Abstract
Cannabis use in the United States is increasing, with highest consumption among men at their peak reproductive years. We previously demonstrated widespread changes in sperm DNA methylation with cannabis exposure in humans and rats, including genes important in neurodevelopment. Here, we use an in vitro human spermatogenesis model to recapitulate chronic cannabis use and assess DNA methylation at imprinted and autism spectrum disorder (ASD) candidate genes in spermatogonial stem cell (SSC)- and spermatid-like cells. Methylation at maternally imprinted genes SGCE and GRB10 was significantly altered in SSC- and spermatid-like cells, respectively, while PEG3 was significantly differentially methylated in spermatid-like cells. Two of ten randomly selected ASD candidate genes, HCN1 and NR4A2, had significantly altered methylation with cannabis exposure in SSC-like cells. These results support our findings in human cohorts and provide a new tool with which to gain mechanistic insights into the association between paternal cannabis use and risk of ASD in offspring.
Keywords: cannabis, DNA methylation, imprinted gene, autism, spermatogenesis
Introduction
Cannabis is considered the most used illicit psychoactive drug in the United States (US) (Chandra et al. 2019). Its use is increasing (US Department of Health and Human Services (SAMHSA) 2018), likely driven by an increasing general perception of safety and expanding legalization of recreational and medicinal use (Cuttler et al. 2016; Hasin 2018; Chandra et al. 2019; Schrott and Murphy 2020). Some mothers- and fathers-to-be are included among cannabis users. Infants born to mothers who used cannabis during pregnancy face serious health concerns, including an increased risk of requiring the NICU, lower birth weight, potential impaired immune system function, neurodevelopmental delay, and autistic-like deficits (Dong et al. 2019; Reece and Hulse 2019a, 2019b). However, the largest group of cannabis consumers in the US are men in their peak reproductive years (Mauro et al. 2018; US Department of Health and Human Services (SAMHSA) 2018). Cannabis use is associated with altered semen quality, including reduced sperm concentration, abnormal morphology, and reduced motility (Rajanahally et al. 2019; Fonseca and Rebelo 2021). Cannabis use is also associated with a reduction in luteinizing hormone which is important for production of testosterone (Fonseca and Rebelo 2021; Meah et al. 2021).
The persistence of these effects is largely unknown. However, cannabis components that enter the bloodstream can impact spermatogonial stem cells, which are located outside of the blood-testis-barrier, in turn altering sperm cell progeny downstream. This raises questions about the impact that cannabis has on the sperm epigenome and how this might contribute to adverse developmental outcomes. Various exposures impact the sperm epigenome at genes important for early life development (Leter et al. 2014; Abbasi 2017; Donkin and Barres 2018), including cannabis (Murphy et al. 2018; Schrott et al. 2020a, 2020b). Our group demonstrated associations between cannabis use in men and widespread changes in sperm DNA methylation (Murphy et al. 2018; Schrott et al. 2020a). We also reported that cannabis use in men is associated with altered sperm DNA methylation at multiple genes implicated in autism, as is tetrahydrocannabinol exposure in rats (Schrott et al. 2020a, 2020b). States with legalized cannabis programs have an increased prevalence of autism diagnoses (Reece A and 2019a), and it is possible that epigenetic changes in gametes mechanistically link such exposures to neurodevelopmental outcomes.
Recently, we established an in vitro model of human spermatogenesis, whereby NIH-approved hESCs derived from a male embryo are differentiated into a mixed population of SSC-like cells, primary spermatocyte-like cells, secondary spermatocyte-like cells, and round spermatid-like cells that have successfully undergone two meiotic divisions (Easley et al. 2012). In addition to providing a novel system for studying spermatogenesis in vitro, this culture system is an elegant model for studying the impact of environmental exposures during the process of spermatogenesis (Easley et al. 2012; Easley et al. 2015; Greeson et al. 2020). This model system is thus useful for experimentally investigating the impact of cannabis exposure on the human sperm epigenome.
The objective of this study was to determine the potential influence of cannabis extract (CE) exposure on DNA methylation at two groups of genes important for early life development, including imprinted genes and autism candidate genes. Imprinted genes are essential for normal embryonic development and play important roles postnatally in neurodevelopment and metabolism (Ferguson-Smith and Bourc'his 2018). DNA methylation at imprinted genes is established at imprint control regions (ICR) in primordial germ cells (PGCs) during embryogenesis (Gkountela et al. 2015; Guo et al. 2015; Tang et al. 2015). During PCG epigenetic reprogramming, parental imprinted marks are erased and then reestablished in cells to reflect the sex of the developing embryo (Gkountela et al. 2015; Guo et al. 2015; Tang et al. 2015). Once established, these imprinted marks are maintained in the gametes and persist in the subsequent generation, escaping post-fertilization reprogramming (Guo et al. 2014). Parental imprinted marks are present in and are propagated across virtually all cell types in a developing embryo. The second group of genes are associated with autism spectrum disorder (ASD). Many ASD candidate genes are marked with bivalent chromatin (Schrott et al. 2020b), a subcomponent of the epigenome that is important for early embryonic development (Bernstein et al. 2006). Bivalent chromatin consists of histone modifications that simultaneously keep a gene silenced while also allowing for that gene to be rapidly activated in response to environmental and developmental cues (Bernstein et al. 2006). We previously found that genes with altered DNA methylation in sperm with cannabis use are enriched for those with bivalent chromatin (Schrott R. et al. 2020). In the present study, we tested the hypothesis that these marks enhance epigenetic vulnerability to environmental exposures.
Results
Cannabis extract exposure impacts DNA methylation at maternally imprinted genes
We used our in vitro spermatogenesis model to determine whether CE exposure alters DNA methylation at this same group of imprinted genes in H1 ESCs. Cells were exposed to 50nM CE or vehicle control during a ten-day differentiation period, reflecting “chronic” cannabis use (Guennewig et al. 2018). Following differentiation, SSC-like cells and haploid spermatid-like cells were isolated using flow cytometry (Figure S3, S4). Given the continual post-pubertal production of sperm from a defined progenitor cell population, the spermatogonia (Neto et al. 2016), we wished to determine whether cannabis exposure impacts spermatogonia, and if so, whether those changes would be propagated to the haploid spermatids.
Paternally imprinted genes
Using bisulfite pyrosequencing, we first analyzed the effect of chronic CE exposure on DNA methylation at paternally imprinted genes in SSC-like cells and haploid spermatid-like cells (n=3). We analyzed the effect of CE on each individual CpG site because it is possible that CE can distinctly impact individual CpG sites. Two-factor ANOVA for CpG site and exposure status revealed no significant effects of exposure for IGF2, MEG3-IG or H19 (Figure S1A-F) in either the SSC-like cells or the haploid spermatid-like cells.
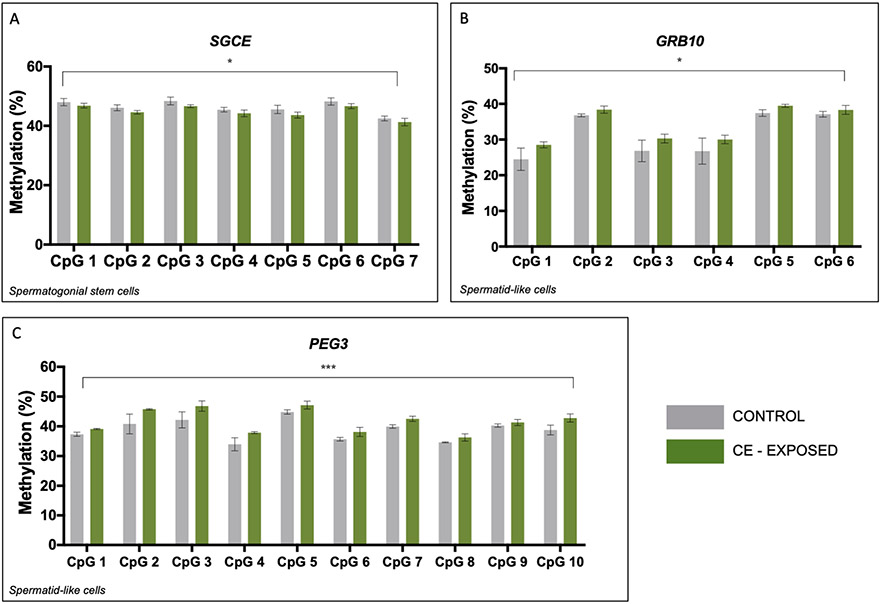
We next performed bisulfite pyrosequencing to analyze the effect of chronic CE exposure on DNA methylation at maternally imprinted genes in SSC-like cells and haploid spermatid-like cells. Two-factor ANOVA for CpG site and exposure status determined no significant effect of exposure for MEST, PLAGL1, and SNRPN (Figure S2A, C, D). Similarly, analysis of NDN revealed no significant effect of exposure in the either cell type (Figure S2B). SGCE revealed a significant main effect of exposure in the SSC-like cells (p<0.05) (Figure 1A), but this effect was not evident in the haploid spermatid-like cells (Figure S2G). GRB10 methylation differences showed a significant effect of exposure in the haploid spermatid-like cells (p=0.02) (Figure 1B) but not for the SSC-like cells (p<0.08) (Figure S2E). PEG3 methylation displayed a significant main effect of exposure in the haploid spermatid-like cells across the region analyzed (p<0.0001) (Figure 1C), but no significant effect in the SSC-like cells (Figure S2F).
Figure 1.
CE significantly impacts spermatogenic cell DNA methylation at three maternally imprinted genes. A-C: Two-factor ANOVA of bisulfite pyrosequencing data for SGCE (A), GRB10 (B), and PEG3 (C) in SSC-like cells (A) and haploid spermatid-like cells (B, C). A: There is a significant effect of exposure on DNA methylation at SGCE in SSC-like cells but not in haploid spermatid-like cells. There is a significant effect CE exposure on DNA methylation in haploid spermatid-like cells at GRB10 (B) and PEG3 (C). Grey=controls, green=CE-exposed. * p<0.05; ** p<0.005; *** p<0.0001
Cannabis extract exposure impacts DNA methylation at neurodevelopmental genes possessing bivalent chromatin
Court & Arnaud published a list of genes that possess bivalent chromatin marks and annotated the CpG nucleotide positions in each gene for the location of the bivalent chromatin markings based on Illumina’s Infinium HumanMethylation450 BeadChip (450k platform) (Court and Arnaud 2017). We then used the Simons Foundation Autism Research Initiative (SFARI) gene list (Simons Foundation Autism Research Initiative 2019) as our baseline list of genes that are important for neurodevelopment and autism. We then divided this into an overlap list consisted of genes in common between the SFARI and bivalent chromatin lists, and our SFARI-only list comprising the remaining SFARI genes. Ten genes from each list were randomly chosen. From each individual gene, the CpG coordinates associated with the gene from the 450K BeadChip manifest were identified, and one site was randomly selected for pyrosequencing assay design. For the overlap list genes, we specifically included only those CpG coordinates that were identified as being associated with bivalent chromatin (Court and Arnaud 2017).
DNA methylation in SFARI-only genes (no bivalent chromatin) in in vitro-derived, CE-exposed spermatogenic cells
We successfully validated bisulfite pyrosequencing assay performance for six of ten randomly selected genes from the SFARI-only gene list (Figure S5A-F).
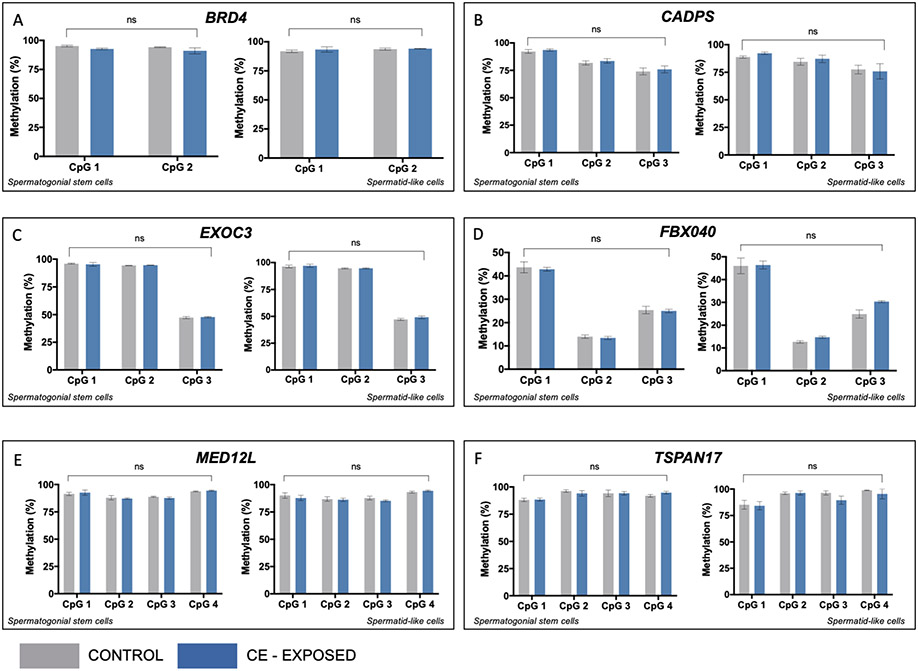
hESCs differentiated into spermatogenic cells were exposed to 50nM CE or vehicle control over the entire ten-day differentiation period. SSC-like and haploid spermatid-like cells were isolated via flow cytometry on day ten. Bisulfite pyrosequencing was used to determine the impact of CE on DNA methylation on SFARI-only genes in SSC-like and haploid spermatid-like cells. There was no significant effect of exposure for any of the six genes analyzed (BRD4, CADPS, EXOC3, FBXO40, MED12L or TSPAN17) in the SSC-like or haploid spermatid-like cells (Figure 2A-F).
Figure 2.
CE does not impact DNA methylation in spermatogenic cells at genes from the SFARI list without bivalent chromatin. In each panel, left graph shows methylation at individual CpG sites in SSC-like cells and right graph shows level in spermatid-like cells. Two-factor ANOVA for bisulfite pyrosequencing data shows no effect of CE exposure on DNA methylation at all genes analyzed, including A: BRD4, B: CADPS, C: EXOC3, D: FBXO40, E: MED12L, F: TSPAN17. Grey=controls, blue=CE exposed. ns=no significance
DNA methylation of SFARI genes possessing bivalent chromatin in in vitro-derived, CE-exposed spermatogenic cells
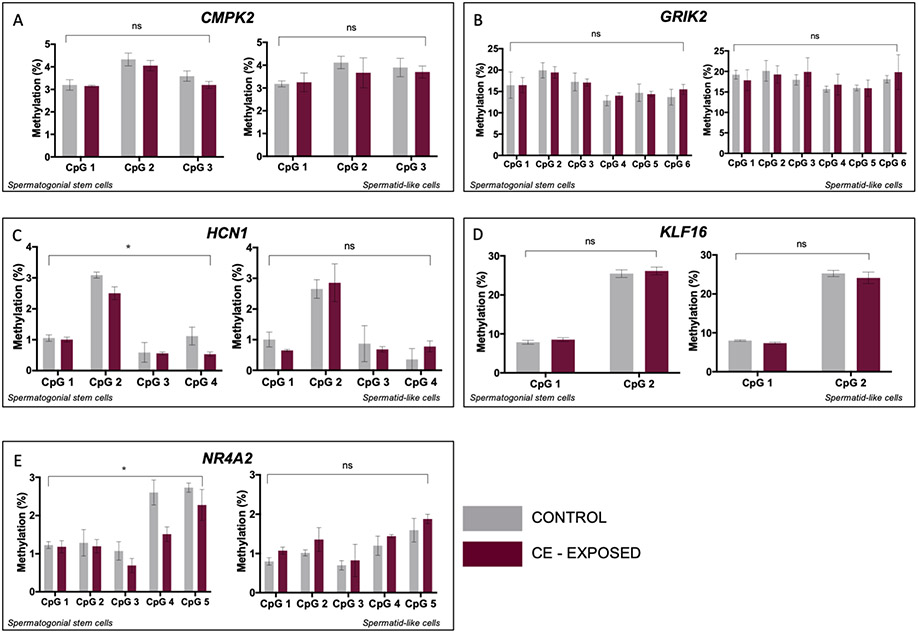
Ten genes were randomly selected for bisulfite pyrosequencing analysis, five of which we were able to successfully validate for assay performance (Figure S6). SSC-like and haploid spermatid-like cells were isolated via flow cytometry from cultures that had been exposed to vehicle control or 50nM CE over a ten-day differentiation period. There was no significant effect of exposure on methylation differences measured for CMPK2, GRIK2, or KLF16 in SSC-like or in haploid spermatid-like cells (Figure 3A, B, D). For HCN1 and NR4A2, however, we observed a significant effect of exposure in the SSC-like cells, (p=0.026 and p=0.014, respectively) (Figure 3C, E). While these changes were not detected in the haploid spermatid-like cells, the effect of exposure did approach significance in haploid spermatid-like cells for NR4A2 (p=0.07, Figure 3E).
Figure 3.
CE exposure impacts DNA methylation in SSC-like cells at genes with bivalent chromatin. In each panel, left graph shows methylation at individual CpG sites in SSC-like cells and right graph shows level in spermatid-like cells. Two-factor ANOVA of bisulfite pyrosequencing data shows no effect of CE exposure on DNA methylation in SSC-like cells or haploid spermatid-like cells for A: CMPK2, B: GRIK2, and D: KLF16. There is a significant effect of exposure on DNA methylation in SSC-like cells for C: HCN1 and E: NR4A2. Grey=controls, purple=CE exposed. * p<0.05; ** p<0.005; *** p<0.0001; ns=no significance
CE differentially impacts DNA methylation at genes possessing bivalent chromatin compared to those that do not
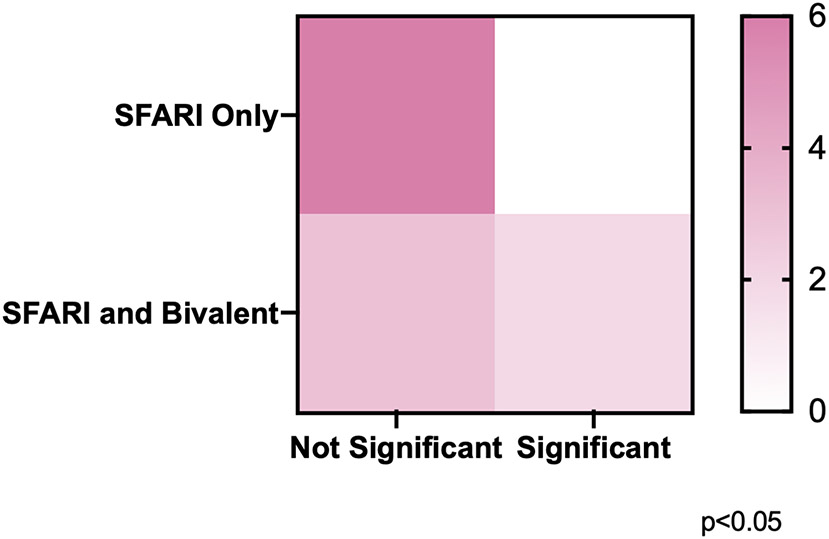
We demonstrated the ability of CE to induce significant methylation changes in chronically exposed SSC-like cells at genes possessing bivalent chromatin, but not at those without this group of epigenetic modifications. Given our a-priori hypothesis that this would be the case, we ran a one-tailed Chi-square test to determine if there was a significant difference between the distribution of significantly impacted genes across the SFARI-only group and the overlap group. We found that among the analyzed genes, there was a significant difference in the distribution of genes whose DNA methylation was affected by CE exposure in the SFARI-only group as compared to the overlap group (p<0.05) (Figure 4).
Figure 4.
Chi-square analysis of the distribution of significant effects of CE exposure on DNA methylation across groups of genes.
Contingency table assignment: SFARI only, non-significant (upper left, n=6), SFARI only, significant (upper right, n=0), SFARI and Bivalent, non-significant (lower left, n=3), SFARI and Bivalent, significant (lower right, n=2). One-tailed Chi-square test demonstrated a significant difference between the distribution of genes whose DNA methylation was affected by CE exposure in the SFARI only compared to the SFARI and Bivalent overlap genes.
Discussion
Cannabis legalization is expanding across the U.S. and around the globe. As of May 2021, there are 36 U.S. states and four territories with some form of legalized cannabis use (National Conference of State Legislatures 2021). With expanding legalization, there is concurrent relaxation of attitudes about cannabis use, with more adolescents and adults perceiving cannabis use to be safe (Hasin 2018; Chandra et al. 2019). The study of the impact of preconception cannabis use on offspring development has been limited. However, the urgency of understanding these consequences continues to grow as access to legalized cannabis increases and men of reproductive age remain the predominant group of cannabis users.
We exposed differentiating cells to vehicle control or 50nM CE, a dose that has been described in the literature as reflective of chronic cannabis use (Guennewig et al. 2018). We then used two distinct flow cytometry methods to isolate SSC-like cells and haploid spermatid-like cells. We analyzed DNA methylation in SSC-like and haploid spermatid-like cells because mature sperm are continuously produced from this stem cell population throughout a man’s post-pubertal life. Thus, it is possible that epigenetic changes present in SSCs themselves may lead to permanent, lifelong changes to sperm DNA methylation in all maturing cells derived from the affected progenitor population.
We first examined imprinted genes in CE- and vehicle-exposed sorted cells. No paternally imprinted genes showed significant effects of exposure in the SSC-like or in the haploid spermatid-like cells. Among the maternally imprinted genes analyzed, we observed a significant effect of CE exposure on DNA methylation at SGCE in the SSC-like cells. SGCE encodes the epsilon member of the sarcoglycan family of transmembrane proteins (Peall et al. 2014). Mutations in SGCE are associated with myoclonus dystonia syndrome (MDS), a rare young-onset movement disorder (Peall et al. 2013; Peall et al. 2014). Maternal imprinting plays a role in regulating the penetrance of this heritable disorder (Peall et al. 2013; Peall et al. 2014). Additionally, the promoter CpG island that houses this SGCE imprint control region also regulates the expression of another imprinted gene, Paternally Expressed Gene 10 (PEG10). PEG10 encodes a retrotransposable element, and deregulation of this region has been reported in cancers (Xie et al. 2018). Thus, alterations in methylation at this regulatory region could have implications for multiple pathologies.
In CE-exposed haploid spermatid-like cells, we observed significant effects of exposure at maternally imprinted PEG3 and GRB10. PEG3 was hypermethylated at every CpG site analyzed in the CE-exposed cells relative to the controls. Importantly, PEG3 is involved in controlling the rate of fetal growth and regulating maternal nurturing behaviors (Huang and Kim 2009; Kim et al. 2013). Mouse models of mutated Peg3 have demonstrated that Peg3 is involved in regulating transcription of placental genes, and epigenetic abnormalities with PEG3 have been associated with multiple forms of cancer and with hydatidiform mole – a pregnancy complication associated with abnormal growth of the trophoblast cells that form the placenta (El-Maarri et al. 2003; Huang JM and Kim 2009; Kim et al. 2013). GRB10 was similarly hypermethylated at each CpG site in the cells exposed to CE relative to those exposed to vehicle control. GRB10’s functions and tissue-specific expression patterns are complex, but it is known to be paternally expressed in sperm and neurons (Hikichi et al. 2003; Plasschaert and Bartolomei 2015). GRB10 has important roles in the regulation of cellular growth and embryonic development (Charalambous et al. 2003; Smith et al. 2007). Knockout of the paternal allele of Grb10 in mice resulted in increased social dominance behaviors, as well as increased anxiety behaviors and general aggression (Garfield et al. 2011; Plasschaert and Bartolomei 2015). Additionally, aberrant imprinting at GRB10 has been associated with the growth retardation phenotypes observed in Silver-Russell Syndrome (Charalambous et al. 2003). While the effect of CE exposure was not significant for these genes in SSCs, it did approach significance. It is, therefore, possible that a small subset of SSCs was affected by CE exposure, and it was from those SSCs that the affected haploid spermatids were derived, as the direction of methylation change was the same. Lineage tracing studies would be necessary to test this hypothesis to follow cells as they differentiate to determine from which progenitor cell they arose.
After using this in vitro model of spermatogenesis to analyze the impact of CE in DNA methylation at imprinted genes, we sought to determine whether CE alters DNA methylation at a group of genes important for neurodevelopment and that possess bivalent chromatin – a relatively unusual epigenetic configuration often found at regulatory regions of genes important for early life development. Many studies investigating the effects of prenatal or preconception exposure to an array of drugs, chemicals, and psychosocial stressors, including our own, similarly report associations with adverse neurodevelopmental outcomes in offspring, or functional roles and pathways associated with neurodevelopmental and early developmental processes that are enriched in their epigenomic datasets (Dolinoy and Jirtle 2008; Wu et al. 2015; Abbasi 2017; Donkin and Barres 2018; Murphy et al. 2018; Greeson et al. 2020; Schrott and Murphy 2020). We therefore questioned what these genes might have in common that makes them vulnerable to multiple types of exposures. Given their roles in early development, we postulated that the presence of bivalent chromatin at these loci might heighten the sensitivity of these genes to their environment, given the ambiguity of the histone modifications present at these regions. Support for this contention came from our prior findings of significant overlap between a curated list of autism candidate genes and genes known to possess bivalent chromatin in hESCs (Schrott et al. 2020b). We further demonstrated enrichment for autism candidate genes and genes possessing bivalent chromatin among genes significantly differentially methylated in sperm of cannabis users (Schrott R. et al. 2020b).
We here further tested our hypothesis using the in vitro model. We analyzed the effect of CE exposure on DNA methylation in differentiating spermatogenic cells for a group of randomly selected genes from the SFARI list of autism candidate genes. For SFARI-only (not marked with bivalent chromatin) genes, we found no significant effects of CE exposure on DNA methylation in SSC-like or haploid spermatid-like cells. Analysis of the overlap genes (with bivalent chromatin) showed a significant effect of exposure on methylation in SSC-like cells at two genes – HCN1 and NR4A2.
HCN1 encodes hyperpolarization-activated cyclic nucleotide-gated voltage-gated ion channels that play a role in regulating neurotransmission and influencing synaptic plasticity in cortical neurons (Huang et al. 2011; Huang et al. 2017; Yang et al. 2018). HCN1 has important roles in controlling dendritic functional integration in hippocampal development and contributing to adult hippocampal function (Seo et al. 2015; Yang et al. 2018). Knockout of Hcn1 in mice results in impaired motor learning and memory and has been associated with drug-induced cognitive dysfunction (Seo et al. 2015; Yang et al. 2018). Importantly, mutations in HCN1 are associated with a range of epileptic phenotypes (Seo et al. 2015; Marini et al. 2018). Interestingly, autism spectrum disorders are associated with epilepsy, with many individuals with autism developing seizure disorders later in life (Tuchman and Cuccaro 2011). Thus, while its role in mental disorders is still being elucidated, possible comorbidities should be considered. It is also known that HCN1 plays an important role in controlling brain development and in regulating the excitability of neurons (Yang et al. 2018). Thus, alterations in how this gene is expressed, including epigenetic changes that influence gene expression, could have profound consequences for neurodevelopment, if the HCN1 methylation changes in sperm are heritable.
NR4A2 encodes a nuclear receptor and transcriptional regulator that functions in the differentiation and maintenance of dopaminergic neurons during neurodevelopment (Blin et al. 2008; Spiers et al. 2015). NR4A2 expression is required for the successful terminal differentiation of mesencephalic dopaminergic neurons during neurogenesis (Blin et al. 2008). Deletions in NR4A2 are associated with intellectual disability, language impairment, and autism (Reuter et al. 2017; Levy et al. 2018). Further, alterations in NR4A2 DNA methylation have been associated with dysregulation of the gene in tissue samples from individuals with Down Syndrome (Zhang et al. 2016). If altered methylation is transmitted from sperm to offspring, it could have serious consequences for offspring neurodevelopment.
A limitation of this study was that our sample sizes were small (n=3) for our CE-exposed and vehicle control experiments, which could have limited our ability to detect significant differences in DNA methylation. Future studies should include additional replicates and doses. We were compelled by the data demonstrating an increased vulnerability of SFARI genes possessing bivalent chromatin to CE exposure compared to those genes without the epigenetic modification. However, larger studies are needed to assess the impact of CE on all CpG sites associated with bivalent chromatin compared to those without to provide more rigorous data in support of this hypothesis.
The use of human embryonic stem cells (hESCs) in research is a powerful scientific tool. Their pluripotent nature renders them a remarkable model for studying early life developmental processes and understanding disease progression (Dvash et al. 2006). These cells have tremendous therapeutic potential, as evidenced by their use in studies involved in understanding and treating diseases such as diabetes, Parkinson’s disease, Alzheimer’s disease, bone marrow failure, cancer therapeutics, and more (Lerou and Daley 2005; Dvash et al. 2006). Much has been characterized in the stem cell lines used for research, such as the mutations they harbor, the diseases they represent, and the karyotypes they possess. However, one area that is less well understood in these cells is the status of their epigenome. This study contributed to the literature characterizing the methylation status of a group of 10 imprinted genes and assessing epigenetic reprogramming in an NIH-approved embryonic stem cell line. Our findings that CE exposure can influence DNA methylation at a subset of imprinted genes and genes possessing bivalent chromatin adds to a growing body of evidence that cannabis exposure can impact the sperm DNA methylome. Future studies are needed to continue to assess the heritability of the effects induced by cannabis.
Methods
Culture of hESCs
NIH-approved WA01 (H1) male-derived hESCs were cultured for expansion on a 3D hESC-quality matrigel matrix (Corning, Corning, NY) in mTeSR Plus media (StemCell Technologies, Cambridge, MA). Cells were acquired through a Duke WiCell Distributor and were approved for use under Duke IRB protocol Pro00087169. Cells were thoroughly validated through STR analysis and full karyotyping by the Duke Sequencing and Genomic Technologies Core and the Cytogenetics Lab at the Duke Medical Center, respectively. Cells were negative for mycoplasma. Media was changed every other day, and cells were passaged once colonies achieved 80% confluence, roughly every 5-7 days, using the ReLeSR passaging reagent (StemCell Technologies, Cambridge, MA).
Differentiation of hESCs in spermatogenic cells
Cells were cultured for 3-5 days in hESC media until 80% confluent prior to initiating differentiation. Once at desired confluency, differentiation media was added (Easley et al. 2012; Easley et al. 2015; Greeson et al. 2020). Differentiation media consisted of MEM-Alpha + L-glutamine (Gibco, Montgomery County, MD), 0.2% bovine serum albumin (Millipore Bio, Burlington, MA), 0.2 mg/ml ascorbic acid (Sigma, St. Louis, MO), 0.2% chemically defined lipid mixture (Sigma, St. Louis, MO), 10μg/ml transferrin in water (Sigma, St. Louis, MO), 5μg/ml insulin in water (Sigma, St. Louis, MO), 20ng/ml recombinant hGDNF (Peprotech, Rocky Hill, NJ), 1ng/ml human recombinant bFGF in 5mM tris, 150mM NaCl buffer at pH 7.5 (Peprotech, Rocky Hill, NJ), 30nM sodium selenite (Sigma, St. Louis, MO), 10 mM HEPES (Gibco, Montgomery County, MD) and 0.5X Penicillin/Streptomycin (Gibco, Montgomery County, MD). The differentiation media was gassed with a blood gas mixture (Airgas, Durham, NC) of 5% CO2, 5% O2, balanced with N2 for 30 seconds, and inverted several times to mix. Media was changed every other day and was gassed for 30 seconds each time prior to use. Differentiation occurred over a ten-day period, where day one was the first day that differentiation media was introduced (Easley et al. 2012; Easley et al. 2015; Greeson et al. 2020).
Cannabis extract exposure
CE was produced by and received from the Research Triangle Institute (RTI) in North Carolina a contracted provider for the National Institute on Drug Abuse. Cells were cultured in differentiation media in the presence of vehicle control (0.1% DMSO) or CE containing 50nM THC in 0.1% DMSO for ten days. Media was changed every other day, and dosing occurred with each media change. For isolation of SSC-like cells, there were n=3 vehicle control replicates and n=3 chronic CE-exposed replicates. For isolation of haploid spermatid-like cells, there were n=3 vehicle control replicates and n=3 chronic CE-exposed replicates. Studies were performed at passage number 48.
Flow cytometry isolation of SSC-like cells and haploid spermatids
Isolation of the SSC-like population was achieved via fluorescence-activated cell sorting (FACS) from a mixed population of SSC-like, primary spermatocyte-like, secondary spermatocyte-like, and spermatid-like cells. Cells were sorted on the SONY cell sorter (Sony Biotechnology, San Jose, CA) at the Duke Cancer Institute’s Flow Cytometry shared resource. Specifically, an APC-conjugated antibody for the ITGA6 protein (R&D Systems, Minneapolis, MN) expressed on the surface of SSCs was used to isolate this population of cells. Following ten days of differentiation, cells were harvested with trypsin and stained with 10μL primary antibody per million cells in 1X PBS with 10% FBS and incubated at room temperature, protected from light, for 30 minutes. Unbound antibody was removed with two 1X PBS washes. Cells were resuspended in 10%FBS in 1X PBS for sorting. ITGA6+ cells were sorted (Figure S3) and collected for subsequent methylation analyses.
For isolation of the haploid spermatids, RedDot 1 (Biotium, Fremont, CA), a nuclear DNA stain, was used to stain live cells and isolate them during flow cytometry based on their DNA content (Figure S4). Following ten days of differentiation, cells were harvested with trypsin and stained with RedDot1 at a 1:100 dilution in differentiation media and incubated for 15 minutes at 37°C, protected from light. Haploid cells were identified based on their DNA content and collected for subsequent methylation analyses.
DNA isolation from cells
DNA was isolated from sorted cells using the Qiagen All Prep DNA/RNA Mini Kit (Qiagen, Germantown, MD). DNA was eluted in 30μL of buffer TE and was quantified on the NanoDrop 2000 (ThermoFisher, Waltham, MA) immediately following extraction. DNA was stored at −20°C until required for use.
Bisulfite pyrosequencing
As previously described (Schrott et al. 2020a, 2020b), the column-based EZ DNA Methylation kit (Zymo Research, Irvine, CA) was used to treat gDNA with sodium bisulfite to convert all unmethylated cytosines to uracils (that ultimately appear as thymines following downstream PCR and sequencing), while all methylated cytosines remain cytosines in the sequence. This resulted in bisulfite-modified DNA (bsDNA) at a final concentration of 10-20ng/μL. BsDNA (10-20 ng) was then used as a template for PCR amplification for bisulfite pyrosequencing. Bisulfite pyrosequencing assay design, validation, and sequencing were performed as described (Bassil et al. 2013).
Gene selection and assay design for imprinted genes
Assay designs for imprinted genes were previously validated and published by our lab (Murphy et al. 2012). Primers and PCR conditions for these genes can be found in (Murphy et al. 2012).
Gene selection and assay design for neurodevelopmental genes
The Simons Foundation Autism Research Initiative (SFARI) gene list contains 913 autism candidate genes based on data in the scientific literature (Simons Foundation Autism Research Initiative 2019). Court & Arnaud’s list of genes that possess bivalent chromatin contains 5379 (Court and Arnaud 2017). There were 293 genes in common between the two lists (the overlap list), and 620 genes present in only the SFARI list (the SFARI-only list). From the overlap list, we assigned each gene a number 1-293 and used a random number generator to randomly select ten genes for follow-up. For the SFARI-only list, we similarly assigned each gene a number 1-620 and used a random number generator to select ten genes at random.
For each gene selected from both lists, there was more than one CpG coordinate associated with bivalent chromatin, and these could not be captured in a single pyrosequencing assay due to too great an intervening distance between bivalent CpG sites. Therefore, for each gene, we then assigned each CpG coordinate a number and then used a random number generator to select the CpG site around which bisulfite pyrosequencing assays would be designed. For all genes, primers and PCR conditions are listed in Table 1.
Table 1.
SFARI and bivalent chromatin gene assay information for bisulfite pyrosequencing
| Gene | F Primer, 5'-3' (*biotin) | R Primer, 5'-3' (*biotin) | Sequencing Primer, 5'-3' | Sequence to Analyze, 5'-3' | Thermocycler Conditions |
|---|---|---|---|---|---|
| BRD4 | BRD4-F | BRD4-R* | BRD4-S | BRD4 | 95°C 15 min |
| 94°C-63.5°C-72°C X 55 ∣30s | |||||
| 72°C 10 min | |||||
| AGGTGGTTTTAGTTTTGAGTGTG | AAATTCAAAAAACACAAACCTCACATTAC | TGTTTTGTTTGTTTGGTTTAT | AGTTTAAAGTYGTGTAYGAG | 4°C ∞ | |
| CADPS | CADPS-F | CADPS-R* | CADPS-S | CADPS | 95°C 15 min |
| 94°C-68°C-72°C X 5 ∣30s | |||||
| 94°C-66°C-72°C X 5 ∣30s | |||||
| 94°C-64°C-72°C X 55 ∣30s | |||||
| 72°C 10 min | |||||
| GTATAGGGGATAAAGTTTTTGTGG | AAAAAACCCTAAACTTCTATCTACTCCT | GATATTATATTGTTAGTGGGAAGTA | YGTTTAGAATTTATATGGYGAAAGGTTTGTTAATGAGTTGYGG | 4°C ∞ | |
| EXOC3 | EXOC3-F | EXOC3-R* | EXOC3-S | EXOC3 | 95°C 15 min |
| 94°C ∣30s | |||||
| 63.5°C ∣30s X55 | |||||
| 72°C ∣30s | |||||
| 72°C 10 min | |||||
| GTTTGGGAGAGTATGGGTGATAT | ACTCCTCATCTAACCTCACACCTAATAA | ATGGGTGATATTTTTTTGAAG | GTATYGTAGTAGGTTAYGGTTTAGGGTAGATTYGT | 4°C ∞ | |
| FBXO40 | FBXO40-F | FBXO40-R* | FBXO40-S | FBXO40 | 95°C 15 min |
| 94°C-66°C-72°C X 5 ∣30s | |||||
| 94°C-63°C-72°C X 5 ∣30s | |||||
| 94°C-60°C-72°C X 50 ∣30s | |||||
| 72°C 10 min | |||||
| TTGAGTGTTGATAAATATAGGGGAAGGAA | CATCTACTTACACCTAAACCCTTAATT | AGGGGAAGGAAAAAG | TYGTAAGTTGAGTTTGGTTATATTGTTTTTTGTGATATYGTTTTATTTTTAATTTYGT | 4°C ∞ | |
| MED12L | MED12L-F | MED12L-R* | MED12L-S2 | MED12L | 95°C 15 min |
| 94°C-59°C-72°C X 5 ∣30s | |||||
| 94°C-57°C-72°C X5 ∣30s | |||||
| 94°C-55°C-72°C X55 ∣30s | |||||
| 72°C 10 min | |||||
| AGTTAGGTATTTGAAAGGAAAGTTTAT | ATCCAATCCTACTACTTTCTAACT | GGAAAGTTTATTTGGAAAGAT | TGGATYGTTTTAAATGTATATATTYGTATATTYGATTGAAGYG | 4°C ∞ | |
| TSPAN17 | TSPAN17-F | TSPAN17-R* | TSPAN17-S | TSPAN17 | 95°C 15 min |
| 94°C ∣30s | |||||
| 62°C ∣30s X55 | |||||
| 72°C ∣30s | |||||
| 72°C 10 min | |||||
| TGTGTTTTATTTTTTTTTTGGAATAGGT | ACCCAAACCTCATACTAATCC | AAAGTTGGAGTTTAATTTTAGTATT | TTTTATTTYGTYGTTTYGTTYGAT | 4°C ∞ | |
| CMPK2 | CMPK2-F | CMPK2-R* | CMPK2-S | CMPK2 | 95°C 15 min |
| 94°C ∣30s | |||||
| 63.5°C ∣30s X55 | |||||
| 72°C ∣30s | |||||
| 72°C 10 min | |||||
| GATGTTGGATTGTGGTAGGTAGA | AAACTTTTCCCTCTTCTTAAAATTACCCT | ATATTTTATTTTAGTGTAGGAAATT | TTAATTTGGTGGAAGAAAGGYGATTTYGAGGTGTTAATTTGGGYGA | 4°C ∞ | |
| GRIK2 | GRIK2-F | GRIK2-R* | GRIK2-S | GRIK2 | 95°C 15 min |
| 94°C-63.5°C-72°C X 5 ∣30s | |||||
| 72°C 10 min | |||||
| TAGTAGTGTAGTAGAATAGGGTTTGGTAAA | AAAAAATCAATAAAAAACCCAACCTTAAT | GGTTTGGTAAAATTTTTGTTAG | TAAAGYGTTTTTTGTGTYGGGTTGTGGTTYGTTATYGATATTTYGTTYGT | 4°C ∞ | |
| HCN1 | HCN1-F | HCN1-R* | HCN1-S | HCN1 | 95°C 15 min |
| 94°C-68°C-72°C X 5 ∣30s | |||||
| 94°C-66°C-72°C X 5 ∣30s | |||||
| 94°C-64°C-72°C X 55 ∣30s | |||||
| 72°C 10 min | |||||
| GAGGTTTTTGGGGTTTAGAAGAAGATTG | CCAAATACAAATTACCCTTCTTTTAGG | TTTTGGGGAGTGTGT | TTAGTTTGTTTTTTTTTATAGGYGTTGGTTATTTTYGTTGTGTTTATTTAGAAATAGYGGTYGG | 4°C ∞ | |
| KLF16 | KLF16-F | KLF16-R* | KLF16-S | KLF16 | 95°C 15 min |
| 94°C-63.5°C-72°C X 55 ∣30s | |||||
| 72°C 10 min | |||||
| GGGGAGGGATGGGTTTGGA | CAACACAACTACATCCACCCCTAA | GGGAAGGTAGTATTGTTAT | TTYGGGATTGTAGTAAGAAATAGGTTTGGGGGTGTTTAGTYGG | 4°C ∞ | |
| NR4A2 | NR4A2-F | NR4A2-R* | NR4A2-S | NR4A2 | 95°C 15 min |
| 94°C-64°C-72°C X 5 ∣30s | |||||
| 94°C-62°C-72°C X 5 ∣30s | |||||
| 94°C-60°C-72°C X 5 ∣30s | |||||
| 94°C-58°C-72°C X 55 ∣30s | |||||
| 72°C 10 min | |||||
| GGGTTTAGGGGAAAGTGAAGT | ACTAACCCTAACCCCCAATATACCTTTAT | GTTAGGTAGGAAATATATTAAAG | YGAGYGYGGGTTAGGAGTTTAGGGAGYGYGG | 4°C ∞ |
Statistical analysis
Statistical analyses were performed in GraphPad Prism Version 9 (GraphPad Software, San Diego, CA). For assay validation, linear regression was used to determine the R2 coefficient. For bisulfite pyrosequencing studies characterizing the imprinted gene status in different cell lines, a One-Factor ANOVA was run to determine the effect of cell line on variation in methylation. For bisulfite pyrosequencing studies assessing the impact of CE exposure on DNA methylation, a Two-Factor ANOVA was run, with one factor being CpG site and the other being Exposure Status. To determine if there was a significant distribution of genes significantly impacted by CE exposure in the overlap list compared to the SFARI-only list, a one-factor Chi-square test was performed.
Supplementary Material
Funding
This work was supported by The John Templeton Foundation under Grant 60957 to SKM.
Abbreviations:
- ASD
autism spectrum disorder
- CE
cannabis extract
- FACS
flow activated cell sorting
- hESC
human embryonic stem cell
- ICR
imprint control regions
- PGC
primordial germ cells
- RTI
Research Triangle Institute
- SFARI
Simons Foundation Autism Research Initiative
- SSC
spermatogonial stem cell
Footnotes
Declaration of Interests
The authors report there are no competing interests to declare.
Disclosure Statement
The authors report no conflict of interest.
Ethics Approval
All stem cell work was performed with NIH-approved cell lines and was conducted with IRB approval.
References
- Abbasi J 2017. The Paternal Epigenome Makes Its Mark. JAMA. 317(20):2049–2051. [DOI] [PubMed] [Google Scholar]
- Bassil CF, Huang Z, Murphy SK. 2013. Bisulfite pyrosequencing. Methods Mol Biol. 1049:95–107. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125(2):315–326. [DOI] [PubMed] [Google Scholar]
- Blin M, Norton W, Bally-Cuif L, Vernier P. 2008. NR4A2 controls the differentiation of selective dopaminergic nuclei in the zebrafish brain. Mol Cell Neurosci. 39(4):592–604. [DOI] [PubMed] [Google Scholar]
- Chandra S, Radwan MM, Majumdar CG, Church JC, Freeman TP, ElSohly MA. 2019. New trends in cannabis potency in USA and Europe during the last decade (2008-2017). Eur Arch Psychiatry Clin Neurosci. 269(1):5–15. [DOI] [PubMed] [Google Scholar]
- Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. 2003. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci U S A. 100(14):8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court F, Arnaud P. 2017. An annotated list of bivalent chromatin regions in human ES cells: a new tool for cancer epigenetic research. Oncotarget. 8(3):4110–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttler C, Mischley LK, Sexton M. 2016. Sex Differences in Cannabis Use and Effects: A Cross-Sectional Survey of Cannabis Users. Cannabis Cannabinoid Res. 1(1):166–175. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Jirtle RL. 2008. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 49(1):4–8. [DOI] [PubMed] [Google Scholar]
- Dong C, Chen J, Harrington A, Vinod KY, Hegde ML, Hegde VL. 2019. Cannabinoid exposure during pregnancy and its impact on immune function. Cell Mol Life Sci. 76(4):729–743. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkin I, Barres R. 2018. Sperm epigenetics and influence of environmental factors. Mol Metab. 14: 10.1016/j.molmet.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvash T, Ben-Yosef D, Eiges R. 2006. Human embryonic stem cells as a powerful tool for studying human embryogenesis. Pediatr Res. 60(2):111–117. [DOI] [PubMed] [Google Scholar]
- Easley CA, Bradner JM, Moser A, Rickman CA, McEachin ZT, Merritt MM, Hansen JM, Caudle WM. 2015. Assessing reproductive toxicity of two environmental toxicants with a novel in vitro human spermatogenic model. Stem Cell Res. 14(3):347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley CA, Phillips BT, McGuire MM, Barringer JM, Valli H, Hermann BP, Simerly CR, Rajkovic A, Miki T, Orwig KE et al. 2012. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2(3):440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Seoud M, Coullin P, Herbiniaux U, Oldenburg J, Rouleau G, Slim R. 2003. Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet. 12(12):1405–1413. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Bourc'his D. 2018. The discovery and importance of genomic imprinting. Elife. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca BM, Rebelo I. 2021. Cannabis and Cannabinoids in Reproduction and Fertility: Where We Stand. Reprod Sci. 10.1007/s43032-021-00588-1 [DOI] [PubMed] [Google Scholar]
- Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, Gilroy K, Baker S, Xia J, Dalley JW, Hurst LD et al. 2011. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 469(7331):534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. 2015. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell. 161(6):1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeson KW, Fowler KL, Estave PM, Kate Thompson S, Wagner C, Clayton Edenfield R, Symosko KM, Steves AN, Marder EM, Terrell ML et al. 2020. Detrimental effects of flame retardant, PBB153, exposure on sperm and future generations. Sci Rep. 10(1):8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guennewig B, Bitar M, Obiorah I, Hanks J, O'Brien EA, Kaczorowski DC, Hurd YL, Roussos P, Brennand KJ, Barry G. 2018. THC exposure of human iPSC neurons impacts genes associated with neuropsychiatric disorders. Transl Psychiatry. 8(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B et al. 2014. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 15(4):447–459. [DOI] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y et al. 2015. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 161(6):1437–1452. [DOI] [PubMed] [Google Scholar]
- Hasin DS. 2018. US Epidemiology of Cannabis Use and Associated Problems. Neuropsychopharmacology. 43(1):195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikichi T, Kohda T, Kaneko-Ishino T, Ishino F. 2003. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 31(5):1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JM, Kim J. 2009. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 442(1-2):18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Li G, Aguado C, Lujan R, Shah MM. 2017. HCN1 channels reduce the rate of exocytosis from a subset of cortical synaptic terminals. Sci Rep. 7:40257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Lujan R, Kadurin I, Uebele VN, Renger JJ, Dolphin AC, Shah MM. 2011. Presynaptic HCN1 channels regulate Cav3.2 activity and neurotransmission at select cortical synapses. Nat Neurosci. 14(4):478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Frey WD, He H, Kim H, Ekram MB, Bakshi A, Faisal M, Perera BP, Ye A, Teruyama R. 2013. Peg3 mutational effects on reproduction and placenta-specific gene families. PLoS One. 8(12):e83359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerou PH, Daley GQ. 2005. Therapeutic potential of embryonic stem cells. Blood Rev. 19(6):321–331. [DOI] [PubMed] [Google Scholar]
- Leter G, Consales C, Eleuteri P, Uccelli R, Specht IO, Toft G, Moccia T, Budillon A, Jonsson BA, Lindh CH et al. 2014. Exposure to perfluoroalkyl substances and sperm DNA global methylation in Arctic and European populations. Environ Mol Mutagen. 55(7):591–600. [DOI] [PubMed] [Google Scholar]
- Levy J, Grotto S, Mignot C, Maruani A, Delahaye-Duriez A, Benzacken B, Keren B, Haye D, Xavier J, Heulin M et al. 2018. NR4A2 haploinsufficiency is associated with intellectual disability and autism spectrum disorder. Clin Genet. 94(2):264–268. [DOI] [PubMed] [Google Scholar]
- Marini C, Porro A, Rastetter A, Dalle C, Rivolta I, Bauer D, Oegema R, Nava C, Parrini E, Mei D et al. 2018. HCN1 mutation spectrum: from neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain. 141(11):3160–3178. [DOI] [PubMed] [Google Scholar]
- Mauro PM, Carliner H, Brown QL, Hasin DS, Shmulewitz D, Rahim-Juwel R, Sarvet AL, Wall MM, Martins SS. 2018. Age Differences in Daily and Nondaily Cannabis Use in the United States, 2002-2014. J Stud Alcohol Drugs. 79(3):423–431. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meah F, Lundholm M, Emanuele N, Amjed H, Poku C, Agrawal L, Emanuele MA. 2021. The effects of cannabis and cannabinoids on the endocrine system. Rev Endocr Metab Disord. eng. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Huang Z, Hoyo C. 2012. Differentially Methylated Regions of Imprinted Genes in Prenatal, Perinatal and Postnatal Human Tissues. PLOS ONE. 7(7):e40924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, Acharya K, Boudreau M-H, Price TM, Raburn DJ et al. 2018. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics. 13(12):1208–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Conference of State Legislatures. 2021. State Medical Marijuana Laws. [updated August 23, 2021; accessed 2021 August 27].
- Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. 2016. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol. 59:10–26. [DOI] [PubMed] [Google Scholar]
- Peall KJ, Kurian MA, Wardle M, Waite AJ, Hedderly T, Lin JP, Smith M, Whone A, Pall H, White C et al. 2014. SGCE and myoclonus dystonia: motor characteristics, diagnostic criteria and clinical predictors of genotype. J Neurol. 261(12):2296–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peall KJ, Smith DJ, Kurian MA, Wardle M, Waite AJ, Hedderly T, Lin JP, Smith M, Whone A, Pall H et al. 2013. SGCE mutations cause psychiatric disorders: clinical and genetic characterization. Brain. 136(Pt 1):294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert RN, Bartolomei MS. 2015. Tissue-specific regulation and function of Grb10 during growth and neuronal commitment. Proc Natl Acad Sci U S A. 112(22):6841–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanahally S, Raheem O, Rogers M, Brisbane W, Ostrowski K, Lendvay T, Walsh T. 2019. The relationship between cannabis and male infertility, sexual health, and neoplasm: a systematic review. Andrology. 7(2):139–147. [DOI] [PubMed] [Google Scholar]
- Reece AS, Hulse GK. 2019a. Effect of cannabis legalization on US autism incidence and medium term projections. Clinical Pediatrics: Open Access. 4(2):17. [Google Scholar]
- Reece AS, Hulse GK. 2019b. Impacts of cannabinoid epigenetics on human development: reflections on Murphy et. al. ‘cannabinoid exposure and altered DNA methylation in rat and human sperm’ epigenetics 2018; 13: 1208-1221. Epigenetics.1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter MS, Krumbiegel M, Schluter G, Ekici AB, Reis A, Zweier C. 2017. Haploinsufficiency of NR4A2 is associated with a neurodevelopmental phenotype with prominent language impairment. Am J Med Genet A. 173(8):2231–2234. [DOI] [PubMed] [Google Scholar]
- Schrott R, Acharya K, Itchon-Ramos N, Hawkey AB, Pippen E, Mitchell JT, Kollins SH, Levin ED, Murphy SK. 2020a. Cannabis use is associated with potentially heritable widespread changes in autism candidate gene DLGAP2 DNA methylation in sperm. Epigenetics.1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrott R, Murphy SK. 2020. Cannabis use and the sperm epigenome: a budding concern? Environmental Epigenetics. 6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrott R, Rajavel M, Acharya K, Huang Z, Acharya C, Hawkey A, Pippen E, Lyerly HK, Levin ED, Murphy SK. 2020b. Sperm DNA methylation altered by THC and nicotine: Vulnerability of neurodevelopmental genes with bivalent chromatin. Sci Rep. 10(1):16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Seol MJ, Lee K. 2015. Differential expression of hyperpolarization-activated cyclic nucleotide-gated channel subunits during hippocampal development in the mouse. Mol Brain. 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Foundation Autism Research Initiative. 2019. SFARI Human Gene Module. [updated June 20, 2019]. www.gene.sfari.org.
- Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ et al. 2007. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol. 27(16):5871–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers H, Hannon E, Schalkwyk LC, Smith R, Wong CC, O'Donovan MC, Bray NJ, Mill J. 2015. Methylomic trajectories across human fetal brain development. Genome Res. 25(3):338–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA. 2015. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell. 161(6):1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchman R, Cuccaro M. 2011. Epilepsy and autism: neurodevelopmental perspective. Curr Neurol Neurosci Rep. 11(4):428–434. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration. 2019. 2018 National Survey on Drug Use and Health: Methodologic summary and definitions. Rockville, MD: Center for Behavioral Health Statistics and Quality, Sustance Abuse and Mental Health Services Administration. Retrieved from https://www.samhsa.gov/data/ [Google Scholar]
- Wu H, Hauser R, Krawetz SA, Pilsner JR. 2015. Environmental Susceptibility of the Sperm Epigenome During Windows of Male Germ Cell Development. Curr Environ Health Rep. 2(4):356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Pan S, Zheng H, Luo Z, Tembo KM, Jamal M, Yu Z, Yu Y, Xia J, Yin Q et al. 2018. PEG10 as an oncogene: expression regulatory mechanisms and role in tumor progression. Cancer Cell Int. 18:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SS, Li YC, Coley AA, Chamberlin LA, Yu P, Gao WJ. 2018. Cell-Type Specific Development of the Hyperpolarization-Activated Current, Ih, in Prefrontal Cortical Neurons. Front Synaptic Neurosci. 10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou W, Liu Y, Li N. 2016. Integrated analysis of DNA methylation and RNAsequencing data in Down syndrome. Mol Med Rep. 14(5):4309–4314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.