sample of TxP_PFAS chemotypes showing percentage increases above the baseline of 37% in going from PFASSTRUCTV4 to V5. Included among these were a number of aromatic chemotypes, either adjacent to fluorinated chains or fluorinated themselves, as well as polyfluorinated chemicals, alternate halogen bonding patterns (Cl, Br) and a few specific functional groups. Missing from this set, and underrepresented in the ∼4000 newly added PFAS to V5, were the more typical PFAS perfluoro capped chains of various lengths and fluorotelomers, presumably because these had already been a specific focus of V4 and earlier curation efforts.
Figure 4.
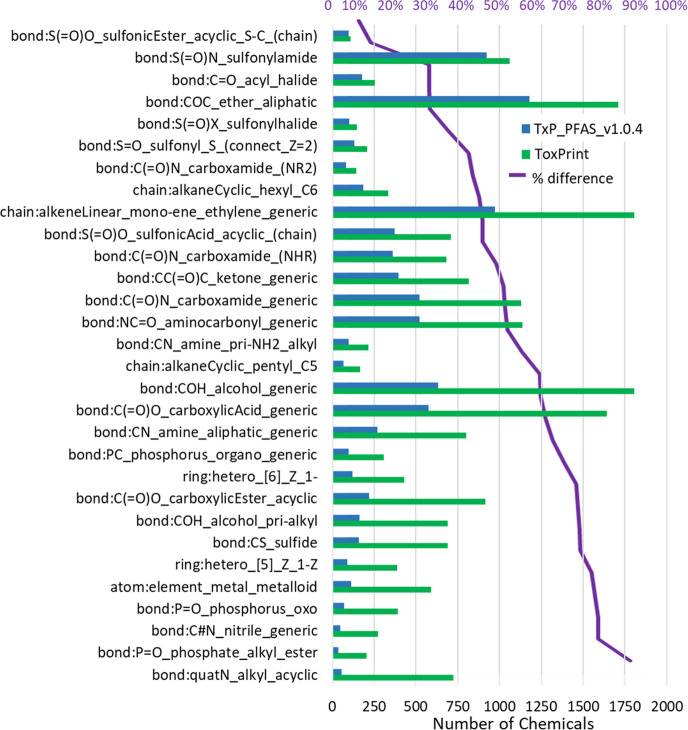
Comparison of structure counts for TxP_PFAS chemotypes in PFASSTRUCTV4 (light blue bars) versus PFASSTRUCTV5 (dark blue bars), with the purple line representing the % increase above the baseline 37% in V5 counts, where the counts of the first 7 chemotypes are reduced by a factor of 3 for graphical display purposes.
Given the large increase in PFASSTRUCTV5 structure counts and clear shifts in the TxP_PFAS chemotype profile, we generated the ToxPrint profile for this new structure set to determine retrospectively whether additional ToxPrints proximate to the fluorinated portion of the chemicals were needed to expand coverage of V5. We re-examined any ToxPrints not previously modified that exceeded 40 hits (increased proportional to the PFASSTRUCTV5 size increase) and confirmed that no new ToxPrints were required to increase the coverage provided by the latest TxP_PFAS set beyond those referenced earlier (i.e., to increase coverage of NoHits).
4.2. ToxPrint vs TxP_PFAS_1.0.4 Coverage of PFASSTRUCTV5
The substantial overlap of the original ToxPrints and the corresponding modified TxP_PFAS fingerprints (see Figure 2 and Table 1) provided an opportunity to compare the relative frequency of the corresponding chemotypes in the PFASSTRUCTV5 file. Our initial hypothesis was that enforcing proximity of the ToxPrint feature to the fluorinated portion of the molecule by addition of a CF group or F atom would yield a substantially smaller number of hits. A total of 56 CF- or F-modified TxP_PFAS chemotypes had a closely corresponding ToxPrint (SI Table S3), and because some were mapped to the same ToxPrint, there were 51 overlapping unique ToxPrints. Figure 5 compares the number of hits for a representative subset of the 51 ToxPrint chemotypes with a corresponding TxP_PFAS chemotype. On one end of the spectrum, the bond:S(=O)O_sulfonicEster_acyclic_S–C_(chain) and bond:S=(O)N_sulfonyamide ToxPrints experienced only a 9% and 13% reduction in counts, respectively, meaning that the ToxPrint feature is almost always (in 91% and 87% of the cases) directly attached to the fluorinated portion of the PFASSTRUCT structure where it occurs. In the case of bond:CS_sulfide or bond:COH_alcohol_pri-alkyl, on the other hand, fewer than 25% of the hits for the ToxPrint remain hits for the corresponding fluorinated TxP_PFAS chemotype, meaning these features most often occur distant from the fluorinated potion of the structure. Overall, the percentage reduction in hits ranged from 8% to 92% across the corresponding set of ToxPrint and TxP_PFAS features, with an average reduction of 54% across the PFASSTRUCT inventory.
Figure 5.
Comparison of structure counts for ToxPrint features (y-axis legend and green bars) versus their corresponding CF- or F-modified TxP_PFAS chemotype (blue bars) across the PFASSTRUCT inventory, sorted from top to bottom by increasing % difference in counts (purple line).
A dramatic illustration of the impact of enforcing proximity of the ToxPrint feature to the fluorinated chain element is provided by the following example. We first performed a substructure search for the carboxylic acid “–CC(=O)OH” (or carboxylate) feature within PFASSTRUCT, which yielded 2747 hits. Next, we performed a substructure search for a linear perfluoroheptyl-C feature (C7F15–C) within the 2747 subset, which yielded 617 hits. Hence, there are 617 structures containing both the carboxylic acid AND the perfluoroheptyl-C feature co-occurring in the same chemicals. Finally, we ask, in how many of these 617 structures were the co-occurring features proximate, or directly bonded to one another? The result is only 24 structures in which the 2 features are bonded, i.e., a nearly 26-fold reduction of hits. This might seem an extreme example since both features are terminal, limiting the outcome. A second example considered a nonterminating secondary alkyl amine functional group (−NHR−) co-occurring with, versus attached to, a linear perfluorooctyl chain. In this example, we found 752 co-occurrences of the two features, but only 2 cases where they were attached, i.e., an even greater 326-fold reduction. We conclude that, whereas co-occurrence of fingerprint features within a small, targeted set of PFAS structures (such as EPA’s test library) might equate with attachment, this is often not the case for the much larger, more structurally diverse PFASSTRUCT inventory, where enforcing proximity of functional group features to fluorinated chains is essential to the accurate detection of attachment.
4.3. Profiling PFASSTRUCT
4.3.1. Perfluorinated Linear Chains
An important subcategory of chemotypes within TxP_PFAS are the perfluoro linear chains of exclusive lengths, both capped (i.e., terminating in CF3) or uncapped (CF2’s on both ends). Figure 6 compares the counts for each type of exclusive perfluoro linear chain, ranging from C2–C11 for cap and C2–C8 for nocap chains, where nocap C7 and C8 exclusives were counted from chemotype combinations (see bottom rows of Table 3). Several trends are apparent: 1) for perfluoro chains of length C4 and above, counts of chemicals with even numbers of perfluoro carbons significantly exceed the counts of the nearest odd numbers of carbons (e.g., C4 ≫ C5, C6 ≫ C7, etc.); 2) there are approximately the same number of chemicals (∼800) with exclusive C4, C6 or C8 capped perfluoro linear chains across the PFASSTRUCT inventory, with a significant drop-off in numbers for > C8 length chains; and 3) the count of cap chains of an exclusive length is greater than that of the corresponding nocap chain of the same length in all cases, most significantly for C3 and longer chains.
Figure 6.
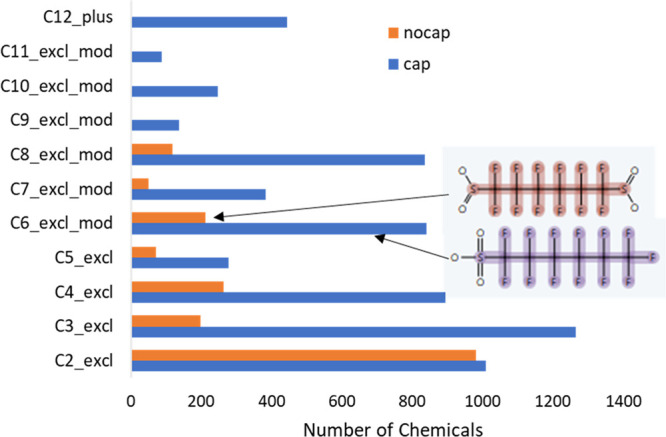
Profile of perfluorinated linear chains across PFASSTRUCT (pfas_chain:perF-linear_...), with cap exclusive counts from C2–C11 (blue) and nocap exclusive counts from C2–C8 (orange), where nocap C7 and C8 exclusive counts were computed with the formulas in Table 3; sample structures are shown indicting presence of a pfas_chain:perF-linear_cap_C6_excl_mod (lower, purple) and pfas_chain:perF-linear_C6_nocap_excl (upper, brown) chemotype.
Not shown in Figure 6 are the C1_excl counts, 5514 for pfas_chain:perF-linear_cap_C1_excl and 1362 for pfas_chain:perF-linear_nocap_C1_excl. Given the large numbers of C1–C3 exclusives, of both cap and nocap linear perfluoro chains, it was of interest to see how often these co-occurred with each other or with the longer perfluoro chains. Given that these chemotypes are fully fluorinated, linear, and exclusive in length, they must be separated by one or more nonfully fluorinated carbons or a heteroatom if co-occurring with another perfluorinated exclusive chemotype. This condition can be satisfied, for example, by a secondary or tertiary branching element on a perfluoro chain, an alternate halogen substitution (e.g., Cl, Br, I), an alkene bond in the chain, or an ether linkage. Using the AND filter condition in the ChemoTyper, we determined that the cap_C1_excl chemotype co-occurred most frequently with the smaller cap_C2,C3,C4_excl chemotypes, i.e., 189, 278, and 43 times, respectively. In contrast, the cap_C1_excl chemotype co-occurred with the cap_C5_excl chemotype only 18 times and with cap_C6_plus only 23 times. Of the 18 co-occurances for the former, we identified 4 branched, 3 alkenes, 2 other halogens, 6 ether linkages, with the remainder having greater nonfluorinated separations between the 2 chemotypes. Similar patterns were seen for co-occurrence of cap_C1_excl with the nocap chemotypes, with slightly higher numbers for the C6 and longer nocap chains. Even fewer overlaps were seen with cap_C2_excl and longer chains. Hence, we conclude that the large majority of the C1_excl counts (5514 cap and 1362 nocap) do not co-occur with longer perfluoro linear chains in the PFASSTRUCT structures.
4.3.2. Perfluorinated Branched Chains
Although most PFAS, including PFOA and PFOS, are typically represented in literature studies as linear perfluoro chains, it is well-known that these and other longer chain PFAS are often accompanied by branched isomers. In recognition of this fact, the Conference of the Parties to the Stockholm Convention on Persistent Organic Pollutants in 2019 listed PFOA, its salts, branched isomers, and PFOA-related compounds in Annex A to the Convention.2 The explicit inclusion of branched isomers of PFOA and related compounds in the listing reflects a growing body of evidence indicating significant levels of PFAS branched isomers in the environment and biota,35 as well as differential properties of linear versus branched isomers of PFOA and PFOS affecting bioaccumulation properties and toxicity.36−38
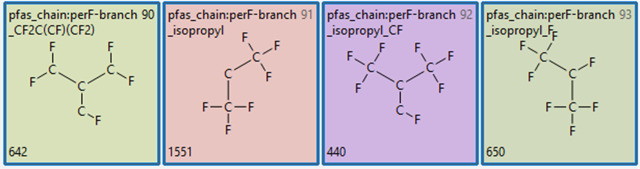
The co-occurrence of the smaller perfluoro chains as noted in the previous section can provide an indirect indication of PFAS branching. More specifically, however, we have included 4 distinct chemotypes in TxP_PFAS that are designed to detect per and poly fluoro branching elements (Figure 7). The first chemotype #90 in Figure 7, present in 642 chemicals in PFASSTRUCT, indicates a branching feature internal to a per or polyfluoro chain. The second chemotype #91 indicates a terminal perfluoro isopropyl branching feature present in 1551 chemicals; given this feature’s high counts, we included two extended features (#92 and #93) in TxP_PFAS to further subset the perfluoro isopropyl branching patterns, each with several hundred counts. The combined coverage of chemotypes #90 and #91 (using the OR filter in the ChemoTyper) yielded a total of 1751 chemicals containing either chemotype #90 or #91, i.e., indicating that 17% of PFASSTRUCT contains a fluorinated branching feature. To further assess how the co-occurrence of smaller perfluoro chains relates to branching, we searched chemotypes #90 AND pfas_chain:perF-linear_cap_C3_excl, which yielded 66 hits. Recalling from the previous section that combining pfas_chain:perF-linear_cap_C1_excl AND pfas_chain:perF-linear_cap_C2_excl yielded a total of 189 hits, additionally combining with the branched chemotype #90 resulted in 59 hits, indicating that nearly a third of the 189 overlaps of those 2 features indeed provide indications of branching.
Figure 7.
TxP_PFAS chemotypes representing branching features, where the number in the lower left corner is the total counts of the chemotype in the PFASSTRUCT inventory.
A recent article by Richard and coauthors proposed a SMILES-based method for detecting PFAS branching for a subset of PFOA-relevant chemicals in PFASSTRUCTV4 containing the formula C7F15–C.39 Given that a manual review was undertaken to validate the results of that study, we sought to determine if a combination of TxP_PFAS chemotypes would successfully detect the 214 confirmed instances of branched isomers of C7F15–C identified within the PFASSTRUCTV4 inventory. The list of 214 DTXSIDs from that earlier study was obtained and confirmed to be a subset of the list of 1750 DTXSIDs containing one or more of the above branched TxP_PFAS chemotypes. However, as discussed in that previous study, a fingerprint method such as presented herein, with distinct independent features, is ill-suited to identifying the exclusive subset of all possible branched isomers corresponding to a particular chemical formula, such as C7F15–C. We encounter the same difficulty here, in that the majority of our perfluoro chain chemotypes are linear. Additionally, due to likely underreporting of branched isomers and greater difficulty discerning branched isomers with typical LC-MS (liquid chromatography mass spectroscopy) monitoring methods, we speculate that the representation of branched isomers actually occurring in the environment is underrepresented within PFASSTRUCT.
4.3.3. Fluorotelomers
Another major subset of chemicals of particular interest to the PFAS community are the so-called fluorotelomers (FT), polyfluorinated compounds typically consisting of a fully fluorinated carbon tail with 1–3 nonfluorinated carbons attached to a functional group. They are commonly named using an “m:n” prefix where m indicates the number of fully fluorinated carbon atoms (m ≥ 2) and n indicates the number of nonfluorinated (usually fully hydrogenated) carbon atoms (n ≥ 1). Several classes of fluorotelomers (FTs) are used in industrial processes and commercial products and have been detected as environmental contaminants.40,41 For example, FT alcohols (FTOH) are raw materials used in the production of FT acrylates and methacrylates, FT sulfonic acids (FTSA) have been associated with aqueous film-forming foam (AFFF) wastewater,42 and FT carboxylic acids (FTCA) are degradation products of FTOHs. Of particular concern, PFOA is a common byproduct of FT production of longer perfluoro chain compounds.43
Given their importance to the PFAS community, and reflecting their prevalence in the PFASSTRUCT inventory, a total of 20 chemotypes in the TxP_PFAS fingerprint set are devoted to the detection of FT-type features. Given the lack of clear community consensus for fluorotelomer nomenclature, our working definition for fluorotelomers in TxP_PFAS is a minimum of 2 perfluoro carbons (CF2CF2) attached to one (n1), two (n2), or three (n3) fully hydrogenated carbons (i.e., CH2, CH2CH2, or CH2CH2CH2), which are then attached to C=O or various heteroatoms that serve as stand-ins for the much broader array of possible functional group attachments.
Shown in Figure 8 is the complete set of FT chemotypes, along with their names, images, representation within the ChemoTyper hierarchy (left side panel) and counts in PFASSTRUCT (number in lower left corner of image box). For each of the three subsets (n1, n2, n3), the pfas_chain:FT_n#_hetero (i.e., not including C) chemotype serves as a generic feature for the set. When combined with the pfas_chain:FT_n#_C=O chemotype, counts for each FT_n# subgroup totaled 734 for n1, 1004 for n2, and 108 for n3, for a total combined count of 1845 fluorotelomer-type chemicals, or 12.5% of PFASSTRUCT.
Figure 8.
TxP_PFAS hierarchy and chemotypes representing fluorotelomer features, where the number in the lower left corner is the total counts of the chemotype in the PFASSTRUCT inventory.
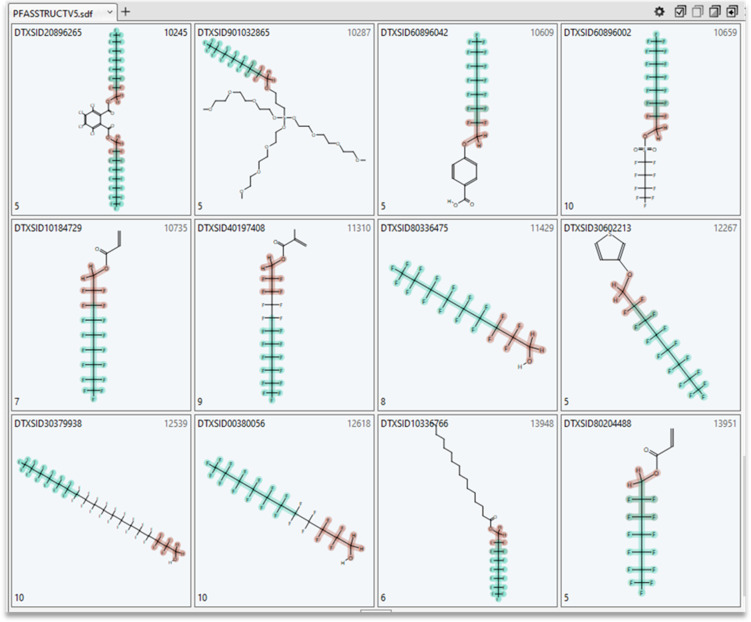
Due to the lack of direct attachment of functional groups to the perfluoro chain in fluorotelomers, TxP_PFAS functional group chemotypes, which each require a direct CF or F attachment, by design, will not be counted, hence requiring the use of heteroatom stand-ins. For the n1 set, FT_n1_O, is the most frequently observed bonding pattern, with 463 hits. Performing substructure searches within this set of hits, we determined that 46 were alcohols (OH), 113 were esters, 99 had O bonded to something other than C or H, and the remaining ∼200 were ethers. Demonstrating the potential for creating PFAS subcategories, combining the chemotypes pfas_chain:FT_n1_O AND pfas_chain:perF-linear_cap_C6_plus in the ChemoTyper yielded 48 hits, all visually confirmed to be co-occurring. A sample of 12 structures from this subset are shown in Figure 9 to illustrate the structural diversity associated with the remaining, distant portion of the chemicals. In this manner, one could construct a large number of meaningful subsets or categories of fluorotelomers for further investigation.
Figure 9.
Subset of 12 chemicals (out of 48 total) resulting from a search for pfas-chain:FT_n1_O (brown highlighted) AND pfas_chain:perF-linear_cap_C6_plus (green highlighted) in the ChemoTyper; the number in the lower left corner of each chemical square indicates the total number of TxP_PFAS chemotypes (not all shown) found in the structure.
Another possible limitation of our fluorotelomer definition, requiring CF2CF2 as the minimum representation of the associated perfluoro chain, is that branching in the perfluoro chain occurring at the first or second carbon attached to the hydrogenated carbon intervening chain will not be detected. A compromise was made in terms of minimizing “false positives” (i.e., incorrectly identifying some structures as fluorotelomers) or “false negatives” (missing chemicals that might be considered fluorotelomers); given the greater opportunity for mis-identification, we chose to minimize false positives. A sample of the PFAS chemicals not recognized by our strict fluorotelomer FT_n1 definition requiring 2 perfluoro carbons (CF2CF2CH2–Q, where Q is a heteroatom, i.e., neither C nor H) that would have been recognized by a less strict definition (CFCFCH2–Q) is shown in Figure 10. The central image is the less strict substructure query used, the 3 structures to the left of it (gray squares) are small structures not generally considered fluorotelomers, the structure above the query structure is a small polyfluorinated PFAS, and the bottom center structure is cyclic (blue squares). Finally, the 2 structures to the right of the query structure (light green squares) are both branched perfluoro structures that our stricter fluorotelomer definition would not perceive as such, i.e., false negatives. Searching across PFASSTRUCT, we compared substructure search counts for CFCFCH2–Q (806) to that for CF2CF2CH2–Q (689), resulting in 117 fewer hits for our stricter FT definition. Visual inspection of the 117 missing structures indicated that only 5 might be considered branched fluorotelomers, whereas the rest are similar to those in the center and left in Figure 10 and would likely not be labeled as fluorotelomers. Hence, the fluorotelomer CF2CF2CH2–Q definition that we adopted appears to be a reasonable compromise.
Figure 10.
Results for a substructure query (yellow center box) of a less strict fluorotelomer FT_n1 representation than the TxP_PFAS version, with the left-most 3 small structures (gray) likely not considered to be fluorotelomers, along with the polyfluoro (top-blue) and cyclic structure (bottom-blue), whereas the 2 right-most green structures could be considered branched fluorotelomers.
We wish to make a final point with respect to the naming of fluorotelomers and the value of structure-based searching. Although the term “fluorotelomer”, or its abbreviation “FT”, is often used within the PFAS community, it is not standard IUPAC (International Union of Pure and Applied Chemistry) nomenclature, nor is it commonly used or understood outside of the PFAS community. In addition, structure-naming algorithms, such as the publicly available OPSIN (Open Parser for Systematic IUPAC Nomenclature) application,44 cannot parse fluorotelomer-type names. An example is DTXSID40880388 whose IUPAC systematic name is 3,3,4,4,5,5,6,6,6-Nonafluorohexane-1-thiol, which can be condensed to the name 2-(Perfluorobutyl)ethanethiol or 4:2 Fluorotelomer thiol. DSSTox expert manual curation has added fluorotelomer-type names either as DSSTox preferred names or synonyms to many PFAS substances,7,11 but there remain hundreds of structures lacking such a synonym. Hence, a simple text search for the term “fluorotelomer” in the DSSTox list of preferred names in PFASSTRUCT (SI Table S5) will retrieve only 242 hits, which is a small fraction of the 1845 hits provided by the structure-based query using the TxP_PFAS FT chemotypes. Even worse is the widespread use of nonstandard acronyms in the PFAS space, particularly for fluorotelomers, which are frequently difficult even for humans to decipher. Examples include FTOHs (FT alcohols), as in 6:2 FTOH, and FTCAs (FT carboxylic acids), among the more common, and FTUCA (FT unsaturated carboxylic acid) and FtTAoS (FT thioether amido sulfonate) among the less common. Because of their widespread use, however, DSSTox curators register many of these acronyms as “ambiguous” synonyms so that a search on the Dashboard will retrieve the most likely record(s) match.
4.3.4. Alternate Halogens, Alkene/Alkyne, Polyfluorination
Additional bonding patterns within the PFAS space not falling under perfluoro chains or fluorotelomers include alternate halogenation (i.e., Cl, Br, I), alkene and alkyne bonds, and polyfluorination, i.e., where one or more fluorines is substituted by a hydrogen on the perfluoro chain. These 16 chemotypes are shown in Figure 11, alongside their ChemoTyper hierarchy placement and showing total counts in PFASSTRUCT. For alternate halogenation, the features pfas_bond:X[notF]CFCX_generic and pfas_bond:X[notF]CFX_generic capture two bonding patterns and each covers all three types of alternate halogens, (Cl, Br, and I) totaling 1199 and 992 counts, respectively. Both bonding patterns were needed to capture the full diversity within PFASSTRUCT as 822 structures are captured in only one or the other group. We find significantly more counts of Cl halogen substitutions within the fluorinated chains than either Br or I, and more Br substitutions than I. These type of substitutions onto a perfluorinated chain create a site of potential reactivity due to the weaker bonds and greater leaving properties of the non-F halogens (I > Br > Cl).
Figure 11.
TxP_PFAS hierarchy and chemotypes representing alternate halogenation, alkene and alkyne bonding patterns, and polyfluorination, where the number in the lower left corner is the total counts of the chemotype in the PFASSTRUCT inventory.
The alkene and alkyne chemotypes that include the direct F bonding pattern were added into TxP_PFAS during final review of the PFASSTRUCT NoHits. An exception was made to include the pfas_bond:alkyne_ethyne_generic_F chemotype, present in only 11 chemicals, due to its prevalence in these NoHits. The alkene version of this generic_F chemotype (#67), however, was well represented in PFASSTRUCT, being present in 915 chemicals.
Finally, 4 chemotypes were included to capture variations in the polyfluorinated bonding patterns observed within PFASSTRUCT; two are terminal “cap” features (#117 and 118) and two are internal “nocap” features (#119, 120). The cap_CHF2CF (#118) and nocap_CFCHFCF (#120) features were the most prevalent within PFASSTRUCT, with a combined total incidence of 1100 hits, whereas the combined total incidence of all 4 chemotypes was 1255, or 8.7% of PFASSTRUCT.
4.3.5. Cyclics, Aromatics, Hetero Rings
Although the term “PFAS” and the majority of PFAS publications and smaller lists focus primarily on alkyl substances, a broader survey of the PFASSTRUCT landscape finds a large number of cyclics (366), aromatics (1298, including fused rings), hetero rings (442), and generic ring systems bound directly to fluorine (2348). Figure 12 lists the 13 TxP_PFAS chemotypes representing these features, resulting in a total coverage of 3311 structures in PFASSTRUCT. Note that 9 of the 13 chemotypes contain fluorine directly bound to carbon within the ring system, whereas the remaining 4 capture a ring system adjacent to an alkyl CF, which indicates likely attachment to a per or polyfluorinated chain system. Note also that only those chain and ring systems found to have sufficient representation within PFASSTRUCT are represented explicitly within the TxP_PFAS chemotype set, whereas others are captured with the pfas_ring:generic_CF chemotype (see Figure 2). Figure 13 shows a sample of the highly diverse PFAS structures captured within this feature space.
Figure 12.
TxP_PFAS hierarchy and chemotypes representing cyclics, aromatics, and fused and hetero rings, where the number in the lower left corner is the total count of the chemotype in the PFASSTRUCT inventory.
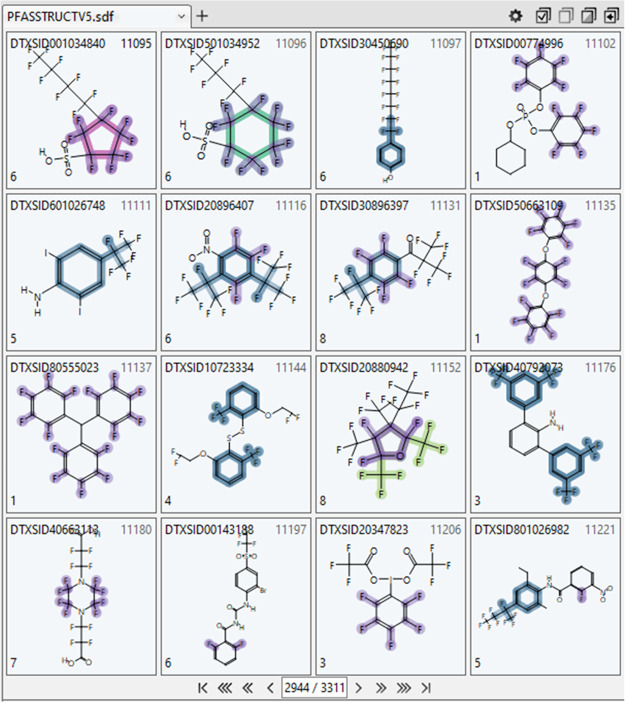
Figure 13.
Subset of 16 chemicals (out of 3311 total) resulting from an OR search in the ChemoTyper upon selecting all chemotypes in the pfas_chain:alkaneCyclic and pfas_ring hierarchy categories; individual TxP_PFAS chemotypes are highlighted on the displayed structures and the number in the lower left corner of each chemical square indicates the total count of unique chemotypes (not all shown) contained in the displayed structure.
Despite the high prevalence of these cyclic and ring chemotypes in PFASSTRUCT, it is noteworthy that several of the unmodified ToxPrint forms of these chemotypes (without enforcing proximity to CF or F) have much greater prevalence, i.e., are most often found distant from the fluorinated portion of the chemical. In all cases, the TxP_PFAS ring chemotypes exceeded 60% reduction from the original ToxPrint counts, meaning that 60% or more of the chemicals containing the nonfluorinated ToxPrint ring chemotypes (i.e., without CF or F attachments) are distant from the fluorinated portion of the PFAS chemical. For the cyclics, chain:alkaneCyclic_butyl_C4 is the one exception as it is almost always found proximate to fluorination in PFASSTRUCT, showing only 8% less incidence with F added to the corresponding ToxPrint, whereas chain:alkaneCyclic_propyl_C3, pentyl_C5, and hexyl_C6 forms show 50%, 59%, and 44% reduction in counts, respectively, with F added.
4.3.6. Bond Type Functional Groups and Categories
Excluding the generic pfas_bond:aromatic_FCc1c chemotype and the alternate halogen (pfas_bond:X[notF]_*) chemotypes already considered, TxP_PFAS contains a total of 51 functional groups within the “bond” hierarchy, most of which have a closely related or directly corresponding ToxPrint as previously discussed. The hierarchy in Figure 3 provides a complete listing of these bond chemotype categories, showing expanded subgroup listings for groups containing more than one chemotype. Shown in Figure 14 are the relative counts of functional group chemotypes in PFASSTRUCT, sorted from high to low (light blue bars), where a representative was chosen for groups containing more than one chemotype. Across the PFASSTRUCT 14,735 chemical landscape, the top ten, most frequently observed functional group chemotypes directly attached to per- or poly- fluoro chains include: a generic heteroatom (C∼Z), carbonyl (C=O), sulfonyl (S=O), ether (COC), sulfur bonded to nitrogen (S∼N), alcohol (COH), carboxylic acid (C(=O)O), carboxamide (C(=O)N), aminocarbonyl (NC=O), and ketone (CC(=O)C). These are a faithful representation of the major functional group categories typically associated with PFAS chemicals.
Figure 14.
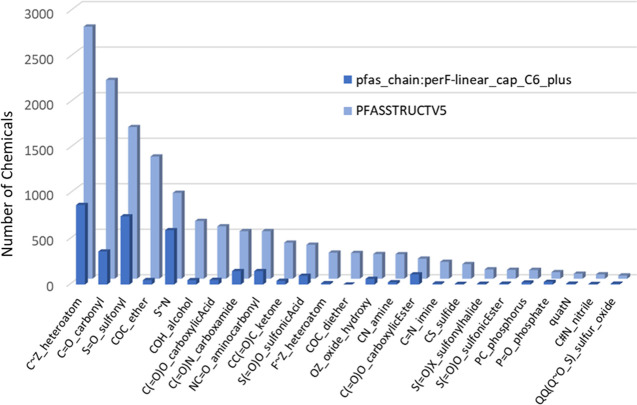
Numbers of chemicals in TxP_PFAS pfas_bond:* categories within PFASSTRUCTV5 in descending order (light blue bars), showing a single representative for groups having multiple subcategories compared with counts within each category of chemicals also containing the pfas_chain:perF-linear_cap_C6_plus chemotype (dark blue bars).
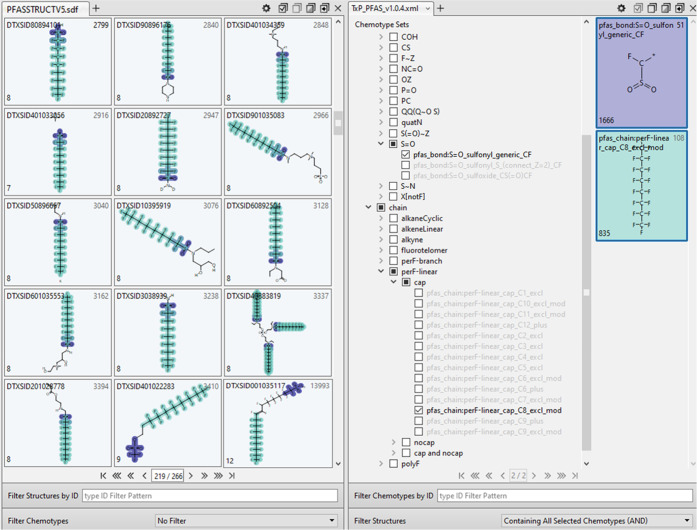
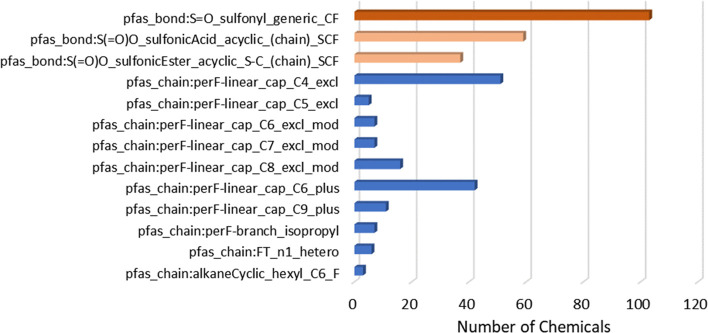
Within the ChemoTyper, a user can construct simple categories by combining two (or more) chemotypes using the “Containing All Selected Chemotypes (AND)” filter. To illustrate, the dark blue bars in the forefront of Figure 14 show the results when a filter is applied separately for each of the listed chemotypes AND pfas_chain:perF-linear_cap_C6_plus. Each of these combined results can serve as a clearly defined structure-based category. For instance, the results indicate that nearly half (750) of the sulfonyl (S=O) containing chemicals are co-occurring with the linear perfluoro capped C6 or greater length chain. Swapping perF-linear_cap_C6_plus for the more specific perF-linear_cap_C8_excl_mod chemotype, in turn, yields fewer results (266) but still a relatively large set of chemicals constituting a category; sample results of this latter search within the ChemoTyper are shown in Figure 15. Hence, a desired degree of specificity can be achieved based on the types of chemotypes chosen for the combinations. In addition, by enforcing proximity to CF or F in the TxP_PFAS functional group (bond type) chemotypes, we have achieved a remarkable degree of success in inferring direct attachment of the fluorinated chain features with an associated functional group. Figure 15 illustrates the point. On visual review of the 265 chemicals containing both the S=O_sulfonyl_generic_CF AND perF-linear_cap_C8_excl_mod chemotypes, we found these chemotypes attached in all but two structures (DTXSID401022283 and DTXSID0001035117, shown in bottom row of Figure 15). Hence, it is important to emphasize that co-occurrence of 2 chemotypes is a necessary but not sufficient condition for attachment, i.e., 2 chemotypes may co-occur in separate parts of the molecule if there are multiple fluorinated moieties within a single chemical, such as in the 2 exceptions in Figure 15. However, as in this example, we have found this nonattached association of the two types of chemotypes (fluorinated alkyl chains and functional groups) to be relatively infrequent across PFASSTRUCT.
Figure 15.
ChemoTyper view resulting from the selection of two co-occurring chemotypes, pfas_bond:S=O_sulfonyl_generic_CF (purple) AND pfas_chain:perF-linear_cap_C8_excl_mod (green), showing a sample of PFASSTRUCT structures containing the two chemotypes highlighted in green and purple.
Although the range of possible chemotype combinations achievable in the ChemoTyper is limited by the available filtering options, the hierarchy and visual representations of the chemotypes alone (right panel in Figure 15) and superimposed on structures (left panel in Figure 15) provide powerful tools to support exploration. The results of any search, such as that in Figure 15, in turn, can be exported as a structure-data (SDF) file or as a tsv or csv fingerprint file. For computational implementation, note that each of the ChemoTyper filtering operations, as well as many more (e.g., including positive and negative conditions), can be achieved by operating directly on the exported fingerprint file, opening the possibility for a wide range of structure-based PFAS category definitions tailored to a particular study.
5. Application: Comparison to OECD PFAS Categories
The goal of creating expert-defined PFAS structure categories, in which each PFAS chemical in an inventory is assigned to a single category, has been the prevailing approach in the PFAS research and regulatory communities. As we attempt to grapple with ever larger and more diverse structure inventories, such as PFASSTRUCT, however, the deficiencies of such an approach become increasingly clear. To illustrate some of the challenges of assigning PFAS categories, either manually or algorithmically, we compare a small subset of the 106 manually assigned PFAS categories accompanying the large OECD Global PFAS list6,9 and consider how well TxP_PFAS chemotypes are able to recapitulate these categories, where the two approaches disagree and why, as well as in what ways the TxP_PFAS chemotypes offer significant advantages over the manual, expert-based approach.
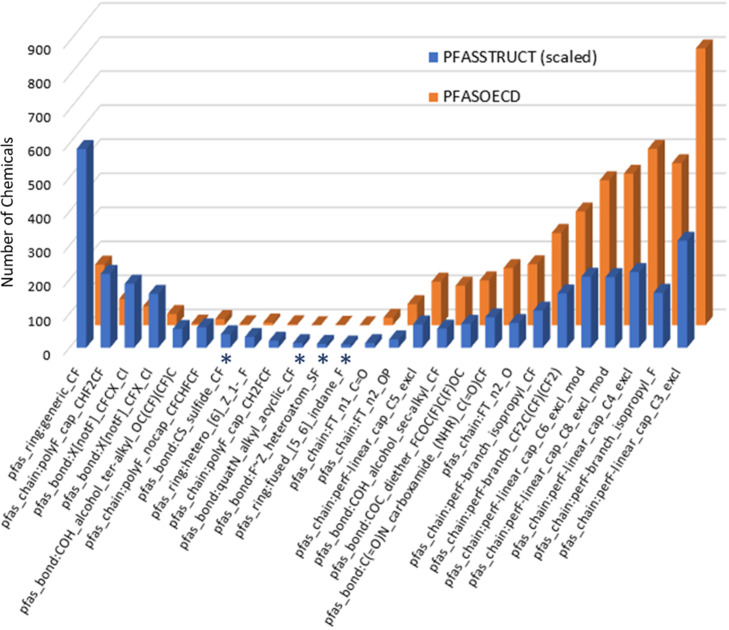
The full OECD Global PFAS list contains both defined structures, as well as polymers and mixtures. We consider here only the 3663 defined structure subset of the OECD Global PFAS list in PFASOECD, which is completely contained within the PFASSTRUCTV5 inventory (SI Table S5). Given that the current PFASSTRUCT inventory is nearly 4 times larger than the PFASOECD inventory, we compared the PFASOECD inventory to a scaled version of the PFASSTRUCT inventory to assess whether there are TxP_PFAS chemotypes significantly enriched in one inventory versus the other. Figure 16 shows a plot of TxP_PFAS counts for PFASSTRUCT (divided by 4) compared to corresponding counts in PFASOECD where we see the largest percentage differences. We found relatively few cases where PFASOECD is deficient in chemotypes relative to PFASSTRUCT (left side of plot), but these interestingly include mostly categories of features that might be considered peripheral to or detracting from the more iconic PFAS structures, such as fluorinated rings and polyfluoro and alternate halogen features. Only 4 chemotypes (starred) have 3 or fewer counts in PFASOECD relative to 39–83 counts in PFASSTRUCT. On the right side of Figure 16, on the other hand, we see PFASOECD containing a greater percentage of the more familiar C3, C4, C6 and C8 perfluorinated chains, as well as n2 fluorotelomers (bonded to O), diethers, and perfluoro branching elements. Overall, however, we find the PFASOECD TxP_PFAS profile to be a good representation of the PFASSTRUCT profile; hence, a category approach for one inventory should reasonably apply to the other.
Figure 16.
Plot of counts of PFASSTRUCTV5 (scaled x 0.25) TxP_PFAS chemotypes (blue bars) versus PFASOECD TxP_PFAS chemotypes (orange bars) for the 26 chemotypes having the highest percentage differences in counts, with the 4 starred (*) chemotypes having 3 or fewer counts in PFASOECD.
Each of the PFASOECD structures was manually assigned to one of 106 OECD PFAS categories by the OECD study authors, and each of the 3663 structures, in turn, has a TxP_PFAS fingerprint. (Note that a polymer category incorrectly assigned to 2 defined structure substances was not included in the 106 total. Also, since chemical structures were not provided with the original OECD Global PFAS list publication, we cannot verify that DSSTox curated structures for this list are the same as were used by the OECD authors to assign categories.) To clearly distinguish between types of categories, TxP_PFAS chemotype names will be italicized and OECD PFAS category names will be bolded throughout this section. A sample of the OECD categories showing relative total counts is provided in Figure 17. Some categories, such as fluorotelomers, are more represented than others but, overall, the categories provide a good representation of the most common PFAS categories of interest and concern, such as perfluorocarboxylic acids (PFCAs) and perfluorosulfonic acids (PFSAs), along with their salts and esters.
Figure 17.
Sample list of OECD PFAS categories with bars indicating the number of OECD list chemicals per category, sorted in descending order.
The OECD PFAS categories range from relatively simple structure-based names, such as perfluoroalkyl iodides (PFAIs) and n:1 fluorotelomer alcohols, to more broadly defined categories, such as perfluoroalkane sulfonic acids (PFSAs), their salts and esters. Additionally, however, are very broad and less well-defined categories, such as n:2 fluorotelomer-based non-polymers, perfluoroalkane sulfonyl-based nonpolymers, and perfluoroalkenes & derivatives. Lastly are the ill-defined categories that are intended to capture multiple features (e.g., n:2 fluorotelomer alcohols (n:2 FTOHs)/thiols and perfluoroalkane sulfonyl amides/amido ethanols (xFASA/Es) and other alcohols) or “other” structures not fitting into existing categories, such as and other per- and polyfluoroalkyl ether-based compounds–monoethers. Because this was an expert, manually compiled list, there are the inevitable misspellings (e.g., perfluoroalkanes & armoatics), inconsistent naming (FT in some places, fluorotelomer in others), and abbreviations that would require a glossary for the average chemist or PFAS researcher (HFE-based silanes, functionalized PTFE, FTAL, n:1 PAPs, n:3 acids, SFAs and derivatives). It is not our intention to diminish the magnitude and value of this OECD effort to provide useful, expert-assigned category labels for one of the largest set of PFAS substances published up to that point. Rather, we wish to emphasize the difficulty of doing so in a manner that is consistent and accompanied by clear structure-based rules defining the categories so as to be computationally feasible and reproducible by others.
Despite its limitations, the OECD PFAS structure list represents the largest available set of chemicals with expert-assigned PFAS categories against which to compare the performance and utility of the TxP_PFAS fingerprint set. In Table 4, we consider a small number of direct category comparisons to illuminate differences and challenges, as well as strengths and limitations of the two approaches. The first 3 OECD categories listed (perfluoroalkyl sulfonyl halides, iodides and amines) are compared to a closely corresponding TxP_PFAS functional group, bond-type chemotype in each case. Of the 14 chemicals categorized by the OECD as perfluoroalkane sulfonyl halides, each has the corresponding pfas_bond:S(=O)X_sulfonylhalide_CF chemotype. However, there are 12 additional OECD PFAS chemicals containing this chemotype (i.e., 26 compared to 14 in the fourth and fifth columns of Table 4), but these were assigned to other OECD categories that were given precedence by the OECD author(s). This particular TxP_PFAS chemotype is found in a larger group of 102 PFAS chemicals across the entire PFASSTRUCT inventory. A similar level of agreement is found in the case of the OECD perfluoroalkyl iodides (PFIAs) category, with only 2/43 chemicals not containing the corresponding TxP_PFAS chemotype (pfas_bond:X[notF]_CFX_I) due to iodine being bonded to a tertiary C, i.e., violating the CF-I condition. We see greater disagreement for the OECD perfluoroalkyl amines category, with 7/19 chemicals not recognized by the pfas_bond:CN_amine_aliphatic_generic_CF chemotype due to the latter being restricted to aliphatic cases (i.e., disallowing double and triple bonded N). For each of these 3 comparisons, the OECD category specifies “perfluoroalkyl”, meaning a fully fluorinated structure is required. The TxP_PFAS chemotype, on the other hand, only specifies that the functional group is bonded to a CF. Only for the larger perfluoroalkyl iodides category did we visually check to confirm whether each of the 43 OECD chemicals in the category were in fact perfluorinated; we found that 8 were not strictly so, i.e., iodine was bonded to a nocap chain (not terminating in CF3).
Table 4. Comparison of OECD PFAS Categories to Analogous or Closely Related TxP_PFAS Chemotype Categories, Noting Discrepancies in Coverage in the Two Approaches and Reasons for the Discrepancies.
For the relatively broad OECD category of side-chain fluorinated aromatics, we find an even larger disagreement with the corresponding pfas_bond:aromatic_FCc1c chemotype category, with the latter missing 75 chemicals assigned by OECD to this category (i.e., 414 minus 339). However, in this case, 37 of the missing chemicals are nonaromatic cyclics mis-categorized by the OECD authors (see, e.g., DTXSID10720484), with the remaining 38 chemicals ambiguous, some having possible resonance structures (see, e.g., DTXSID10293289).
The OECD combined category of perfluoroalkane sulfonic acids (PFSAs), their salts and esters provides an example of a broad category encompassing multiple types of features. We achieved nearly 100% coverage with the pfas_bond:S=O_sulfonyl_generic_CF chemotype, missing only a single chemical no longer registered in DSSTox as a defined structure (DTXSID50880591). Interestingly, this chemotype has much broader representation across the OECD inventory outside of the OECD category, i.e., 445 total instances, meaning 342 OECD chemicals containing this chemotype (sulfonyl bonded to CF) are assigned to other OECD categories. We also use this example to demonstrate the profiling capability of the TxP_PFAS chemotypes, with two more specific TxP_PFAS bond-type chemotypes listed in Figure 18 that identify the distinct sulfonic acid and ester subgroup counts (orange bars) within the broader OECD category. We can extend profiling of this PFSAs OECD category even further using the perfluoro chain chemotypes, with the resulting counts (blue bars) shown in Figure 18.
Figure 18.
TxP_PFAS chemotype profile of the OECD PFAS perfluoroalkane sulfonic acids (PFSAs), their salts and esters category containing 104 total members within the OECD PFAS list, where the top row, pfas_bond:S=O_sulfonyl_generic_CF (dark orange bar), represents the main category counts and subsequent rows represent separate subcategory counts within the main category: the light orange bars in rows 2 and 3 represent bond-type subcategory counts and the remaining blue bars represent pfas_chain:... subcategory counts.
Fluorotelomers constitute nearly a quarter of the OECD categories (25/106), covering 1309 of the 3663 OECD PFAS list chemicals. The total numbers of structures specific to n:1 (6 categories) and n:2 fluorotelomers (15 categories), respectively, are 270 and 995, indicating significantly more n:2 categories and structures. The last OECD category considered in Table 4 is a super category consisting of all 25 of the OECD categories containing the term “fluorotelomer” or “FT” combined. We constructed a similar super category by combining all 20 of the pfas_chain:FT* TxP_PFAS chemotypes for comparison. In this case, we find a large deficit in coverage of the TxP_PFAS FT chemotypes, only identifying 616/1309 of the OECD FT-categorized chemicals, i.e., missing 693. This lack of correspondence is due to the stricter definition of fluorotelomer adopted for the TxP_PFAS set, requiring CF2CF2-(CH2)n-Q, where n = 1–3 and Q = heteroatom (not C or F) or Q = (C=O). Shown in Figure 19 are several types of structures considered by the OECD as FTs that are not included under the more restrictive TxP_PFAS FT definition. Consistent with their greater relative counts, the majority of these 693 structures are categorized by OECD as n:2 FTs. Despite not being categorized as an FT by the TxP_PFAS, each of these 693 OECD PFAS structures contains one or more pfas_chain:* type chemotypes, and 273 of the 693 contain one or more pfas_bond:* functional group chemotypes.
Figure 19.
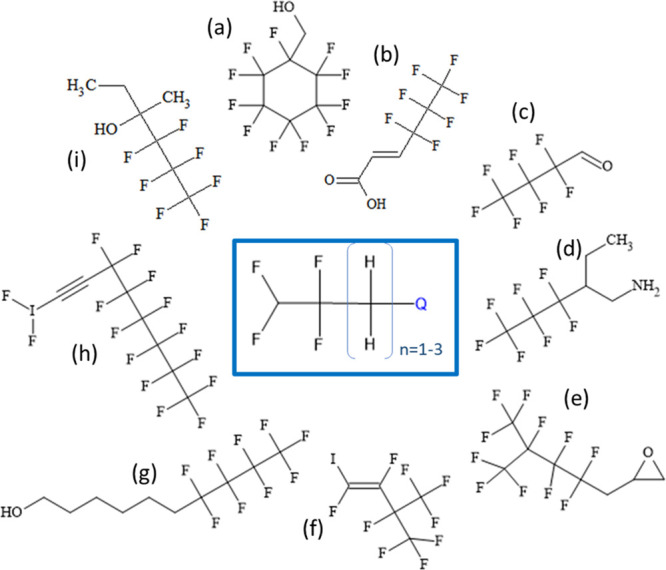
Sample structures (a–i) in the 25 OECD PFAS fluorotelomer categories that fall outside the TxP_PFAS fluorotelomer chemotype definitions (center blue box), where Q = a non-C,F heteroatom or a C=O.
The above OECD category comparison exercise points to several challenges in the PFAS field, including lack of community nomenclature standards applied to terms such as fluorotelomer, widespread use of acronyms and nonstandard names, and lack of clear guidance as to which PFAS features should take precedence over others in constructing PFAS categories, and for what purposes. Furthermore, the OECD categories focus exclusively on functional group attachments to fluorinated portions of chemicals, without any distinctions based on the length or types of fluorinated chains or rings. In spite of these limitations, the exercise serves to highlight several types of possible missing features within TxP_PFAS, particularly for the FT chemicals labeled as such by the OECD, as represented in Figure 19.
Lastly, recall that the full set of 129 TxP_PFAS features when mapped onto chemicals in PFASSTRUCTV5 yielded only 25 NoHits (i.e., chemicals without a single chemotype) out of the 14,735 total, implying excellent overall coverage. However, we did not distinguish the combined set of chain-type and aromatic chemotypes, which reproduce this degree of coverage on their own, from the group of chemotypes containing one or more heteroatoms (i.e., not C, H, or F). A coverage assessment of all 87 TxP_PFAS containing heteroatoms, i.e., bond-type, FT-type, metals, and heterocycle chemotypes, was undertaken to shed light on this issue. In total, these 87 heteroatom chemotypes were present in 11,191 chemicals. We then used molecular formula to identify 1289 chemicals in PFASSTRUCTV5 that contained no heteroatoms (i.e., only C, F and H) meaning that 13,446 structures out of the 14,735 total in PFASSTRUCTV5 contain a heteroatom compared to the 11,191 predicted by TxP_PFAS. The difference of 2254 chemicals where the heteroatom is undetected by TxP_PFAS is most likely due it being distant from the fluorinated portion of the structure, as in Figure 19g. However, other possibilities include structures such as in Figure 19a, b, d, e, and h, where the nearby heteroatom is not perceived by a TxP_PFAS chemotype. Depending on the number of representative cases, these types of exceptions could represent an area for future expansion of the TxP_PFAS chemotypes.
6. Summary and Conclusions
We have presented herein the first publicly available, structure-fingerprint set tailored to the large and growing PFAS chemical domain, currently exceeding 14,000 structures within EPA’s latest PFASSTRUCTV5 inventory. In relating the history of development of the TxP_PFAS fingerprint set, which used the public ToxPrints as a starting point, we have also related many of the underlying assumptions, along with the heuristic and expert-informed process used to iterate in on the final selection of 129 TxP_PFAS chemotypes (Figure 2). The chemotypes are split into two large groups, the first containing a selection of mostly ToxPrint bond-type functional groups that were modified to incorporate attachment to either a CF group or F atom to enforce proximity to the fluorinated portion of the chemical. For the 51 ToxPrints where a direct comparison could be made, we showed that this resulted in a dramatic reduction in chemotype-chemical counts (averaging 54%) in going from the unmodified ToxPrint to the corresponding modified TxP_PFAS chemotype, thus achieving the desired result of focusing only on those functional groups proximate to the per or polyfluorinated portions of the PFAS chemicals as well as demonstrating the need to do so (Figure 5). The remaining TxP_PFAS chemotypes consist of some additional fluorinated functional (bond) groups as well as various lengths and types of per and polyfluorinated chains and bonding patterns, including branching, polyfluorination, alternate halogenation, and fluorotelomer-type bonding patterns. Using the publicly available ChemoTyper application, we additionally showed, by means of several examples, how the TxP_PFAS fingerprints can be visualized, filtered, and used to profile the PFASSTRUCT inventory in various ways, including using chemotypes alone or in combinations to construct structure-based PFAS categories. Lastly, we used a small set of the OECD expert-defined PFAS categories, manually assigned by OECD authors to over 3600 PFAS structures in the OECD Global PFAS database, to evaluate agreement, or lack thereof, with a set of analogous structure-based TxP_PFAS categories. Overall, the results demonstrated the ability of the TxP_PFAS chemotypes to recapitulate several of the expert-based PFAS category concepts, albeit with clearly defined structure rules that can be consistently, transparently, and reproducibly applied to automatically process the entire PFASSTRUCT inventory without need to consult an expert.
We have demonstrated considerable potential of the TxP_PFAS fingerprint set to support research into PFAS properties, environmental fate, and toxicity. Not only do the fingerprints provide a computationally feasible, standardized, structure-based means for partitioning the PFAS space into local chemical domains of interest, but they do so in a way that is flexible, chemically meaningful, and builds on prior knowledge and PFAS-specific features of interest to the PFAS research community. That being said, limitations of the TxP_PFAS fingerprint approach, many of which would be shared by any binary bit-based fingerprint-type approach, include the following:
-
(1)
Only a single instance of a chemotype is reported, even if many instances are present in the chemical, e.g., in multi-ether chain fragments or when multiple perfluoro chains are attached to a central phosphorus.
-
(2)
It is difficult to capture or delineate some types of chemical categories, such as branching of defined length chains, or subcategories nested within larger categories when the core atom has multiple possible valence states (e.g., sulfinic acids nested within sulfonic acids).
-
(3)
Fingerprint bits are independent, i.e., co-occurrence of two chemotypes in a chemical is a necessary but not sufficient condition for assuming the chemotypes are attached. However, by virtue of attaching bond-type TxP_PFAS chemotypes to a CF or F, we in effect enforced an overlap to the per- and poly fluoro chain-type chemotypes should they co-occur, thus increasing the probability that they are in fact attached.
-
(4)
By focusing TxP_PFAS features only on the per- or polyfluorinated local regions of PFAS chemicals, we have chosen to ignore features of PFAS chemicals located distant from the fluorinated region. This limitation is deliberate and by design as the TxP_PFAS fingerprint set has a focused objective. One way to broaden the perceptual scope, however, would be to couple the TxP_PFAS fingerprint profile with the PFAS-agnostic ToxPrint fingerprint profile. The latter would perceive a broader array of features, including those distant from the fluorinated portion of the molecule. These would provide other possible inputs to support categorization efforts, particularly when distant features are believed to exert a strong effect on the chemical leaving ability or reactivity of the fluorinated chain.
-
(5)
Finally, structure-based features or fingerprints, in and of themselves, can be useful descriptors for purposes of modeling PFAS properties, but global, electronic, and physicochemical properties pertinent to PFAS characterization, such as vapor pressure, dipole moment, size, polar surface area, and energy of frontier orbitals, to name but a few, will be required to capture more detailed and deterministic drivers of activity within defined domains of PFAS chemical space. These global properties coupled with similarity and clustering algorithms, such as employed in the PFAS-Map approach mentioned previously,21 should be considered complementary to the present approach, with the TxP-PFAS features helping to structure-profile similarity clusters, illuminate underlying structure correlates, and further restrict the domain of investigation to allow for more mechanistic drivers of activity to be revealed.
It is not possible with a set of only 129 defined features to recapitulate all the nuance and knowledge that an expert might bring to bear on the challenge of PFAS categorization and different needs may dictate that different features take precedence in assigning chemicals to categories, e.g., one study might focus on the effect of variations of perfluoro chain lengths while keeping the terminal functional group constant, whereas another might do the opposite, fixing the chain length and varying the functional group. Hence, categorization should not be considered a fixed, singular objective since many complex chemicals could fall into multiple structural categories. In addition, the level and type of category specificity required to support a study can vary significantly and a range of chemical properties and attributes can inform chemical selection prior or subsequent to application of TxP_PFAS structure-based profiling. A category should serve the objectives of the study, whether it be a structure–activity study of bioactivity, modeling of environmental fate, or prioritizing chemicals for testing. A case in point is EPA’s 2021 National PFAS Testing Strategy45 in which a tiered combination of structural filters, the PFAS-Map structural categories,19 physicochemical properties (e.g., vapor pressure), and data-availability (e.g., in vivo or in vitro toxicity), coupled with structure-based clustering, were used to create chemical categories to inform selection of potential candidates for additional tiered testing. Future iterations and expansions of this approach could potentially benefit from application of TxP_PFAS, e.g., to profile more specific structure features within and across the current broadly defined category clusters.
In employing the TxP_PFAS features moving forward, researchers are encouraged to report the TxP_PFAS feature names, whether used alone or in combinations to construct new categories, to document reproducible and unambiguous structural definitions. Looking to the future, as the community gains experience using the TxP_PFAS fingerprint set and research priorities change, it is likely there will be the need to modify or add new chemotypes to the TxP_PFAS set. The public availability of the ChemoType Editor (available for download from http://chemotyper.org) provides users with an easy-to-use tool for creating new chemotypes, which can then be validated and tested within the public ChemoTyper. The public accessibility, visualization tools, and extensibility of CSRML provide distinct advantages over SMARTS (http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html), which have been more commonly implemented in cheminformatics tools and fingerprint methods to date. However, public tools for interconversion of CSRML and SMARTS, such as provided by the ChemoType Editor, will be useful for promoting use of CSRML. Significant advantages of ToxPrint and TxP_PFAS features coded in CSRML are that they provide a clear and intuitive correspondence to a structure-defined chemical domain. EPA researchers have collaborated with CSRML developers within MN/AM to incorporate the ability to download ToxPrint fingerprint files for any DSSTox inventory or list of chemicals from the Dashboard. This capability has been expanded within a new online suite of EPA tools, known as Cheminformatics Modules (https://www.epa.gov/chemical-research/cheminformatics). ToxPrints can be selected as one of several possible fingerprint methods within the Search module, allowing structure/substructure/similarity searching across the DSSTox inventory with a number of chemical filtering options. In addition, a dedicated ToxPrint module enables users to generate, export, and visualize ToxPrint fingerprints contained in a list of DSSTox or imported structures. ToxPrints found to be statistically enriched within ToxCast assay actives, in turn, are incorporated into the “Alerts” module for several assay categories. Given that the ToxPrint CSRML functionality is already incorporated, it is feasible and straightforward to extend the same functionality for the TxP_PFAS fingerprints in future updates of the Cheminformatics Modules.
The newly created TxP_PFAS fingerprint CSRML with the accompanying PFASSTRUCT file and public ChemoTyper are for the first time enabling robust exploration and organization of PFAS space in a way that can be clearly communicated and understood by chemists and nonchemists alike. Using these tools, we were able to systematically explore the structural composition of the large PFASSTRUCT structure space, shedding light on the types and frequency of PFAS features. Given the lack of bioactivity, toxicity, and environmental transformation and fate data on the vast majority of these chemicals, it is our hope that the TxP_PFAS fingerprint set and accompanying tools will spur modeling studies to investigate the impacts of both local and distant chemical features on the reactivity of PFAS structures, the ability of the fluorinated portion of the chemical to dissociate and react, and properties that are conferred onto the larger molecule by the addition of the PFAS moiety. Lastly, it is our hope that the availability of these public resources will encourage the use of systematic, structure-based means for organizing and communicating PFAS features, offering greatly expanded opportunities for the larger PFAS community to create, explore, and model structure-based categories within PFAS inventories.
Acknowledgments
The authors wish to acknowledge Risa Sayre and Kelly Carstens for helpful comments in review of the manuscript, Daniel Chang for the support and encouragement to pursue this project, and the invaluable contribution of Valery Tkachenko of Science Data Experts to the development of the ToxPrint module capabilities within EPA’s Cheminformatics Modules on-line application. We also acknowledge the incredible dedication, talent, and productivity of members of the DSSTox chemical curation team (Indira Thillainadarajah, Brian Meyer, Saku Sivasupramaniam, and Vicente Samano) for expert review and registration of chemicals in each of the various DSSTox PFAS lists referenced and used in the present study.
Glossary
Abbreviations
- CT
chemotype
- CSRML
chemical subgraphs and reactions markup language
- DMSO
dimethyl sulfoxide
- DSSTox
Distributed Structure-Searchable Toxicity Data Network
- EPA
United States Environmental Protection Agency
- FT
fluorotelomers
- PFAS
per- and polyfluorinated alkyl substances
- PFOA
perfluorooctanoic acid
- PFOS
perfluorooctanesulfonic acid
- OECD
Organisation for Economic Cooperation and Development
- OPSIN
Open Parser for Systematic IUPAC Nomenclature
- TxP
ToxPrint
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.2c00403.
Table S1 FP ToxPrint fingerprint matrix exported from the ChemoTyper for PFASSTRUCTV5 containing 14,735 rows indexed by DTXSID substance identifier and 729 columns indexed by ToxPrint names (alphabetized); Table S2 TxP_PFAS_v1.0.4 fingerprint matrix exported from the ChemoTyper for PFASSTRUCTV5 containing 14,735 rows indexed by DTXSID substance identifier and 129 columns indexed by TxP_PFAS_v1.0.4 chemotype names (alphabetized); Table S3 Chemotype count totals for TxP_PFAS_v1.0.4 mapped to PFASSTRUCTV5 compared to counts for PFASSTRUCTV4, as well as corresponding ToxPrint names where a close correspondence exists; Table S4 Chemotype count totals for ToxPrints mapped to PFASSTRUCTV5 compared to counts for PFASSTRUCTV4, as well as indications of which ToxPrints have a closely corresponding TxP_PFAS chemotype; Table S5 DSSTox chemical identifiers (DTXSID, SMILES, name, CASRN, formula) for PFASSTRUCTV5 list, indicator column for overlapping content in PFASSTRUCTV4 (10,586) and PFASOECD (3662) lists, indicator columns for the 7 TxP_PFAS chemotypes used in the OECD Category analysis of Section 5, indicator columns for chemicals containing one or more of the 20 TxP_PFAS fluorotelomer (FT) chemotypes or designated as an OECD FT, and assigned OECD Structure-Category Name for the 3662 overlapping OECD vs PFASSTRUCTV5 chemicals, with the last separated column listing the 106 unique OECD Structure Categories; PFASSTRUCTV5_20221101.sdf containing 14,735 structures, also described and available for viewing and download at: https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCTV5; TxP_PFAS_v1.0.4.xml CSRML file containing coding for 129 TxP_PFAS chemotypes and their hierarchy index. (ZIP)
Author Present Address
⊥ R.L., Simulacra and Simulation, New York City, New York, United States
Author Present Address
¶ H.H., Project Enhancement Corp., Contractor to the U.S. Dept. of Energy.
Author Contributions
A.M.R. is lead author, helped to compile and procure EPA’s PFAS testing inventory, heads the DSSTox project and curation efforts to compile the various PFASSTRUCT lists, and led development and testing of the final TxP_PFAS CSRML file; R.L. initiated this project while an ORISE student contractor at EPA and coded many of the original TxP_PFAS chain chemotypes in CSRML; M.A. took over helping with the project after R.L. left EPA and coded most of the modified ToxPrints in CSRML; H.H. helped in the transition of the project from R.L. to M.A. and created an initial version of the TxP_PFAS hierarchical index; C.Y. and J.R. provided indispensable support and scientific inspiration for this project with development of the public ChemoTyper and ToxPrints, and are providing a public forum for distribution of the TxP_PFAS CSRML file on the ChemoTyper Web site; T.M. provided early help with CSRML coding issues, and B.B. is the main developer of the publicly available ChemoType Editor application that was used to create some of the TxP_PFAS CSRML chemotypes; A.W. was instrumental in compiling PFAS lists for curation and registration in DSSTox and publishing these lists on EPA’s CompTox Chemicals Dashboard; G.P. heads up “read-across” research within EPA’s computational toxicology program, constructed expert-based PFAS categories to initially prioritize and select PFAS chemicals for assay testing to support read-across and modeling efforts, and was instrumental in motivating development of the TxP_PFAS fingerprints and in promoting the use of structure-based tools for further prioritization and analysis efforts of PFAS. CRediT: Ann M Richard conceptualization, data curation, formal analysis, methodology, project administration, resources, software, supervision, visualization, writing-original draft, writing-review & editing; Ryan Lougee conceptualization, formal analysis, investigation, methodology, software; Matthew Adams investigation, methodology, software; Hannah Hidle investigation, methodology, software; Chihae Yang methodology, software, visualization, writing-review & editing; James Rathman methodology, software, visualization; Tomasz Magdziarz methodology, software, visualization; Bruno Bienfait software, visualization; Antony John Williams data curation, writing-review & editing; Grace Patlewicz conceptualization, validation, writing-review & editing.
The work presented in this manuscript was solely supported by appropriated funds of the U.S. Environmental Protection Agency, National Institutes of Health, and U.S. Food and Drug Administration.
The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency or the U.S. Food and Drug Administration. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
The authors declare no competing financial interest.
Special Issue
Published as part of the Chemical Research in Toxicology virtual special issue “AI Meets Toxicology”.
Supplementary Material
References
- Technical Fact Sheet – Perfluorooctane Sulfonate (PFOS) and Perfluorooctanoic Acid (PFOA); U.S. EPA, 2017. https://19january2021snapshot.epa.gov/sites/static/files/2017-12/documents/ffrrofactsheet_contaminants_pfos_pfoa_11-20-17_508_0.pdf (accessed 02-28-2023).
- The new POPs under the Stockholm Convention: List perfluorooctanoic acid (PFOA), its salts and PFOA-related compounds in Annex A with specific exemptions; Stockholm Convention POPRC, 2019. http://www.pops.int/TheConvention/ThePOPs/TheNewPOPs/tabid/2511/Default.aspx (accessed 12-16-2021).
- Sunderland E. M.; Hu X. C.; Dassuncao C.; Tokranov A. K.; Wagner C. C.; Allen J. G. A review of the pathways of human exposure to poly-and perfluoroalkyl substances (PFASs) and present understanding of health effects. J. Expos. Sci. Environ. Epidemiology 2019, 29, 131–147. 10.1038/s41370-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA’s Per- and Polyfluoroalkyl Substances (PFAS) Action Plan; U.S. EPA, 2019. https://www.epa.gov/sites/default/files/2019-02/documents/pfas_action_plan_021319_508compliant_1.pdf (accessed 09-15-2022).
- PFAS Strategic Roadmap: EPA’s Commitments to Action 2021–2024; U.S. EPA, 2021. https://www.epa.gov/pfas/pfas-strategic-roadmap-epas-commitments-action-2021-2024 (accessed 09-15-2022).
- OECD Global PFAS List . Toward a new comprehensive global database of per- and polyfluoroalkyl substances (PFASs); OECD, 2018. http://www.oecd.org/chemicalsafety/risk-management/global-database-of-per-and-polyfluoroalkyl-substances.xlsx (accessed 11-25-2021).
- Grulke C. M.; Williams A. J.; Thillainadarajah I.; Richard A. M. EPA’s DSSTox database: History of development of a curated chemistry resource supporting computational toxicology research. Comput. Toxicol. 2019, 12, 100096 10.1016/j.comtox.2019.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. J.; Grulke C. M.; Edwards J.; McEachran A. D.; Mansouri K.; Baker N. C.; Patlewicz G.; Shah I.; Wambaugh J. F.; Judson R. S.; Richard A. M. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J. Cheminf. 2017, 9, 61. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA PFAS OECD . PFAS: Listed in OECD Global Database; U.S. EPA, 2018. https://comptox.epa.gov/dashboard/chemical-lists/PFASOECD (accessed 08-25-2022).
- Cosgrove D. A.; Green K. M.; Leach A. G.; Poirrette A.; Winter J. A system for encoding and searching Markush structures. J. Cheminf. Modeling 2012, 52, 1936–1947. 10.1021/ci3000387. [DOI] [PubMed] [Google Scholar]
- Williams A. J.; Gaines L. G. T.; Grulke C.; Lowe C.; Sinclair G.; Samano V.; Thillainadarajah I.; Meyer B.; Patlewicz G.; Richard A. M. Assembly and curation of a list of per- and polyfluoroalkyl substances (PFAS) to support environmental science research. Frontiers Environ. Sci. 2022, 209, 1. 10.3389/fenvs.2022.850019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines L.G. T.; Sinclair G.; Williams A. A Proposed approach to defining per- and polyfluoroalkyl substances (PFAS) based on molecular structure and formula. Integ. Environ. Assess. Management 2023, 1–15. 10.1002/ieam.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Buser A.; Cousins I.; Demattio S.; Drost W.; Johansson O.; Ohno K.; Patlewicz G.; Richard A.; Walker G.; White G.; Leinala E. A New OECD Definition for Per- and Polyfluoroalkyl Substances. Environ. Sci. Technol. 2021, 55, 15575–15578. 10.1021/acs.est.1c06896. [DOI] [PubMed] [Google Scholar]
- Worth A. P.; Bassan A.; De Bruijn J.; Gallegos Saliner A.; Netzeva T.; Pavan M.; Patlewicz G.; Tsakovska I.; Eisenreich S. The role of the European Chemicals Bureau in promoting the regulatory use of (Q) SAR methods. SAR QSAR in Environ. Res. 2007, 18, 111–125. 10.1080/10629360601054255. [DOI] [PubMed] [Google Scholar]
- Patlewicz G.; Ball N.; Booth E. D.; Hulzebos E.; Zvinavashe E.; Hennes C. Use of category approaches, read-across and (Q) SAR: general considerations. Regul. Toxicol. Pharmacol. 2013, 67, 1–12. 10.1016/j.yrtph.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Buck R. C.; Franklin J.; Berger U.; Conder J. M.; Cousins I. T.; De Voogt P.; Jensen A. A.; Kannan K.; Mabury S. A.; van Leeuwen S. P. Perfluoroalkyl and polyfluoroalkyl substances in the environment: terminology, classification, and origins. Integ. Environ. Assess. Management 2011, 7, 513–541. 10.1002/ieam.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD . Reconciling Terminology of the Universe of Per- and Polyfluoroalkyl Substances: Recommendations and Practical Guidance, OECD Series on Risk Management, No. 61; OECD Publishing: Paris, 2021. https://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV/CBC/MONO(2021)25&docLanguage=En.
- OECD . Fact Cards of Major Groups of Per- and Polyfluoroalkyl Substances (PFASs), Series on Risk Management No. 68; OECD, 2022. https://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/cbc/mono(2022)1&doclanguage=en (accessed 08-24-2022).
- Sha B.; Schymanski E. L.; Ruttkies C.; Cousins I. T.; Wang Z. Exploring open cheminformatics approaches for categorizing per- and polyfluoroalkyl substances (PFASs).. Environ. Sci. Processes Impacts 2019, 21, 1835–1851. 10.1039/C9EM00321E. [DOI] [PubMed] [Google Scholar]
- Djoumbou Feunang Y.; Eisner Y. R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminf. 2016, 8, 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.; Rajan K. A database framework for rapid screening of structure-function relationships in PFAS chemistry. Nature Sci. Data 2021, 8, 1–10. 10.1038/s41597-021-00798-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap C. W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- Yang C.; Tarkhov A.; Marusczyk J.; Bienfait B.; Gasteiger J.; Kleinoeder T.; Magdziarz T.; Sacher O.; Schwab C. H.; Schwoebel J.; Terfloth L.; et al. New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J. Chem. Inf. Model. 2015, 55, 510–528. 10.1021/ci500667v. [DOI] [PubMed] [Google Scholar]
- Patlewicz G.; Helman G.; Pradeep P.; Shah I. Navigating through the minefield of read-across tools: A review of in silico tools for grouping. Comp. Toxicol. 2017, 3, 1–18. 10.1016/j.comtox.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida C.; Barton-Maclaren T.; Benfenati E.; Caloni F.; Chandrasekera P. C.; Chesne C.; Cronin M. T.; De Knecht J.; Dietrich D. R.; Escher S. E.; Fitzpatrick S. Internationalization of read-across as a validated new approach method (NAM) for regulatory toxicology. Altex 2020, 37, 579. 10.14573/altex.1912181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R.; Chandler K.; Houck K.; Hunter S.; Judson R.; Kleinstreuer N.; Knudsen T.; Martin M.; Padilla S.; Reif D.; Richard A.; Rotroff D.; Sipes N.; Dix D. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 2012, 25, 1287–302. 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Patlewicz G.; Richard A. M.; Williams A. J.; Grulke C. M.; Sams R.; Lambert J.; Noyes P. D.; DeVito M. J.; Hines R. N.; Strynar M.; Guiseppi-Elie A.; et al. A chemical category-based prioritization approach for selecting 75 per-and polyfluoroalkyl substances (PFAS) for tiered toxicity and toxicokinetic testing. Environ. Health Perspec. 2019, 127, 014501 10.1289/EHP4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereto-Massagué A.; Ojeda M. J.; Valls C.; Mulero M.; Garcia-Vallvé S.; Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods 2015, 71, 58–63. 10.1016/j.ymeth.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Durant J. L.; Leland B. A.; Henry D. R.; Nourse J. G. Reoptimization of MDL Keys for Use in Drug Discovery. Chem. Inf. Model. 2002, 42, 1273–1280. 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- Bolton E. E.; Wang Y.; Thiessen P. A.; Bryant S. H. PubChem: Integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem. 2008, 4, 217–241. 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- Morgan H. L. The generation of a unique machine description for chemical structures – a technique developed at chemical abstracts service. J. Chem. Doc. 1965, 5, 107–113. 10.1021/c160017a018. [DOI] [Google Scholar]
- A Consensus Study Report of National Academies of Sciences, Engineering, and Medicine. A Class Approach to Hazard Assessment of Organohalogen Flame Retardants 2019, 1. 10.17226/25412. [DOI] [PubMed] [Google Scholar]
- Patlewicz G.; Richard A.; Williams A.; Judson R.; Thomas R. Towards reproducible structure-based chemical categories for PFAS to inform and evaluate toxicity and toxicokinetic testing. Comput. Toxicol. 2022, 24, 100250 10.1016/j.comtox.2022.100250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C. N.; Williams A. J. Enabling high-throughput searches for multiple chemical data using the US-EPA CompTox chemicals dashboard. J. Chem. inf. Model. 2021, 61, 565–570. 10.1021/acs.jcim.0c01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzaro A.; Zaggia A.; Fant M.; Conte L.; Falletti L. Identification and quantification of linear and branched isomers of perfluorooctanoic and perfluorooctane sulfonic acids in contaminated groundwater in the veneto region. J. Chromatogr. A 2018, 1533, 143–154. 10.1016/j.chroma.2017.12.036. [DOI] [PubMed] [Google Scholar]
- Liu H. S.; Wen L. L.; Chu P. L.; Lin C. Y. Association among total serum isomers of perfluorinated chemicals, glucose homeostasis, lipid profiles, serum protein and metabolic syndrome in adults: NHANES, 2013–2014. Environ. Pollut. 2018, 232, 73–79. 10.1016/j.envpol.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Liang Y.; Gao K.; Wang Y.; Wang C.; Fu J.; Wang Y.; Jiang G.; Jiang Y. Levels, spatial distribution and isomer profiles of perfluoroalkyl acids in soil, groundwater and tap water around a manufactory in China. Chemosphere 2019, 227, 305–314. 10.1016/j.chemosphere.2019.04.027. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zeng X. W.; Bloom M. S.; Qian Z.; Hinyard L. J.; Belue R.; Lin S.; Wang S. Q.; Tian Y. P.; Yang M.; Chu C.; et al. Renal function and isomers of perfluorooctanoate (PFOA) and perfluorooctanesulfonate (PFOS): Isomers of C8 Health Project in China. Chemosphere 2019, 218, 1042–1049. 10.1016/j.chemosphere.2018.11.191. [DOI] [PubMed] [Google Scholar]
- Richard A. M.; Hidle H.; Patlewicz G.; Williams A. J. Identification of Branched and Linear Forms of PFOA and Potential Precursors: A User-Friendly SMILES Structure-based Approach. Frontiers in Environ. Sci. 2022, 10, 271. 10.3389/fenvs.2022.865488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt C. M.; Muir D. C.; Mabury S. A. Biotransformation pathways of fluorotelomer-based polyfluoroalkyl substances: A review. Environ. Toxicol. Chem. 2014, 33, 243–267. 10.1002/etc.2407. [DOI] [PubMed] [Google Scholar]
- Field J. A.; Seow J. Properties, occurrence, and fate of fluorotelomer sulfonates. Critical Reviews in Environ. Sci. Technol. 2017, 47, 643–691. 10.1080/10643389.2017.1326276. [DOI] [Google Scholar]
- McDonough C. A.; Choyke S.; Barton K. E.; Mass S.; Starling A. P.; Adgate J. L.; Higgins C. P. Unsaturated PFOS and other PFASs in human serum and drinking water from an AFFF-impacted community. Environ. Sci. Technol. 2021, 55, 8139–8148. 10.1021/acs.est.1c00522. [DOI] [PubMed] [Google Scholar]
- Washington J. W.; Jenkins T. M. Abiotic hydrolysis of fluorotelomer polymers as a source of perfluorocarboxylates at the global scale. Environ. Sci. Technol. 2015, 49, 14129–14135. 10.1021/acs.est.5b03686. [DOI] [PubMed] [Google Scholar]
- Lowe D. M.; Corbett P. T.; Murray-Rust P.; Glen R. C. Chemical name to structure: OPSIN, an open source solution. J. Chem. Inf. Model. 2011, 51, 739–753. 10.1021/ci100384d. [DOI] [PubMed] [Google Scholar]
- National PFAS Testing Strategy; U.S. EPA, 2021. https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/national-pfas-testing-strategy; https://www.epa.gov/system/files/documents/2021-10/pfas-natl-test-strategy.pdf (accessed 11-07-2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- OECD Global PFAS List . Toward a new comprehensive global database of per- and polyfluoroalkyl substances (PFASs); OECD, 2018. http://www.oecd.org/chemicalsafety/risk-management/global-database-of-per-and-polyfluoroalkyl-substances.xlsx (accessed 11-25-2021).