Abstract
Background:
Indoor environments contain a broad diversity of non-pathogenic Basidiomycota yeasts, but their role in exacerbating adverse health effects has remained unclear.
Objective:
To understand the role of Vishniacozyma victoriae exposure and its impact on human health.
Methods:
A qPCR assay was developed to detect and quantify an abundant indoor yeast species, Vishniacozyma victoriae (syn. Cryptococcus victoriae), from homes participating in the New York City Neighborhood Asthma and Allergy Study (NAAS). We evaluated the associations between V. victoriae, housing characteristics, and asthma relevant health endpoints.
Results:
V. victoriae was quantified in 236 of the 256 bedroom floor dust samples ranging from less than 300 to 45,918 cell equivalents/mg of dust. Higher concentrations of V. victoriae were significantly associated with carpeted bedroom floors (P=0.044), mean specific humidity (P=0.004), winter (P<0.0001) and spring (P=0.001) seasons, and the presence of dog (P=0.010) and dog allergen Can f 1 (P=0.027). V. victoriae concentrations were lower in homes of children with asthma vs. without asthma (P=0.027), an association observed only among the non-seroatopic children.
Significance:
These findings add to the inverse associations observed between microbial exposures in homes and asthma outcomes in children, specifically the contribution of non-pathogenic Basidiomycota yeasts.
Keywords: Allergy, Asthma, Cryptococcus, ITS, qPCR, Yeast
II. INTRODUCTION
Microbial bioaerosols contribute to personal exposure within the built environment and have been associated with adverse health effects such as respiratory morbidities and allergic sensitization (1, 2). However, microbial exposures early in life may confer protective health effects such as decreased asthma symptoms (3), highlighting the dichotomy of microbial exposures and their role in health. Recently, contemporary molecular methods have resolved a much broader assemblage of endogenous and exogenous microbial sources that contribute to personal exposure (4). In addition to bacteria, fungi are ubiquitous and are commonly encountered within the built environment (5).
Traditional methods to detect and identify fungi have been primarily restricted to viable culture and light microscopy. Culture-based microbial assessments have identified yeasts as the most prevalent fungi in some indoor environments (4, 6). These methodological approaches have limited the identification of potential sources of fungal exposure such as unicellular yeast species that are unable to be identified (7, 8). Contemporary sequencing methods developed over the last decade have enabled the identification of fungi via the Internal Transcribed Spacer (ITS) region of DNA, known as the fungal barcode (9). This molecular approach has provided much needed insight to the complete spectrum of fungal assemblages in indoor air and dust samples (1, 10, 11). Yeast species were previously overlooked or unable to be identified in indoor environments (12–14), especially the presence of yeasts placed in the phylum Basidiomycota.
Cryptococcus species are members of the phylum Basidiomycota placed in the order Tremellales (15). Species placed within this order can include pathogenic and non-pathogenic yeast species (16, 17). Non-pathogenic Cryptococcus species such as Cryptococcus victoriae, which has since been renamed Vishniacozyma victoriae (15, 18), have been recently resolved in a variety of indoor environmental assessments that have utilized contemporary ITS sequencing methods (11, 14). V. victoriae is a slow growing polar budding yeast originally isolated in the Antarctic (19), but has since been shown to adapt to multiple conditions and inhabit a broad geographic range (15). The abundance of non-pathogenic Cryptococcus species is associated with either increased or decreased asthma severity, suggesting the potential involvement of multiple pulmonary immunological mechanisms following exposure. Decreased levels of Cryptococcus species diversity are significantly associated with increased asthma risk (1), which highlights the potential clinical significance of personal exposure to environmental Basidiomycota yeast species. In contrast, exposures to yeast and other microorganisms early in life can result in decreased allergy related symptoms, suggesting that yeast may confer protective effects against asthma in exposed children (10, 20, 21). Given the high prevalence of V. victoriae and the fact that its contribution to human health is not understood, it was chosen for this study.
Asthma has been an ongoing public health concern, particularly in cities such as New York City (22, 23). Asthma prevalence in school aged children (age 0–13 years) ranges from 5.1% to 17.6% and varies by neighborhood in New York City (24). Furthermore, children from lower socio-economic neighborhoods are approximately three times as likely to develop asthma as those living in wealthier neighborhoods (25). Many studies have examined the impact of detectable allergens and air quality on asthma prevalence in New York City, but the prevalence of fungi, specifically Basidiomycota yeast, has not been measured.
Although an inverse relationship between relative abundance of yeasts in indoor environments and asthma prevalence has been previously reported (1, 20), the specific quantification of V. victoriae in the homes of asthmatic children and associations with health outcomes has not been determined. To facilitate a better understanding of the burden of non-pathogenic Cryptococcus yeasts in indoor U.S. environments, species-specific primers, probes, and a quantitative polymerase chain reaction (qPCR) method were developed to provide the detection and quantification of V. victoriae (10). In this study, we provide the first quantification of V. victoriae in dust samples from homes participating in the New York City Neighborhood Asthma and Allergy Study. Furthermore, the influence of housing factors and health effects associated with exposures to V. victoriae in homes of children from low and high asthma prevalence neighborhoods in the New York metropolitan area were examined. This is the first study to utilize quantitative measurements and correlative statistics to support the findings of previous studies examining the relationship between yeast exposure and allergic airway disease.
III. MATERIALS AND METHODS
3.1. Fungal Strains and Growth Conditions
We used 11 fungal strains in this study (Supplementary Table 1). The Vishniacozyma victoriae type strain was purchased from the American Type Culture Collection (ATCC) (Strain ID: MYA-305). This strain is an aerobic polar budding yeast with a Biosafety Level 1 classification. V. victoriae was grown in Potato Dextrose Broth (BD Bioscience, San Jose, CA) and shaken at 15°C for 4 days. V. victoriae cultures entered the logarithmic growth phase at 50 hours post inoculation and developed a creamy pink color over time. V. victoriae yeast cells that entered the logarithmic growth phase were utilized in subsequent method development experiments. Fungi tested for cross reactivity in the qPCR assay included Ascomycota species Alternaria alternata, Aspergillus fumigatus, Candida albicans, Cladosporium cladosporioides, Cyberlindnera fabianii, Epicoccum nigrum, and Penicillium purpurogenum from the National Institute for Occupational Safety and Health (NIOSH) culture collection (Supplementary Table 1). Cross reactivity was additionally determined against Basidiomycota yeast species including Naganishia globosa and Rhodotorula glutinis from the Agricultural Research Service collection and NIOSH culture collection, respectively (Supplementary Table 1). Culture and growth parameters for each tested species can be referred to in Supplementary Table 1.
3.2. DNA Extraction
DNA was extracted from liquid culture (standards) and environmental dust samples utilizing the DNEasy PowerLyzer PowerSoil kit (Qiagen, Valencia, CA) following the manufacturer’s protocol with a few modifications. An aliquot of dust up to 25 mg was taken from each dust sample and processed for DNA extraction. Additional beads were added to each tube prior to DNA extraction, including 1 gram of garnet beads, 0.1 grams of 0.5 mm glass beads, and 0.3 grams of 0.1 mm glass beads (Biospec, Bartlesville, OK) (26). Samples were eluted in 50 μL of Tris-EDTA Buffer and stored at 4 °C. All DNA samples were diluted ten-fold to limit potential inhibition. DNA concentrations were reported per mg of dust in order to standardize differences in mass of collected dust samples.
3.3. Primer and Probe Design
The Internal Transcribed Spacer (ITS) 2 region of V. victoriae (Genbank accession number MH809977), was targeted for the primer and probe development. The ITS regions, known as the fungal barcode, are regions of high heterogeneity situated between ribosomal RNA genes that can be utilized for species identification of fungi (9). Compared to the ITS1 region, the ITS2 region is more heterogeneous at the Vishniacozyma genus level and provides better differentiation of Basidiomycota with reduced over-estimation (27). Therefore, this was the region targeted for the primer and probe development. Integrated DNA Technology’s (IDT) PrimerQuest tool (Coralville, IA) was utilized to design five primers and probes specific for V. victoriae (Supplementary Table 2). The full ITS region of V. victoriae was entered into the software, and parameters such as approximate melting temperatures (Tm) of 62 °C, amplicon length of 100 base pairs, and GC content approximately 50% were selected. Probes were designed with carboxyfluorescein (FAM) dyes and tetramethylrhodamine (TAMRA) quenchers (IDT). Primer and probe pairs were initially tested in silico through The National Center for Biotechnology Information (NCBI) Blast (blast.ncbi.nlm.nih.gov) analyses to confirm species-specific amplification of the targeted ITS2 region.
3.4. V. victoriae qPCR Assay and Standard Curve Development
V. victoriae cells were harvested during the logarithmic growth phase and quantified using a hemocytometer for use in the development of a standard curve. DNA extraction tubes containing 250 mg of sterile Coarse A4 Arizona Test Dust (Powder Technology Inc, Schofield, WI) were spiked with known concentrations of V. victoriae cells starting at 107 cells in 100 μl of Potato Dextrose broth (BD #254290). Course A4 Arizona Test Dust was utilized as it is an International Organization for Standardization (ISO) standard for house dust. The DNA was extracted as specified above and stored at 4 °C. DNA standards were serially diluted ten-fold in Tris-EDTA (TE) buffer and used to develop a standard curve ranging from 1 to 106 cell equivalents/sample of V. victoriae. Cell equivalents units were used as they relate the quantification cycle (Cq) of the qPCR reaction back to the standard curve that correlates with the known concentration of cells added in order to account for copy number variation.
Quantitative real-time polymerase chain reactions (qPCR) for all extracted DNA samples, standards, and primer-probe pairs were run on a QuantStudio 6 Flex machine (Applied Biosystems, Foster City, CA) under the following conditions: 95 °C for 3 minutes, then 40 cycles of 95 °C for 5 seconds and 57 °C for 30 seconds. Quantification cycles were all normalized to 0.1. Each qPCR reaction was performed containing 3.5 μl of DNA template, 200 nM probes, 500 nM of each primer, and IDT PrimeTime Gene Expression Master Mix (IDT) with ROX reference dye for a final reaction volume of 10 μl. All samples were run in duplicate. Cell equivalent measurements were extrapolated from the Cq and aligned to the number of cells per reaction that corresponded with the cell-based standard. The standard curve reaction efficiency of each primer-probe pair was determined utilizing the slope of the linear portion of the log transformed cells per reaction.
3.5. V. victoriae qPCR Assay Specificity and Inhibition Testing
Supplementary Table 1 lists the fungal strains used to test the specificity of the developed primers and probes evaluated in this study. Cultures were inoculated on the selected nutrient media and temperature indicated and grown for 72 hours. Cells were counted via hemocytometer, and 106 cells in 100 μL of media (Potato Dextrose, Malt Extract and Yeast Malt Broth) were spiked into tubes of sterile Coarse A4 Arizona Test Dust (Powder Technology Inc). DNA was extracted as above and stored utilizing the same protocol as the environmental dust and standard samples. Inhibition tests were performed by spiking an aliquot of each environmental sample with a high concentration of V. victoriae DNA (from approximately 106–107 cells), running qPCR, and looking for significantly increased cycle threshold (Ct) values across all the samples. A significantly increased Cq value would result in decreased quantifications of V. victoriae suggesting polymerase inhibition. No samples were significantly inhibited (data not shown).
3.6. Study Subject Data Collection
The NYC Neighborhood Asthma and Allergy Study is a case-control study of asthma for which 7–8 year old children were recruited through a health care provider used primarily by a middle-income population (28). Recruitment targeted children living in higher asthma prevalence neighborhoods (HAPN, >11.0%) and lower asthma prevalence neighborhoods (LAPN, <9.0%) in the Bronx, Queens, Brooklyn and Manhattan boroughs as previously described (28). Children with complete data on V. victoriae exposure were included in the analysis. This study was approved by Columbia University’s Institutional Review Board and the portion conducted at The Ohio State University was determined exempt by The Ohio State University Institutional Review Board under study 2017E0224.
During home visits, parents answered a detailed questionnaire pertaining to the child’s health, demographics and environmental exposures as previously described (23). Asthma cases were defined by questionnaire, including the modules from the International Study of Asthma and Allergy in Childhood (ISAAC) (23, 25). Children were classified as asthmatic if their caregiver reported at least one of the following for the child in the past 12 months: 1) wheeze, 2) being awakened at night by cough without having a cold, 3) wheeze with exercise or 4) report of medication use for asthma. Children not meeting the asthma definition were classified as non-asthmatic. Serum immunoglobulin E (IgE) levels to common inhalant allergens (German cockroach, mouse urine proteins, Dermatophagoides farinae, cat dander, dog dander, common ragweed, and mixed tree pollen) were measured by ImmunoCAP (Phadia, Uppsala, Sweden) as previously described (29, 30). Children with measurable IgE (>0.35 IU/ml) to at least one allergen tested were considered seroatopic.
3.7. Environmental Sample Collection
Study participant home dust samples were collected from the child’s bedroom floor by vacuuming approximately 2 square meters surrounding the bed for 3 minutes using a vacuum cleaner and Dustream® collector (Indoor Biotechnologies, Charlottesville, VA). Samples were collected between March 2008 and June 2011 and were stored at −20 °C before being shipped. Aliquoted dust was stored at −20 °C for approximately 7 months until total DNA was extracted. Samples from 275 homes were utilized in this study. Bed dust samples were also collected during the home visits for measurement of inhalant allergens as previously described (23). Temperature, relative humidity, and specific humidity were measured at 5-minute intervals using HOBO H08-003-02 data loggers (Onset Computer Corporation, Bourne, MA). Monitors were installed for periods of 6–13 days (average 9.1 days, standard deviation 2.1 days) after the dust sample was collected. Monitors were preferentially placed in the living room, 1.5 m above the floor and away from windows and drafts, as described previously (31).
3.8. Statistical Analysis
The quantifications of V. victoriae were determined from dust samples from homes participating in the NYC Neighborhood Asthma and Allergy Study. Given that V. victoriae and allergen concentrations in dust were approximately log-normally distributed, log transformed values were used for all statistical tests. Differences in geometric means were tested between housing and built environment data along with health outcomes. Variables conventionally considered as potential confounders were included in the multivariable model (32). Linear regression models were tested with logarithmically transformed V. victoriae as the dependent variable. Prevalence ratios (PR) for risk of asthma were calculated using a generalized estimating equation model with United Hospital Fund (UHF) used as a cluster variable because some of the neighborhood level data were available at the UHF level. Data were analyzed in SPSS version 26 (Chicago, IL) and visualized in R version 3.3.1 (Vienna, Austria).
IV. RESULTS
4.1. Specificity of qPCR Assay
The amplified ITS2 regions by the designed primer-probe pairs shown in Supplementary Table 2 were analyzed in silico and aligned with 100% identity to V. victoriae sequences, uncultured fungus, or other unidentified Cryptococcus sequences deposited in NCBI. To further test primer and probe specificity, ten other strains derived from the phyla Ascomycota and Basidiomycota (Supplementary Table 1) were tested with each primer-probe pair in the V. victoriae qPCR assay. Only the sample with V. victoriae DNA resulted in amplification over the limit of detection in qPCR when evaluated with all five of the primer-probe pairs, confirming the species-specificity of the assay.
4.2. Primer-Probe Pair Selection
All five primer-probe pairs were tested with the developed standards for V. victoriae. The efficiencies within the limit of detection were calculated for each primer-probe pair utilizing ThermoFisher’s qPCR Efficiency Calculator (Thermo Fisher Scientific, Waltham, MA). Supplementary Table 2 lists the efficiencies and correlation coefficients for each of the developed standard curves. Primer-probe pair VV05 had the highest correlation coefficient and greatest efficiency without polymerase inhibition and was therefore selected for downstream analyses to quantify V. victoriae in NYC Neighborhood Asthma and Allergy Study environmental samples (Figure 1A).
Figure 1: V. victoriae Standard Curve Development and Distribution.
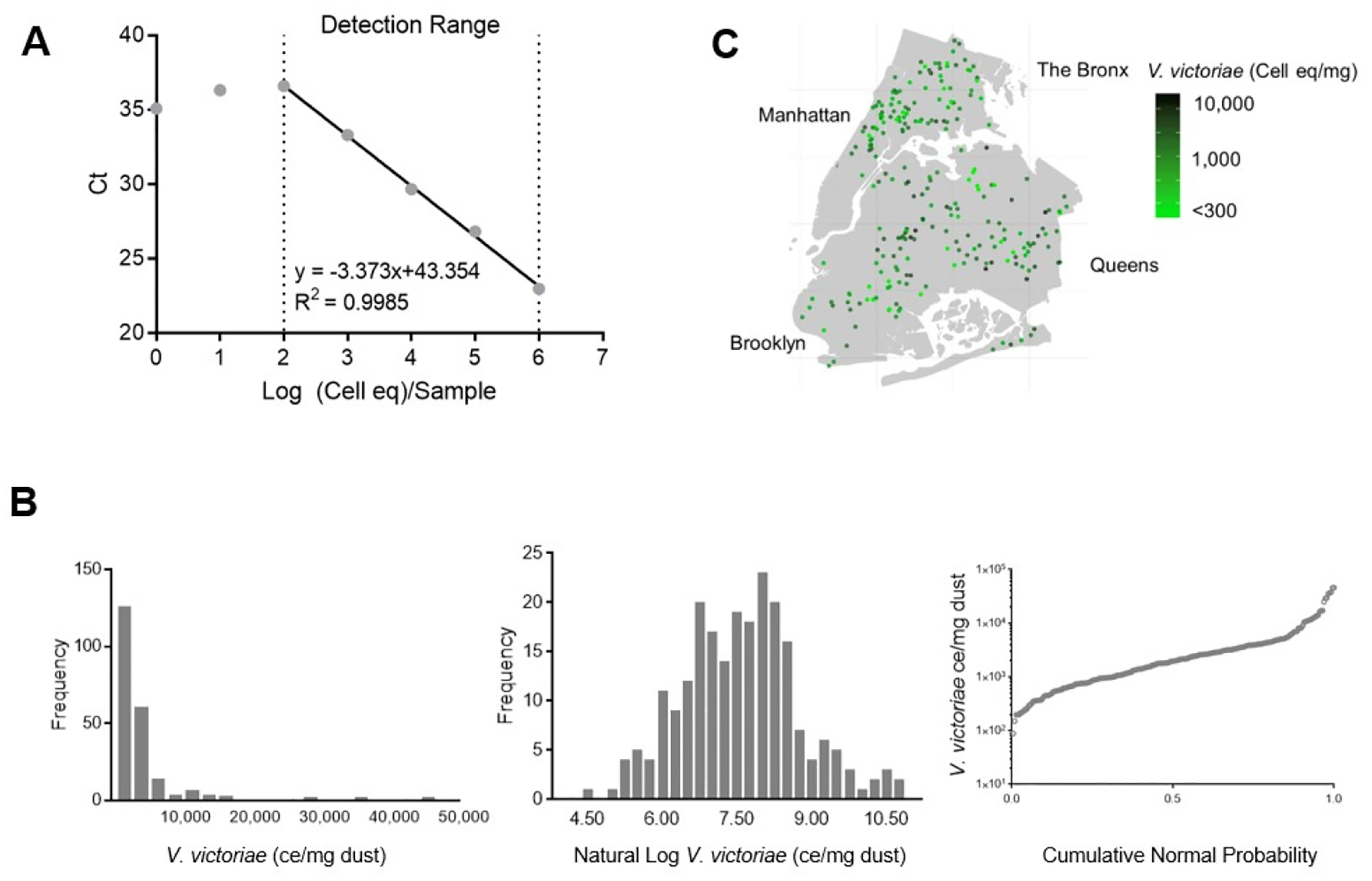
V. victoriae was quantified in samples utilizing a specific primer and probe set. A: The developed standard curve and range of detection. B: Numerical frequency distribution of V. victoriae quantifications in NYC Neighborhood Asthma and Allergy Study samples (left), log transformed quantifications (middle), and cumulative normal probability (right). C: Geographical distribution of V. victoriae ce/mg dust over the New York metropolitan area.
4.3. Sensitivity of qPCR Assay
The limit of quantification for the V. victoriae qPCR assay utilizing the primer-probe pair VV05 was 36 cell equivalents (cell eq) of V. victoriae per reaction, which equates to 100 cell equivalents of V. victoriae in the total dust sample processed assuming extraction efficiencies were comparable across all samples. The limit of quantification was determined as it was the lowest concentration of cell equivalents which fell within the linear range of the standard curve for the assay. The limit of detection was set to Ct values ≤ 36.085, which aligned to the lowest concentration of V. victoriae consistently detected over the quantification cycle that did not have undetermined Cq values. Inhibition tests confirmed that none of the samples had significant polymerase inhibition (data not shown).
4.4. Detection and Quantification of V. victoriae in Environmental Samples
Environmental samples (n=275) from the NYC Neighborhood Asthma and Allergy Study cohort were evaluated for the presence and quantification of V. victoriae. Samples with <5.0 mg of dust were excluded (n=19) from the analyses. Of the remaining 256 samples, 20 had quantifications of V. victoriae below the limit of detection (300 V. victoriae cell eq/mg of dust (ce/mg)) and were excluded from the analyses. The samples with quantifications of V. victoriae within the limit of detection (n=236) were approximately log normally distributed (Figure 1B, middle), ranging from <300 to 45,918 ce/mg with a geometric mean [95% C.I.] of 1,622 [1,370–1,971] ce/mg (Figure 1B, left). Spatial analysis of the distribution of V. victoriae determined in the qPCR assay revealed a heterogeneous distribution of V. victoriae across neighborhoods that was dependent on a number of individual housing characteristics with no apparent clustering of low or high concentrations by neighborhood (Figure 1C).
4.5. Associations with Housing Characteristics
The presence of V. victoriae in homes was higher in homes with carpeted bedroom floors (P=0.044), a dog in home (P=0.010), and from samples collected during the winter and spring seasons (P<0.001) (Table 1). V. victoriae was also positively correlated with mean specific humidity (P=0.004) (Table 2). Among homes without a dog, the geometric mean concentrations of V. victoriae were similar for those that had never had a dog (1,456 [1,197–1,772] ce/mg) and those that previously owned a dog (1,625 [1,006–2,624] ce/mg) (data not shown). Furthermore, V. victoriae prevalence was associated with homes with the presence of dog allergen Can f 1 (P=0.027) (Table 2).
Table 1:
Geometric mean of V. victoriae in bedroom floor dust by home and neighborhood characteristics and household behavior. Data in bold indicate statistically significant associations.
| Characteristics | n | Geometric mean [95% C.I.] ce/mg | P value for differenceI | |
|---|---|---|---|---|
| Neighborhood asthma prevalence | HAPN | 129 | 1,791 [1,394–2,301] | 0.21 |
| Housing Type | Single | 46 | 2,125 [1,395–3,236] | 0.10 |
| Carpeted bedroom floor | No | 111 | 1,349 [1,073–1,696] | 0.044 |
| Report of mold odor | No | 210 | 1,623 [1,352–1,948] | 0.95 |
| Report of visible mold | No | 144 | 1,550 [1,261–1,905] | 0.51 |
| Report of unrepaired water damage | No | 184 | 1,548 [1,285–1,865] | 0.29 |
| Report of leaky pipes | No | 211 | 1,669 [1,394–1,999] | 0.34 |
| Report of adding water to air | No | 143 | 1,642 [1,316–2,048] | 0.89 |
| Remove shoes entering home | No | 146 | 1,720 [1,387–2,133] | 0.40 |
| Basement in building | No | 37 | 1,672 [1,098–2,545] | 0.89 |
| Wet mop | No | 186 | 1,643 [1,362–1,982] | 0.80 |
| Cat in home | No | 208 | 1,658 [1,385–1,984] | 0.49 |
| Dog in home | No | 202 | 1,481 [1,235–1,775] | 0.010 |
| Season of collection | Winter | 71 | 2,562 [1,932–3,397] | <0.001 |
ANOVA for geometric means
Table 2:
Correlation between V. victoriae in bedroom floor dust and neighborhood characteristics and allergens. Data in bold indicate statistically significant associations.
| Characteristic | Correlation coefficient* | P value |
|---|---|---|
| Neighborhood asthma prevalence† | −0.11 | 0.096 |
| Year home built | −0.002 | 0.98 |
| Median neighborhood household income (500 meters) in $10K‡ | 0.089 | 0.17 |
| Family household income in $10K | 0.080 | 0.23 |
| Number people living in home | 0.13 | 0.052 |
| Der f 1(ug/g)∥ | 0.074 | 0.26 |
| Bla g 2 (ug/g)∥ | −0.072 | 0.27 |
| Mus m 1 (ug/g)∥ | −0.038 | 0.57 |
| Fel d 1 (ug/g)∥ | 0.053 | 0.42 |
| Can f 1 (ug/g)∥ | 0.14 | 0.027 |
| Airborne PM2.5§ | 0.017 | 0.80 |
| Airborne black carbon§ | −0.057 | 0.39 |
| Mean specific humidity** | 0.20 | 0.004 |
Spearman rank correlation coefficient.
School based prevalence of asthma among 5-year-old children for the child’s United Hospital Fund Neighborhood (several zip codes).
GIS census-based variable of the median income of the household in the surrounding radian 500 meters.
Allergens measured in bed dust.
Airborne fine particulate matter (PM2.5) and black carbon fraction of PM2.5 were measured in the air of the home during the week following dust collection.
Specific humidity in home measured during the week following dust collection.
V. victoriae was not significantly associated with neighborhood asthma prevalence, housing type, reports of mold odor or visible mold, unrepaired water damage, leaky pipes, adding water to air (such as via a humidifier), removing shoes when entering home, presence of basement in building, wet mop cleaning, or having a cat in the home (all P>0.05, Table 1). When examining correlations between V. victoriae in bedroom floor dust and home and neighborhood characteristics, V. victoriae was not significantly (P>0.05) associated year home built, median neighborhood or family household income, or number of people living in the home (Table 2).
Associations between V. victoriae prevalence and environmental characteristics were additionally examined. V. victoriae was not associated with the presence of perennial allergens (Table 2). V. victoriae was also not associated with airborne PM2.5 (P=0.80) or black carbon (P=0.39) (Table 2). V. victoriae was associated with the number of people living in home (P=0.041), presence of dog in home (P=0.023), winter (P <0.001) and spring seasons (P=0.001) (Table 3).
Table 3:
Multivariable model for concentration of V. victoriae (n=221).* Data in bold indicate statistically significant associations.
| Characteristic | Beta [95% C.I.] | P value | |
|---|---|---|---|
| Apartment building | −0.15 [−0.53,0.22] | 0.43 | |
| Carpet | 0.28 [−0.062, 0.63] | 0.18 | |
| Neighborhood asthma prevalence† | −0.005 [−0.044,0.033] | 0.78 | |
| Number of people living in home | 0.12 [0.005, 0.23] | 0.041 | |
| Dog in home | 0.55 [0.078, 1.0] | 0.023 | |
| Mean specific humidity∥ | 0.013 [−0.072, 0.098] | 0.76 | |
| reference | N/A | ||
| 0.94 [0.46, 1.4] | <0.001 | ||
| 1.0 [0.43, 1.7] | 0.001 | ||
| Summer | −0.065 [−0.58, 0.45] | 0.81 | |
Multivariable regression model with logarithmically transformed V. victoriae as the dependent variable and all variables listed in the table and a variable indicating if the sample collected was <10mg as independent variables.
School based prevalence of asthma among 5-year-old children for the child’s United Hospital Fund Neighborhood (several zip codes).
Specific humidity in home measured during the week following dust collection.
4.6. Associations with Asthma Related Health Effects
Associations between V. victoriae and asthma symptoms were tested in a model that adjusted for sex, race, Hispanic ethnicity, season, neighborhood asthma prevalence, smoker in home, ever dog ownership, and collected sample >10 mg (Figure 2). Lower V. victoriae concentrations were observed in homes with an asthmatic child as compared with homes without an asthmatic child (P=0.027) (Table 4). In addition, concentrations of V. victoriae were lower in the homes of asthmatic children with more frequent symptoms as compared to homes of children without asthma (P=0.041) (Table 4). The inverse association with asthma was only observed among the non-seroatopic (P=0.007) and not the seroatopic children (P=0.35) (Table 4). Of the children classified as seroatopic, the vast majority (77%) were sensitized to at least two allergens (data not shown). Furthermore, the geometric mean of the total IgE was 33.2 IU/mL among the non-seroatopics as compared to 208 IU/mL among the seroatopic children.
Figure 2: Association of V. victoriae with Asthma.
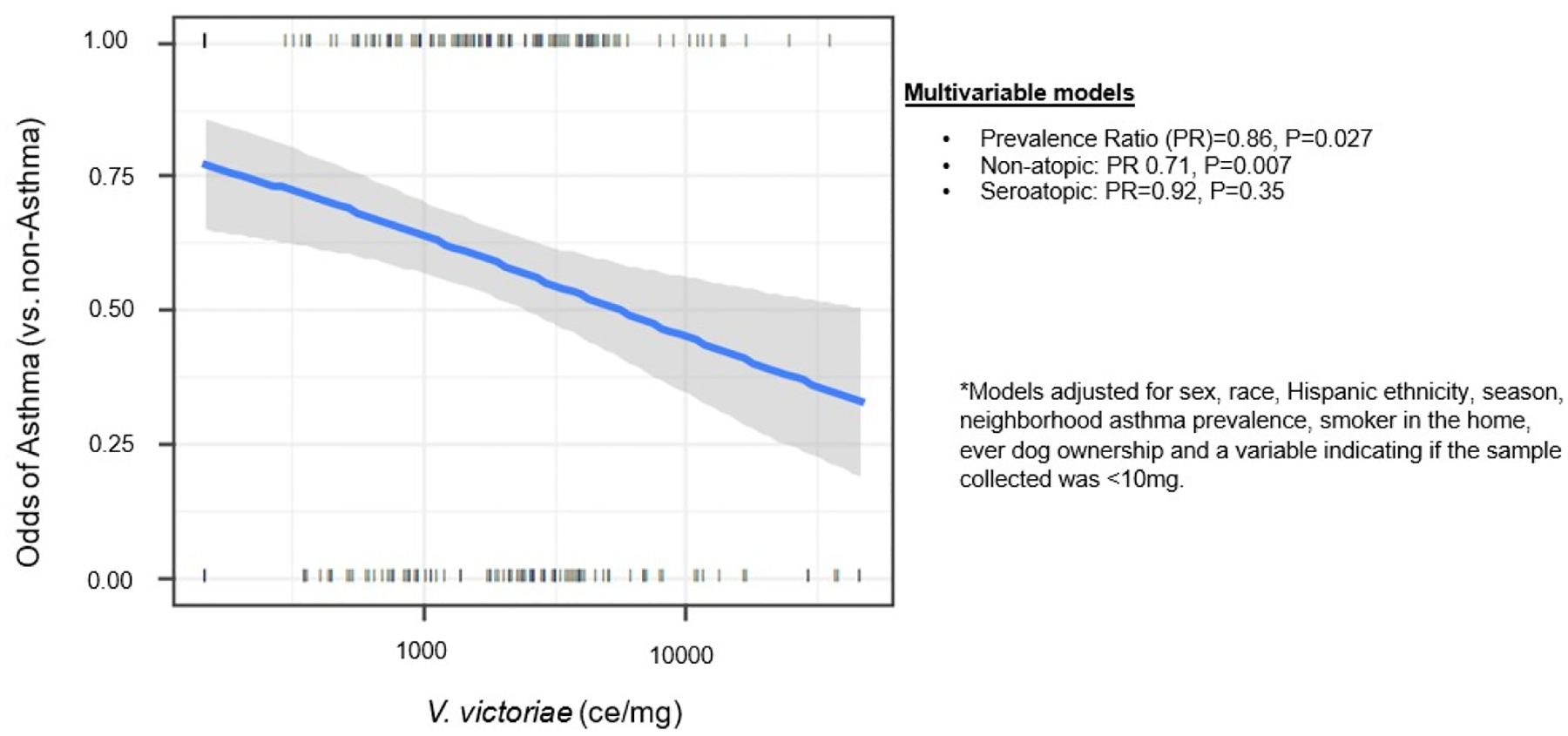
Multivariable model examining asthmatic cases vs. controls, cross-sectional, adjusted for sex, race, Hispanic ethnicity, season, neighborhood asthma prevalence, smoker in the home, ever dog ownership and a variable indicating if the sample collected was <10 mg, n = 227.
Table 4:
Associations* between V. victoriae in bedroom floor dust and asthma symptoms. Data in bold indicate statistically significant associations.
| Factor | n | Overall |
|---|---|---|
| Asthma (case vs. control) | n=227 | PR =0.86 [0.75–0.98], P=0.027 |
| Frequent asthma symptoms (vs. no asthma symptoms) | n=138 | PR= 0.79 [0.63–0.99], P=0.041 |
| Seroatopy | n=216 | PR=0.96 [0.83–1.11], P=0.60 |
| Among non-seroatopic asthma (case vs. control) | n=105 | PR=0.71 [0.55–0.91], P=0.007 |
| Among seroatopic asthma (case vs. control) | n=111 | PR= 0.92 [0.77–1.1], P=0.35 |
Models adjusted for sex, race, Hispanic ethnicity, season, neighborhood asthma prevalence, smoker in the home, ever dog ownership and a variable indicating if the sample collected was <10mg.
V. DISCUSSION
5.1. Environmental Yeast Exposures
To date, environmental yeasts have remained a largely overlooked source of fungal exposure in indoor environments (1). Popular fungal qPCR methods, such as the Environmental Relative Moldiness Index (ERMI) panel (33), do not include primers that are specific for individual Basidiomycota yeast species. Therefore, there are substantial knowledge gaps about the role exposures to these yeast species play in allergic airway disease. This study provides insight into the prevalence, distribution, and associations with housing characteristics of the non-pathogenic Basidiomycota yeast V. victoriae in dust samples from the NYC Neighborhood Asthma and Allergy study.
Although studies have suggested that pulmonary exposure to pathogenic Cryptococcus species, such as Cryptococcus neoformans, exacerbates asthma (34), non-pathogenic Cryptococcus species have not been examined until this study. Non-pathogenic Cryptococcus species, including V. victoriae, are abundant and contribute to personal exposure in indoor environments (1, 8, 10, 12, 14). Exposures to non-pathogenic Cryptococcus species have varying impacts on asthma severity (1, 34). However, it has been shown that decreased fungal diversity has been associated with increased asthma risk (10). This suggests that a diverse mycobiome including environmental yeasts (specifically the non-pathogenic Cryptococcus yeasts such as V. victoriae) may potentially be protective, and this study contributes to this theory. However, it is important to note that environmental microbial exposures comprise a mixed exposure rather than just one species. There are synergistic and antagonistic effects among various microbes which may play an important role in the formation of an immune response to exposure (35).
5.2. Detection of V. victoriae
Prior to the development of this qPCR assay, the specific identification and quantification of V. victoriae in environmental samples was not easily evaluated. In the present study, V. victoriae was prevalent and detected in 85.8% of tested NYC Neighborhood Asthma and Allergy Study homes. Previous studies that employed molecular methods and pan fungal primers to assess fungal exposure typically utilized standard curves developed with one fungal species to estimate total fungi in a sample (4, 36). This presents a potential confounding variable as fungal species are likely to contain varying copy numbers of the region of interest. In this study, V. victoriae was shown to be associated with mean specific humidity and is predominantly identified in the spring and winter seasons. The type culture was originally isolated from soil in the Antarctic (15, 19), therefore associations with lower temperatures indicated the species’ ecological and environmental requirements for lower temperatures to facilitate growth. V. victoriae was found to be associated with several housing characteristics including carpets, number of people living in the home, and mean specific humidity, which are commonly associated with increased fungal growth (37). Studies have suggested that elevated levels of yeast in bedroom floor dust are associated with reduced wheeze and asthma development (20), which corroborates the finding that the presence of V. victoriae was associated with reduced asthma cases compared to control.
5.3. Association Between V. victoriae and Dog
V. victoriae was identified for the first time to be associated with the presence of dog allergen Can f 1. However, HAPN and LAPN homes in the New York City Neighborhood Asthma and Allergy Study had similar levels of Can f 1 (23). Yeasts comprise a significant portion of the dog microbiome (38, 39), and these data suggest that V. victoriae may be associated with the dog endogenous flora or that dogs disturb soil and then transfer the species into the built environment. Associations between the presence of a dog in home and decreased asthma cases and symptoms have been recently reported (40, 41). Microbiomes associated with dogs have been shown to confer similar protective immunological responses (3, 42, 43). Fujimara and colleagues have shown that mice exposed to dust from homes with a dog had increased levels of Lactobacillus johnsonii. When challenged in an ovalbumin animal model, L. johnsonii supplemented mice had decreased allergic airway response (42). However, it was noted that L. johnsonii was not responsible for all the protective effects seen in the dust from homes with dogs, suggesting that other unexplored organisms associated with dogs, potentially yeasts, could contribute to the airway protection observed.
Similar responses have been reported following Streptococcus pneumoniae and certain helminth infections, in which T regulatory cells were induced resulting in a suppression of the allergic airway response (44–46). Comparable trends have been reported when examining allergic airway disease in Amish and Hutterite Farm children, who share similar genetic backgrounds but vary in the microbial composition of their indoor environments (47). Compared to Hutterite children, Amish children with more abundant indoor microbiomes associated with animals had less allergic sensitization, serum IgE levels, eosinophils, and Th2 responses typical of allergic airway disease (47), further suggesting that microbial exposures within the indoor environment can influence allergic airway disease.
The associations of V. victoriae with dog and dog allergen in the NYC Neighborhood Asthma and Allergy Study, and the associations with decreased asthma cases, indicate for the first time, that study participants with asthma were less likely to have non-pathogenic Cryptococcus species present in their environment compared to control subjects. Although the immunological mechanisms are not understood, future animal models could help provide mechanistic insight into repeated exposure of V. victoriae and the species’ influence on adverse respiratory health effects. The results of this study highlighting the prevalence of V. victoriae in the study homes as well as the associations between this species and potential protective effects are in agreement with other recent studies indicating that certain yeast taxa confer protection against allergic airway disease (20). The results of this study highlight the contributions of V. victoriae to NYC Neighborhood Asthma and Allergy Study homes and may indicate that exposure to this yeast, especially in environments with a dog, potentially confers protection or attenuates the response to adverse health effects such as allergic airway disease. This association should continue to be evaluated in future studies.
5.4. Study Limitations
Although cell equivalent measurements of V. victoriae were calculated, the copy number of the ITS region for V. victoriae currently remains unknown and represents a potential limitation of this study. Furthermore, as with all qPCR assay based methods, the potential for extraction and amplification biases must be considered as a potential study limitation (36). For example, there may be highly similar sequences to organisms not tested for cross reactivity in the samples that are being amplified resulting in the over estimation of the quantification of V. victoriae. Some of the developed standard curves had efficiencies greater than 100%, indicating mild polymerase inhibition. Presence of PCR inhibitors in the DNA samples is a potential limitation that could have been introduced into the assay (48). Furthermore, the standard curve approach assumes that the efficiency of the extraction and amplification is the same for the standards as it is for the environmental samples (36). Typically, the standards are created from pure cultures, as with our approach, therefore the sample template may differ slightly resulting in a different efficiency. However the aforementioned limitations apply to any qPCR based approach, and this methodology has been utilized and accepted by the scientific community as a reliable tool for the quantification of indoor fungal species (49, 50).
The environmental parameters of the homes such as humidity were monitored in the living room of the homes, however dust samples were collected from children’s bedrooms. Given that the distance from the bedroom to living room or size of the house was not measured, it is possible that the environmental parameters would be different in the bedroom. The finding that V. victoriae was more strongly associated with non-seroatopic than seroatopic children is striking. Non-seroatopic asthmatics included those participants who did not have IgE reactivity to the panel of tested allergens. It is possible that these children could have IgE reactivity to allergens that were not tested and thus be a false negative. However, given that 77% of the seroatopic children were sensitized to at least 2 allergens and the differences in the mean of total IgE between the two groups (33.2 IU/mL in non-seroatopic versus 208 IU/mL in seroatopic participants), it is likely that these groups are distinct.
Lastly, given the cross-sectional nature of this study, it must be noted that we cannot infer causality and the direction of the associations cannot be proven. However, it seems unlikely that yeast growth would cause some of the associated factors to be true, such as cause carpeted flooring to be put in place, raise the specific humidity, or lead to dog ownership. It is possible that the behavioral practices of families with asthmatic children could have led to changes in domestic fungal growth (such as use of a fungicide or humidifier) or that there is an unmeasured confounder causing the observation of an association between V. victoriae and the control, non-asthmatic case. Still, these findings are in keeping with other studies observing inverse associations between fungal species and fungal diversity in general and asthma outcomes.
5.5. Conclusion
In conclusion. V. victoriae was quantified in 236 of the 275 NYC Neighborhood Asthma and Allergy Study samples within the limit of detection, and was associated with the presence of carpet, mean humidity, the presence of dog allergen and dog in home, and with the non-asthmatic control compared to the asthmatic case. These data suggest a potentially protective role of non-pathogenic Cryptococcus species, such as V. victoriae, in indoor repeated fungal exposures. Moreover, our data suggest that V. victoriae is associated with the dog mycobiome and provides evidence that personal exposure to this species could potentially confer protective effects against allergic airway disease. Future studies aim to validate these associations in an in vivo model in order to elucidate the mechanisms behind these associations and environmental yeast exposures. This study provides novel insight into the contribution of often overlooked environmental yeast species towards the indoor microbiome, and more specifically, the role exposure to these species plays in protection against allergic airway disease.
Supplementary Material
VI. ACKNOWLEDGMENTS
The authors would like to acknowledge Angela Lemons for her assistance with techniques and manuscript preparation. We would like to thank the NYC Neighborhood Asthma and Allergy Study field team for their hard work. We would also like to thank the families who have participated on the study. The findings and conclusions in this study are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
VIII. FUNDING
This study was supported in part by an interagency agreement between NIOSH and NIEHS (AES 12007001-1-0-6) as a collaborative National Toxicology Program research activity. This study was supported by the National Institute of Environmental Health Sciences (NIEHS) [grant #’s R01 ES014400, P30 ES09089] the Department of Housing and Urban Development (HUD) Healthy Homes and Lead Technical Study [HUD NYHHU0003-11, NYHHU0021-13]).
Abbreviations:
- ATCC
American Type Culture Collection
- BD
Becton, Dickinson
- CBS
Westerdijk Fungalbio Diversity Institute
- Cq
Quantification Cycle
- ISO
International Organization for Standardization
- ITS
Internal Transcribed Spacer
- NCBI
National Center for Biotechnology Information
- NIOSH
National Institute for Occupational Safety and Health
- NO
Nitric oxide
- PR
Prevalence Ratios
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supplementary information is available at Journal of Exposure Science & Environmental Epidemiology’s website.
IX. REFERENCES
- 1.Dannemiller KC, Gent JF, Leaderer BP, Peccia J. Indoor microbial communities: Influence on asthma severity in atopic and nonatopic children. J Allergy Clin Immunol. 2016;138(1):76–83.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green BJ, Summerbell RC. Aerosolized fungal fragments. In: Samson RA, Adan OCG (eds). Fundamentals of mold growth in indoor environments and strategies for healthy living. The Netherlands: Wageningen Academic Publishers; 2011. [Google Scholar]
- 3.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–9. [DOI] [PubMed] [Google Scholar]
- 4.Adams RI, Miletto M, Taylor JW, Bruns TD. The diversity and distribution of fungi on residential surfaces. PLoS One. 2013;8(11):e78866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpe RA, Bearman N, Thornton CR, Husk K, Osborne NJ. Indoor fungal diversity and asthma: a meta-analysis and systematic review of risk factors. J Allergy Clin Immunol. 2015;135(1):110–22. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, Cox-Ganser JM, Kreiss K, White SK, Rao CY. Hydrophilic fungi and ergosterol associated with respiratory illness in a water-damaged building. Environ Health Perspect. 2008;116(1):45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmadi B, Mirhendi H, Shidfar MR, Nouripour-Sisakht S, Jalalizand N, Geramishoar M, et al. A comparative study on morphological versus molecular identification of dermatophyte isolates. J Mycol Med. 2015;25(1):29–35. [DOI] [PubMed] [Google Scholar]
- 8.Rittenour WR, Ciaccio CE, Barnes CS, Kashon ML, Lemons AR, Beezhold DH, et al. Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environ Sci Process Impacts. 2014;16(1):33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. 2012;109(16):6241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dannemiller KC, Mendell MJ, Macher JM, Kumagai K, Bradman A, Holland N, et al. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air. 2014;24(3):236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemons AR, Hogan MB, Gault RA, Holland K, Sobek E, Olsen-Wilson KA, et al. Microbial rRNA sequencing analysis of evaporative cooler indoor environments located in the Great Basin Desert region of the United States. Environ Sci Process Impacts. 2017;19(2):101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson B, Zhou Y, Bautista EJ, Urch B, Speck M, Silverman F, et al. Characterization of the bacterial and fungal microbiome in indoor dust and outdoor air samples: a pilot study. Environ Sci Process Impacts. 2016;18(6):713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaprakash B, Adams RI, Kirjavainen P, Karvonen A, Vepsalainen A, Valkonen M, et al. Indoor microbiota in severely moisture damaged homes and the impact of interventions. Microbiome. 2017;5(1):138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitkaranta M, Meklin T, Hyvarinen A, Paulin L, Auvinen P, Nevalainen A, et al. Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl Environ Microbiol. 2008;74(1):233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Garcia V, Zalar P, Brizzio S, Gunde-Cimerman N, van Broock M. Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol Ecol. 2012;82(2):523–39. [DOI] [PubMed] [Google Scholar]
- 16.Idnurm A, Lin X. Rising to the challenge of multiple Cryptococcus species and the diseases they cause. Fungal Genet Biol. 2015;78:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khawcharoenporn T, Apisarnthanarak A, Mundy LM. Non-neoformans cryptococcal infections: a systematic review. Infection. 2007;35(2):51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mondini A, Donhauser J, Itcus C, Marin C, Perșoiu A, Lavin P, et al. High-throughput sequencing of fungal communities across the perennial ice block of Scărișoara Ice Cave. Annals of Glaciology. 2018;59(77):134–46. [Google Scholar]
- 19.Montes MJ, Belloch C, Galiana M, Garcia MD, Andres C, Ferrer S, et al. Polyphasic taxonomy of a novel yeast isolated from antarctic environment; description of Cryptococcus victoriae sp. nov. Syst Appl Microbiol. 1999;22(1):97–105. [DOI] [PubMed] [Google Scholar]
- 20.Behbod B, Sordillo JE, Hoffman EB, Datta S, Webb TE, Kwan DL, et al. Asthma and allergy development: contrasting influences of yeasts and other fungal exposures. Clin Exp Allergy. 2015;45:154–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman DL, Huffnagle GB. Potential contribution of fungal infection and colonization to the development of allergy. Medical mycology. 2009;47(5):445–56. [DOI] [PubMed] [Google Scholar]
- 22.Lin S, Fitzgerald E, Hwang SA, Munsie JP, Stark A. Asthma hospitalization rates and socioeconomic status in New York State (1987–1993). J Asthma. 1999;36(3):239–51. [DOI] [PubMed] [Google Scholar]
- 23.Olmedo O, Goldstein IF, Acosta L, Divjan A, Rundle AG, Chew GL, et al. Neighborhood differences in exposure and sensitization to cockroach, mouse, dust mite, cat, and dog allergens in New York City. J Allergy Clin Immunol. 2011;128(2):284–92.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Environment and Health Data Portal NYC.gov: The City of New York; 2020 [Asthma Prevalence - Children ever diagnosed with asthma (ages 0–13 years) - Percentage, 2017, Neighborhood (NYC KIDS)]. Available from: http://a816-dohbesp.nyc.gov/IndicatorPublic/VisualizationData.aspx?id=2392,4466a0,11,Map,Percentage,2017.
- 25.Mainardi TR, Mellins RB, Miller RL, Acosta LM, Cornell A, Hoepner L, et al. Exercise-induced wheeze, urgent medical visits, and neighborhood asthma prevalence. Pediatrics. 2013;131(1):e127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto N, Bibby K, Qian J, Hospodsky D, Rismani-Yazdi H, Nazaroff WW, et al. Particle-size distributions and seasonal diversity of allergenic and pathogenic fungi in outdoor air. Isme j. 2012;6(10):1801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang RH, Su JH, Shang JJ, Wu YY, Li Y, Bao DP, et al. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS One. 2018;13(10):e0206428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornell AG, Chillrud SN, Mellins RB, Acosta LM, Miller RL, Quinn JW, et al. Domestic airborne black carbon and exhaled nitric oxide in children in NYC. J Expo Sci Environ Epidemiol. 2012;22(3):258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donohue KM, Al-alem U, Perzanowski MS, Chew GL, Johnson A, Divjan A, et al. Anti-cockroach and anti-mouse IgE are associated with early wheeze and atopy in an inner-city birth cohort. J Allergy Clin Immunol. 2008;122(5):914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C, Gauvey-Kern K, Johnson A, Kelvin EA, Chew GL, Perera F, et al. Cord blood versus age 5 mononuclear cell proliferation on IgE and asthma. Clin Mol Allergy. 2010;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamerius JD, Acosta LM, Jacobson JS, Goldstein IF, Quinn JW, Rundle AG, et al. Socioeconomic and Outdoor Meteorological Determinants of Indoor Temperature and Humidity in New York City Households. Weather, Climate and Society. 2013;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355(21):2226–35. [DOI] [PubMed] [Google Scholar]
- 33.Vesper S, McKinstry C, Haugland R, Wymer L, Bradham K, Ashley P, et al. Development of an Environmental Relative Moldiness index for US homes. J Occup Environ Med. 2007;49(8):829–33. [DOI] [PubMed] [Google Scholar]
- 34.Goldman DL, Davis J, Bommarito F, Shao X, Casadevall A. Enhanced allergic inflammation and airway responsiveness in rats with chronic Cryptococcus neoformans infection: potential role for fungal pulmonary infection in the pathogenesis of asthma. The Journal of infectious diseases. 2006;193(8):1178–86. [DOI] [PubMed] [Google Scholar]
- 35.Siggins A, Gunnigle E, Abram F. Exploring mixed microbial community functioning: recent advances in metaproteomics. FEMS Microbiol Ecol. 2012;80(2):265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol. 2012;78:4481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dannemiller KC, Gent JF, Leaderer BP, Peccia J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air. 2016;26(2):179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster ML, Dowd SE, Stephenson C, Steiner JM, Suchodolski JS. Characterization of the fungal microbiome (mycobiome) in fecal samples from dogs. Vet Med Int. 2013;2013:658373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chermprapai S, Ederveen THA, Broere F, Broens EM, Schlotter YM, van Schalkwijk S, et al. The bacterial and fungal microbiome of the skin of healthy dogs and dogs with atopic dermatitis and the impact of topical antimicrobial therapy, an exploratory study. Vet Microbiol. 2019;229:90–9. [DOI] [PubMed] [Google Scholar]
- 40.Campo P, Kalra HK, Levin L, Reponen T, Olds R, Lummus ZL, et al. Influence of dog ownership and high endotoxin on wheezing and atopy during infancy. J Allergy Clin Immunol. 2006;118(6):1271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. Jama. 2002;288(8):963–72. [DOI] [PubMed] [Google Scholar]
- 42.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111(2):805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2):410–2, 2.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitagaki K, Businga TR, Racila D, Elliott DE, Weinstock JV, Kline JN. Intestinal helminths protect in a murine model of asthma. J Immunol. 2006;177(3):1628–35. [DOI] [PubMed] [Google Scholar]
- 45.Parande Shirvan S, Ebrahimby A, Dousty A, Maleki M, Movassaghi A, Borji H, et al. Somatic extracts of Marshallagia marshalli downregulate the Th2 associated immune responses in ovalbumin-induced airway inflammation in BALB/c mice. Parasit Vectors. 2017;10(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston JA, Thorburn AN, Starkey MR, Beckett EL, Horvat JC, Wade MA, et al. Streptococcus pneumoniae infection suppresses allergic airways disease by inducing regulatory T-cells. Eur Respir J. 2011;37(1):53–64. [DOI] [PubMed] [Google Scholar]
- 47.Ober C, Sperling AI, von Mutius E, Vercelli D. Immune development and environment: lessons from Amish and Hutterite children. Current opinion in immunology. 2017;48:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hedman J, Radstrom P. Overcoming inhibition in real-time diagnostic PCR. Methods Mol Biol. 2013;943:17–48. [DOI] [PubMed] [Google Scholar]
- 49.Vesper SJ, Wymer LJ, Meklin T, Varma M, Stott R, Richardson M, et al. Comparison of populations of mould species in homes in the UK and USA using mould-specific quantitative PCR. Lett Appl Microbiol. 2005;41(4):367–73. [DOI] [PubMed] [Google Scholar]
- 50.Haugland RA, Brinkman N, Vesper SJ. Evaluation of rapid DNA extraction methods for the quantitative detection of fungi using real-time PCR analysis. J Microbiol Methods. 2002;50(3):319–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


