Abstract
Background
To learn about the molecular etiology of strabismus, we are studying the genetic basis of 'congenital fibrosis of the extraocular muscles' (CFEOM). These syndromes are characterized by congenital restrictive ophthalmoplegia affecting muscles in the oculomotor and trochlear nerve distribution. Individuals with the classic form of CFEOM are born with bilateral ptosis and infraducted globes. When all affected members of a family have classic CFEOM, we classify the family as a CFEOM1 pedigree. We have previously determined that a CFEOM1 gene maps to the FEOM1 locus on chromosome 12cen. We now identify additional pedigrees with CFEOM1 to determine if the disorder is genetically heterogeneous and, if so, if any affected members of CFEOM1 pedigrees or sporadic cases of classic CFEOM harbor mutations in ARIX, the CFEOM2 disease gene.
Results
Eleven new CFEOM1 pedigrees were identified. All demonstrated autosomal dominant inheritance, and nine were consistent with linkage to FEOM1. Two small CFEOM1 families were not linked to FEOM1, and both were consistent with linkage to FEOM3. We screened two CFEOM1 families consistent with linkage to FEOM2 and 5 sporadic individuals with classic CFEOM and did not detect ARIX mutations.
Conclusions
The phenotype of two small CFEOM1 families does not map to FEOM1, establishing genetic heterogeneity for this disorder. These two families may harbor mutations in the FEOM3 gene, as their phenotype is consistent with linkage to this locus. Thus far, we have not identified ARIX mutations in any affected members of CFEOM1 pedigrees or in any sporadic cases of classic CFEOM.
Background
Congenital fibrosis of the extraocular muscles (CFEOM) and Duane syndrome (DS) are complex strabismus disorders that present with congenital restrictive ophthalmoplegia with or without ptosis. These disorders were traditionally believed to reflect primary structural extraocular muscle (EOM) anomalies and have been referred to as 'congenital fibrosis syndromes' [1]. Neuropathology studies of DS [2,3] and one form of CFEOM (CFEOM1) [4], and the identification of ARIX as the gene mutated in a second form of CFEOM (CFEOM2) [5], however, support our hypothesis that CFEOM results from maldevelopment of the oculomotor (nIII) and/or trochlear (nIV) nuclei and DS results from maldevelopment of the abducens (nVI) nucleus. The continued definition of these phenotypes and identification of the underlying disease genes will assist clinical diagnostics and lead to a better understanding of the unique developmental features of the oculomotor lower motor neuron unit.
Although several distinct CFEOM phenotypes have been defined [6-8], each likely resulting from maldevelopment of a unique combination of alpha motor neurons in nIII and/or nIV, most reports of CFEOM families describe a stereotypical clinical phenotype. The affected members of these pedigrees are born with bilateral ptosis and restrictive ophthalmoplegia. The primary vertical position of each eye is downward and cannot be elevated above the midline. On forced duction testing there is resistance to passive movement of the globe. Although the primary position of both eyes is infraducted, there is variability in the secondary position of each eye (i.e. exotropic, esotropic, or neutral), and the degree of residual horizontal movement within the lower quadrants (full to completely restricted). This CFEOM phenotype was first described in the medical literature in 1840 [9] and was recognized to occur as a familial trait in 1879 [10]. Subsequently, families segregating this phenotype have been published under myriad names [1,4,6,11-21]. We now refer to individuals with this phenotype as having "classic CFEOM" and to families in which all affected members have this phenotype as " CFEOM1 pedigrees". We previously mapped a CFEOM1 locus, referred to as FEOM1, to a ≤ 3 cM region spanning the centromere of chromosome 12, flanked by D12S1584 (AFM136xf6) on the p-arm and D12S1668 (AFMb320wd9) on the q-arm [6,15].
In addition to families with CFEOM1, we have identified several less common familial CFEOM phenotypes (CFEOM2 and CFEOM3). These phenotypes are classified as CFEOM based on the presence of affected members with congenital restrictive ophthalmoplegia affecting extraocular muscles in the nIII/nIV distribution. By definition, however, one or more affected family members do not have the classic CFEOM phenotype. In families with CFEOM2, the eyes of affected family members are fixed in an exotropic, or outward, position. Thus far, this phenotype segregates as an autosomal recessive trait and maps to the FEOM2 locus on chromosome 11q13 [8], and affected individuals carry homozygous mutations in ARIX[5]. ARIX encodes a homeodomain transcription factor required for nIII and nIV development in mice and zebrafish [22,23]. In families with CFEOM3, the CFEOM phenotype is variably expressed. Some affected members have classic CFEOM. Others, however, are unilaterally affected, the primary position of the eye is orthotropic rather than infraducted, and/or the eye can be raised into the upper quadrants. Thus far, this phenotype segregates as an autosomal dominant trait and maps to either FEOM3 on 16qter [7] or to FEOM1[24].
In our attempt to understand phenotype-genotype correlations between specific CFEOM phenotypes and FEOM loci, we noted that the CFEOM1 phenotype in all pedigrees reported to date maps to the FEOM1 locus. To determine if CFEOM1 is indeed genetically homogeneous, we identified all unpublished CFEOM1 pedigrees in our database, analyzed them for linkage to the FEOM loci, and found that most but not all were consistent with linkage to FEOM1. The two small pedigrees not linked to FEOM1 were consistent with linkage to FEOM3. In addition, to further define the spectrum of human ARIX mutations, we identified all CFEOM1 families consistent with linkage to FEOM2 or sporadic individuals with classic CFEOM and determined that none harbored mutations in the ARIX gene.
Results
From our database, 33 pedigrees were of sufficient size and had sufficient clinical data and 14 sporadic individuals had sufficient clinical data to qualify for the study. Of these, 20 pedigrees met CFEOM1 and 5 sporadic individuals met classic CFEOM inclusion criteria. Although not an inclusion criterion for the study, the CFEOM1 phenotype in all 20 families was inherited as an autosomal dominant trait with full penetrance. The phenotype in 9 of the 20 pedigrees was previously demonstrated to map to the FEOM1 locus [6,15,25,26] (Table 1). Therefore, the remaining 11 families were included in this study (Figs. 1 &2, Table 1, and see additional files 1–3 [Additional File 1, Additional File 2, Additional File 3].
Table 1.
Summary of the genetic analysis of CFEOM1 pedigrees
| Pedigree | CFEOM Phenotype | Inheritance | Forced Ductions | Cytogenetic analysis | FEOM1 | FEOM3 | FEOM2 | ARIX mutations | Publication |
| A | CFEOM1 | AD | + | normal | LINKED | refs. [6,15] | |||
| B | CFEOM1 | AD | + | normal | LINKED | refs. [6,15] | |||
| C | CFEOM1 | AD | + | not done | LINKED | ref. [15] | |||
| H | CFEOM1 | AD | + | normal | LINKED | ref. [15] | |||
| AA | CFEOM1 | AD | + | normal | LINKED | ref. [15] | |||
| AC | CFEOM1 | AD | + | normal | LINKED | ref. [15] | |||
| AD | CFEOM1 | AD | + | normal | LINKED | ref. [15] | |||
| CD | CFEOM1 | AD | + | not done | LINKED | ref. [25] | |||
| CB | CFEOM1 | AD | not done | not done | c/w linkage | nc/w linkage | NOT LINKED | ref. [26] | |
| BJ | CFEOM1 | AD | + | normal | LINKED | current | |||
| CZ | CFEOM1 | AD | + | normal | LINKED | current | |||
| AG | CFEOM1 | AD | + | normal | c/w linkage | NOT LINKED | NOT LINKED | current | |
| AJ | CFEOM1 | AD | + | not done | c/w linkage | nc/w linkage | nc/w linkage | current | |
| AH | CFEOM1 | AD | + | normal | c/w linkage | nc/w linkage | nc/w linkage | current | |
| T | CFEOM1 | AD | + | not done | c/w linkage | nc/w linkage | nc/w linkage | current | |
| CT | CFEOM1 | AD | + | normal | c/w linkage | nc/w linkage | nc/w linkage | current | |
| BC | CFEOM1 | AD | + | not done | c/w linkage | nc/w linkage | (c/w linkage) | none | current |
| E | CFEOM1 | AD | + | normal | c/w linkage | c/w linkage | NOT LINKED | current | |
| K | CFEOM1 | AD | + | normal | nc/w linkage | c/w linkage | (c/w linkage) | none | current |
| BT | CFEOM1 | AD | + | normal | NOT LINKED | c/w linkage | nc/w linkage | current |
AD = autosomal dominant; + = positive forced duction testing for restriction; c/w linkage = consistent with linkage; nc/w li nkage = not consistent with linkage.
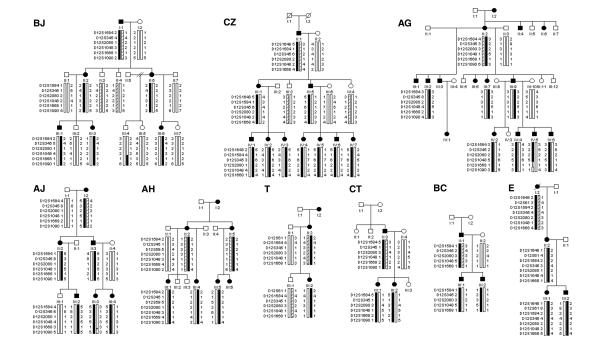
Figure 1.
Haplotype analysis of pedigrees BJ, CZ, AG, AJ, AH, T, CT, BC, and E at the FEOM1 locus. Black symbols denote those individuals who are clinically affected with classic CFEOM. Genotyping data and schematic segregating haplotype bars for chromosome 12cen markers are shown below the symbol for each study participant. Allele sizes here and in figure 2 were assigned as linkage studies were performed are not equivalent when compared between families. Black bars denote the potential disease-associated region. Diagonally hatched or white bars highlight the inheritance of the non-disease-associated haplotypes. References to specific individuals within the text refer to the generation number (Roman numeral) and position within generation (Arabic numeral). In all 9 pedigrees each family's disease-associated haplotype is inherited by all CFEOM1 individuals and by no asymptomatic individuals.
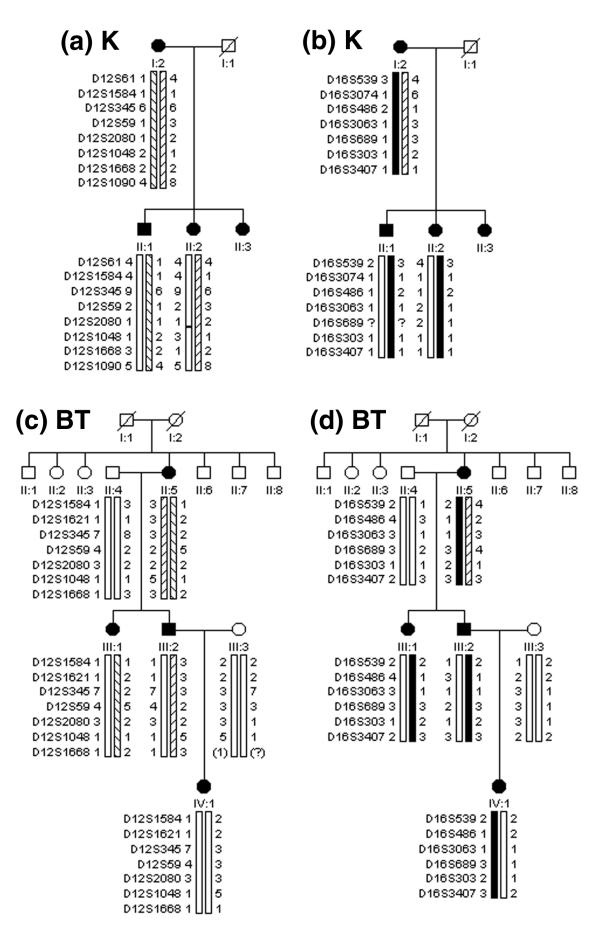
Figure 2.
Haplotype analysis for pedigrees K at the (a)FEOM1 and (b)FEOM3 loci and BT at the (c)FEOM1 and (d)FEOM3 loci. Symbols are defined in the legend to figure 1. In each family the CFEOM1 phenotype is co-inherited with FEOM3 markers and not with FEOM1 markers.
The 11 families are ethnically diverse, not consanguineous and, to the best of our knowledge, unrelated. Eight of the families reside in the US and are of mixed European ancestry (BJ, AG, AJ, AH, CT, E, K, BT), while families CZ, T, and BC are of Italian, Irish, and Japanese ancestry, respectively. Family history of CFEOM in several previous generations was documented in two pedigrees (AG, T), and a previous family history was recounted but could not be corroborated in five others (BJ, AJ, AH, E, K). In contrast, in pedigrees CZ, BC, CT and BT, neither the parents nor more distant relatives of the eldest affected family member were reportedly affected. Examination of individuals I–1 and I–2 of pedigrees BC and CT confirmed their unaffected status. Individuals I–1 and I–2 in pedigrees CZ and BT were deceased. These data suggest that the CFEOM1 mutation rate is not negligible. Cytogenetic analyses, when performed, did not reveal abnormalities (Table 1).
Nine of the 11 CFEOM1 pedigrees contain too few participants to establish linkage to a specific locus. Haplotype analysis of these families using multiple markers that span the critical region of a given locus can, however, eliminate linkage to the locus, determine genetic heterogeneity, and guide future mutation analyses. If the phenotype results from a mutation at a given locus, haplotype analysis at that locus will be consistent with linkage. If the phenotype does not result from a mutation at a given locus, however, haplotype data from a small family may be consistent or inconsistent with linkage. Thus, haplotype data in a small family that is consistent with linkage can result either from a disease mutation at that locus or by chance. Haplotype data in a small family that is inconsistent with linkage strongly suggests that the family's phenotype is not linked to the locus.
Linkage to FEOM1
Genetic analyses of the two largest families (BJ, CZ) established linkage of their phenotype to the FEOM1 locus (Fig. 1, Table 1 & see Additional File 1). Maximum lod scores of 3.01 were obtained at a theta value of zero for the fully informative markers D12S59 and D12S1048 in family BJ, and the fully informative markers D12S1648, D12S345, and D12S59 in family CZ.
Linkage to FEOM1 was ruled out in family BT (see Additional file 1). A lod score of -2 was obtained at a theta value of 0.04 for the fully informative markers D12S1621, D12S59 and D12S1668. The FEOM1 critical region is ≤ 3 cM, and a theta of 0.04 corresponds to a genetic distance of approximately 4 cM, thus eliminating linkage of this family's CFEOM disease gene to the entire FEOM1 critical region. Exclusion of linkage to the FEOM1 locus is further supported by haplotype analysis of this family (Fig. 2c). The affected sister and brother inherit different FEOM1 haplotypes from their affected mother, and the brother's affected daughter inherits her unaffected paternal grandfather's FEOM1 haplotype, thus proving non-association between the FEOM1 haplotype and the disease phenotype.
The eight remaining CFEOM pedigrees were too small to produce statistically significant lod scores; however, seven of the eight families displayed haplotype and linkage data consistent with linkage to the FEOM1 locus (Fig. 1, see Additional File 1). Genetic and haplotype analysis of all nine families consistent with linkage to the FEOM1 locus did not reveal any recombination events within the previously defined FEOM1 critical region.
The smallest family, K, revealed haplotype and linkage data that was inconsistent with linkage to the FEOM1 locus (Fig. 2a, see Additional File 1). A lod score of -2 was obtained at a theta value of 0.002 for the only two informative markers, D12S59 and D12S1090. These theta values eliminate linkage to only 0.8 cM of the < 3 cM FEOM1 critical region and, therefore, this locus cannot be formally ruled out. Nevertheless, the minimum number of recombination events in this family occurs only if the affected son and daughter inherit different FEOM1 haplotypes from their affected mother, thus strongly suggesting that the disease gene in this family does not map to the chromosome 12 locus.
Genetic heterogeneity was tested taking into account the eleven new families. Admixture analysis of the two-point data with the HOMOG program showed evidence for linkage to FEOM1 with heterogeneity for both markers tested (D12S345 and D12S1048). Chi-squares of 34.601 and 40.377 were obtained for D12S345 and D12S1048 respectively which resulted in significant likelihood ratios of 3.26 × 107 and 5.86 × 108. Alpha (the proportion of linked families) was 0.90.
Linkage to FEOM3
All families except BJ and CZ were analyzed for linkage to the 5.6 cM FEOM3 locus flanked by D16S486 and 16qter. Of the seven small families consistent with linkage to FEOM1, only the largest (AG) can be definitively excluded from linkage to FEOM3 (see Additional File 2). Five of the remaining families (AJ, AH, T, BC, CT) showed haplotype data inconsistent with linkage to FEOM3, but the theta values obtained at lod scores of -2 were insufficient to rule out the entire FEOM3 critical region.
The two families whose phenotype did not map to FEOM1 (K, BT) had haplotype and linkage data consistent with linkage to FEOM3 (Fig. 2b &2d, see Additional File 2). In addition, one of the small families consistent with linkage to the FEOM1 locus (E) had haplotype and linkage data that was also consistent with linkage to the FEOM3 locus (see Additional File 2).
Linkage to FEOM2
All families whose phenotype was not linked to FEOM1 or FEOM3 were tested for linkage to FEOM2. Assuming autosomal dominant inheritance with complete penetrance, two families (AG, E) are not linked and five families are inconsistent with linkage (AJ, AH, T, CT, BT) to the FEOM2 locus (see Additional File 3). Family BC is consistent with linkage to both the FEOM2 and FEOM1 loci (maximum lod score 0.3 at both loci), and family K is consistent with linkage to both the FEOM2 and FEOM3 loci (maximum lod score 0.3 at both loci).
ARIX mutation analysis
Genomic DNA samples from affected member of pedigree BC and K and 5 sporadic individuals with classic CFEOM were used as templates to sequence the three ARIX exons and flanking introns. No mutations were identified.
Discussion
We have established clinical criteria for classic CFEOM and CFEOM1, and have identified 5 sporadic individuals with classic CFEOM and 20 pedigrees with CFEOM1. Of these 20 pedigrees, 18 are linked, or consistent with linkage, to the FEOM1 locus. Two small CFEOM1 pedigrees are not consistent with linkage, however, establishing that CFEOM1 is genetically heterogeneous.
Eleven of the 20 pedigrees are large enough to establish linkage to a specific locus; we previously reported that the CFEOM1 phenotype in 9 of these pedigrees maps to the FEOM1 locus [6,15,25,26] and we now demonstrate that the remaining two also map to FEOM1. Our analysis of the remaining 9 CFEOM1 pedigrees demonstrates that 6 most likely result from mutations in the FEOM1 gene. Five of these 6 are consistent with linkage to FEOM1 and are either not linked or not consistent with linkage to FEOM2 and FEOM3. One is consistent with linkage to FEOM1 and not FEOM3 and, although consistent with linkage to FEOM2, ARIX mutations were not identified. Therefore, although not proved, the CFEOM1 phenotype in these 6 families seems likely to result from mutations in the FEOM1 gene. The phenotype of a seventh family, E, is consistent with linkage to both FEOM1 and FEOM3 and will be screened for mutations at both these loci.
In contrast to the 18 pedigrees whose CFEOM1 phenotype is consistent with linkage to FEOM1, pedigree BT is not linked to FEOM1 and pedigree K is inconsistent with linkage to this locus. It is notable that haplotype analysis of both these small CFEOM1 families demonstrates co-inheritance with the FEOM3 locus. In the reported family whose autosomal dominant CFEOM3 phenotype maps to the FEOM3 locus, 9 of the 17 affected members had classic CFEOM. Our current data now suggests that, at least in small pedigrees, CFEOM1 can also map to the FEOM3 locus. It will require the identification of additional large CFEOM1 families to determine if they too can map to this locus.
ARIX, which encodes a transcription factor critical to nIII and nIV development in mice and zebrafish [22,23], was recently identified as the FEOM2 gene mutated in affected members of CFEOM2 families [5]. It was unknown, however, if classic CFEOM may also result from mutations in this gene. We now find that we are unable to identify ARIX mutations underlying classic CFEOM in either sporadic cases or in individuals from CFEOM1 families.
This finding is consistent with our prediction that, compared to ARIX, the genes mutated in CFEOM1 may have a more restricted function in the development of nIII and that their expression may actually be regulated by ARIX. This prediction is based on the CFEOM1 phenotype and on the neuropathological study of an affected member of a CFEOM1 pedigree whose disease gene maps to the FEOM1 locus. This study revealed absence of the superior division of the oculomotor nerve and the corresponding central caudal and medial nIII subnuclei, and marked abnormalities of the levator palpebrae superioris and superior rectus muscles [4]. These findings suggest that while ARIX is necessary for both nIII and nIV development, the CFEOM1 genes may be necessary for the development of only these two specific nIII subnuclei.
Conclusions
The genetic analysis of the 11 CFEOM1 pedigrees in this report demonstrates that this disorder is genetically heterogeneous. While the phenotype of all large CFEOM1 pedigrees analyzed thus far map to the FEOM1 locus, smaller CFEOM1 pedigrees may harbor mutations in the FEOM3 gene. The CFEOM1 phenotype does not, however, appear to result from mutations in ARIX. The CFEOM1 families identified in this study contribute critical alleles toward the identification of the mutated FEOM1 and FEOM3 genes. Once identified, we anticipate that the study of the function of these genes will contribute to our understanding of midbrain motor neuron development.
Materials and Methods
Subjects
We established inclusion criteria for this study as follows. First, we established criteria for a "classic CFEOM individual" as an individual with congenital nonprogressive bilateral ophthalmoplegia and ptosis, an infraducted primary position of each eye with the inability to raise either eye above the midline, and forced duction testing positive for restriction, if testing was performed. We then established criteria for a "CFEOM1 pedigree" as a family in which every affected member met the criteria for "classic CFEOM". Second, we reviewed all participants enrolled in our ongoing CFEOM study and determined which individuals and which pedigrees met these two criteria. For a pedigree to be considered, we required clinical examination records and/or photographs/videos of the primary positions of gaze for all affected study participants. Third, from these CFEOM1 pedigrees we identified those with a family structure sufficient for linkage analysis. Pedigrees were required to have affected study participants in three or more generations, or affected study participants in two generations with at least two participating offspring of affected individuals, or two or more affected study participants within one generation. In this way we did not assume a mode of inheritance. Lastly, sporadic individuals with classic CFEOM were screened for ARIX mutations. The study was approved by the Children's Hospital institutional review board, and all study participants signed informed consent forms. Our methods adhered to the Declaration of Helsinki for research involving human subjects.
Molecular studies
Blood samples were obtained from all participating family members, and lymphocyte DNA was extracted using the Puregene kit (Gentra, Research Triangle Park, NC). Chromosome analyses of GTG banded metaphase cells at a 400 band level minimum resolution were performed on one or more affected family members of each family whenever possible to rule out cytogenetic abnormalities. All families were analyzed for linkage to the FEOM1 locus. Families not linked to the FEOM1 locus were also analyzed for linkage to the FEOM2 and FEOM3 loci. Linkage studies were conducted using three or more locus specific polymorphic DNA micro satellite markers for each family. The FEOM1 markers included D12S1648, D12S61, D12S1584, D12S1621, D12S345, D12S59, D12S2080, D12S1048, D12S1668, and D12S1090[6,15]. The FEOM2 markers included D11S1337, D11S4162, D11S4196, D11S1314, and D11S1369[8]. The FEOM3 markers included D16S539, D16S3077, D16S498, D16S486, D16S476, D16S3063, D16S689, D16S2621, D16S303, and D16S3407[7]. The primer sequences for these polymorphisms are available from the Genome Database http://gdbwww.gdb.org. Unlabeled primers were purchased from Genosys Biotechnologies, Inc. http://www.genosys.com. Fluorescently labeled primers were purchased from Research Genetics, Inc. http://www.researchgenetics.com. Radioactive products were made by 30 cycles of PCR amplification of 10-μl reaction volumes containing 10–30 ng of genomic DNA, 40 ng of each primer, 200 μM each of dATP, dTTP, dGTP, and dCTP, 1 μCi α-32P-dCTP (3,000 Ci mmol-1) and 0.5 U Taq polymerase (Perkin Elmer). The radioactively labeled PCR products were separated on 6% denaturing polyacrylamide sequencing gels, and the alleles were visualized by autoradiography [6]. For the fluorescently labeled products, α-32P-dCTP was omitted, fluorescent primers were used, and the products were analyzed in an ABI PrismTM 377 DNA Sequencer (Perkin Elmer) following the manufacturer's specifications.
Lod score calculations
An individual was scored as affected based on clinical examination records and/or photographs of primary position of gaze. Lod scores were calculated using the Fastlink version 3.0 package of programs [27], assuming autosomal dominant inheritance with complete penetrance, and a disease incidence of 1 in 1,000,000 births, as described previously [7,8]. Because of the absence of specific allele frequencies for each of the ethnic groups represented in the study, we assumed ten marker alleles of equal frequency. To assess nonallelic heterogeneity linkage data from two FEOM1 markers spanning the FEOM1 critical region (D12S345 and D12S1048) were analyzed using a HOMOG version 3.35 program [28].
ARIX mutation detection
ARIX was PCR amplified from genomic DNA using five primer sets and sequenced on an automated ABI 377 DNA seqeuncer (PE-Applied Biosystems) as described previously [5].
Supplementary Material
Table 2: Two-point linkage data between CFEOM1 and the FEOM1 locus
Table 3: Two-point linkage data between CFEOM1 and the FEOM3 locus
Table 4: Two-point linkage data between CFEOM1 and the FEOM2 locus
Acknowledgments
Acknowledgements
This work was supported by National Eye Institute EY12498 and EY13583 and by the Children's Hospital Mental Retardation Research Center (P30 HD18655).
Contributor Information
Elizabeth C Engle, Email: engle@enders.tch.harvard.edu.
Nathalie McIntosh, Email: mcintosh@brandeis.edu.
Koki Yamada, Email: kamada@enders.tch.harvard.edu.
Bjorn A Lee, Email: bjornl@mail.vet.upenn.edu.
Roger Johnson, Email: rhjohnson@aol.com.
Michael O'Keefe, Email: mokeefe@materprivate.ie.
Robert Letson, Email: letso001@tc.umn.edu.
Arnold London, Email: londo005@tc.umn.edu.
Evan Ballard, Email: eaballard@associatedeyecare.com.
Mark Ruttum, Email: mruttum@mcw.edu.
Naomichi Matsumoto, Email: naomat@net.nagasaki-u.ac.jp.
Nakamichi Saito, Email: nksaito@med.kyushu-u.ac.jp.
Mary Louise Z Collins, Email: mcollins@gbmc.org.
Lisa Morris, Email: lisamor@umich.edu.
Monte Del Monte, Email: madm@umich.edu.
Adriano Magli, Email: magli@cds.unina.it.
Teresa de Berardinis, Email: debera@libero.com.
References
- Brown HW. Congenital structural muscle anomalies. In: JH Allen, editor. Strabismus Ophthalmic Symposium. St. Louis: C. V. Mosby Co.; 1950. pp. 205–236. [Google Scholar]
- Hotchkiss MG, Miller NR, Clark AW, Green WG. Bilateral Duane's retraction syndrome: A clinical-pathological case report. Arch Ophth. 1980;98:870–874. doi: 10.1001/archopht.1980.01020030864013. [DOI] [PubMed] [Google Scholar]
- Miller NR, Kiel SM, Green WR, Clark AW. Unilateral Duane's retraction syndrome (type 1). Arch Ophth. 1982;100:1468–1472. doi: 10.1001/archopht.1982.01030040446016. [DOI] [PubMed] [Google Scholar]
- Engle EC, Goumernov B, McKeown CA, Schatz M, Johns DR, Porter JD, Beggs AH. Oculomotor nerve and muscle abnormalities in congenital fibrosis of the extraocular muscles. Annals of Neurology. 1997;41:314–325. doi: 10.1002/ana.410410306. [DOI] [PubMed] [Google Scholar]
- Nakano M, Yamada K, Fain J, Sener EC, Selleck CJ, Awad AH, Zwaan J, Mullaney PB, Bosley TM, Engle EC. Homozygous mutations in ARIX(PHOX2A) result in congenital fibrosis of the extraocular muscles type 2. Nat Genet. 2001;29:315–320. doi: 10.1038/ng744. [DOI] [PubMed] [Google Scholar]
- Engle EC, Kunkel LM, Specht LA, Beggs AH. Mapping a gene for congenital fibrosis of the extraocular muscles to the centromeric region of chromosome 12. Nature Genetics. 1994;7:69–73. doi: 10.1038/ng0594-69. [DOI] [PubMed] [Google Scholar]
- Doherty E, Macy M, Wang S, Dykeman C, Melanson M, Engle E. CFEOM3: a new extraocular congenital fibrosis syndrome that maps to 16q24.2-q24.3. Investigative Ophthalmology and Visual Science. 1999;40:1687–1694. [PubMed] [Google Scholar]
- Wang S, Zwaan J, Mullaney P, Jabak M, Al-Awad A, Beggs A, Engle E. Congenital fibrosis of the extraocular muscles type 2 (CFEOM2), an inherited exotropic strabismus fixus, maps to distal 11q13. American Journal of Human Genetics. 1998;63:517–525. doi: 10.1086/301980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten M. Erfahrungen uber den strabismus und die Muskeldurchschneidung am Auge in physiologischpathologischer und therapeutischer Beziehung. Monatsschr Med Augenheilkd Chir. 1840;3:474–499. [Google Scholar]
- Heuck G. Ueber angeborenen vererbten Beweglichkeits – Defect der Augen. Klin Monatsbl Augenheilkd. 1879;17:253–278. [Google Scholar]
- Black G, Perveen R, Hatchwell E, Reck A, Clayton-Smith J. Locus heterogeneity in autosomal dominant congenital external ophthalmoplegia. Journal of Medical Genetics. 1998;35:985–988. doi: 10.1136/jmg.35.12.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradburn AA. Hereditary ophthalmoplegia in five generations. Trans Ophthalmol Soc. 1912;32:142–153. [Google Scholar]
- Catford GV. A familial musculo-fascial anomaly. Trans Ophthal Soc UK. 1966;86:19–36. [PubMed] [Google Scholar]
- Cibis GW. Congenital familial external ophthalmoplegia with co-contraction. Ophthalmic Paediatrics and Genetics. 1984;4:163–168. doi: 10.3109/13816818409006116. [DOI] [PubMed] [Google Scholar]
- Engle EC, Marondel I, Houtman WA, de Vries B, Lowenstein A, Lazar M, Ward DC, Kucherlapati R, Beggs AH. Congenital fibrosis of the extraocular muscles (autosomal dominant congenital external ophthalmoplegia): genetic homogeneity, linkage refinement, and physical mapping on chromosome 12. American Journal of Human Genetics. 1995;57:1086–1094. [PMC free article] [PubMed] [Google Scholar]
- Harley RD, Rodrigues MM, Crawford JS. Congenital fibrosis of the extraocular muscles. Journal of Pediatric Ophthalmology and Strabismus. 1978;15:346–358. doi: 10.3928/0191-3913-19781101-04. [DOI] [PubMed] [Google Scholar]
- Houtman WA, vanWeerden TW, Robinson PH, deVries B, Hoogenraad TU. Hereditary congenital external ophthalmoplegia. Ophthalmologica. 1986;193:207–218. doi: 10.1159/000309712. [DOI] [PubMed] [Google Scholar]
- Laughlin RC. Congenital fibrosis of the extraocular muscles; a report of six cases. Am J Ophthalmology. 1956;41:432–438. doi: 10.1016/0002-9394(56)91259-4. [DOI] [PubMed] [Google Scholar]
- Lees F. Congenital static familial ophthalmoplegia. J Neurol Neurosurg Psychiatry. 1960;23:46–51. doi: 10.1136/jnnp.23.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA. Mendelian inheritance in man: catalogs of autosomal dominant, autosomal recessive, and X-linked phenotypes, ninth. Baltimore: The Johns Hopkins University Press;; 1990. [Google Scholar]
- Traboulsi E, Jaafar M, Kattan H, Parks M. Congenital fibrosis of the extraocular muscles: Report of 24 cases illustrating the clinical spectrum and surgical management. American Orthoptic Journal. 1993;43:45–53. [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Sener EC, Lee BA, Turgut B, Akarsu AN, Engle EC. New clinical variation of a fibrosis syndrome in a Turkish family maps to the CFEOM1 locus on chromosome 12. Arch Ophth. 2000;118:1090–1097. doi: 10.1001/archopht.118.8.1090. [DOI] [PubMed] [Google Scholar]
- Flaherty MP, Grattan-Smith P, Steinberg A, Jamieson R, Engle EC. Congenital fibrosis of the extraocular muscles associated with cortical dysplasia and maldevelopment of the basal ganglia. Ophthalmology. 2001;108:1313–1322. doi: 10.1016/S0161-6420(01)00582-6. [DOI] [PubMed] [Google Scholar]
- Hart B, Carlow T, Engle E. Congenital fibrosis of the extraocular muscles type 1: MRI and clinical findings. Proceedings of the American Society of Neuroradiology, 38th Annual Meeting; Atlanta. 2000. p. 317.
- Cottingham RW, Jr, Idury RM, Schäffer AA. Faster Sequential Genetic Linkage Computations. American Journal of Human Genetics. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- Ott J. Analysis of Human Genetic Linkage, Rev ed edn. Baltimore: The Johns Hopkins University Press;; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 2: Two-point linkage data between CFEOM1 and the FEOM1 locus
Table 3: Two-point linkage data between CFEOM1 and the FEOM3 locus
Table 4: Two-point linkage data between CFEOM1 and the FEOM2 locus