Abstract
Major impediments to conveyance of intravenously administered drugs to tumors are biofouling, opsonization, and rapid clearance from the circulation by macrophages and reticuloendothelial phagocytes. Cloaking nanoparticles with stealth epilayers partly overcomes these hurdles but it also foils interactions with tumor cells. Here, we describe the synthesis, characterization, and validation of smart gold nanords (GNRs) that spontaneously transform from inert passengers in the blood stream to active cell-penetrating nanoparticles within tumors to potently sensitize tumors to radiation therapy. Intrinsically cationic and cell-penetrating GNRs were shielded from phagocytosis with a cloaking polyethylene glycol epilayer containing an intervening cleavable peptide. In the absence of an external trigger, this epilayer is clipped off by the tumor microenvironmental protease, cathepsin B, in colorectal cancers to uncloak and expose the free-circulating native unPEGylated GNR that is readily internalized by cancer cells and turn into immovable small clusters of GNRs. Selective uncloaking of GNRs in the tumor reduced off-target toxicity confirmed by hematologic, biochemical, and histopathological analysis of blood, serum, and normal organs, respectively. Subsequent irradiation led to significant tumor growth delay and improved survival of mice. By addressing multiple barriers to efficient transport and cellular internalization of nanoparticles, our results demonstrate that clinically meaningful radiosensitization can be achieved with rationally designed GNRs.
Keywords: cathepsin B, gold nanorod, cleavable peptide, radiation therapy, colorectal cancer
Introduction
An increasing appreciation of the challenges with selective and efficient delivery of therapeutic payloads to tumors via intravenous administration of nanocarriers has fueled the quest for novel approaches to tackle this vexing problem. The journey from the intravenous injection site to the tumor is riddled with obstacles. First, the extraneous ‘foreign’ nanoparticle is tagged with opsonins that form a protein corona around the particle, flagging it for complement activation, engulfment by circulating macrophages, and rapid clearance from circulation. Second, upon encountering resident phagocytic cells in the liver and spleen, the nanoparticle is again efficiently eliminated from the circulation. Third, the nanoparticle may ride within the center of the advancing laminar flow column of blood in a blood vessel such that it does not contact the endothelial lining of tumor neovasculature to facilitate tumbling through the wide interendothelial fenestrations and pores that are the hallmark of leaky chaotic immature neoangiogenic blood vessels, the enablers of the enhanced permeability and retention effect. Fourth, upon extravasating from the tumor vasculature, the nanoparticle faces a harsh tumor microenvironment that is not conducive for interactions with cancer cells. Not surprisingly, an appraisal of published reports of nanoparticle delivery to tumors concluded that on average only 0.7% of intravenously administered nanoparticles reach the tumor as a consequence of these pharmacokinetic impediments.[1] Yet, nanoparticles have shown promise in terms of minimizing systemic exposure to drugs and thus reducing toxicity of some chemotherapeutic agents. Clearly, newer techniques are needed to improve tumor-specific delivery of nanoparticles to tumors and to subcellular compartments where they exert their therapeutic effects.
One such strategy is to exploit the intrinsic difference between the extracellular microenvironment of tumors and that of the blood stream or normal tissues to transform an inert non-interactive nanoparticle into a cell-penetrant nanoparticle. A hallmark of cell penetrating peptides, exemplified by the transactivator of transcription (TAT) protein of the human immunodeficiency virus (HIV), is the positive charge density on their surface that is thermodynamically favorable for interaction with negatively-charged sialic acids on the outer membrane of cells.[2] And a hallmark of tumor microenvironments is the abundance of extracellular proteases secreted by proliferating dysplastic cancer cells to loosen the intracellular matrix, degrade and invade the basement membrane, and disseminate distantly via vascular or lymphatic channels.[3] Coupling these two hallmarks together, we sought to design a nanoparticle that is inherently cationic and cell-penetrant but cloaked in an armor that shields it from non-specific cellular interactions outside the tumor (in the blood steam, with RES cells, or normal tissue cells) but spontaneously shed this cloak when it encounters the proteases in the tumor microenvironment and thus penetrates tumor cells. In doing so using a cylindrical nanoparticle, we address each of the four aforementioned barriers to conveying nanoparticles to tumors.
In the initial embodiment of this paradigm, we chose colorectal cancer, a disease where extant standard-of-care for rectal cancer is preoperative chemoradiation therapy followed by surgery. Unfortunately, only about 20% of patients achieve a pathologic complete response (pCR) and could have undergone organ-preserving treatment without the need for radical surgery. In this disease, a preponderance of data suggests that radiation dose escalation can improve pCR rates but the ability to adequately escalate the radiation dose is limited by the tolerance of adjacent normal tissues. Among strategies to escalate radiation dose, there is a convergence of literature that confirms that infusing a tumor with high atomic number (Z) element-containing nanoparticles greatly increases the microscopic radiation dose deposited within the vicinity of the nanoparticle, via an increase in secondary electron showers, and thereby increases treatment response.[4–15] We therefore chose a gold (Z=79) nanoparticle as our prototype nanoparticle for delivery to colorectal cancers.
The tumor microenvironment of colorectal cancers is characterized by the abundance of the cysteine protease cathepsin B secreted by their invading tumor cells to degrade components of the extracellular matrix. Cathepsin B, coded by the gene CTSB, is located at chromosome 8p22 and is expressed physiologically by the human body. This enzyme is responsible for antigen processing, hormone activation, and bone turnover. Pro-cathepsin B, a proenzyme form of cathepsin B tethers to cell surface in a calcium dependent manner via the Annexin II tetramer complex. However, in the tumor microenvironment, the pro-cathepsin B no longer binds to Annexin II during its conversion to cathepsin B causing protein overexpression in the extracellular matrix.[16] Expression of cathepsin B in the tumor matrix aggravates tumorigenesis and is responsible for cancer invasion and metastasis.[17] This is observed in cancers of the breast, cervix, colorectum, stomach, bladder, ovary, thyroid, and prostate.[18]
In this study, we report the synthesis, characterization, optimization, and validation of a gold nanorod (GNR) construct that is decorated with a cathepsin B cleavable peptide sequence interspersed within its stealth polyethylene glycol (PEG) epilayer. In controlled experiments in vivo, we demonstrate that this construct is non-toxic, displays excellent pharmacokinetic and biodistribution properties, and potently sensitizes colorectal cancer xenografts to radiation therapy via tumor microenvironment-specific cleavage of the peptide to dePEGylate the GNR, exposure of the native cationic GNR surface, and internalization into tumor cells.
Materials and Methods
Synthesis and characterization of peptides
The following proteins were purchased from Peptide 2.0 (Chantilly, VA) with >97% purity: GFGFVG, CR9GGFGFVG, and GGFGFVGGR9WC, stored at −20 °C and used without further purification. p-Nitrophenyl carbonate polyethylene glycols (HS-PEG20k-OMe and NPC-PEG20k-OMe) was purchased from Laysan Bio (Arab, AL).
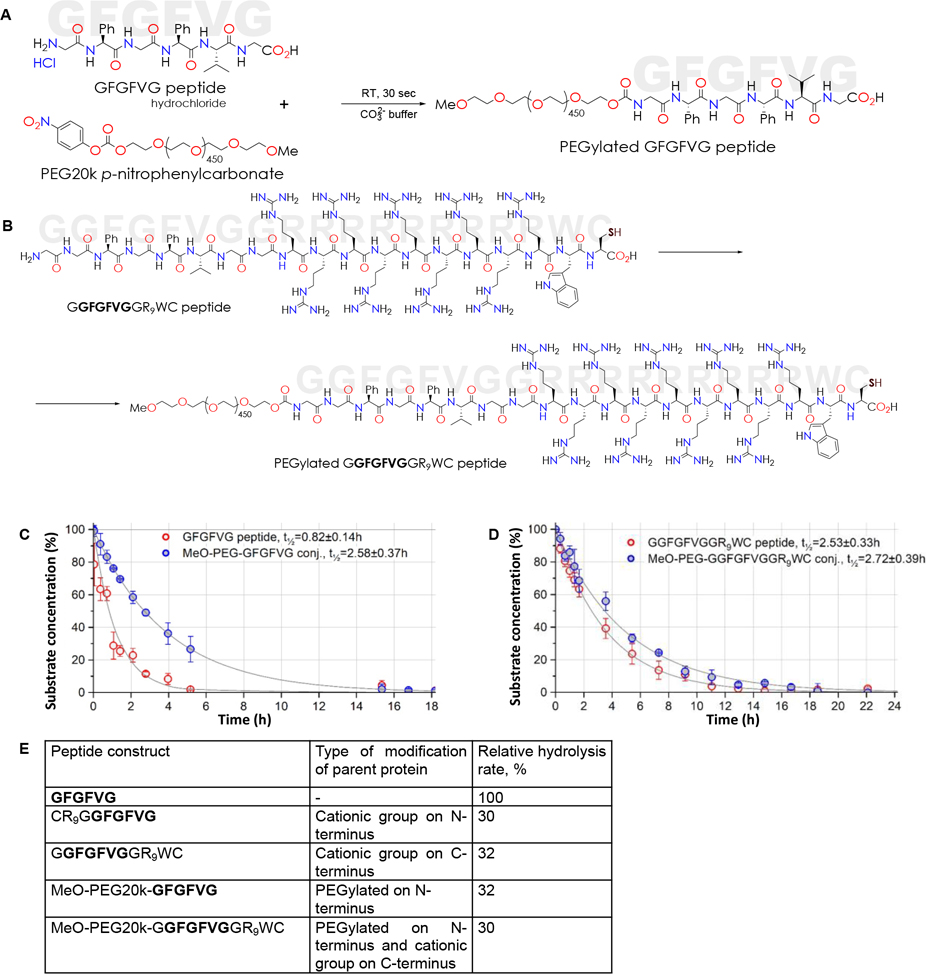
Building on the highly specific hexapeptide GFGFVG that is cleaved by cathepsin B, we first added a cationic spacer comprised of 9 arginine moieties (the guanidinium groups of an arginine residue maintains a positive charge) and the cysteine (for future attachment to GNRs) to either the N-terminus or the C-terminus to see the effect of this spacer on hydrolysis rate of GFGFVG. When hydrolysis rate at 25 °C was quantified by HPLC measurement of substrate concentration at serial time points up to 24 hr after addition of cathepsin B, the hydrolysis rate was noted to be somewhat similar between the constructs with N-terminus cationic spacer and C-terminus cationic spacer. We chose the construct with the cationic spacer on the C-terminus for future experiments since N-terminus derivatization with 20 kDa PEG could be readily performed using “good leaving group” chemistry. As such, two constructs were created: a PEGylated peptide (MeO-PEG20k-GFGFVG) and a PEGylated peptide with a cationic spacer on the C-terminus of the peptide (MeO-PEG20k-GGFGFVGGR9WC). Briefly, 10 μM of each protein was dissolved in 1 mL of 0.1M carbonate–bicarbonate buffer solution (pH=8.5) and cooled to 4 °C. 1 mL of DMF solution of NPC-PEG20k-OMe cooled to 4 °C was added in 15 sec. Reaction mixture was left at 4 °C for 30 min, then allowed to warm up to RT. PEGylated product isolation was performed using Teledyne ISCO CombiFlash Rf+ Lumen LC system (Lincoln, Nebraska) with an integrated evaporative light scattering detector (ELSD). MeO-PEG20k-GFGFVG separation was performed with RediSep Rf Gold Silica Gel Disposable columns (particle size 20 – 40 μm, pore size 60Å, bed weight 4 g) with ELSD and UV-Vis detectors (at 253 nm) simultaneously. ELSD conditions were optimized to achieve maximum sensitivity: the spray chamber temperature was set at 60 °C, the nitrogen flow was maintained at 2.5 SLPM and the gain was set to 1. The mobile phase composition was a mixture of methanol (A) and 0.015M aqueous ammonium formate (B) in a gradient elution at a flow rate of 10 ml/min. The gradient started with a methanol–water phase (80:20, v/v) for 6 column volumes (CV). At 14 CV the composition of the mobile phase was methanol–water 40:60 (v/v). At 18 CV it changed to a methanol–water mixture of 25:75 (v/v) and at 20 CV to 10:90 (v/v). MeO-PEG20k-GGFGFVGGR9WC separation were performed with RediSep Rf SCX column (particle size 40–63 μm, pore size 100Å, bed weight 5 g) with ELSD and UV-Vis detectors (at 253 nm) simultaneously. The mobile phase composition was a mixture of 0.015M aqueous ammonium formate, pH=3.8 (A) and 0.015M ammonium formate, pH=8 in 15% acetonitrile (B) in a gradient elution at a flow rate of 10 ml/min. The gradient started with 100% A for 2 column volumes (CV). At 6 CV the composition of the mobile phase was A:B 90:10 (v/v). At 12 CV it changed to 100% phase B. PEGylated peptide conjugates were lyophilized and stored at −20 °C. Purity was verified by HPLC analysis. Figure 1A and 1B show the synthetic steps, for MeO-PEG20k-GFGFVG and MeO-PEG20k-GGFGFVGGR9WC conjugates, respectively.
Figure 1: Synthesis and characterization of peptide constructs.
(A) Conjugation schema for synthesis of MeO-PEG20k-GFGFVG. (B) Conjugation schema for synthesis of MeO-PEG20k-GGFGFVGGR9WC. (C) Hydrolysis kinetics of native GFGFVG hexapeptide and PEGylated GFGFVG when suspended in DI water and treated with Cathepsin B (1.34 × 10−13 M) at 25 °C. Progress curves with pseudo-first order kinetic half-life expressed as mean ± SD. (D) Hydrolysis kinetics of GGFGFVGGR9WC and PEGylated GGFGFVGGR9WC when dissolved in DI water and treated with Cathepsin B (1.34 × 10−13 M) at 25 °C. Progress curves with pseudo-first order kinetic half-life expressed as mean ± SD. (E) Table of hydrolysis rates of constructs demonstrating that hydrolysis rates of PEGylated or cationic spacer conjugated are slower than parent hexapeptide but that of cationic spacer + PEG construct is not any slower than PEGylated or cationic spacer conjugated constructs.
Synthesis and characterization of gold nanorods (GNRs)
Cetyltrimethylammonium bromide (CTAB, >99%), hydrogen tetrachloroaurate trihydrate (HAuCl4·3H2O, ≥49% gold basis), silver nitrate (AgNO3, ≥99%), hydroquinone (C6H6O2, ≥99.5%), and sodium borohydride (NaBH4, 99%) were purchased from Sigma-Aldrich. All solutions were freshly prepared before use, except for HAuCl4 (stored at 4 °C) and CTAB solutions (stored at 28 °C). Seedless synthesis of gold nanorods (GNRs) was performed according to methods described before, with slight modifications.[19–21] The typical synthesis had the following steps. 25 mL of CTAB (100mM) and 25 mL HAuCl4 (1mM) solutions were transferred to a 250 mL Erlenmeyer flask and the mixture was hand-stirred. After 15 min, 37.5 μL of 0.1M AgNO3 solution was added and hand-stirred followed by the addition of 1.25 mL of 50mM hydroquinone solution. Next, during magnetic stirring at a rate of 600 rpm, 75 μL of ice cold 10mM NaBH4 (aq.) was added in a single injection to the flask. After 1–2 min, the stirring was stopped and the reaction mixture was kept at 27 °C overnight. The as-synthesized GNRs were purified twice by centrifugation at 15,000 g for 30 min and redispersed into a small volume of 10 mM CTAB. The stock solution derived from the supernatant was analyzed by UV-Vis spectrophotometry. Transmission electron microscopy (TEM) was performed on a JEOL 1230 instrument operating at 80 kV. GNR size distribution calculations (for >200 nanorods) were performed using ImageJ (National Institutes of Health).
Synthesis and characterization of PEG GNR and peptide GNR conjugates
PEG GNR and peptide GNR conjugates were prepared by combining 1 mL of suspension of CTAB-stabilized GNRs with OD=2 with 1 mL of PEG solution, prepared with 2 mg of HS-PEG20k-OMe or PEG20k-GGFGFVGGR9WC in 0.05M sodium phosphate buffer, pH=6.9. The GNR suspensions were dialyzed using 50 kDa biotech grade cellulose ester membrane (Spectra/Por G235034) in 18 MΩ.cm DI water (Millipore) until phosphate and CTAB were completely removed, which was traced by measuring the electrical conductivity. The zeta potential values (ζ, mV) and average hydrodynamic particle size corresponding to the rotational diffusion (dh, nm) of resulting GNRs were determined using a ZEN 3600 Malvern Zetasizer equipped with a 633 nm laser (Worcestershire, UK). The surface potential and conductivity of all formulations and samples suspended in DI water were measured 3–5 times, with data presented as averages and standard deviations. Particles in DI water placed in 10 mm quartz cuvettes were analyzed on a Varian CARY-300 BIO UV-Vis spectrophotometer for their characteristic optical absorbance spectra measured over a range of 200 to 900 nm, with DI water used as the reference.
FTIR spectra were obtained with a Thermo Nicolet 6700 spectrometer (Thermo Scientific, Waltham, MA) equipped with a Smart iTR accessory for ZnSe crystal ATR sampling. 0.5 3L of each sample was placed on the crystal and allowed to dry (PIKE Technologies, WI) using a N2-purged sample chamber. Data were collected between 4000 and 600 cm−1, 256 scans, 2 cm−1 spectra resolution at RT.
Mapping peptide cleavage by cathepsin B by HPLC
Chromatographic measurements were performed on an Agilent 1200 series HPLC system (Agilent Technologies, Santa Clara, CA, USA) with G1315C diode array detector. The system was equipped with a Jupiter 5 μm C18 300Å guard column (Phenomenex, Torrance, CA) and a Gemini C18 column 5 μm 110Å, 50×2 mm (Phenomenex). A gradient of A [0.1% trifluoroacetic acid (Alfa Æsar) in water] and B [0.1% trifluoroacetic acid in acetonitrile (HPLC gradient grade, VWR)] with increasing organic proportion was used for peptide separation (main separation ramp: 1.5% B to 85% B from 0 to 9 min, flow rate 750 μL/min).
In all experiments, each repeated three or more times, an equal amount of 1·10−7 M of protein substrate was used, 1.34·10−13 M Cathepsin B was activated using 1 mM dithiothreitol (DTT), and the reaction volumes were adjusted to 1 mL with 50 mM sodium citrate, pH=6.0. The solutions were sterilized by filtration using Corning 4 mm Syringe Filters, 0.2 μm pore RC Membrane, directly into a C4000–1W Target DP sample vessel (National Scientific Co., Duluth) and left in G1367C Autosampler (Agilent) in the dark at 25 °C. One μL volumes were drawn with a needle assembly from the vial and injected into the column; UV signal was recorded at 220 nm with 380 nm reference wavelength and peak areas integrated using ACD/Labs 2015 Pack 2 Spectrus Processor (ACDLabs, Toronto, ON). In each experiment injections were performed iteratively over a period at least 6 times longer than reaction half-life; each chromatogram was processed individually. The retention time of each protein substrate was determined before the experiment and remained constant during the set of HPLC runs.
The reaction progress curves were plotted as reaction time vs. % substrate concentration. Experimental peak areas were compared with the peak areas obtained by direct injection of a known amount of protein substrate considered as 100%.
Cell culture
Human colorectal adenocarcinoma cell lines (HCT116, HT29, SW480, DLD1), pancreatic adenocarcinoma cell lines (PANC1, BXPC3) and the breast cancer cell line (MDA MB231) were obtained from the American Type Culture Collection and cultured in complete media containing 10% FBS (Fetal Bovine Serum, Sigma Aldrich) and 1% penicillin-streptomycin (Sigma Aldrich). HCT116 and HT29 cell lines were cultured in McCoy’s media, SW480 and MDA MB231 in DMEM 1×, and DLD1, PANC1 and BXPC3 in RPMI 1640. Cells were incubated at 37 °C in 5% CO2.
Western blot analysis
Colorectal, pancreatic, and breast cancer cell lines were cultured in a T75 flask at 37 °C. Once the cells were 70% confluent, they were washed twice in ice-cold PBS, lysed using RIPA buffer (Sigma Aldrich), and protein concentration was determined using bicinchoninic acid (BCA) assay protein kit (Pierce, Thermo Fisher Scientific). 30μg of protein lysates were loaded onto a precast polyacrylamide gel and separated by electrophoresis after which they were transferred onto a PVDF membrane by wet electrophoresis. The membrane was blocked with 5% powdered milk in PBS and 0.05% Tween-20 at room temperature for 1 hr following which it was incubated overnight at 4 °C with cathepsin B recombinant rabbit monoclonal antibody (Invitrogen, ThermoFischer Scientific, 1:500). Anti-rabbit vinculin secondary antibody (1:3000) was later added and incubated for 1 hr at room temperature. Following antibody incubation, the membrane was washed thrice at 15 min intervals with PBS containing 0.05% Tween-20. The membrane was developed using Pierce chemiluminescent reagents. The blots were quantified using Image J.
Extracellular cathepsin B protein estimation
To estimate the levels of extracellular cathepsin B, semiconfluent colorectal, pancreatic, and breast cancer cells were serum deprived for 24 hrs.[17] The media was then collected and centrifuged at 1300 rpm for 5 min to remove dead cells and debris. 5 mL of this media was collected and concentrated using an Amicon® Ultra 10kDa centrifugal filter (Millipore Sigma). The protein concentration of the cell lysates was determined using the BCA assay kit. A volume of media proportional to the protein concentration of the lysates was used for electrophoretic separation and Western blot analysis.
Enzyme-linked immunosorbent assay (ELISA)
Cathepsin B human ELISA kit was purchased from Abcam (ab119584) and stored at −20 °C. Cancer cells were cultured at 37 °C until semi-confluent. The cells were then serum deprived for 24 hr following which 5 mL media was collected, centrifuged at 1500 rpm at 4 °C for 10 min. All reagents and working standards were prepared according to ab119584 cathepsin B human ELISA kit protocol. The protein assay was performed as per the manufacturer’s instructions. Briefly, 100 μL of standards and samples were added to each of the 96-well plates and incubated at room temperature for 2 hr. After washing three times with 0.01M PBS, the conjugates were added and incubated at room temperature for 2 hr. The plates were washed thrice with washing buffer and 90 μL of substrate solution was added and incubated at room temperature for 30 min. The absorbance was measured using BioTek Cytation5 plate reader at 450 nm.[22]
In vitro uptake of PEG GNR and peptide GNR
To measure the cellular internalization of PEG GNR and peptide GNR, colorectal, breast, and pancreatic cancer cell lines were cultured in a 35 mm petri dish with 1 mL of culture media. Once they were 70% confluent, 10 μM of CA074, a specific inhibitor of cathepsin B, was added and incubated for 12 hr following which cells were incubated in serum free media containing 0.05OD of PEG GNR (Au: 0.73 μg/mL) and peptide GNR for 24 hr.[23] Cells were washed thrice with PBS, trypsinized with 0.25% trypsin-EDTA (ThermoFisher), neutralized in 1 mL of complete media, and centrifuged at 1300 rpm for 5 min. The pellets were resuspended in 1 mL of PBS after which they were transferred to liquid scintillation vials (Sigma Aldrich) and allowed to evaporate at 56 °C. Cells were digested using 1 mL of aqua regia (prepared by mixing hydrochloric and nitric acids, Fisher Scientific) for 4 hr at 60 °C and diluted in 1% hydrochloric acid. The elemental gold content in each of these samples was measured using inductively coupled plasma mass spectrometry (ICP-MS) (Agilent 7900) after calibration with serial dilutions of TraceCERT® gold standards (Sigma-Aldrich, 38168) and normalized to cell count.[24]
Immunostaining
Cells were fixed in 100% ice cold methanol for 3 mins at room temperature following which they were blocked using 2% bovine serum albumin (BSA) for 20 mins at room temperature. Cells were then washed thrice with PBS and incubated overnight at 4 °C with primary antibody (1:100) in blocking buffer. Cells were washed three times with PBS and incubated with secondary antibody (AlexaFlour 594, Thermo Fisher, 1:200) in blocking buffer for 30 mins in a dark room at room temperature. Cells were rewashed thrice with PBS, mounted using mounting medium with DAPI (abcam), and visualized under Leica DMI 6000 microscope.
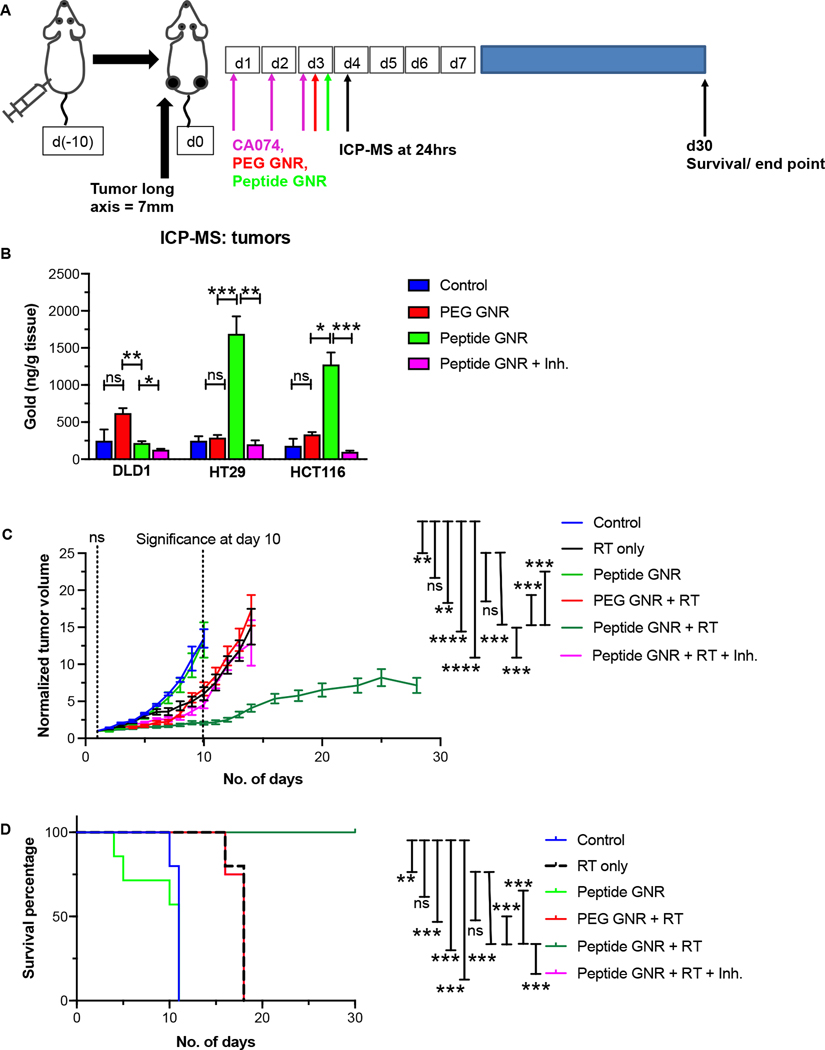
In vivo tumor inoculation and treatment with GNR
All mice used in these studies were purchased from The University of Texas MD Anderson Cancer Center Experimental Radiation Oncology Mouse Facility and maintained in accordance with the protocol and guidelines of Institutional Animal Care and Use Committee (IACUC). Adult male Swiss nude mice bearing bilateral subcutaneous HT29, HCT116, and DLD1 xenograft tumors (100 μL of 1×106 cells in PBS) were used for this study. Once the tumors reached approximately 7–8 mm in diameter, mice were randomized into four groups (untreated, PEG GNR, peptide GNR, peptide GNR + inhibitor). 10 mg/Kg body weight of CA074 (MedChem Express LLC, NJ) was given intraperitoneally for 5 consecutive days, followed by 8 OD of PEG and peptide GNR (Au: 117 μg/mL) given intravenously via the tail vein on day 3 of CA074 injection.[23, 25]
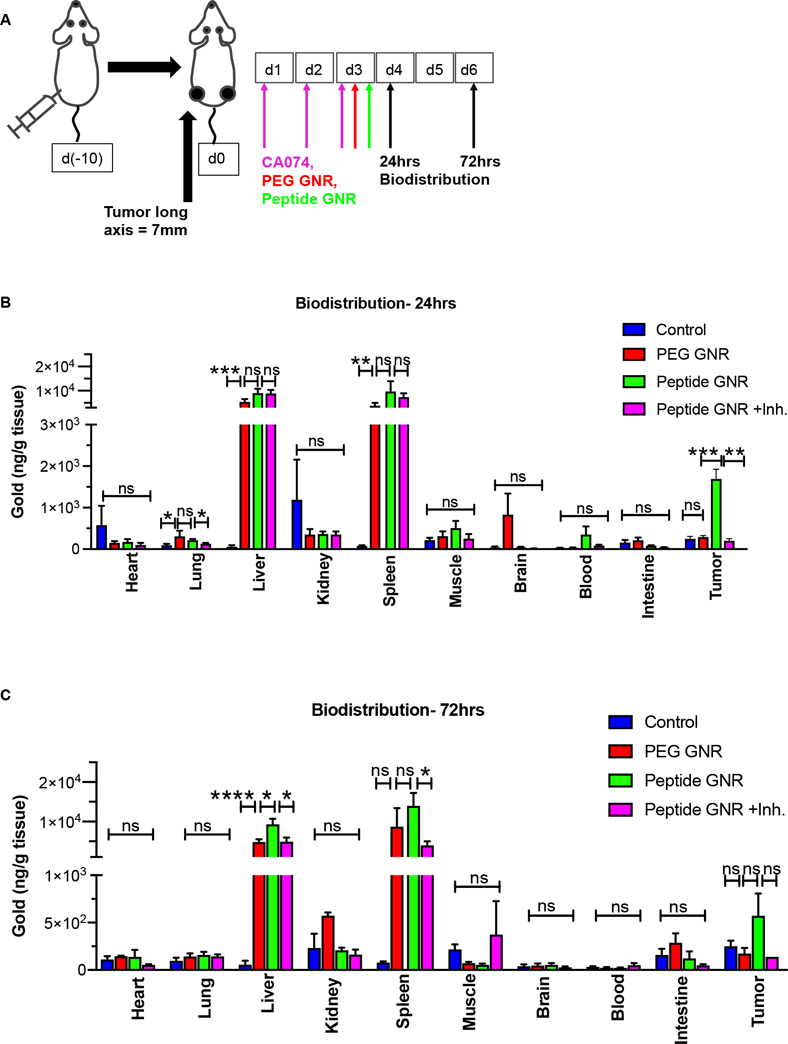
In vivo biodistribution and pharmacokinetics
24 hr and 72 hr post-treatment, mice were euthanized, and tissues (heart, lung, liver, spleen, kidney, blood, intestine, tumor, brain, muscle) were harvested, weighed, lyophilized, digested in aqua regia, and quantified using ICP-MS to measure the elemental amount of gold (ppb/g) present per gram of dry tissue.
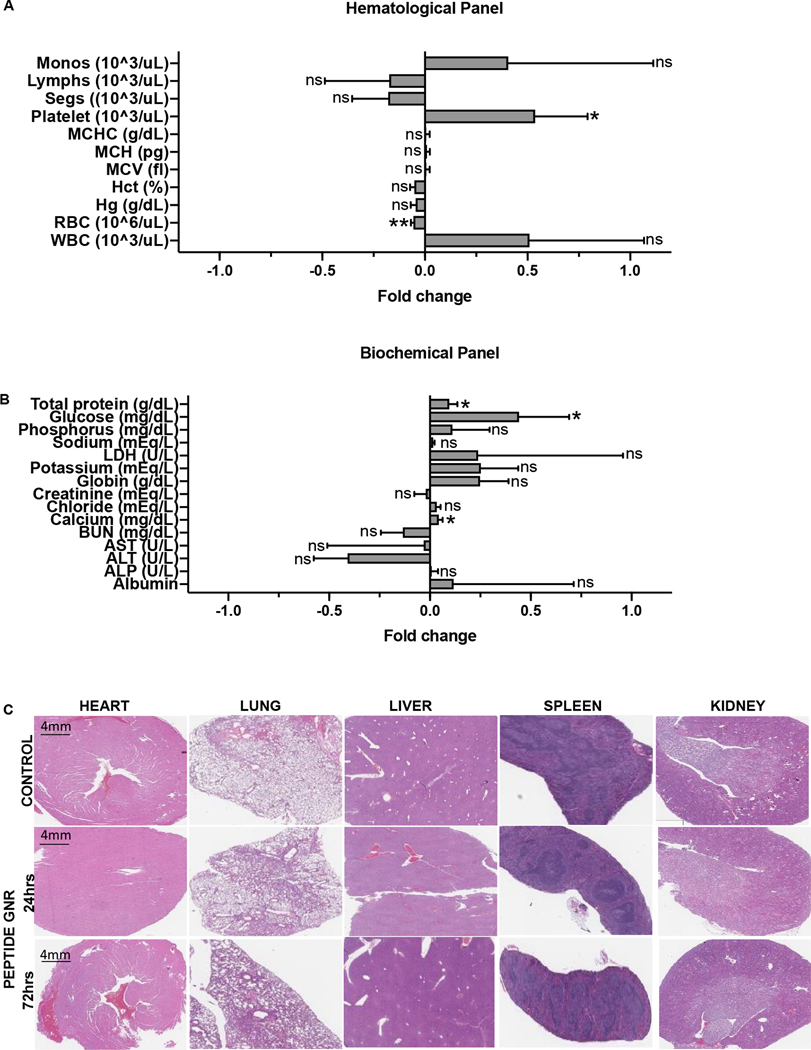
In vivo toxicity study
This experiment was performed with the same mice used for the biodistribution study. Mice were sacrificed 24 hr and 72 hr after treatment with PEG GNR and peptide GNR and samples were collected for hematological, biochemical, and histopathological analysis. A panel of 11 hematologic parameters; red blood cell count (RBC), hemoglobin (Hg), white blood cell count (WBC), platelet count, monocyte count (Monos), lymphocyte count (Lymph), segmented neutrophils (SEGS), hematocrit levels (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), and mean corpuscular hemoglobin concentration (MCHC), and 16 biochemical parameters; glucose, total protein, albumin, globulin, bilirubin, aspartate aminotransferase (AST), alkaline phosphatase (ALP), sodium, potassium, calcium, phosphorus, chloride, lactate dehydrogenase (LDH), blood urea nitrogen (BUN), and creatinine were measured using an autoanalyzer. Blood was drawn from the facial vein of mice and collected in potassium EDTA tubes and lithium heparin collection tubes for hematological and biochemical analysis, respectively. For the histopathological toxicity study, major organs (heart, lung, liver, spleen, kidney) were collected and fixed in 10% buffered formalin at 4 °C overnight following which they were embedded in paraffin. Tissues were sectioned at 4 μm thickness and stained with hematoxylin and eosin (H&E), imaged using a Leica DMI750 microscope at 400X, and examined by a veterinary pathologist.
In vivo tumor growth inhibition and survival
Twenty-four hours post-GNR treatment, mice were irradiated with a single dose of 10 Gy using a Philips RT-250 orthovoltage unit. Tumors were measured using a digital Vernier caliper along their long axis (L) and short axis (S) one day prior to radiation for baseline volume estimation using the formula L × S2/2. Subsequently, post-treatment tumor dimensions were measured roughly thrice weekly and calculated tumor volumes for each mouse were normalized to the baseline volumes. Mean tumor volumes were calculated for each group and plotted against time until all the mice had tumors measuring 20 mm after which they were euthanized according to institutional policies. Survival data was plotted as a Kaplan-Meier curve with survival times measured from the date of GNR treatment to the date of euthanasia.
Clinical database analysis of cathepsin B association with survival
To demonstrate the association between CTSB and disease-specific survival in patients, we analyzed clinical data and RNASEQ data of ~1650 patients downloaded from UCSC Xena and performed survival analysis across 3 major cancer types: breast, colorectal, and pancreatic cancer.[26] The clinical data and RNASEQ data were downloaded from UCSC Xena. The expression data was divided into high CTSB and low CTSB based on quartile CTSB gene expression. Disease-specific survival analysis was plotted collectively and individually for these cancers using the Kaplan-Meier method.
Statistical analysis
All the results are presented as mean with its corresponding standard error of mean (SEM). The Student’s t-test was used to analyze differences between treatment groups. Values were considered statistically significant if the two-tailed p-value was <0.05. Differences between groups in the Kaplan-Meier survival curves were analyzed using the log rank test, with the results being considered significant if the two-tailed p-value was < 0.05. All statistical tests were performed using R statistical software version 3.3.1.
Results
PEGylation and cationic spacer conjugation reduce cathepsin B-mediated hydrolysis of GFGFVG but dual conjugation does not reduce it further
The degradation reaction half-lives (t½) of native GFGFVG hexapeptide and PEGylated GFGFVG when suspended in DI water and treated with Cathepsin B (1.34 × 10−13 M) at 25 °C, computed by the least-squares regression method, are shown in Figure 1C. The mere modification of the peptide with PEG increases the half-life nearly three-fold and reduces the hydrolysis rate to ~32% of hydrolysis rate of peptide alone (100%). Similarly, addition of cationic spacer to either the N-terminus or the C-terminus of the peptide reduces the hydrolysis rate to ~30% and ~32% of hydrolysis rate of peptide alone. We, therefore, chose the C-terminus for addition of the cationic spacer and the ‘good leaving group’ chemistry for PEGylation on the N-terminus. The MeO-PEG20k-GGFGFVGGR9WC construct had a hydrolysis rate of ~30% and a half-life about three-fold higher than native peptide (Figure 1D); and importantly not very different from those of either the PEGylated peptide or the cationic spacer conjugated peptide. Based on these results (Figure 1E) we conclude that a construct with PEG on the N-terminus and a cationic spacer on the C-terminus will allow us to now conjugate a cathepsin B-cleavable epilayer to the surface of a GNR such that once cleaved by the enzyme, the residual cationic spacer will cause internalization of the nanoparticle into cells.
Peptide GNRs are dePEGylated efficiently by cathepsin B
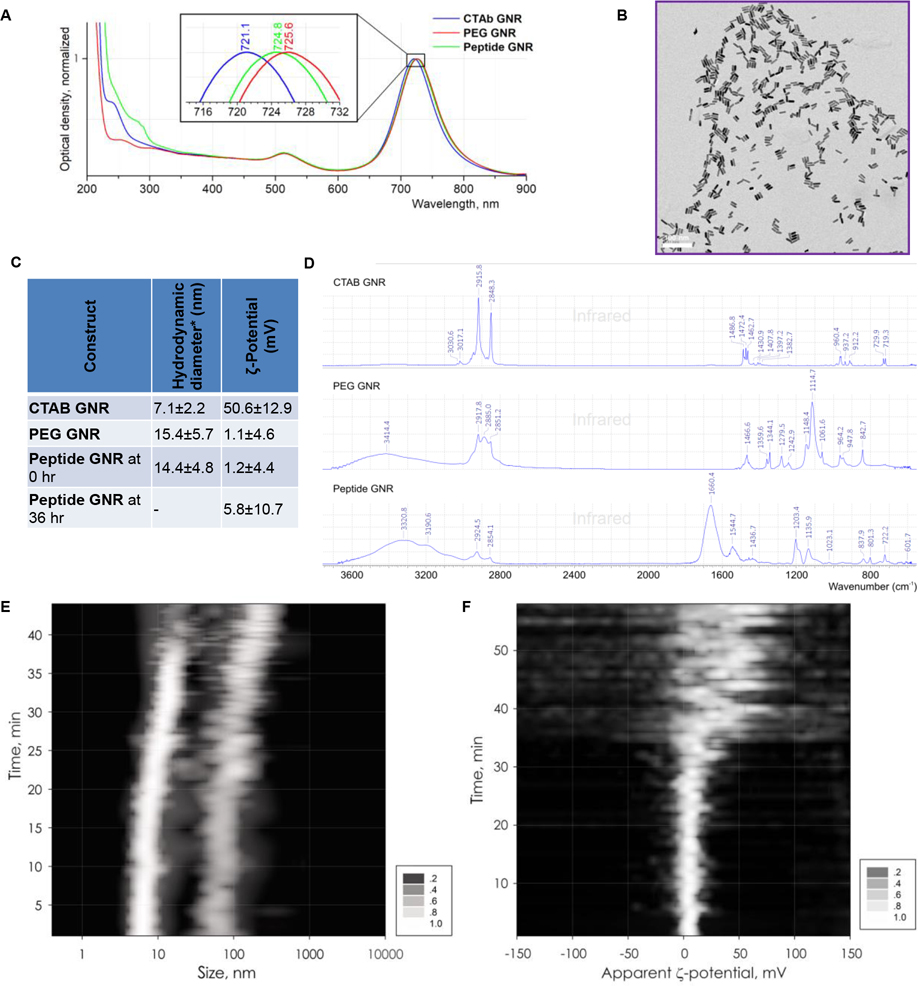
When the above constructs (PEGylated cationic spacer linked peptide GGFGFVGGR9WC and HS-PEG-OMe) were conjugated to GNRs, the resulting constructs were stable and retained characteristics of GNRs. They are hereafter referred to as peptide GNR and PEG GNR, respectively. As seen in Figure 2A, decoration of GNRs with PEG or peptide results in a red shift of the peak wavelength (of the second peak in the classical bimodal distribution of GNRs) from 721 nm for CTAB GNRs to 726 nm for PEG GNRs and 725 nm for peptide GNRs. TEM images showed uniform size and shape of the GNRs measuring 20.38±2.15 nm in length, 5.95±0.45 nm in width/diameter for an aspect ratio of 3.46±0.56 (Figure 2B).
Figure 2: Synthesis and characterization of GNR constructs.
(A) UV spectra of CTAB stabilized GNRs, PEG GNRs, and peptide GNRs in DI water at 25 °C demonstrate bimodal GNR spectral absorbance with short axis near 510 nm and long axis near 720 nm with red-shifting upon PEG/peptide conjugation. (B) Transmission electron microscopy image of GNRs. (C) Table shows hydrodynamic particle size (* peak corresponding to the rotational diffusion of a bimodal distribution of particles) and ζ-potential of CTAB stabilized GNRs, PEG GNRs, and peptide GNRs at 0 hr and 36 hr after Cathepsin B treatment at RT (1.34 × 10−13 M). Peak, corresponding to the rotational diffusion (D) FTIR spectra of CTAB stabilized GNRs, PEG GNRs, and peptide GNRs. (E) Composite plot of dynamic light scatter at different time points after Cathepsin B treatment showing aggregation of GNR at ~40 min due to dePEGylation. (F) Composite plot of ζ-potential at different time points after Cathepsin B treatment showing aggregation of GNR at ~40 min due to dePEGylation.
Similarly, dynamic light scattering (DLS) size distribution plots of CTAB GNRs, PEG GNRs, and peptide GNRs showed a bimodal apparent hydrodynamic diameter distribution, with a lower size peak corresponding to the translational diffusion mode (Γtr), and the second peak arising from mixed diffusions (Γmix = Γtr + Γrot, rotational and translational),[27] as seen in Supplementary Figure S1. As expected, size increased slightly with PEGylation and peptide conjugation (Figure 2C). Similarly, the surface potential also reduced from highly positively charged for CTAB GNRs to near neutral for PEG GNRs and peptide GNRs (Figure 2C). To document the aggregation process triggered by cathepsin B-mediated dePEGylation of peptide GNRs we stacked a series of individual DLS plots of GNRs taken at 30 sec intervals to create a composite plot of hydrodynamic diameter dynamics over a ~40 min interval. Figure 2D shows the spectrum of CTAB GNR on the top with distinctive features of the cationic surfactant: asymmetric and symmetric CH2 stretchings at 2916 and 2848 cm−1, respectively; peaks in CH2 wagging region (1383, 1408, 1431 cm−1); symmetric N–CH3 and C–N bands at 1397 and 912 cm−1, respectively; and CH2 scissoring vibration and asymmetric bending mode of the head [N(CH3)3] methyl groups between 1460 and 1490 cm−1. The middle spectrum of PEG GNR has all distinctive features of polyethylene glycol: asymmetric CH2 stretchings at 2918 cm−1 and symmetric at 2885 and 2851 cm−1; CH2 scissoring stretching at 1467 cm−1; C-O-C asymmetric and symmetric stretchings at 1115 (highest intensity) and 843 cm−1, respectively. The bottom spectrum of peptide GNR has characteristic features of both PEG and protein, including peaks typically observed in peptide bonds: C=O stretching at 1660 cm−1 and N-H in-plane bending at 1545 cm−1. The catalytic hydrolysis experiments were repeated multiple times, and Figure 2E shows a result of superposition of seven such experiments as a single image, displaying how both rotational and translational diffusion-based hydrodynamic diameters change after addition of 1.34·10−13 M of DTT-activated Cathepsin B to a suspension of peptide GNRs at 37 °C. These changes were not detected by the naked eye over a short period of time, but confirm the process of formation of larger aggregates, which is initiated by catalytic loss of the PEG envelope of GNRs. We view this as indicative of a change in particle behavior from free-circulating PEG GNRs to immovable small clusters of GNRs which are cell penetrant and can enhance radiosensitivity.
Surface charge distribution measurements, performed on the same samples by ζ-potentiometry, demonstrated a shift from a distinct positive charge of +50.6±12.9 mV of CTAB GNRs to near neutral charge of PEG GNRs and peptide GNRs after removal of the surfactant (CTAB) (Figure 2C, Supplementary Figure S1B, D, F). To get a more detailed view of how the surface charge of peptide GNRs evolves during the process of catalytic dePEGylation, composite ζ-potential plots were obtained in the same manner as the time course DLS plots, described above. Figure 2F shows the result of four stacked measurements at 37 °C demonstrating evidence of the GNR dePEGylation progress, resulting in ζ-potential change from nearly neutral to a distinctly positive over a ~50 min interval, when cathepsin B cleaves the peptide to expose the cationic spacer and facilitate cellular uptake.
Lastly, similar to the hydrolysis experiments with peptide alone, we performed HPLC-based analysis of cathepsin B cleavage kinetics of peptide GNRs where the degradation product concentrations were recorded instead of substrate concentrations. Since this was performed at 25 °C, the kinetics of cleavage was slower than that revealed indirectly by DLS and ζ-potentiometry; yet, this provides more objective evidence of hydrolysis (Supplementary Figure S2).
Extracellular cathepsin B facilitates in vitro uptake of peptide GNRs.
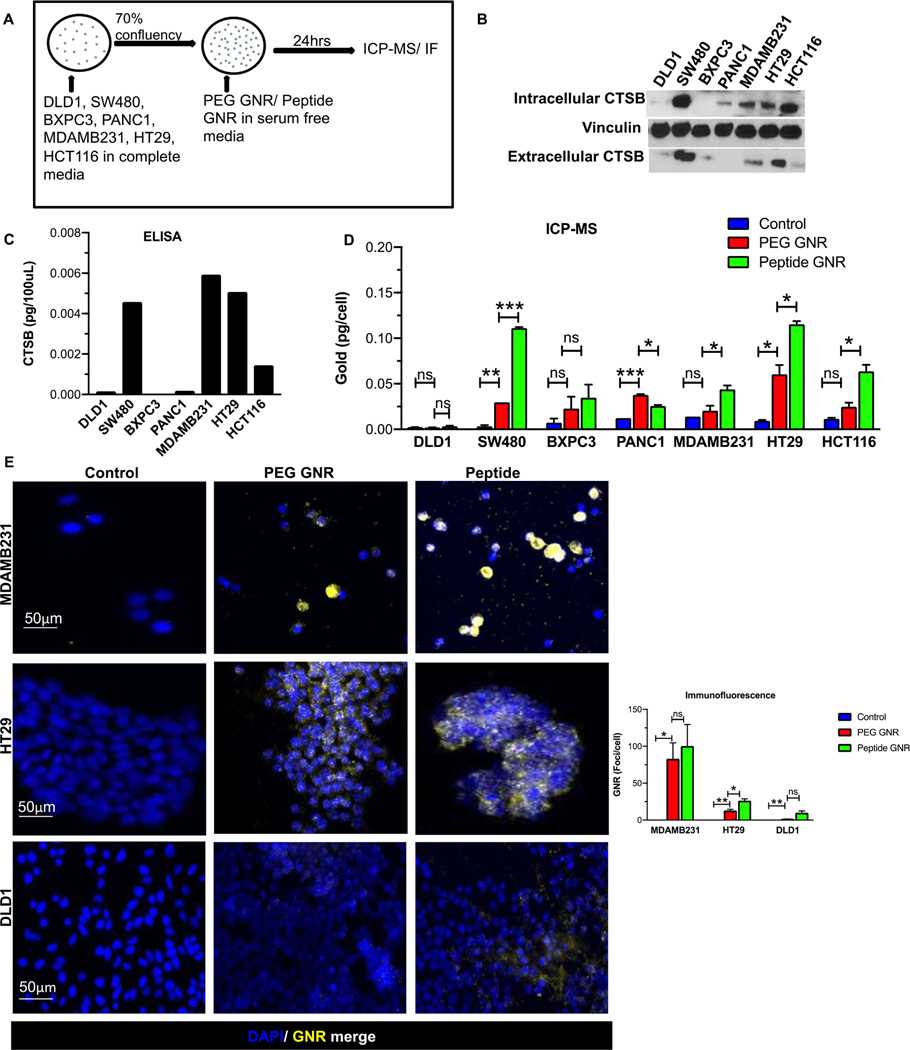
To demonstrate that cathespin B facilitates cellular internalization of GNRs, we chose a panel of human colorectal, breast, and pancreatic cancer cell types with differential cathepsin B expression namely, SW480, HCT116, HT29, DLD1, MDAMB231, BXPC3, and PANC1. A schematic of the experimental approach is displayed in Figure 3A. The protein levels of cathepsin B in each of these cell lines were analysed using Western blots (Figure 3B) and ELISA (Figure 3C). These data show the presence of abundant extracellular cathepsin B in SW480, MDAMB231, and HT29 cells, modest expression of extracellular cathepsin B in HCT116 cells, and low to no expression in DLD1, PANC1, and BXPC3 cells (Figure 3B & 3C). Although these treatment conditions used serum-free media, as noted later, the in vivo treatment conditions mimicked a possible clinical scenario where GNRs were administered intravenously and were therefore suspended in serum-rich conditions.
Figure 3: Cathepsin B accelerates cellular uptake of peptide-GNRs.
(A) Schematic representation of experimental setup. (B-C) Cathepsin B protein expression in different human cancer cells (Colorectal cancer - HT29, HCT116, SW480, DLD1; Pancreatic cancer - BXPC3, PANC1; Breast cancer - MDAMB231) obtained by (B) western blotting and (C) ELISA. (D) ICP-MS analysis of gold content in human cancer cell lines incubated with media, PEG GNRs, and peptide GNRs. Error bars represent standard error of the mean (SEM). (E) Representative images from fluorescence microscopy showing PEG GNR and peptide GNR uptake in high Cathepsin B expressing cell lines MDAMB231 (top panel) and HT29 (middle panel), and low Cathepsin B expressing cell line DLD1(bottom panel). Bar chart on the right depicts number of GNR foci per cell. * p<0.05, ** p<0.01, *** p<0.001, ns - not significant. Data were analyzed by the unpaired two-tailed t-test and expressed as mean ± SEM.
Next, to show that extracellular cathepsin B was responsible for cellular uptake of peptide conjugated GNR, we analysed the amount of gold present in various cancer cell lines qualitatively and quantitatively using dark field fluorescence microscopy and ICP-MS, respectively. Our ICP-MS analysis demonstrated a significantly higher uptake of peptide GNR when compared to PEG GNR in SW480, BXPC3, MDAMB231, HT29, and HCT116 cells (p < 0.05) (Figure 3D). Additionally, dark field microscopic images demonstrate selective uptake of peptide GNR in cancers cells that have high extracellular cathepsin B expression. Cell lines MDAMB231 and HT29, that express high cathepsin B, showed abundant GNR internalization by the cells when compared to PEG GNR whereas DLD1, which has no extracellular cathepsin B expression, showed no GNR uptake (Figure 3E). This indicated that enhanced uptake of peptide GNR corresponds to the expression levels of extracellular cathepsin B which facilitates its internalization.
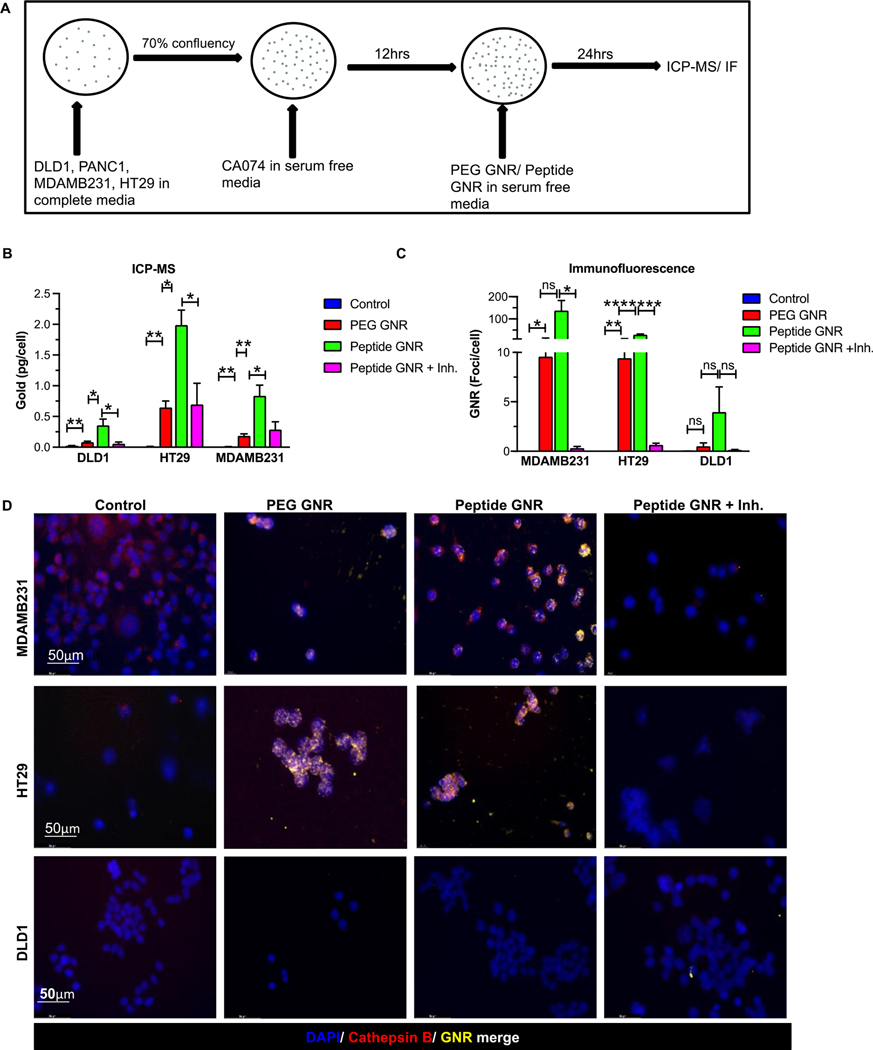
Inhibition of cathepsin B abrogates cellular internalization of peptide GNRs
To show that extracellular cathepsin B facilitates selective uptake of peptide GNRs, cells were treated with CA074, a selective cathepsin B inhibitor, and analysed using ICP-MS and immunofluorescence (Figure 4A). ICP-MS demonstrated that HT29, MDAMB231, and PANC1 cells, when treated with peptide GNR along with the cathepsin inhibitor CA074, showed a two-fold reduction in the GNR uptake as compared to cells treated with peptide GNRs (Figure 4B). Further, immunofluorescence imaging identified intracellular GNRs in the presence of cathepsin B but low to no GNR uptake when treated with the cathepsin inhibitor, CA074 (Figure 4C & 4D). Taken together, these findings confirm that cathepsin B facilitates selective uptake of peptide GNRs by cancer cells and the cathepsin B inhibitor CA074 abrogates this uptake.
Figure 4: Inhibition of cathepsin B abrogates in vitro uptake of GNRs.
(A) Schematic representation of experimental design. (B) Quantitative analysis of gold uptake by ICP-MS showing increased uptake of peptide GNR constructs that is abrogated by treatment with the Cathepsin B inhibitor, CA074. (C-D) Quantification of GNR foci per cell (C) and representative dark field microscopic images (D) of MDAMB231 (top panel), HT29 (middle panel) and DLD1 (bottom panel) cells treated with PEG GNRs, peptide GNRs, or peptide GNRs with inhibitor. These again confirm excellent internalization of GNRs by cells expressing Cathepsin B and abrogation of this uptake by treatment with the inhibitor. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns - not significant. Data were analyzed by the unpaired two-tailed t-test and expressed as mean ± SEM.
In vivo uptake of peptide GNRs
Adult nude mice with bilateral subcutaneous HT29, HCT116, and DLD1 xenograft tumors were randomized into four groups (untreated controls, PEG GNR, peptide GNR, and peptide GNR+ CA074). Once the tumors were 7–8 mm in size (around day 10 post-tumor injection) intraperitoneal injection of CA074 at 10 mg/kg body weight was given for 3 consecutive days followed by 100 μL of 8 OD PEG GNR and peptide GNR intravenously via the tail vein. 24 hrs later, mice were euthanized, and tumors were harvested to measure gold concentration using ICP-MS. (Figure 5A)
Figure 5: In vivo tumor growth inhibition and survival.
(A) Schematic representation of experimental approach. (B) ICP-MS analysis of gold content in HT29, HCT116, and DLD1 xenograft tumors in athymic Swiss nude mice (n=4 mice/group) 24 hr after GNR administration shows excellent uptake of peptide GNRs by high extracellular cathepsin B-expressing HT29 cells, followed by moderate extracellular cathepsin B-expressing HCT116 cells, and minimal uptake of peptide GNRs by low extracellular cathepsin B-expressing DLD1 cells. Treatment with inhibitor abrogates the increased uptake seen in HT29 and HCT116 tumors. Data represents mean ± S.E.M. Unpaired two-tailed t-test. (C) Tumor regrowth curves following treatment with radiation (RT) in the presence of vehicle, PEG GNRs, peptide GNRs, and peptide GNRs with cathepsin B inhibitor (CA074) (n=10 mice/group). Difference in regrowth rates of RT-treated and peptide GNR + RT-treated tumors was highly significant with p <0.0005. By day 10, there was a two-fold reduction in the size of normalized tumor volumes in the peptide GNR + RT group when compared to the controls. This regrowth delay was abrogated by treatment with the inhibitor. (D) Mice treated with peptide GNR + RT remained alive at day 30 whereas none of the mice in the other treatment groups survived. Curves for PEG GNR +RT and Peptide GNR + RT + Inh. coincide with one another. Log-rank Mantel-Cox test. n=10 mice/group. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns - not significant. Other than the survival data, data were analyzed by the unpaired two-tailed t-test and expressed as mean ± SEM.
Quantitative analysis of tumors using ICP-MS showed that the presence of GNRs corresponded to the levels of cathepsin B expressed in each of these tumor types (Figure 5B). HT29 which had the highest cathepsin B levels demonstrated the highest uptake of GNRs followed by HCT116 (p < 0.005). DLD1 which expressed very low to no cathepsin B had very minimal GNR uptake. Additionally, the tumor samples that received peptide GNR treatment along with CA074 did not show any GNR uptake. These results suggest that peptide GNR uptake is dependent on cathepsin B levels and that CA074 blocks uptake of peptide GNRs.
Peptide GNRs potently sensitize tumors to radiation in vivo and improve survival of mice
To evaluate whether peptide GNRs retain their radiosensitizing properties in vivo, we perfomed a tumor regrowth delay experiment with adult nude mice harboring subcutaneous HT29 xenograft tumors. Once tumors were established, mice were randomized into six groups (untreated, RT only, peptide GNR, PEG GNR + RT, peptide GNR + RT, peptide GNR + RT + CA074) and tumor volumes were measured thrice weekly. After intraperitoneal injection of CA074 at 10 mg/Kg body weight for 5 consecutive days, 8 OD of PEG or peptide GNR were injected intravenously. 24 hrs after GNR injection, the mice were radiated with a single dose of 10 Gy radiation. Tumor volumes were normalized to pretreatment starting volumes and plotted against days after treatment (Figure 5C). Treatment effectiveness was assessed by comparing the slopes of curves generated by plotting log relative tumor volume to time. Peptide GNR + RT had a highly significant (p <0.0005) decrease in the rate of tumor regrowth after day 14 of treatment when compared to the untreated mice as well as the RT only group. Notably, this translates to an improvement in survival (Figure 5D). Mice treated with peptide GNR + RT + CA074 showed no reduction in tumor growth when compared to peptide GNR + RT. This indicates that cathepsin B facilitates uptake of GNR which radiosensitizes cancer cells, inhibits tumor regrowth, and consequently improves survival.
Invivo biodistribution confirms GNR uptake by tumors
To determine the biodistribution and pharmacokinetics of GNRs, we conducted ICP-MS analysis on different organs (heart, lung, liver, spleen, kidney, muscle, brain, blood, intestine) at 24 hrs and 72 hrs post GNR injection (Figure 6A). Adult nude mice with subcutaneous colorectal cancer xenografts were randomized into four groups (untreated controls, PEG GNR, peptide GNR, and peptide GNR+CA074). Following intraperitoneal injection of CA074 at 10 mg/kg body weight for 3 consecutive days, 100 μL of PEG GNR or peptide GNR at 8 OD was given intravenously via the tail vein. Elemental gold concentration per unit dry weight of normal organs, measured by ICP-MS, confirmed tumor uptake at both time points, and the expected high levels of uptake by the RES in the liver and spleen at 24 hrs (Figure 6B) and 72 hrs (Figure 6C).
Figure 6: In vivo biodistribution of GNRs.
Athymic Swiss nude mice bearing bilateral subcutaneous HT29, HCT116, and DLD1 xenograft tumors were randomized into 4 groups and treated with vehicle, CA074, PEG GNRs, or peptide GNRs (n = 4 mice/group). Mice were euthanized 24 hr and 72 hr after GNR administration and organs extracted for ICP-MS analysis of elemental gold content. (A) Schematic representation of experimental approach. (B) Biodistribution of GNRs in major organs at 24 hr. (C) Biodistribution of GNRs in major organs at 72 hr. Data represents mean ± S.E.M. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns - not significant. Data were analyzed by the unpaired two-tailed t-test and expressed as mean ± SEM.
Peptide GNR treatment is well tolerated with no appreciable toxicity
To assess biocompatibility of GNR constructs, we performed hematological, biochemical, and histopathological analysis of samples at 24 hrs and 72 hrs after intravenous infusion in mice (Figure 7). The hematological (Figure 7A) and biochemical (Figure 7B) analysis showed no significant changes following peptide GNR administration when compared to the controls. Presence of very small amounts of gold in the kidneys with no significant difference in the renal function test indicates that the particles are easily eliminated through the kidneys and are not toxic. Histopathological analysis of the liver showed mild to moderate Kupffer cell hyperplasia with aggregates of mononuclear cells (Figure 7C) and the spleen showed lymphoid aggregates and increased multilineage hematopoiesis. Additionally, there were no histopathological changes in the heart, lungs, and kidneys of mice that received peptide GNRs when compared to the untreated control.
Figure 7: Safety and biocompatibility analysis of GNRs.
(A) Hematological analysis 24 hr after peptide GNR administration. (B) Serum biochemical analysis 24 hr after GNR administration. (C) Histopathological analysis of major organs 24 hr and 72 hr after GNR administration. * p<0.05, ** p<0.01, ns - not significant. Data were analyzed by the unpaired two-tailed t-test and expressed as mean ± SEM.
Cathepsin B expression by human tumors is associated with worse survival outcomes
To understand the association between CTSB and overall survival in human subjects, we analysed tumor samples across 3 different cancer types: colorectal, breast, and pancreatic cacners, using the TCGA database. The TCGA database contains 1650 patients with clinically and genomically characterized tumors across these three cancer types. The difference in CTSB expression across various cancer types was plotted as a boxplot (Supplementary Figure S3A). We then compared the survival of these cancer patients stratified by high (upper quartile) and low (lower quartile) of CTSB gene expression. Analysis of 413 and 412 samples in high CTSB and low CTSB groups, respectively, revealed that the patients with high expression of CTSB had poorer overall survival than patients with low expression of CTSB (p ≤ 0.0001) (Supplementary Figure S3B). However, datasets with smaller numbers such as individual colorectal, breast, and pancreatic cancer cohorts (Supplementary Figure S3C–H) did not show a statistically significant difference in survival between high- and low-CTSB expressing tumors. Nonethess, the pooled analysis (Supplementary Figure S4A & 4B) is in agreement with our preclinical results that high CTSB expression is often associated with worse survival of patients.
Discussion
In this study, we demonstrate that protease cleavable GNR constructs can evade RES capture, transform into cell-penetrant GNRs within the tumor micorenvironment, and potently sensitize colorectal cancers to radiation therapy. These findings are consistent with previous studies demonstrating that cellular internalization facilitates radiosensitization.[28–31] Unconjugated GNPs predominantly accumulate in the “leaky” tumor microenvironment due to enhanced permeability and retention effect. [32–34] However, from our study, we can conclude that GNPs when conjugated with a tumor targeting moiety results in its increased accumulation at the tumor site, thereby enhancing radiosensitivity and inhibiting tumor growth following radiotherapy.
Cathepsin B, a cysteine protease belonging to the papain family, is physiologically present as inactive pro-cathepsin B in the rough endoplasmic reticulum and is trafficked via the Golgi apparatus and vesicles to the late endosome/lysosome and to the cell membrane and extracellular space via secretory lysosomes. The acidic environment inside the lysosome activates the enzyme which functions as both an endopeptidase and exopeptidase to mediate intracellular proteolysis and protein turnover.[35] Overexpression of cathepsin B in tumors, beginning at the premalignant stages and extending to the leading edge of invasive tumors, is associated with an imbalance between cathepsins and their endogenous inhibitors (stefins, cystatins, and kininogens). Extralysosomal cathepsin B has pleiotropic functions including degradation of tumor microenvironmental extracellular matrix proteins such as laminin, fibronectin, and collagen IV to facilitate tumor cell migration, invasion, and metastasis. Consistent with the notion that cathepsin B in the extracellular space drives aggressiveness of invasive cancers, our analysis also confirmed that patients with high cathepsin B expressing tumors had a significantly poorer survival when compared to the patients with low cathepsin B expressing tumors. (Figure S1) We therefore exploited this inherent characteristic of colorectal cancer to transform an otherwise cell-impenetrant nanoparticle into a cell-penetrant nanoparticle that is internalized by cells to mediate radiosensitization.
Passive accumulation of a cathepsin B responsive nanoconstruct in the extracellular matrix of the colorectal cancer microenvironment results in proteolytic conversion of the PEGylated neutrally charged construct to a dePEGylated cationic gold nanoparticle that is readily taken up by tumor cells. This relies on the active site of cathepsin B docking specifically and preferentially to the scissile peptide GFGFVG to trigger hydrolysis.[36] However, having a bulky constituent like PEG or a positively charged polyarginine moiety, in proximity to the GFGFVG group may result in reduced sensitivity to the hydrolytic effect of cathepsin B due to steric hindrance or electrostatic interactions. To build an optimal PEG-peptide-gold nanoconstruct, we evaluated several structures, based on the parent protein moiety GFGFVG, shown in Figure 1A for their degradation half-lives at 25 °C. The final construct retained excellent sensitivity to hydrolytic cleavage by cathepsin B in solution. Peptide hydrolysis leading to GNR internalization was confirmed in vitro by increased quantitative and qualitative uptake by cells as a function of cathepsin B expression and abrogation of this uptake by pretreatment with the selective cathepsin B inhibitor, CA074.
Extension of our findings to an in vivo setting is a key validation step in nanoparticle delivery to tumors, activation within the tumor microenvironment, and functional utility in increasing therapeutic efficacy of an anticancer treatment modality. Our in vivo experiments demonstrate excellent accumulation of our particles in tumors and potent radiosensitization. 24 hrs after intravenous injection of 11.7 μg of gold per mouse, we achieve an uptake of 1.7 μg/g tumor which translates to ~14.5% ID/g. Also, the dramatic reduction in tumor growth in mice that received the peptide GNR treatment and radiation resulted in better survival of these mice compared to the mice that received PEG GNR or peptide GNR with CA074. Collectively, these results show that peptide GNR conjugates have a high affinity towards cancer cells that express cathepsin B which leads to its increased intracellular retention sufficient to produce an improved response to radiotherapy and survival. Our toxicity studies revealed no appreciable changes in hematological, biochemical, or histopathological changes in normal organs of mice treated with peptide GNRs, further supporting continued advancement of a clinical translational strategy. The mild to moderate Kupffer cell hyperplasia and aggregates of mononuclear cells (probably histiocytes) seen in the liver histopathology specimens is likely to be a consequence of entrapment of a nanomaterial by the reticuloendothelial system.
We note an important distinction between our use of cathepsin B for cleavage, dePEGylation, and internalization of peptide GNRs into colorectal cancers for improved radiosensitization and the uncertain role of cathepsin B inhibitors as anticancer agents. Whereas the latter need to have high specificity for tumors, good bioavailability, potent activity in the extracellular space without abrogating the protein turnover function of cathepsin B, and some level of reversibility, these are not requirements for stimulus responsive nanoparticles that merely use tumor microenvironmental extracellular cathepsin B to cleave a peptide on their surface to expose a cationic outer scaffold. By not interfering with cathepsin B function, such nanoconstructs have less potential to have their activity be influenced by pleiotropism of cathepsin B function in cancer and merely rely on increased extracellular cathepsin B expression by aggressive cancers to mediate their therapeutic effect.
In conclusion, several attempts have been made to deliver gold nanoparticles into the tumors to improve therapeutic efficacy of radiotherapy, with limited success. In this study, we exploit the therapeutic vulnerability of cathepsin B secreting tumor cells to drive preferential uptake and cellular internalization of peptide GNRs in colorectal cancers to potently increase radiosensitization. Since cathepsin B overexpression is a characteristic feature of many tumors beyond colorectal cancer, we expect that the same construct could have therapeutic utility for other cancers too. We designed a peptide construct that is cleaved by the tumors secreting cathepsin B. When these GNRs reach the tumor ECM, they are cleaved, dePEGylated and internalizaed into the tumors which, when combined with radiotherapy, lead to tumor growth inhibition and improved survival in mice models with heterotopic colorectal cancer.
Supplementary Material
Acknowledgements
We acknowledge support from the MD Anderson Cancer Center Support Grant P30CA92261 and the Mayo Clinic Cancer Center Support Grant P30CA015083 from the National Institutes of Health (NIH). This work was also supported in part by NIH research grant R01DE028105 (to S.K.), and R01CA257241 and R01CA155446 (to S.K. and S.H.C.).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Analysis of nanoparticle delivery to tumours, Nature Reviews Materials 1(5) (2016) 16014. [Google Scholar]
- [2].Kim GC, Cheon DH, Lee Y, Challenge to overcome current limitations of cell-penetrating peptides, Biochim Biophys Acta Proteins Proteom 1869(4) (2021) 140604. [DOI] [PubMed] [Google Scholar]
- [3].Le T.a.B., Vikas, First Aid for the USMLE Step 1 2019, 2018. https://mhebooklibrary.com/doi/book/10.1036/9781260143683. [Google Scholar]
- [4].Chithrani DB, Jelveh S, Jalali F, van Prooijen M, Allen C, Bristow RG, Hill RP, Jaffray DA, Gold nanoparticles as radiation sensitizers in cancer therapy, Radiat Res 173(6) (2010) 719–28. [DOI] [PubMed] [Google Scholar]
- [5].Geng F, Song K, Xing JZ, Yuan C, Yan S, Yang Q, Chen J, Kong B, Thio-glucose bound gold nanoparticles enhance radio-cytotoxic targeting of ovarian cancer, Nanotechnology 22(28) (2011) 285101. [DOI] [PubMed] [Google Scholar]
- [6].Geng F, Xing JZ, Chen J, Yang R, Hao Y, Song K, Kong B, Pegylated glucose gold nanoparticles for improved in-vivo bio-distribution and enhanced radiotherapy on cervical cancer, J Biomed Nanotechnol 10(7) (2014) 1205–16. [DOI] [PubMed] [Google Scholar]
- [7].Hainfeld JF, Dilmanian FA, Zhong Z, Slatkin DN, Kalef-Ezra JA, Smilowitz HM, Gold nanoparticles enhance the radiation therapy of a murine squamous cell carcinoma, Phys Med Biol 55(11) (2010) 3045–59. [DOI] [PubMed] [Google Scholar]
- [8].Hainfeld JF, Slatkin DN, Smilowitz HM, The use of gold nanoparticles to enhance radiotherapy in mice, Phys Med Biol 49(18) (2004) N309–15. [DOI] [PubMed] [Google Scholar]
- [9].Khoshgard K, Hashemi B, Arbabi A, Rasaee MJ, Soleimani M, Radiosensitization effect of folate-conjugated gold nanoparticles on HeLa cancer cells under orthovoltage superficial radiotherapy techniques, Phys Med Biol 59(9) (2014) 2249–63. [DOI] [PubMed] [Google Scholar]
- [10].Polf JC, Bronk LF, Driessen WH, Arap W, Pasqualini R, Gillin M, Enhanced relative biological effectiveness of proton radiotherapy in tumor cells with internalized gold nanoparticles, Appl Phys Lett 98(19) (2011) 193702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rahman WN, Bishara N, Ackerly T, He CF, Jackson P, Wong C, Davidson R, Geso M, Enhancement of radiation effects by gold nanoparticles for superficial radiation therapy, Nanomedicine 5(2) (2009) 136–42. [DOI] [PubMed] [Google Scholar]
- [12].Roa W, Zhang X, Guo L, Shaw A, Hu X, Xiong Y, Gulavita S, Patel S, Sun X, Chen J, Moore R, Xing JZ, Gold nanoparticle sensitize radiotherapy of prostate cancer cells by regulation of the cell cycle, Nanotechnology 20(37) (2009) 375101. [DOI] [PubMed] [Google Scholar]
- [13].Chatterjee DK, Wolfe T, Lee J, Brown AP, Singh PK, Bhattarai SR, Diagaradjane P, Krishnan S, Convergence of nanotechnology with radiation therapy-insights and implications for clinical translation, Transl Cancer Res 2(4) (2013) 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schuemann J, Berbeco R, Chithrani DB, Cho SH, Kumar R, McMahon SJ, Sridhar S, Krishnan S, Roadmap to Clinical Use of Gold Nanoparticles for Radiation Sensitization, Int J Radiat Oncol Biol Phys 94(1) (2016) 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schuemann J, Bagley AF, Berbeco R, Bromma K, Butterworth KT, Byrne HL, Chithrani BD, Cho SH, Cook JR, Favaudon V, Gholami YH, Gargioni E, Hainfeld JF, Hespeels F, Heuskin AC, Ibeh UM, Kuncic Z, Kunjachan S, Lacombe S, Lucas S, Lux F, McMahon S, Nevozhay D, Ngwa W, Payne JD, Penninckx S, Porcel E, Prise KM, Rabus H, Ridwan SM, Rudek B, Sanche L, Singh B, Smilowitz HM, Sokolov KV, Sridhar S, Stanishevskiy Y, Sung W, Tillement O, Virani N, Yantasee W, Krishnan S, Roadmap for metal nanoparticles in radiation therapy: current status, translational challenges, and future directions, Phys Med Biol 65(21) (2020) 21RM02. [DOI] [PubMed] [Google Scholar]
- [16].Mai J, Waisman DM, Sloane BF, Cell surface complex of cathepsin B/annexin II tetramer in malignant progression, Biochim Biophys Acta 1477(1–2) (2000) 215–30. [DOI] [PubMed] [Google Scholar]
- [17].Bian B, Mongrain S, Cagnol S, Langlois MJ, Boulanger J, Bernatchez G, Carrier JC, Boudreau F, Rivard N, Cathepsin B promotes colorectal tumorigenesis, cell invasion, and metastasis, Mol Carcinog 55(5) (2016) 671–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ruan H, Hao S, Young P, Zhang H, Targeting Cathepsin B for Cancer Therapies, Horiz Cancer Res 56 (2015) 23–40. [PMC free article] [PubMed] [Google Scholar]
- [19].Liopo A, Derry PJ, Ermolinsky B, Su R, Conjusteau A, Ermilov S, Zubarev ER, Oraevsky A, Controlled bacteria-gold nanorod interactions for enhancement of optoacoustic contrast, Photons Plus Ultrasound: Imaging and Sensing 2014, International Society for Optics and Photonics, 2014, p. 894368. [Google Scholar]
- [20].Requejo KI, Liopo AV, Zubarev ER, Synthesis of Gold Nanorods Using Poly (vinylpyrrolidone) of Different Molecular Weights as an Additive, ChemistrySelect 3(43) (2018) 12192–12197. [Google Scholar]
- [21].Requejo KI, Liopo AV, Zubarev ER, Gold nanorod synthesis with small thiolated molecules, Langmuir 36(14) (2020) 3758–3769. [DOI] [PubMed] [Google Scholar]
- [22].Rong X, Xiang L, Li Y, Yang H, Chen W, Li L, Liang D, Zhou X, Chronic Periodontitis and Alzheimer Disease: A Putative Link of Serum Proteins Identification by 2D-DIGE Proteomics, Front Aging Neurosci 12 (2020) 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Matarrese P, Ascione B, Ciarlo L, Vona R, Leonetti C, Scarsella M, Mileo AM, Catricala C, Paggi MG, Malorni W, Cathepsin B inhibition interferes with metastatic potential of human melanoma: an in vitro and in vivo study, Mol Cancer 9 (2010) 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Khoo AM, Cho SH, Reynoso FJ, Aliru M, Aziz K, Bodd M, Yang X, Ahmed MF, Yasar S, Manohar N, Cho J, Tailor R, Thames HD, Krishnan S, Radiosensitization of Prostate Cancers In Vitro and In Vivo to Erbium-filtered Orthovoltage X-rays Using Actively Targeted Gold Nanoparticles, Sci Rep 7(1) (2017) 18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Van Acker GJ, Saluja AK, Bhagat L, Singh VP, Song AM, Steer ML, Cathepsin B inhibition prevents trypsinogen activation and reduces pancreatitis severity, Am J Physiol Gastrointest Liver Physiol 283(3) (2002) G794–800. [DOI] [PubMed] [Google Scholar]
- [26].Cancer N Genome Atlas Research, Comprehensive molecular characterization of clear cell renal cell carcinoma, Nature 499(7456) (2013) 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Glidden M, Muschol M, Characterizing gold nanorods in solution using depolarized dynamic light scattering, The Journal of Physical Chemistry C 116(14) (2012) 8128–8137. [Google Scholar]
- [28].Wolfe T, Chatterjee D, Lee J, Grant JD, Bhattarai S, Tailor R, Goodrich G, Nicolucci P, Krishnan S, Targeted gold nanoparticles enhance sensitization of prostate tumors to megavoltage radiation therapy in vivo, Nanomedicine 11(5) (2015) 1277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gargioni E, Schulz F, Raabe A, Burdak-Rothkamm S, Rieckmann T, Rothkamm K, Targeted nanoparticles for tumour radiotherapy enhancement-the long dawn of a golden era?, Ann Transl Med 4(24) (2016) 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kang B, Mackey MA, El-Sayed MA, Nuclear targeting of gold nanoparticles in cancer cells induces DNA damage, causing cytokinesis arrest and apoptosis, J Am Chem Soc 132(5) (2010) 1517–9. [DOI] [PubMed] [Google Scholar]
- [31].Jones BL, Krishnan S, Cho SH, Estimation of microscopic dose enhancement factor around gold nanoparticles by Monte Carlo calculations, Med Phys 37(7) (2010) 3809–16. [DOI] [PubMed] [Google Scholar]
- [32].Liu CJ, Wang CH, Chen ST, Chen HH, Leng WH, Chien CC, Wang CL, Kempson IM, Hwu Y, Lai TC, Hsiao M, Yang CS, Chen YJ, Margaritondo G, Enhancement of cell radiation sensitivity by pegylated gold nanoparticles, Phys Med Biol 55(4) (2010) 931–45. [DOI] [PubMed] [Google Scholar]
- [33].Maeda H, Wu J, Sawa T, Matsumura Y, Hori K, Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review, J Control Release 65(1–2) (2000) 271–84. [DOI] [PubMed] [Google Scholar]
- [34].Xie H, Diagaradjane P, Deorukhkar AA, Goins B, Bao A, Phillips WT, Wang Z, Schwartz J, Krishnan S, Integrin alphavbeta3-targeted gold nanoshells augment tumor vasculature-specific imaging and therapy, Int J Nanomedicine 6 (2011) 259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Turk D, Podobnik M, Popovic T, Katunuma N, Bode W, Huber R, Turk V, Crystal structure of cathepsin B inhibited with CA030 at 2.0-A resolution: A basis for the design of specific epoxysuccinyl inhibitors, Biochemistry 34(14) (1995) 4791–7. [DOI] [PubMed] [Google Scholar]
- [36].Biniossek ML, Nagler DK, Becker-Pauly C, Schilling O, Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S, J Proteome Res 10(12) (2011) 5363–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.