Abstract
BACKGROUND:
New ultrasensitive methods for detecting residual disease after surgery are needed in human papillomavirus–associated oropharyngeal squamous cell carcinoma (HPV+OPSCC).
METHODS:
To determine whether the clearance kinetics of circulating tumor human papillomavirus DNA (ctHPVDNA) is associated with postoperative disease status, a prospective observational study was conducted in 33 patients with HPV+OPSCC undergoing surgery. Blood was collected before surgery, postoperative days 1 (POD 1), 7, and 30 and with follow-up. A subcohort of 12 patients underwent frequent blood collections in the first 24 hours after surgery to define early clearance kinetics. Plasma was run on custom droplet digital polymerase chain reaction (ddPCR) assays for HPV genotypes 16, 18, 33, 35, and 45.
RESULTS:
In patients without pathologic risk factors for recurrence who were observed after surgery, ctHPVDNA rapidly decreased to <1 copy/mL by POD 1 (n = 8/8). In patients with risk factors for macroscopic residual disease, ctHPVDNA was markedly elevated on POD 1 (>350 copies/mL) and remained elevated until adjuvant treatment (n = 3/3). Patients with intermediate POD 1 ctHPVDNA levels (1.2–58.4 copies/mL) all possessed pathologic risk factors for microscopic residual disease (n = 9/9). POD 1 ctHPVDNA levels were higher in patients with known adverse pathologic risk factors such as extranodal extension >1 mm (P = .0481) and with increasing lymph nodes involved (P = .0453) and were further associated with adjuvant treatment received (P = .0076). One of 33 patients had a recurrence that was detected by ctHPVDNA 2 months earlier than clinical detection.
CONCLUSIONS:
POD 1 ctHPVDNA levels are associated with the risk of residual disease in patients with HPV+OPSCC undergoing curative intent surgery and thus could be used as a personalized biomarker for selecting adjuvant treatment in the future.
Keywords: cell-free DNA, circulating tumor DNA, head and neck cancer, human papillomavirus (HPV), oropharyngeal cancer
LAY SUMMARY:
• Human papillomavirus–associated oropharyngeal squamous cell carcinoma (HPV+OPSCC) is increasing at epidemic proportions and is commonly treated with surgery.
• This report describes results from a study examining the clearance kinetics of circulating tumor HPV DNA (circulating tumor human papillomavirus DNA [ctHPVDNA]) following surgical treatment of HPV+OPSCC.
• We found that ctHPVDNA levels 1 day after surgery are associated with the risk of residual disease in patients with HPV+OPSCC and thus could be used as a personalized biomarker for selecting adjuvant treatment in the future.
• These findings are the first to demonstrate the potential utility of ctHPVDNA in patients with HPV+OPSCC undergoing surgery.
INTRODUCTION
Human papillomavirus–associated oropharyngeal squamous cell carcinoma (HPV+OPSCC) is increasing at epidemic proportions, surpassing cervical cancer to become the most common HPV-associated malignancy in the United States.1 By the year 2030, HPV+OPSCC will account for approximately 30,000 new cancer diagnoses per year.2 Compared to patients with “traditional” carcinogen-induced OPSCC, patients with HPV+OPSCC are younger, healthier, and more responsive to treatment and therefore are more likely to be cured and have longer life expectancy after cure.3–5 Current treatment algorithms for HPV+OPSCC lead to significant long-term morbidity and decreased quality of life.6–8 Interest is now focused on deintensifying treatment to decrease treatment-induced morbidity, whereas maintaining survival outcomes. One approach to deintensification is using surgery to eliminate or reduce the dose of radiation.9–11 Transoral robotic surgery (TORS) to remove the primary tumor, paired with a cervical lymphadenectomy, is now the most common approach to treating early-stage HPV+OPSCC in the United States.11 Unfortunately, most patients who undergo TORS go on to receive radiation (58%) or chemoradiation (CRT) (31%) due in part to the lack of effective methods for identifying patients with residual disease (RD), and, thus, those in need of adjuvant treatment.10,12 The inability to accurately stratify patients into risk groups postoperatively results in practitioners defaulting to overtreatment.7 Current decision-making for who is in need of adjuvant treatment, and what type, is nonstandardized and based on indirect clinicopathologic risk factors at both the primary site (perineural invasion [PNI], lymphovascular invasion [LVI], and margin status) and neck (number and size of lymph nodes involved and presence and extent of extranodal extension [ENE]).13 These clinicopathologic risk factors have poor individualized predictive and prognostic capacity. Because the vast majority of surgeries result in removal of all visible tumor and lymph nodes, accurate detection of occult RD after surgery is the primary limiting factor in more effectively tailoring treatment and personalizing care in HPV+OPSCC.
Circulating tumor DNA (ctDNA) is released into the blood by cancer cells, contributing a small fraction of total cell-free DNA.14 HPV-associated cancers also release ctHPVDNA.15–26 Most recently, droplet digital polymerase chain reaction (ddPCR)–based approaches, which have the benefit of high sensitivity and fidelity, have been used to demonstrate that 1) most patients have detectable ctHPVDNA at the time of diagnosis (sensitivity of 89%–98% and specificity of 97%–100% compared to p16 immunohistochemistry [IHC]), 2) ctHPVDNA levels trend down slowly during definitive CRT in responding patients, and 3) ctHPVDNA is a marker of recurrence in the post-CRT setting with levels re-elevating in patients who suffer recurrence.20–24,26 Taken together, ctHPVDNA detection appears to be a promising noninvasive approach to determining the presence or absence of HPV+OPSCC. However, existing work has focused on patients receiving CRT. The kinetics of ctHPVDNA in the surgical setting, its relationship to surgical pathology risk factors for recurrence, and capacity to detect RD have not been established in HPV+OPSCC nor any HPV-associated cancer.
We describe results from a prospective observational biomarker study of longitudinal ctHPVDNA monitoring in a cohort of patients with HPV+OPSCC undergoing definitive surgery using custom ddPCR assays for detection of high-risk HPV DNA. We tested the hypothesis that clearance kinetics of ctHPVDNA after surgical treatment of HPV+OPSCC would be associated with risk of RD.
MATERIALS AND METHODS
Study Design and Enrollment
All patients presenting for treatment of newly diagnosed HPV+OPSCC at Massachusetts Eye and Ear/Massachusetts General Hospital from January 21, 2020, to March 31, 2021, were approached for informed consent and consecutively enrolled in this prospective observational biomarker study. Patients gave written informed consent to a protocol approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (DFCI 18–653). The study was conducted in accordance with the US Common Rule. Eligibility criteria for inclusion in the study included 1) age older than 18 years, 2) willingness to contribute blood samples for research purposes, 3) biopsy-proven, untreated HPV+OPSCC clinical stage T0–4, N1–3, M0 American Joint Committee on Cancer, 8th edition, 4) having undergone definitive TORS and cervical lymphadenectomy or CRT, and 5) no history of head and neck cancer.
Diagnosis, Treatment, and Post-treatment Monitoring
HPV+OPSCC was defined by a positive p16 IHC (70% of carcinoma cells showing nuclear reactivity). All patients underwent cross-sectional imaging of the head, neck, and chest before treatment, at 3 months after treatment and 1 year. Clinical examinations were performed at 2- to 3-month intervals after treatment. Determination of disease recurrence required confirmatory tissue biopsy. Surgery involved resection of the primary tumor using a transoral robotic oropharyngectomy with selective neck lymphadenectomy levels 2 to 4. Patients receiving CRT were treated with 70-Gy intensity modulated radiotherapy and weekly intravenous cisplatin 40 mg/m2 or carboplatin AUC 1.5 and paclitaxel 30 mg/m2.
Surgical Pathology
The following pathology features were defined and measured: 1) ENE was defined as none, ≤1 mm, >1 mm, or gross (extensive soft tissue infiltration seen intraoperatively), 2) margins were defined as negative (>1 mm), microscopically positive/close (≤1 mm), focally positive (1 defined region/edge of tumor positive without obvious disease remaining in patient), or grossly positive. Risk factors for microscopic RD were PNI, LVI, ENE ≤1 mm, focally positive margins, and >2 lymph nodes involved. Risk factors for macroscopic RD were gross ENE, gross positive margins, and distant metastases. RD was defined as microscopic or macroscopic RD.
Blood Collection and Processing
Ten to twenty milliliters of blood was collected in DNA BCT tubes (Streck, La Vista, Nebraska) according to the following schema: 1) pretreatment, 2) 6:00 am on postoperative day (POD) 1, 3) POD 7, 4) POD 30, 5) 3 months after treatment completion, and 6) 1 year after treatment completion. Additional blood collections were performed in some patients on the basis of their clinical course. A subcohort of 12 patients underwent frequent collections in the immediate postoperative period to examine early clearance kinetics. In addition to the above blood collections, in this early kinetics cohort, blood was collected at the following times: 1) immediately after resection (tumor removed and patient still under general anesthesia in the operating room) and 2) 6, 12, 18, and 24 hours postoperatively. A cohort of patients treated with CRT was used as a comparison. In this cohort, blood was collected before treatment, weekly during treatment (6 time points), and with 3-month and 1-year imaging. Blood was double-spun at room temperature for 10 minutes at 1600g and then 3000g and frozen until extraction at −80°C. Total cell-free DNA was extracted using the QIAamp circulating nucleic acid kit (Qiagen, Valencia, California) and quantified with a Qubit fluorometer (Thermo Fisher Scientific).
ddPCR and Assay Characteristics
Five ddPCR assays were developed and optimized to detect conserved regions of E7 from HPV 16, 18, 33, 35, and 45.26 Probes were confirmed to have no cross-genotype reactivity, to be linear (R2 > 0.96 for all genotypes), and to have limits of detection ≤0.1% fractional abundance. Detailed assay metrics and performance can be found in Siravegna et al.26
Hypothesis and Statistical Analysis
The primary objective of this study was to test the hypothesis that postoperative ctHPVDNA levels would decrease to undetectable within 24 hours in patients without clinicopathologic risk factors for recurrence (ie, those patients who would not require adjuvant treatment), would remain marginally elevated in those patients with risk factors for microscopic RD (ie, those who would need adjuvant radiation therapy), and would remain significantly elevated in patients with known high-risk features (ie, those needing adjuvant CRT). Wilcoxon rank sum tests were used to make comparisons between groups.
RESULTS
Patient Characteristics
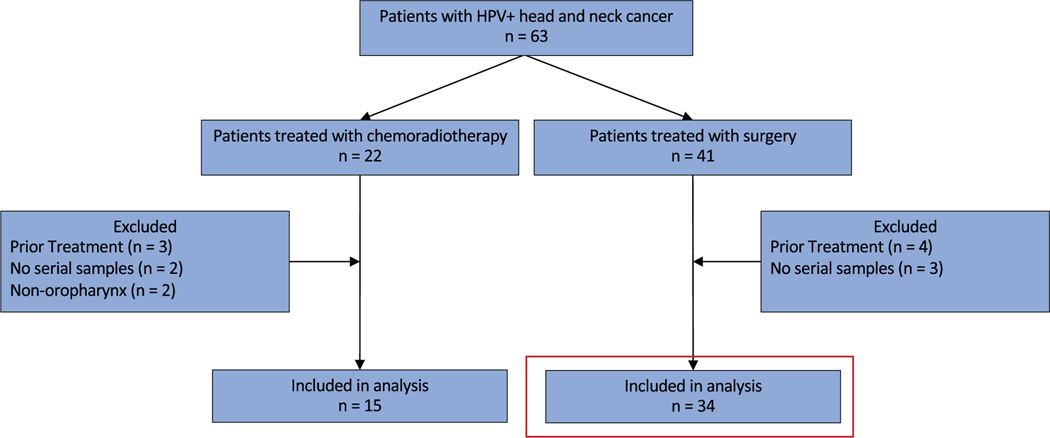
From January 21, 2020, to March 30, 2021, excluding a 4-month block of time due to cessation of research during the coronavirus pandemic, 63 patients presenting for treatment of an HPV+OPSCC were enrolled. Forty-one patients were treated surgically, and 22 patients were treated with CRT. Forty-nine patients were included in the final analysis: 34 patients treated surgically and 15 patients treated with CRT (Fig. 1; Table 1). All patients had HPV+OPSCC as defined by p16 overexpression. 86% of patients (n = 42) had confirmatory direct HPV testing by DNA PCR or RNA in situ hybridization (ISH).
Figure 1.
Consolidated Standards of Reporting Trials diagram of patient cohorts. The red box is the primary surgical cohort. HPV indicates human papillomavirus.
TABLE 1.
Patient Characteristics at Time of Treatment (n = 33)
| Characteristic | Surgical Patients (n = 34) | CRT Patients (n = 15) |
|---|---|---|
|
| ||
| Mean age (range) | 62 (49–85) | 59 (49–70) |
| Sex | ||
| Male | 30 | 13 |
| Female | 4 | 2 |
| TNM statusa | ||
| T | pT | cT |
| 0 | 0 | 1 |
| 1 | 18 | 5 |
| 2 | 15 | 7 |
| 3 | 1 | 1 |
| 4 | 0 | 1 |
| N | pN | cN |
| 0 | 4 | 0 |
| 1 | 27 | 11 |
| 2 | 2 | 1 |
| 3 | 1 | 3 |
| M | ||
| 0 | 33 | 15 |
| 1 | 0 | 0 |
| Primary site | ||
| Palatine tonsil | 20 | 7 |
| Base of tongue | 15 | 5 |
| Overlapping | 0 | 2 |
| Unknown primary | 0 | 1 |
Abbreviation: CRT, chemoradiation.
Pathologic criteria when available.
ctHPVDNA at Presentation and Association With Clinical Risk Factors
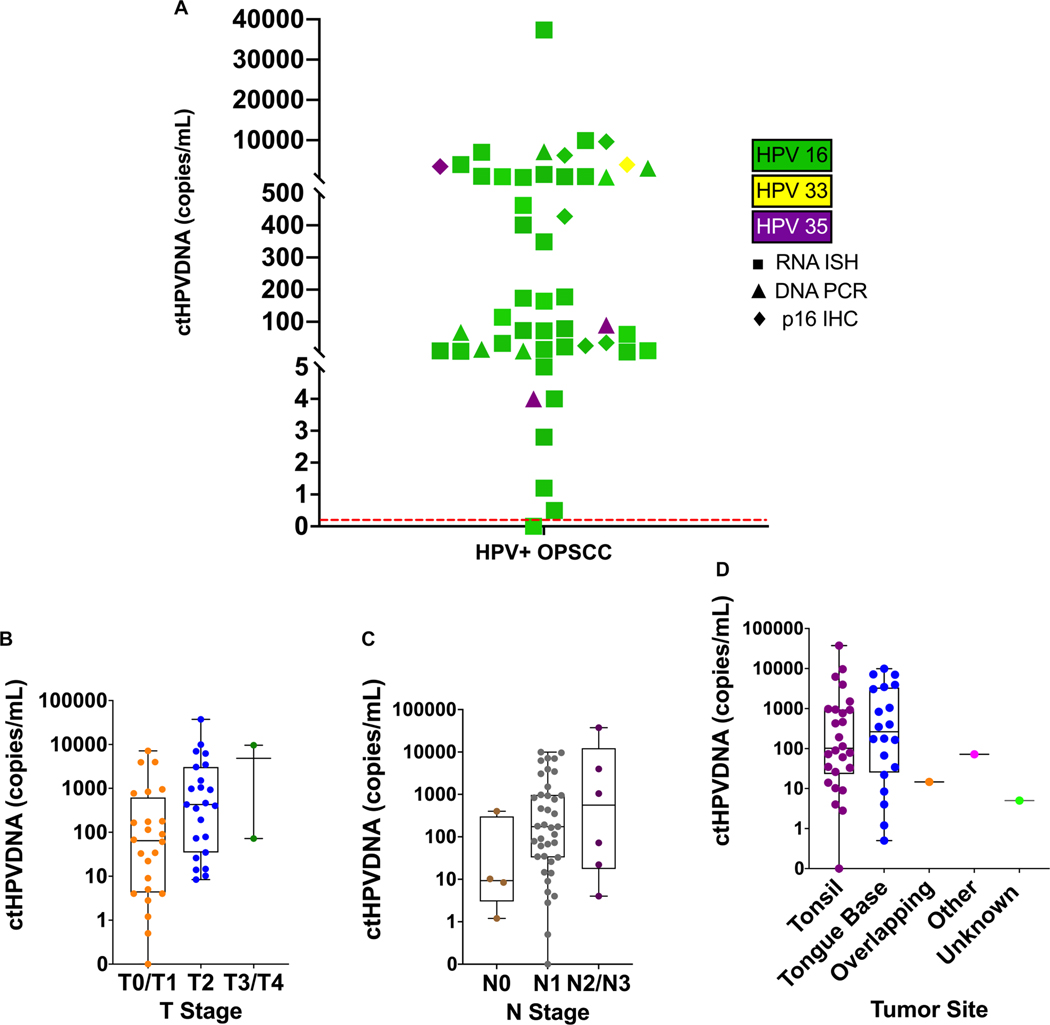
Forty-eight of 49 patients included in the analysis had detectable ctHPVDNA at presentation (sensitivity 98%). Mean ctHPVDNA at presentation was 2076 copies/mL, the median was 164 copies/mL, and the range was 0 to 37,350 copies/mL (Fig. 2A). T stage and N stage were borderline associated with ctHPVDNA levels at presentation (P = .0453; P = .0506), whereas tumor anatomic site within the oropharynx was not (P = .58) (Fig. 2B,C,D). HPV genotype on tissue testing matched HPV genotype by ctHPVDNA in all cases.
Figure 2.
ctHPVDNA detection at the time of diagnosis and correlation with tumor stage and site. (A) ctHPVDNA detection at the time of diagnosis. The color of the shapes corresponds to genotype detected, whereas shape corresponds to the test used on biopsy to confirm HPV+OPSCC. All patients with direct HPV testing (RNA ISH and DNA PCR) also had p16 IHC. The red dotted line represents the test threshold for a positive versus negative test, demonstrating 1 patient below threshold. (B) T stage and (C) N stage were borderline associated with ctHPVDNA levels at presentation (P = .0453; P = .0506), whereas (D) the tumor anatomic site was not (P = .58). The median and interquartile range are shown for each graph (box). P values are based on a 2-sided Mann-Whitney U test. ctHPVDNA indicates circulating tumor human papillomavirus DNA; HPV, human papillomavirus; HPV+OPSCC, human papillomavirus (HPV)–associated oropharyngeal squamous cell carcinoma; IHC, immunohistochemistry; ISH, in situ hybridization; OPSCC, oropharyngeal squamous cell carcinoma; PCR, polymerase chain reaction.
Surgical Cohort
Forty-one patients presenting for surgical treatment of HPV+OPSCC were enrolled. Seven patients were excluded due to having been partially treated before accrual (for example, diagnostic tonsillectomy performed before presentation) or for missing key serial samples related to cessation of research during the coronavirus pandemic, leaving 34 patients for analysis. Mean ctHPVDNA at presentation was 1070 copies/mL, the median was 139 copies/mL, and the range was 0 to 9922 copies/mL. The ctHPVDNA level for one patient was below the pre-specified cut off for test positivity, leaving 33 patients for longitudinal analysis. Interestingly, HPV was detectable in the saliva of this patient, which cleared following surgery, on POD 1 (Supporting Fig. 1). The mean and median follow-up for the cohort was 12 months, and the range was 6 to 20 months.
Surgical Clearance Kinetics in the First 24 Hours After Surgery
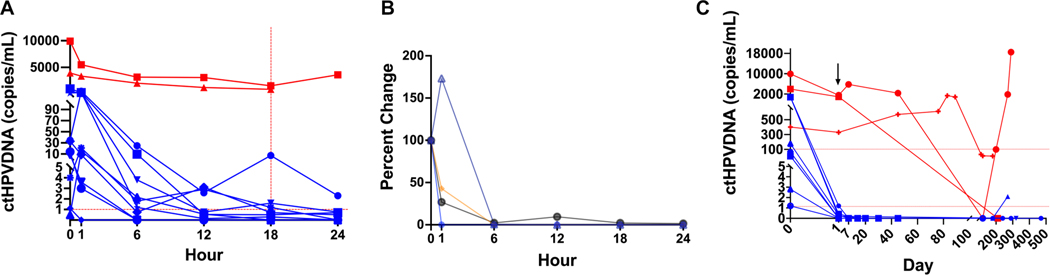
Twelve patients underwent frequent blood collections in the immediate postoperative period (Fig. 3A; Supporting Fig. 2). Four of 12 patients had surgical pathology without any pathologic risk factors for recurrence and went on to observation without recurrence. These patients were considered ideal for determining early clearance kinetics with assumed no RD (Fig. 3B). All 4 patients had ctHPVDNA levels that decreased precipitously within 6 hours and stayed below 1 copy/mL by 6:00 am on POD 1 (18 hours postoperatively). Thus, <1 copy/mL on POD 1 was associated with no RD after surgery. Three additional patients had POD 1 ctHPVDNA levels below 1 copy/mL, all of whom had risk factors of unclear significance—1 with LVI and 2 with PNI (Supporting Figs. 3 and 10BB,CC,DD).
Figure 3.
ctHPVDNA kinetics after surgery. (A) Early kinetics cohort demonstrating clear separation within 6 hours of patients with risk factors for persistent macroscopic disease (red lines) and remainder of cohort (blue lines). Samples were collected immediately before surgery, immediately after tumor removal, and every 6 hours for 24 hours. Total cohort size was 12 patients. Vertical red dotted line represents 6:00 AM postoperative day (POD) 1. (B) Patients undergoing early kinetics lacking all known clinicopathologic risk factors (n = 4) demonstrated a rapid decrease to <1 copy/mL in 1 to 18 hours after surgery. Data were normalized to the pretreatment level. (C) Patients with risk factors for macroscopic RD (red lines) and no risk factors (blue lines) demonstrated clear separation within 18 hours of surgery (POD 1) in full cohort. The black arrow indicates POD 1 values on the x-axis scale. ctHPVDNA indicates circulating tumor human papillomavirus DNA.
Five patients had ctHPVDNA ≥1 copy/mL on POD 1, all of whom had adverse pathology risk factors. Two of these patients had ctHPVDNA levels that remained markedly elevated at >850 copies/mL. Both of these patients had ENE: 1 with gross ENE (Supporting Fig. 10Y) and 1 with >1 mm ENE (Supporting Fig. 10X). Additionally, the patient with >1 mm ENE was found to have distant metastases soon after surgery, which likely contributed to the elevated levels. Thus, significantly elevated POD 1 ctHPVDNA associated with macroscopic RD after surgery. Adverse pathology features of the remaining 3 patients with ctHPVDNA ≥1 copy/mL on POD 1 included >2 lymph nodes positive (2) and focally positive surgical margins + ≤1 mm ENE (1). Notably, these 3 patients’ POD 1 ctHPVDNA levels were between patients with no pathology risk factors and those with risk factors for macroscopic RD (range, 1.2–7.2 copies/mL), consistent with the clinical assumption that this group of patients would have microscopic RD. Based on these results, and receiver operating characteristic (ROC) analysis, we set 1 copy/mL (AUC, 0.84) and 100 copies/mL (AUC, 1.0) as thresholds for the remainder of the analysis to test the hypothesis that ctHPVDNA as early as POD 1 may differentiate patients with no RD (<1 copy/mL), microscopic RD (1–100 copies/mL), and macroscopic RD (>100 copies/mL).
POD 1 ctHPVDNA Levels Associate With Residual Disease Risk
Based on the early kinetics analysis, POD 1 levels of 1 and 100 copies/mL were applied to the full surgical cohort (Supporting Fig. 4). We first compared known pathologic risk factors for recurrence to postsurgical kinetics (Table 2). Examining patients with high-risk pathology features for macroscopic RD, 2 patients had pathology consistent with gross RD: 1 with gross positive margins (Supporting Fig. 10M) and 1 with gross ENE (Supporting Fig. 10Y). In both patients, POD 1 ctHPVDNA remained significantly elevated (>300 copies/mL). When combining these patients with a third patient (Supporting Fig. 10X), discussed above, with distant metastases identified soon after surgery, patients with assumed macroscopic RD after surgery demonstrated significantly elevated POD 1 ctHPVDNA levels (range, 329–1554 copies/mL) (Fig. 3C).
TABLE 2.
Clinicopathologic Risk Factors Based on Surgical Pathology
| Pathology | No. (%) | <1 Copy/mL | 1–100 Copies/mL | >100 Copies/mL |
|---|---|---|---|---|
|
| ||||
| Distant metastasis | 1 (3) | 0 | 0 | 1 |
| Gross positive margins | 1 (3) | 0 | 0 | 1 |
| Gross ENE | 1 (3) | 0 | 0 | 1 |
| ENE >1 mm | 4 (12) | 0 | 3 | 1 |
| Focal positive margins | 3 (9) | 1 | 2 | 0 |
| Close margins | 4 (12) | 3 | 1 | 0 |
| PNI | 6 (18) | 3 | 2 | 1 |
| LVI | 8 (24) | 4 | 2 | 2 |
| ENE ≤1 mm | 2 (6) | 1 | 1 | 0 |
| Lymph nodes | ||||
| 0 | 4 (12) | 4 | 0 | 0 |
| 1 | 17 (51) | 10 | 7 | 0 |
| 2 | 7 (21) | 6 | 1 | 0 |
| >2 | 4 (12) | 1 | 2 | 1 |
ENE, extranodal extension; LVI, lymphovascular invasion; PNI, perineural invasion.
We next examined patients without any pathologic risk factors for recurrence. Eight of 9 patients had POD 1 ctHPVDNA levels <1 copy/mL (Fig. 3C). Notably, the only patient without pathologic risk factors for recurrence with a POD 1 ctHPVDNA level ≥1 copy/mL was found to have a second primary HPV+OPSCC 5 months later, which we hypothesize may be the cause of their delayed clearance kinetics (Supporting Fig. 10A). Patients with risk factors for macroscopic RD had significantly higher POD 1 ctHPVDNA levels compared to patients without pathology risk factors (P = .0037).
While some clinicopathologic risk factors have been established as markers of aggressive biology and increased risk of recurrence/persistence in HPV+OPSCC, the prognostic capacity of other pathologic features is less clear. Using cutoffs of 1 and 100 copies/mL on POD 1, we next examined the relationship between these pathologic risk factors and POD 1 ctHPVDNA levels.
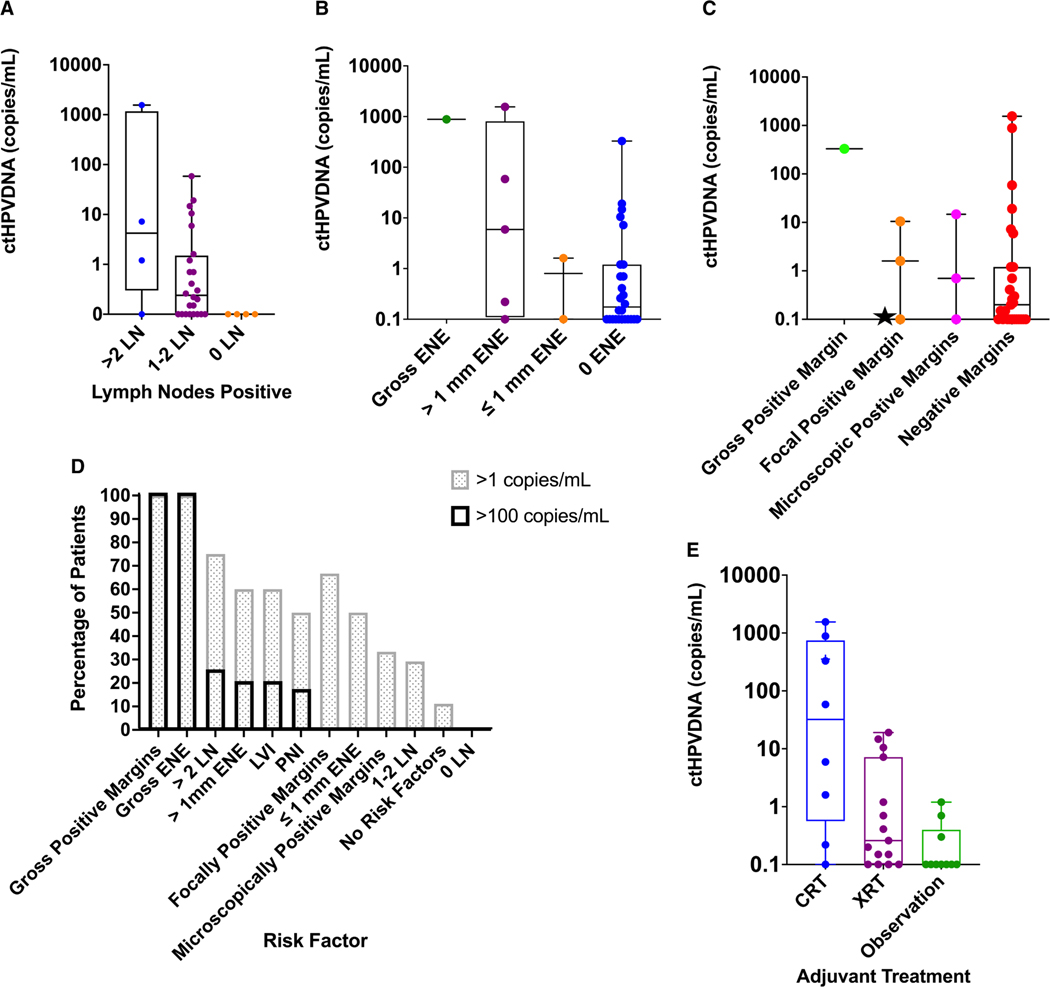
Both number of lymph nodes involved and growth outside of the lymph node capsule (ENE) are prognostic indicators in HPV+OPSCC.27 Here, increasing lymph node burden was associated with higher ctHPVDNA levels on POD 1 (Fig. 4A,D) (P = .0453). ENE >1 mm has been used in the literature and, in prospective trials, as a feature predictive of aggressive biology. POD 1 ctHPVDNA was higher in patients with >1 mm ENE compared to those with ≤1 mm (P = .0481) (Fig. 4B,D).
Figure 4.
Postoperative ctHPVDNA levels and clinicopathologic risk factors. Patients were stratified by postoperative day (POD) 1 ctHPVDNA: (A) lymph node burden, (B) ENE, and (C) surgical margins. The star represents patient with positive surgical margin on initial evaluation, but no tumor cells were identified on reresection; this suggests false-positive histopathology. (D) The percentage of patients with pathologic risk factors with >1 and >100 copies/mL on POD 1 demonstrating higher POD 1 ctHPVDNA with higher risk features. (E) POD 1 ctHPVDNA is predictive of the adjuvant treatment received. The adjuvant treatment received correlates with POD 1 ctHPVDNA levels. The median and interquartile range are shown for each graph. CRT indicates chemoradiation; ctHPVDNA, circulating tumor human papillomavirus DNA; ENE, extranodal extension; LN, lymph node; LVI, lymphovascular invasion; PNI, perineural invasion; XRT, radiation therapy.
Surgical margins were clinically categorized as grossly positive (n = 1), focally positive (n = 4), close/microscopically positive (≤1 mm) (n = 3), and negative (n = 26). POD 1 ctHPVDNA levels trended down from grossly positive margins to negative margins but were not statistically different (P = .21) (Fig. 4C). Only 1 patient with grossly or focally positive margins had <1 copy/mL on POD 1. Notably, this patient underwent re-resection of the focally positive margin without evidence of cancer in the specimen and went on to be observed with no evidence of recurrence, suggesting this case may not have had focally positive margins but rather a false positive on histomorphologic examination (Fig. 3C, starred data point).
The prognostic significance of PNI and LVI in HPV+OPSCC is unknown. Here, PNI or LVI was seen in 38% of the overall cohort, and neither was associated with POD 1 ctHPVDNA levels (P = .52 and P = .23, respectively).
Last, pathologic tumor stage, anatomic site, and presenting ctHPVDNA level were examined. T stage, N stage, tumor site, and pretreatment ctHPVDNA levels did not correlate with clearance to <1 copy/mL on POD 1 (P = .18, P = .30, and P = .70, respectively), suggesting the adequacy and completeness of the surgery at removing all disease is what drives POD 1 ctHPVDNA levels as opposed to these initial variables.
POD 1 ctHPVDNA Is Associated With Postoperative Adjuvant Treatment
Ten patients (29%) received observation after surgery, 16 (47%) received adjuvant radiation, and 8 (24%) received CRT. When comparing POD 1 ctHPVDNA levels to adjuvant treatments received, 9 of 10 patients who went on to observation had POD 1 ctHPVDNA <1 copy/mL. The mean POD 1 ctHPVDNA level was 0.2 copies/mL, the median was 0 copies/mL, and the range was 0 to 1.2 copies/mL (Supporting Fig. 6). As mentioned above, the sole patient whose level remained above 1 copy/mL was later found to have a second HPV+OPSCC primary. Ten of 16 patients treated with adjuvant radiation had levels that decreased below 1 copy/mL on POD 1. The mean POD 1 ctHPVDNA level for the cohort was 3.4 copies/mL, the median was 0.2 copies/mL, and the range was 0 to 19.1 copies/mL (Supporting Fig. 7). Two of 8 patients who received CRT had POD 1 levels <1 copy/mL. The mean POD 1 ctHPVDNA level for the cohort was 404 copies/mL, the median was 58 copies/mL, and the range was 0.2 to 1554 copies/mL (Supporting Fig. 8). POD 1 ctHPVDNA was markedly different between patients who received observation, radiation, and CRT (P = .0076) (Fig. 4E; Supporting Fig. 9). In total, 12 patients with POD 1 ctHPVDNA <1 copy/mL received adjuvant radiation or CRT.
Detection of Recurrent/Persistent Disease
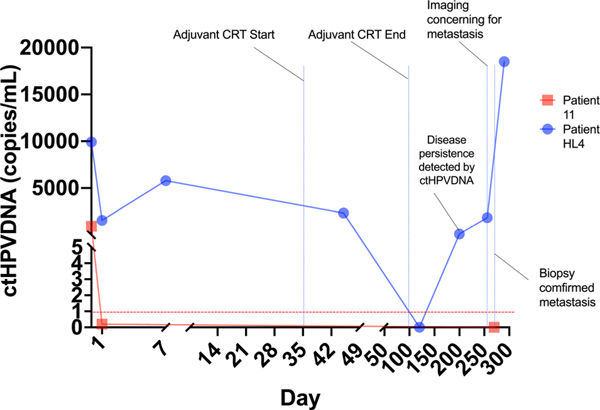
Before final data analysis, 1 patient was known to have biopsy-proven recurrence or disease persistence. This patient received surgery followed by CRT for ENE >1 mm. ctHPVDNA levels remained markedly elevated after surgery, transiently decreased after adjuvant CRT and immediately rebounded, indicating persistent disease (Fig. 5). The patient was found to have lung metastases on post-treatment imaging, which was confirmed with biopsy. Notably, pretreatment chest CT did not show evidence of metastasis. The patient subsequently developed clinical progression of their metastases with increasing ctHPVDNA levels. ctHPVDNA detection of persistent disease occured 56 days before clinical detection.
Figure 5.
ctHPVDNA detects recurrence earlier than standard of care monitoring. This figure contrasts 2 patients with the same stage presentation prior to surgery (cT1N1) who had significant differences in postoperative circulating tumor human papillomavirus DNA (ctHPVDNA) levels indicating differences in their residual disease (RD) burden. Patient HL4 (blue line) had significant elevation on postoperative day (POD) 1, downtrending only after adjuvant chemoradiotherapy (blue vertical lines) and immediate re-elevation signifying persistence. Persistence was detected by ctHPVDNA 2 months before clinical detection. Patient 11 (red line) had an immediate decrease in ctHPVDNA to zero after surgery. ctHPVDNA remained undetectable for the remainder of the study with no clinical evidence of recurrence.
Two additional patients had ctHPVDNA levels >0 at final data analysis. The first patient underwent surgery followed by observation, with ctHPVDNA levels decreasing to <1 on POD 1. ctHPVDNA levels remained zero until POD 270 at which time an elevation was detected (Supporting Figs. 4 and 10C). At the time of publication, no recurrence had been clinically detected (follow-up 15 months), but we continue to monitor the patient to understand whether this is a true positive, or technical artifact. In a second patient, ctHPVDNA levels remained markedly elevated despite surgery and adjuvant CRT, never reaching zero, before data analysis (Supporting Figs. 4 and 10M). At the time of analysis, no persistence/recurrence had been clinically detected; however, during the intervening time of manuscript submission, a new neck metastasis was identified (day 270), confirming disease persistence, as suggested by the lack of ctHPVDNA clearance.
CRT Cohort Clearance Kinetics
As a comparison cohort for surgical patients, 15 patients undergoing definitive CRT were followed longitudinally during and after treatment. The mean and median follow-up for the cohort was 12 months, and the range was 7 to 15 months. All patients had detectable ctHPVDNA at presentation with a mean of 4356 copies/mL, a median of 428 copies/mL, and a range of 9 to 37,350 copies/mL. Using a threshold of 95% decrease in ctHPVDNA levels, as described by Chera et al.,20 93% of patients cleared ≥95% ctHPVDNA by the last week of treatment and 73% by week 4 (Supporting Fig. 11A,C). The mean time to ≥95% clearance was 3.4 weeks on treatment, and the median was 4.0 weeks. Using a more stringent threshold of clearance, <1 copy/mL remaining, 73% of patients cleared ctHPVDNA to <1 copy/mL by the last week of treatment and 33% by 4 weeks. The mean time to clearance <1 copy/mL was 4.5 weeks on treatment, and the median was 5.0 weeks (Supporting Fig. 11B,C,D). T stage, N stage, anatomic site, and pretreatment ctHPVDNA level were not associated with time to clearance (all P > .05).
DISCUSSION
We have conducted a prospective observational biomarker study of ctHPVDNA clearance kinetics in patients with HPV+OPSCC undergoing curative intent surgery. We found that POD 1 ctHPVDNA levels are associated with the risk of residual disease. These findings are the first to demonstrate the potential utility of ctHPVDNA as a tool for personalized treatment decision-making in patients with HPV+OPSCC undergoing surgery and add to a growing body of literature which support ctHPVDNA use as a diagnostic test and a biomarker of treatment response and recurrence after CRT.20,21,26
Few studies across all cancer types have examined ctDNA in the immediate postoperative period (first 24 hours), and none in HPV-associated cancers. To our knowledge, only 1 study has systematically examined viral ctDNA in the surgical setting. To et al28 examined a heterogeneous group of 11 patients with recurrent nasopharyngeal carcinoma, reporting a rapid decline in ctEB-VDNA levels to undetectable after surgery in patients who did not recur and residual elevation in those who recurred. The authors found the half-life of ctEBVDNA to be 139 minutes, suggesting early postsurgical monitoring of viral ctDNA could be an accurate biomarker of RD. Although postsurgical monitoring of ctEBVDNA has not become commonplace due to the infrequency with which surgery is used in nasopharyngeal carcinoma, surgery is common in HPV+OPSCC. Other studies of ctHPVDNA have included small numbers of patients treated surgically.29
To determine early clearance kinetics in HPV+OPSCC, we first used a cohort of 12 patients presenting for surgical treatment with clinical staging consistent with early-stage disease (cT1N1M0). These patients were chosen to optimize our chances of identifying “ideal” patients with no RD after surgery. Demonstrating the inaccuracy of current clinical staging, which is based on cross-sectional imaging and physical examination, only 4 of 12 patients had no risk factors on surgical pathology and subsequently went on to observation without a recurrence.30 All 4 of these patients cleared ctHPVDNA within 6 hours, demonstrating the short half-life and rapid clearance of ctHPVDNA in patients with no RD. Contrary to this, patients with risk factors for macroscopic RD had markedly elevated ctHPVDNA levels after surgery, which did not appreciably decline until after adjuvant treatment. Both the rapid decline to <1 copy/mL in patients without risk factors and the marked elevation in patients with assumed or known macroscopic RD were validated in the full surgical cohort using POD 1 ctHPVDNA thresholds of <1 and >100 copies/mL. Cut points were set by data visualization and ROC analysis. ROC analysis of the full surgical cohort suggested similar cut points (0.7 and 329 copies/mL) with nearly equivalent performance to 1 copy/mL (AUC, 0.84) and 100 copies/mL (AUC, 1). Taken together, our findings suggest that after additional validation ctHPVDNA could potentially be used to identify patients who could be safely observed after surgery, and those patients who require adjuvant treatment for macroscopic RD.
Of great interest was the finding that, in addition to ctHPVDNA appearing to accurately stratify patients at lowest and highest risk for RD (8/8 patients without risk factors had <1 copy/mL, and 3/3 patients with macroscopic RD risk factors had >100 copies/mL), POD 1 ctHPVDNA also demonstrated an ability to stratify patients according to intermediate risk factors. For example, presence and extent of ENE and lymph node burden, which are both known prognostic risk factors, were associated with elevated POD 1 ctHPVDNA. However, these data must be interpreted with caution due to the low number of patients in each prognostic risk group and the presence of outlier samples, which can have disproportionate impacts on statistical evaluations when sample sizes are small.
When we examined emerging trends in risk stratification in HPV+OPSCC, ctHPVDNA accurately mirrored these as well. For example, 0, 1, and 2 lymph nodes involved have all been suggested to be cutoffs for observation after surgery, with 0 lymph nodes being the most conservative/traditional approach and 2 lymph nodes reflecting an emerging understand of the nonaggressive biology of most HPV+OPSCCs. Interestingly, all patients with 0, 1, or 2 lymph nodes involved, who lacked additional risk factors, had POD 1 levels <1 copy/mL (Supporting Fig. 5). Median ctHPVDNA values on POD 1 in ≤2 versus >2 nodes were 0.2 and 4.2 copies/mL; this suggests 2 positive lymph nodes may be an appropriate cutoff for observation after surgery. Pathology risk factors of unknown significance in HPV+OPSCC, such as PNI and LVI, were not associated with POD 1 ctHPVDNA, and we found no difference between close margins and negative margins. This coincides with emerging literature to suggest wide margins are not necessary to achieve disease control in HPV+OPSCC, unlike traditional head and neck squamous cell carcinomas where >3- to 5-mm surgical margins are established to be necessary for local disease control.31
We also found that POD 1 ctHPVDNA levels were markedly different between patients who went on to receive observation, radiation, and CRT, suggesting that ctHPVDNA may accurately perform as a composite biomarker, stratifying patients into risk groups for adjuvant treatment. Currently used clinicopathologic risk factors for choosing adjuvant treatment only approximate risk and are nonspecific on a patient-by-patient basis, leading to overtreatment (adjuvant treatment received when not needed) in many patients. Here, 12 patients (36% of the cohort) with POD 1 ctHPVDNA <1 copy/mL received radiation or CRT. We posit that these patients represent the cohort of patients who could be safely de-escalated (ie, those patients who are overtreated based on clinicopathologic criteria). Although it is not possible to test this hypothesis in this observational cohort, future prospective trials are needed to assess whether POD 1 ctHPVDNA could be used alone or as an adjunct to pathologic risk stratification in HPV+OPSCC.
Although this observational study is the largest cohort of surgical patients available to date, it has a number of limitations, including that it is underpowered to assess disease recurrence because of the infrequent nature of this outcome. Significantly longer follow-up of these patients will be required to identify additional recurrences and assess the ability of ctHPVDNA to both predict these recurrences and identify them earlier than standard monitoring. It is encouraging, nonetheless, that the 2 biopsy confirmed cases of recurrence/persistence that occurred in this study were identified by ctHPVDNA months before clinical detection. Because there is no gold standard for detection of RD immediately after surgery, the performance and utility of ctHPVDNA must be inferred from clinicopathologic risk factors. Such inference presents challenges due to the number of risk factors, the fact that many patients possess multiple risk factors, and risk factors have gradations within them. Analyses presented here, such as the association between number of lymph nodes and ctHPVDNA, are univariate and do not account for the affect of additional risk factors the patient may possess. Furthermore, because the number of patients in any given risk category is low, outlier samples can drastically impact statistical findings. Thus, validation of the findings from this cohort and the cutoff values used for analyses are necessary in a second, larger independent cohort.
In summary, POD 1 ctHPVDNA levels were found to be associated with the risk of residual disease in patients with HPV+OPSCC undergoing curative intent surgery and thus could be used as a personalized biomarker for selecting adjuvant treatment in the future. ctHPVDNA should be evaluated as a tool to guide real-time treatment decision-making after surgery in future prospective studies to validate the findings of this observational study.
Supplementary Material
FUNDING SUPPORT
Funding for this work came from Sundry funds (Daniel L. Faden). Daniel L. Faden receives salary support from the National Institute of Dental and Craniofacial Research of the National Institutes of Health. Ryan B. Corcoran receives salary support from National Cancer Institute of the National Institutes of Health.
Giulia Siravegna is a consultant/advisory board member for Eli Lilly. Ryan B. Corcoran has received consulting or speaking fees from AbbVie, Amgen, Array Biopharma/Pfizer, Asana Biosciences, Astex Pharmaceuticals, AstraZeneca, Avidity Biosciences, BMS, C4 Therapeutics, Chugai, Elicio, Erasca, Fog Pharma, Genentech, Guardant Health, Ipsen, Kinnate Biopharma, LOXO, Merrimack, Mirati Therapeutics, Natera, Navire, N-of-one/Qiagen, Novartis, nRichDx, Remix Therapeutics, Revolution Medicines, Roche, Roivant, Shionogi, Shire, Spectrum Pharmaceuticals, Symphogen, Tango Therapeutics, Taiho, Warp Drive Bio, and Zikani Therapeutics; holds equity in Avidity Biosciences, C4 Therapeutics, Erasca, Kinnate Biopharma, nRichDx, Remix Therapeutics, and Revolution Medicines; and has received research funding from Asana, AstraZeneca, Lilly, Novartis, and Sanofi. A. John Iafrate holds equity in Invitae. Daniel L. Faden has received research funding from Bristol-Myers Squibb and Foundation Medicine, holds equity in Illumina, and receives consulting fees from Merck, Noetic, and Focus on Boston.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The other authors made no disclosures.
Additional supporting information may be found in the online version of this article.
REFERENCES
- 1.Number of HPV-associated cancer cases per year. Centers for Disease Control and Prevention. Updated August 2, 2019. Accessed May 19–20, 2020. https://www.cdc.gov/cancer/hpv/statistics/cases.htm
- 2.Tota JE, Best AF, Zumsteg ZS, Gillison ML, Rosenberg PS, Chaturvedi AK. Evolution of the oropharynx cancer epidemic in the United States: moderation of increasing incidence in younger individuals and shift in the burden to older individuals. J Clin Oncol. 2019;37:1538–1546. doi: 10.1200/jco.19.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011 [DOI] [PubMed] [Google Scholar]
- 4.D’Souza G, Cullen K, Bowie J, Thorpe R, Fakhry C. Differences in oral sexual behaviors by gender, age, and race explain observed differences in prevalence of oral human papillomavirus infection. PLoS One. 2014;9:e86023. doi: 10.1371/journal.pone.0086023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3235–3242. doi: 10.1200/jco.2015.61.6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudell JJ, Schaner PE, Desmond RA, Meredith RF, Spencer SA, Bonner JA. Dosimetric factors associated with long-term dysphagia after definitive radiotherapy for squamous cell carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 2010;76:403–409. doi: 10.1016/j.ijrobp.2009.02.017 [DOI] [PubMed] [Google Scholar]
- 7.Kelly JR, Husain ZA, Burtness B. Treatment de-intensification strategies for head and neck cancer. Eur J Cancer. 2016;68:125–133. doi: 10.1016/j.ejca.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsen ML, Mady LJ, Hodges J, Wasserman-Wincko T, Johnson JT. Burden of treatment: reported outcomes in a head and neck cancer survivorship clinic. Laryngoscope. 2019;129:E437–E444. doi: 10.1002/lary.27801 [DOI] [PubMed] [Google Scholar]
- 9.Wirth LJ, Burtness B, Nathan C-AO, Grégoire V, Richmon J. Point/counterpoint: do we de-escalate treatment of HPV-associated oropharynx cancer now? And how? Am Soc Clin Oncol. 2019;39:364–372. doi: 10.1200/edbk_238315 [DOI] [PubMed] [Google Scholar]
- 10.Ferris RL, Flamand Y, Weinstein GS, et al. Phase II randomized trial of transoral surgery and low-dose intensity modulated radiation therapy in resectable p16+ locally advanced oropharynx cancer: an ECOG-ACRIN Cancer Research Group trial (E3311). J Clin Oncol. 2022;40:138–149. doi: 10.1200/jco.21.01752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cracchiolo JR, Baxi SS, Morris LG, et al. Increase in primary surgical treatment of T1 and T2 oropharyngeal squamous cell carcinoma and rates of adverse pathologic features: National Cancer Data Base. Cancer. 2016;122:1523–1532. doi: 10.1002/cncr.29938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan KY, Puram SV, Li MM, et al. National treatment trends in human papillomavirus-positive oropharyngeal squamous cell carcinoma. Cancer. 2020;126:1295–1305. doi: 10.1002/cncr.32654 [DOI] [PubMed] [Google Scholar]
- 13.Head and Neck Cancers (version 3.2021) National Comprehensive Cancer Network (NCCN). Accessed July 7, 2021, https://www.nccn.org/login?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf
- 14.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 15.Dahlstrom KR, Li G, Hussey CS, et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer. 2015;121:3455–3464. doi: 10.1002/cncr.29772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao H, Banh A, Kwok S, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys. 2012;82:e351–e358. doi: 10.1016/j.ijrobp.2011.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn SM, Chan JY, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140:846–854. doi: 10.1001/jamaoto.2014.1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chera BS, Kumar S, Beaty BT, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2019;25:4682–4690. doi: 10.1158/1078-0432.Ccr-19-0211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chera BS, Kumar S, Shen C, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol. 2020;38:1050–1058. doi: 10.1200/jco.19.02444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanna GJ, Lau CJ, Mahmood U, et al. Salivary HPV DNA informs locoregional disease status in advanced HPV-associated oropharyngeal cancer. Oral Oncol. 2019;95:120–126. doi: 10.1016/j.oraloncology.2019.06.019 [DOI] [PubMed] [Google Scholar]
- 23.Hanna GJ, Sridharan V, Margalit DN, et al. Salivary and serum HPV antibody levels before and after definitive treatment in patients with oropharyngeal squamous cell carcinoma. Cancer Biomark. 2017;19:129–136. doi: 10.3233/cbm-160071 [DOI] [PubMed] [Google Scholar]
- 24.Damerla RR, Lee NY, You D, et al. Detection of early human papillomavirus-associated cancers by liquid biopsy. JCO Precis Oncol. 2019;3:PO.18.00276. doi: 10.1200/po.18.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riaz N, Sherman E, Pei X, et al. Precision radiotherapy: reduction in radiation for oropharyngeal cancer in the 30 ROC trial. J Natl Cancer Inst. 2021;113:742–751. doi: 10.1093/jnci/djaa184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siravegna G, O’Boyle CJ, Varmeh S, et al. Cell-free HPV DNA provides an accurate and rapid diagnosis of HPV-associated head and neck cancer. Clin Cancer Res. Published online: December 2, 2021. doi: 10.1158/1078-0432.Ccr-21-3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lydiatt WM, Patel SG, O’Sullivan B, et al. Head and neck cancers—major changes in the American Joint Committee on Cancer eighth edition Cancer Staging Manual. CA Cancer J Clin. 2017;67:122–137. doi: 10.3322/caac.21389 [DOI] [PubMed] [Google Scholar]
- 28.To EW, Chan KC, Leung SF, et al. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res. 2003;9:3254–3259. [PubMed] [Google Scholar]
- 29.Lopez EM, Tanner AM, Du E, et al. Decline in circulating viral and human tumor markers after resection of head and neck carcinoma. Head Neck. 2021;43:27–34. doi: 10.1002/hed.26444 [DOI] [PubMed] [Google Scholar]
- 30.McMullen CP, Garneau J, Weimar E, et al. Occult nodal disease and occult extranodal extension in patients with oropharyngeal squamous cell carcinoma undergoing primary transoral robotic surgery with neck dissection. JAMA Otolaryngol Head Neck Surg. 2019;145:701–707. doi: 10.1001/jamaoto.2019.1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holcomb AJ, Herberg M, Strohl M, et al. Impact of surgical margins on local control in patients undergoing single-modality transoral robotic surgery for HPV-related oropharyngeal squamous cell carcinoma. Head Neck. 2021;43:2434–2444. doi: 10.1002/hed.26708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.