Abstract
Bacterial cell wall biosynthesis is the target of many important antibiotics. Its spatiotemporal organization is closely coordinated with cell division. However, the role of peptidoglycan synthesis within cell division is not fully understood. Even less is known about the impact of antibiotics on the coordination of these two essential processes. Visualizing the essential cell division protein FtsZ and other key proteins in Staphylococcus aureus, we show that antibiotics targeting peptidoglycan synthesis arrest cell division within minutes of treatment. The glycopeptides vancomycin and telavancin completely inhibit septum constriction in all phases of cell division. The beta-lactam oxacillin stops division progress by preventing recruitment of the major peptidoglycan synthase PBP2 to the septum, revealing PBP2 as crucial for septum closure. Our work identifies cell division as key cellular target of these antibiotics and provides evidence that peptidoglycan synthesis is the essential driving force of septum constriction throughout cell division of S. aureus.
Peptidoglycan synthesis inhibiting antibiotics critically impair cell division progress within minutes of treatment.
INTRODUCTION
The increasing threat of antibiotic resistance requires innovation in the research and development process. However, our understanding of how even clinically successful antibiotics kill bacteria is still limited. A deeper insight into cellular effects preceding and contributing to cell death can certainly help improve anti-infective strategies. Peptidoglycan synthesis (PGS) is the central pathway of constructing the bacterial cell wall. The principle of this process is similar in all bacteria that have a cell wall; however, differences exist in the set of proteins comprising the cell wall biosynthesis machinery (CWBM) and in its spatiotemporal organization (1, 2). The clinically highly relevant Gram-positive pathogen Staphylococcus aureus is a well-established model organism for investigating PGS due to a comparably small set of CWBM proteins and the lack of an elongasome machinery or comparable peripheral PGS activity, resulting in spatially focused PGS at the cell division septum (1). PGS in S. aureus begins with a sequence of intracellular biochemical steps leading to the formation of the ultimate precursor molecule lipid II. The lipid II building block consists of a disaccharide-pentapeptide (GlcNAc-MurNAc-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala) bound to an undecaprenyl-pyrophosphate (C55PP) carrier lipid on the inner leaflet of the cytoplasmic membrane. Lipid II translocation to the outer leaflet of the cytoplasmic membrane is performed by specific flippases, most notably MurJ (3, 4). Transglycosylase (TGase) enzymes polymerize the disaccharide units to peptidoglycan strands and release the carrier lipid. Transpeptidases (TPases) cross-link pentapeptide side chains of neighboring strands to form a dense molecular cell wall mesh. This reaction is performed by the penicillin-binding proteins (PBPs) PBP1 to PBP4, of which PBP1 and PBP2 are essential in S. aureus (1, 5, 6). PBP2 is the only bifunctional PBP catalyzing both TGase and TPase reactions and is accordingly described as the main peptidoglycan synthase of S. aureus (7).
Components of the CWBM are key targets for a number of antibiotics, including some of the clinically most relevant classes, such as beta-lactams and glycopeptides (8). Beta-lactam antibiotics directly inhibit PBPs by inhibiting enzyme function through irreversibly binding to the TPase active site (9). In contrast, glycopeptide antibiotics bind to the d-Ala-d-Ala terminus present in lipid II and nascent peptidoglycan, thereby hindering TGase and TPase substrate access (10, 11). Despite their different molecular targets, the cellular effects induced by glycopeptides and beta-lactams are frequently compared as they both interfere with the final stage of cell wall biosynthesis. Consequently, cells overall share common fates resulting from destabilization of the cell wall after treatment with either class of antibiotics (12, 13). Similarly, treatment with these agents also leads to delocalization of PBP2, demonstrating a massive impact on the spatiotemporal organization of the CWBM in S. aureus (14).
Bacterial cell wall biosynthesis is closely coordinated with cell division (1, 7, 15–21). In multiple species including S. aureus, the cell division septum is the main region of PGS (1). The key protein of bacterial cell division is the cytoplasmic FtsZ (filamenting temperature-sensitive mutant z), which polymerizes into filaments. These filaments condense into a Z-ring at mid-cell through guanosine triphosphatase (GTPase) activity of individual FtsZ molecules. The dynamic front assembly and end disassembly of these filaments lead to a circular motion of FtsZ filaments along the Z-ring circumference, which is referred to as treadmilling (20, 21). This process enables Z-ring condensation, which subsequently provides a scaffold for septal cell wall biosynthesis (7, 21–25). In S. aureus, FtsZ treadmilling has been shown to be essential for early septum constriction. However, it was shown to be redundant after full assembly of the CWBM at the septum (7). This clearly shows how strongly the processes of cell division and PGS are interconnected. Corroboratively, several CWBM components have been shown to be essential for proper cell division (5, 26–28). However, the precise role of PGS throughout cell division remains indistinctly defined.
Even less is known about how CWBM targeting antibiotics interfere with cell division. Here, we describe the impact of the antibiotics vancomycin, telavancin, and oxacillin, which differentially target PGS, on the spatiotemporal organization of cell division of S. aureus in unprecedented detail. We show that these antibiotics have drastic effects on bacterial cell division within minutes of treatment through their inhibitory activity toward PGS. We provide evidence that FtsZ treadmilling cannot drive septum constriction without a functioning PGS, showing that PGS is essential for septum constriction throughout cell division of S. aureus. In this regard, our results highlight the special role of the main peptidoglycan synthase protein PBP2 as a driver of septum closure. We show that the first cellular effects of glycopeptides and beta-lactams are directed toward cell division and more specific and intricate than previously anticipated. Overall, our work contributes to a deeper understanding of the cellular effects of antibiotics targeting cell wall biosynthesis beyond their primary targets.
RESULTS
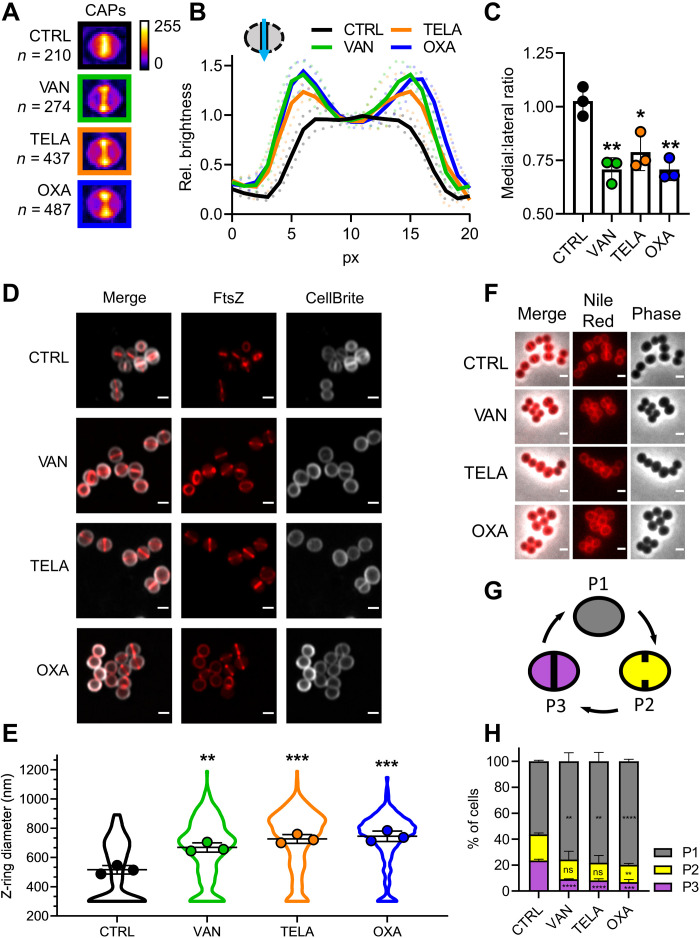
PGS targeting antibiotics drastically increase FtsZ-ring diameter
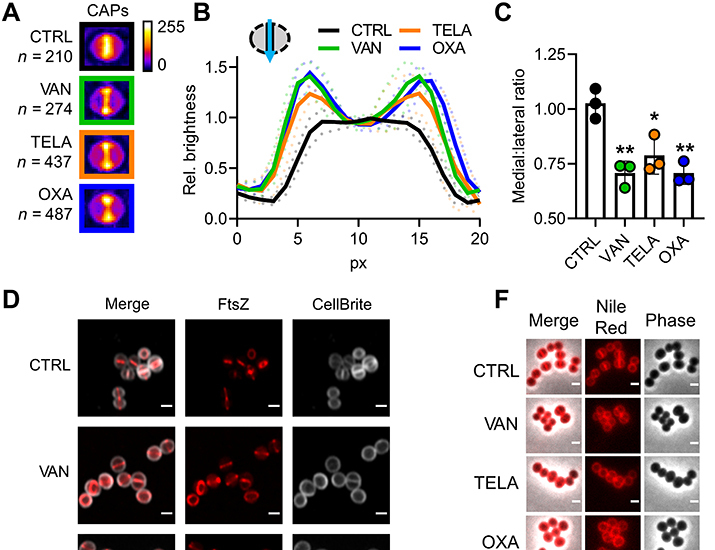
Currently, it is unclear how inhibition of PGS by antibiotics affects bacterial cell division despite the close interconnection of these two processes. To gain insight into the immediate effects of cell wall biosynthesis inhibitors on cell division, we treated S. aureus with the glycopeptide vancomycin, its semisynthetic derivative telavancin, and the beta-lactam oxacillin. To visualize the impact of antibiotic treatment on FtsZ localization, we developed convolved average projections (CAPs) as an analysis tool. CAPs are generated by condensing the fluorescence micrographs of all individual cells of an experiment after rotation into a vertical orientation into one single image. This image serves as a comprehensive representation of the entire sample population (Supplementary Materials, fig. S1). The CAP of an untreated control showed a uniform brightness distribution along the septum, which is in line with the expected even distribution of cells in different stages of septum closure (Fig. 1, A to C). Treatment with either vancomycin, telavancin, or oxacillin resulted in a pronounced shift of the intensity distribution toward the lateral edges of the CAP septum (Fig. 1, A to C), indicating that more cells with large Z-rings were found in these samples. As a control, we quantified how many cells maintained Z-ring structure and membrane integrity during treatment. We found no significant differences between control and treated samples, showing that the antibiotics did not affect Z-ring integrity (Supplementary Materials, fig. S2).
Fig. 1. Cell wall targeting antibiotics interfere with cell division.
(A) Representative CAPs of S. aureus RN4220 pCQ11-FtsZ-SNAP cells treated with vancomycin, telavancin, or oxacillin and an untreated control. SNAP-tags were labeled with SNAP-Cell TMR Star. n corresponds to the number of individual cells used for CAP generation. (B) Quantification of CAP brightness along the septal axis, plotted relative to the medial brightness value. Mean of three replicates ± SD. (C) Medial:lateral ratio of CAP brightness profiles. Mean of three replicates ± SD. (D) AiryScan superresolution micrographs of S. aureus RN4220 pCQ11-FtsZ-SNAP treated with vancomycin, telavancin, or oxacillin and an untreated control. SNAP-tag was labeled with SNAP-Cell 505-Star. Cell membrane was stained with CellBrite Fix 640. Scale bars, 1 μm. (E) Z-ring diameter distribution after antibiotic treatment. Violin plots show individual cell data, and points show replicate means. (F) Representative micrographs of S. aureus RN4220 cells after antibiotic treatment, stained with the membrane dye Nile Red. Scale bars, 1 μm. (G) Scheme of S. aureus cell cycle membrane morphologies. (H) Cell cycle phase distribution in S. aureus RN4220 after antibiotic treatment. Mean of three replicates ± SD. Gray: P1 cells; yellow: P2 cells; violet: P3 cells. The number of individual cells per replicate are shown in table S2 for all datasets. Unpaired two-tailed Student’s t tests (95% confidence interval) against the untreated control. ns, not significant, *P = 0.05 to 0.01; **P = 0.01 to 0.001; ***P = 0.001 to 0.0001; ****P ≤ 0.0001.
Given the changes in FtsZ localization as observed by CAP analyses, we quantified the impact of the CWBM-targeting antibiotics on Z-ring diameter on the individual cell level using confocal Airyscan superresolution microscopy (Fig. 1, D and E). We used the improved resolution for precise measurements of individual Z-ring diameters with a comfortable lower threshold of 300 nm. The precise evaluation of antibiotic impact on Z-ring size confirmed the indications of the CAP analysis: Mean Z-ring diameters were drastically increased by 30% (vancomycin), 41% (telavancin), and 44% (oxacillin) compared to the untreated control with a notable shift in the distribution toward larger diameters (Fig. 1, D and E). Notably, we found diameters well above 1100 nm in treated cells, while in the untreated control, the FtsZ ring diameter did not exceed 893 nm, corroborating reports of cell swelling during treatment with either glycopeptides or beta-lactams (12, 28).
Together, the severe effects of CWBM-targeting antibiotics on Z-ring sizes indicated a considerable disruption of the cell division process. As a complementary type of analysis, we analyzed cell cycle phase distribution using the membrane stain Nile Red (Fig. 1, F to H), an established approach to study staphylococcal cell cycle dynamics (29). In line with earlier reports (29), we found approximately 55% of the cells without membrane invagination (P1), 20% of the cells with initiated septum constriction (P2), and the remaining 25% of the cells in the late stage of cell division (P3). Treatment with either antibiotic had considerable impact on these dynamics as the proportion of cells in P3 was significantly reduced to approximately 8%, while the proportion of cells in P1 increased significantly (Fig. 1H).
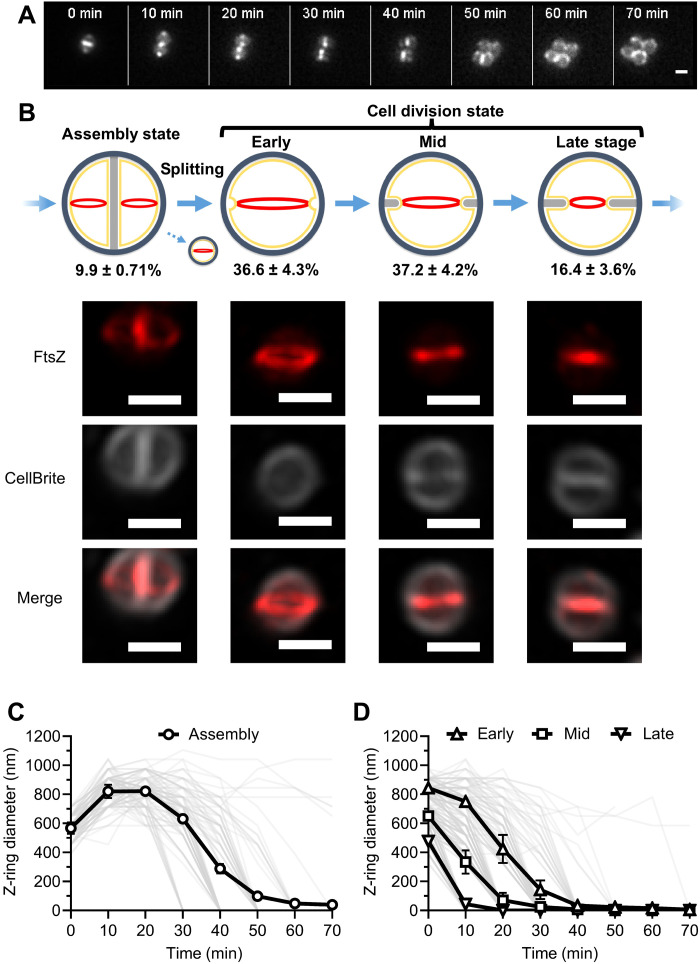
Visualization of S. aureus Z-ring dynamics throughout the cell cycle
The Z-ring size is a measure of septum progression (7, 21, 24, 25). The increased size after antibiotic treatment suggested massive impairment of cell division progress caused by the CWBM-targeting antibiotics. This was further supported by the changes in cell cycle phase distribution. To elucidate speed and extent of antibiotic impact, we visualized Z-ring behavior over time using live-cell microscopy with highly inclined and laminated optical sheet (HiLo) illumination and analyzed Z-ring size dynamics at the individual cell level. Therefore, we first visualized dynamics of untreated cells throughout the cell cycle (Fig. 2, A and B). As Z-rings were frequently assembled before division of the parent cell, we evaluated Z-ring sizes starting from this assembly state (Fig. 2C). Within the first minutes, Z-ring diameters increased because of cell splitting and the resulting rapid transformation of the new cells into a spherical shape. Both observations are in line with earlier reports (29, 30). Subsequent cell division began with a short phase of stagnation, followed by a fast constriction resulting in septum closure and cell separation. Because the cultures were not synchronized, we observed cells in different stages of the cell cycle at every image acquisition. We therefore categorized the cells depending on their Z-ring diameter at t = 0 min according to their progression in the cell cycle into early (large Z-rings), mid (medium Z-rings), and late (small Z-rings) stages of cell division (Fig. 2B). All three categories—early, mid and late division stage cells—showed a rapid septum constriction and subsequent cell division (Fig. 2D).
Fig. 2. Live-cell dynamics of Z-ring size over time in S. aureus.
(A) Representative HiLo microscopy images of Z-ring dynamics over time. Scale bar, 1 μm. (B) Pictogram illustrating the cell cycle states and cell division stages in S. aureus with corresponding Z-ring phenotypes and representative Airyscan superresolution micrographs of S. aureus RN4220 pCQ11-FtsZ-SNAP. SNAP-tag was labeled with SNAP-Cell 505-Star. Cell membrane was stained with CellBrite Fix 640. Percentages show the relative abundance of each phenotype at t = 0 min as the mean of three independent biological experiments ± SD. Coloring of the pictogram: red: Z-ring; light gray: new cell wall; dark gray: old cell wall; yellow: cell membrane. Micrographs are z-stack maximum projections. Scale bars, 1 μm. (C) Quantification of Z-ring dynamics over the course of one complete cell cycle of S. aureus. Per replicate, 30 cells in the assembly state were chosen at t = 0 min, and Z-ring size was quantified over 70 min. Individual cell data are plotted as thin semitransparent lines. Black thick lines with open circles show the mean of three independent biological experiments ± SD. (D) Quantification of Z-ring dynamics over time after classification of cells into cell division stages according to Z-ring size at t = 0 min. Individual cell data are plotted as thin semitransparent lines. Black thick lines with open symbols show the mean of three independent biological experiments ± SD. The number of individual cells per replicate are shown in table S2 for all datasets.
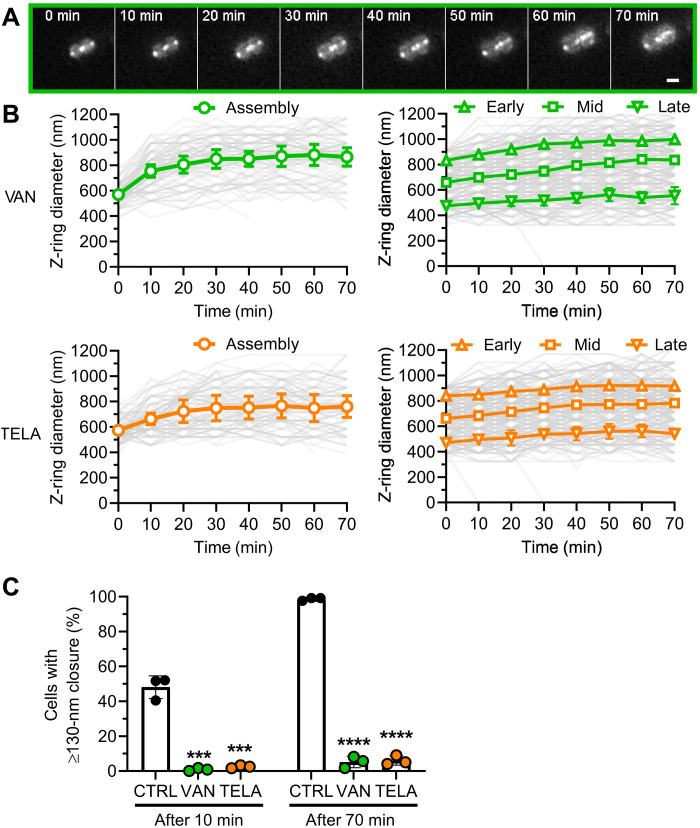
Lipid II binding glycopeptides inhibit septum constriction within minutes of treatment
Next, we visualized the live-cell dynamics of Z-rings during treatment with vancomycin and telavancin. Treatment with both glycopeptides drastically affected Z-ring dynamics. Vancomycin and telavancin nearly completely abolished septum constriction within minutes of treatment (Fig. 3, A and B). Both glycopeptides reduced the percentage of cells with active septum progression to less than 3% within the first 10 min of treatment (Fig. 3C). The effect was independent of the Z-ring size at the time point of the antibiotics application and resulted in a complete arrest of nearly all septum progression. Consequently, only 6% of cells treated with either glycopeptide showed septum progression within 70 min of treatment (Fig. 3C). In contrast, more than 98% of untreated cells showed septum progression in this time. Treated cells in the assembly state at the time of exposure still showed parent cell splitting, revealing that divisome components responsible for daughter cell separation remained functional. This is in line with the reduced proportion of cells in P3 in the cell cycle dynamics analysis. Formed daughter cells, however, were immediately arrested in their cell cycle as they were unable to perform septum constriction (Fig. 3B). Merely 3 of the 735 analyzed cells were able to divide after addition of one of the glycopeptides, while 98% of untreated cells proceeded to cell division. These data demonstrate the fast and massive impact of glycopeptide action on cell division.
Fig. 3. Glycopeptides vancomycin and telavancin lead to immediate arrest of septum progression.
(A) Representative HiLo microscopy images of Z-ring dynamics over time during vancomycin treatment. Scale bar, 1 μm. (B) Quantification of Z-ring dynamics over time after classification of cell cycle states and cell division stages according to Z-ring size at t = 0 min. Green: cells treated with vancomycin; orange: cells treated with telavancin. Individual cell data are plotted as thin semitransparent lines. Black thick lines with open symbols show the mean of three independent biological experiments ± SD. Open symbols correspond to the time points when the cells were imaged. (C) Quantification of active septum progression. Points correspond to values of individual replicates, and bar shows the mean of three independent biological experiments ± SD. The number of individual cells per replicate are shown in table S2 for all datasets. Statistical significance was determined against the untreated control with unpaired two-tailed Student’s t tests with a 95% confidence interval. Statistical significance was denoted ***P = 0.001 to 0.0001; ****P ≤ 0.0001.
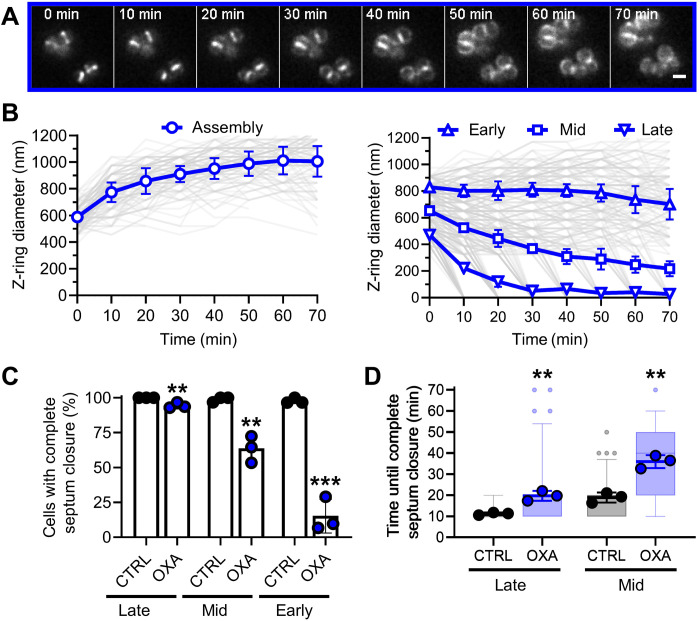
Oxacillin interferes with septum constriction in a cell division stage–dependent manner
We proceeded to visualize the effect of oxacillin on Z-ring dynamics. Similar to the glycopeptides, the beta-lactam had substantial impact on septum progression. The effect was more intricate, as it depended on the stage of cell division. Mid- and late-stage subpopulations were still able to perform septum closure. In contrast, constriction was almost completely inhibited in early-stage cells with large Z-rings (Fig. 4, A and B). The percentage of cells performing septum closure was 95%, 63%, and 15% for late-stage, mid-stage, and early-stage cells, respectively (Fig. 4C). Cells in the assembly state still showed parent cell splitting but subsequently stopped in the early stage of cell division (Fig. 4B) as observed during glycopeptide treatment. We observed a significant increase in the time needed to perform septum closure in the cells that were still able to divide (Fig. 4D). For the category of early-stage cells, the sample size was too small for a reliable analysis given the nearly complete halt of septum progression. Late-stage and mid-stage cells took significantly longer to close the septum by reducing the septum closure velocity (Fig. 4, A and D).
Fig. 4. Oxacillin differentially affects septum constriction depending on the cell division stage.
(A) Representative HiLo microscopy images of Z-ring dynamics over time during oxacillin treatment. Scale bar, 1 μm. (B) Quantification of Z-ring dynamics over time after classification of cells into cycle states and cell division stages according to Z-ring size at t = 0 min. Individual cell data are plotted as thin semitransparent lines. Black thick lines with open symbols show the mean of three independent biological experiments ± SD. Open symbols correspond to the time points, when cells were imaged, and z-ring sizes were analyzed. (C) Quantification of septum closure activity as % of cells with complete septum closure within 70 min of treatment. Points correspond to values of individual replicates, and bars show the mean of three independent biological experiments ± SD. (D) Quantification of time needed for complete septum closure. Semitransparent boxplots show the distribution of individual cell data with 9% to 95% confidence interval. Semitransparent small points represent outlier of individual cell data. Large points correspond to values of individual replicates, and thick lines show the mean of three independent biological experiments ± SD. The number of individual cells per replicate are shown in table S2 for all datasets. Statistical significance was determined against the untreated control with unpaired two-tailed Student’s t tests with a 95% confidence interval. Statistical significance was denoted as **P = 0.01 to 0.001; ***P = 0.001 to 0.0001.
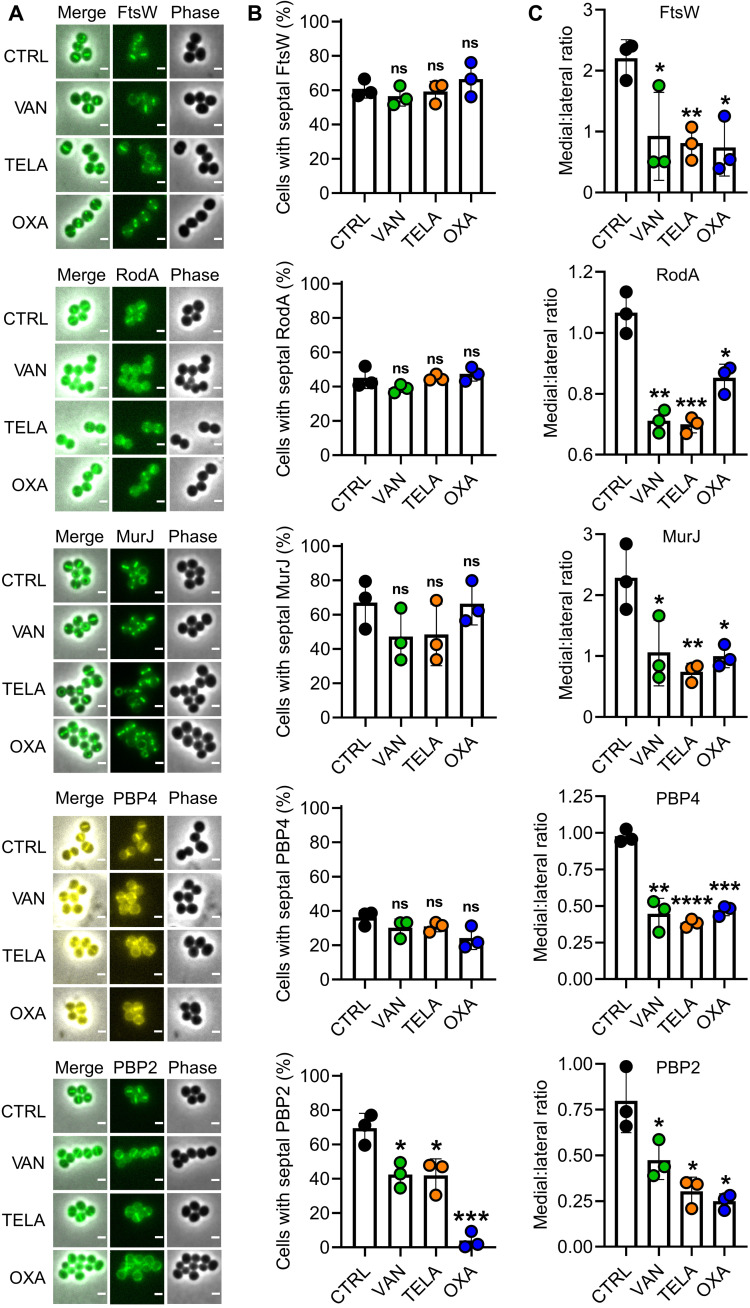
Subcellular localization of CWBM components is unaffected with the exception of PBP2 during oxacillin treatment
We analyzed the impact of the antibiotics on the subcellular localization of important proteins of the CWBM that are known to localize to the septum (7). The members of the shape, elongation, division, and sporulation (SEDS) protein family FtsW and RodA that localize in the early phase of cell division to the septum and the lipid II flippase MurJ, the recruitment of which represents the turning point of cell division, were investigated. The penicillin binding proteins 2 and 4 that localize through different mechanisms to the septum in the late stage of cell division were likewise studied. Despite the differences in structure, function, and temporal localization, a general pattern was observed for all proteins with the notable exception of PBP2 (Fig. 5). FtsW, RodA, MurJ, and PBP4 were still septally localized after antibiotic treatment to the same degree as in the untreated control (Fig. 5B), and CAP analyses reproduced the pattern observed for FtsZ (see Fig. 1, A to C), revealing no effect of the antibiotic treatment on the subcellular localization of these CWBM components (Fig. 5C). In contrast, the impact of antibiotics on PBP2 was more intricate. Vancomycin and telavancin had only moderate effects on septal localization of PBP2. In contrast, the impact of oxacillin was more drastic, as almost no cell showed a septal PBP2 signal (Fig. 5B).
Fig. 5. Impact of antibiotic treatment on CWBM components localizing to the septum.
(A) Representative micrographs of S. aureus RN4220 FtsW-gfp, S. aureus RN4220 RodA-gfp, S. aureus RN4220 pCQ11-MurJ-mNeongreen, S. aureus RN4220 pCQ11-PBP4-YFP, and S. aureus RN4220 RNpPBP2-31 after 25 min of treatment with vancomycin, telavancin, or oxacillin and an untreated control. (B) Relative abundance of cells with septal FtsW, RodA, MurJ, PBP4, and PBP2 localization as % of total population after 25 min of treatment with vancomycin, telavancin, or oxacillin and an untreated control. Boxes and thin lines report the mean of three independent biological experiments ± SD, respectively. Points indicate the values of the individual replicates. (C) Medial:lateral ratios of CAP brightness profiles of FtsW, RodA, MurJ, PBP4, and PBP2 localization after 25 min of treatment with vancomycin, telavancin, or oxacillin and an untreated control. Ratios were determined as the ratio of medial maximum to the mean of both lateral maxima. CAPs and brightness profiles are shown in fig. S3. Boxes and thin lines report the mean of three independent biological experiments ± SD, respectively. Points indicate the values of the individual replicates. The number of individual cells per replicate are shown in table S2 for all datasets. Statistical significance was determined against the untreated control with unpaired two-tailed Student’s t tests with a 95% confidence interval. Statistical significance was denoted as ns, P > 0.05; *P = 0.05 to 0.01; **P = 0.01 to 0.001; ***P = 0.001 to 0.0001; ****P ≤ 0.0001.
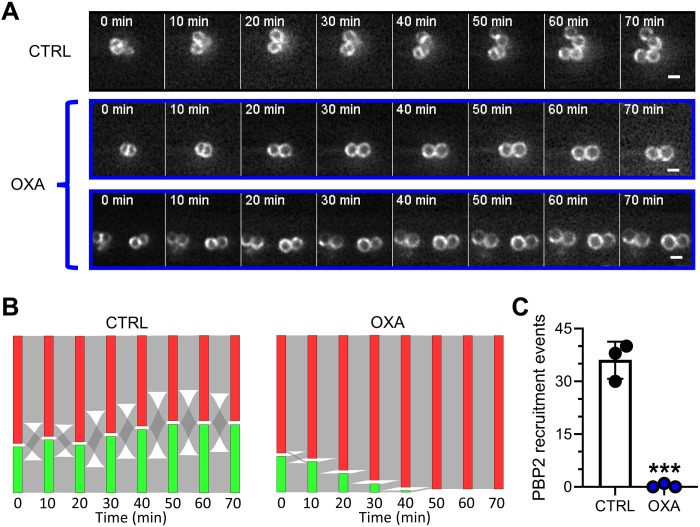
Oxacillin inhibits recruitment of PBP2 to the septum
This striking difference prompted us to investigate the dynamics of PBP2 in our live-cell setup. Untreated cells showed a “biphasic” behavior of oscillation between septal and nonseptal PBP2 localization (Fig. 6, A and B, and fig. S4). Cells with PBP2 at the septum were able to divide within 22 min on average, leaving the daughter cells with a nonseptal PBP2 localization. After subsequent progression through their cell cycle, PBP2 was recruited to the new division septum, and cells divided again. This resulted in an oscillation between the two states of septal and nonseptal PBP2 localization over time (Fig. 6B). Again, the cells were not synchronized in their cell cycle. Therefore, we observed respective subpopulations of both states at every time point.
Fig. 6. Oxacillin specifically prevents PBP2 recruitment to the septum.
(A) Representative HiLo microscopy images of PBP2 dynamics over time during oxacillin treatment and an untreated control. Scale bars, 1 μm. (B) Fluxes of cells with specific PBP2 localization as a function of time in an untreated control (left) and during oxacillin treatment (right). Red: cells without septal PBP2 localization; green: cells with septal PBP2 localization. Red-to-green fluxes indicate cells that recruit PBP2 to the septum. Green-to-red fluxes indicate cells that divide and thereby lose septal PBP2 localization in the daughter cells. Bar height represents relative abundance of cells with the respective PBP2 localization. Sum of both bars is always 100%. Mean of three independent biological experiments is shown (for individual replicate data, see fig. S4). For each replicate and condition, n = 30. (C) Number of events of PBP2 recruitment to the septum, quantified from data shown in (C). Points correspond to values of individual replicates, and bars show the mean of three independent biological experiments ± SD. The number of individual cells per replicate are shown in table S2 for all datasets. Statistical significance was determined against the untreated control with unpaired two-tailed Student’s t tests with a 95% confidence interval. Statistical significance was denoted as ***P = 0.001 to 0.0001.
During treatment with oxacillin, this oscillatory dynamic was massively disturbed within a few minutes (Fig. 6, B to C, and fig. S4). While cells with septal PBP2 still divided, cells were unable to recruit PBP2 to the septum from the moment oxacillin was added (Fig. 6C). We observed only a single event of PBP2 recruitment after addition of oxacillin in a total of 90 analyzed cells. Consequently, oxacillin treatment resulted in a constant decrease of cells with septal PBP2 localization. Oxacillin did not delocalize PBP2 already localized at the septum at the moment of exposure. All cells with septal PBP2 localization at t = 0 min were able to perform septum constriction and cell division. These results are in agreement with the effect on FtsZ-ring dynamics (see Fig. 4), where mid- and late-stage cells were still able to divide. Cells with nonseptal PBP2 at t = 0 min correspond to early-stage cells unable to divide.
Live-cell PBP2 dynamics during glycopeptide treatment explained the moderate effect on septal localization we observed earlier (see Fig. 5). Both vancomycin and telavancin had a moderate effect on PBP2 localization as a downstream effect of the cell cycle arrest and the cell splitting of the cells in the assembly state at t = 0 (Fig. 3B and fig. S4).
DISCUSSION
Bacterial cell wall biosynthesis is closely coordinated with cell division, which, in the case of S. aureus, is initiated by the assembly of the Z-ring structure composed of treadmilling filaments of the tubulin-like FtsZ protein in the center of the coccoid cell. The dynamic Z-ring then serves as a scaffold for stepwise recruitment of the divisome composed of proteins required for PGS and cell division. While the effects of beta-lactam and glycopeptide antibiotics on PGS are very well established, substantially less is known about the impact of these antibiotics on cell division. Differential effects of beta-lactam and glycopeptides antibiotics on septum structure have been recently observed using electron microscopy (12). Using these two classes of antibiotics targeting the final steps of PGS, we show here that inhibition of PGS in S. aureus has fast and drastic effects on cell division. The remarkable speed and extent, by which the glycopeptides and oxacillin inhibit septum constriction, reveals cell division interference as an essential part of these antibiotics’ cellular action. The effects presented here precede the well-documented changes in PG architecture, cell lysis, and oxidative stress (12, 13, 31–33).
Neither antibiotic affected cell splitting and Z-ring integrity. Vancomycin and telavancin effectively stopped septum constriction within minutes, trapping cells at the division stage they were in at the time of exposure. However, we could not observe any drastic effect on subcellular localization of key CWBM components, which could explain this impairment by itself. We therefore propose that cell division arrest directly emerges from the glycopeptides molecular mechanism of action, i.e., inhibition of both TGase and TPase reactions in PGS via substrate binding (10, 11). Our results show that FtsZ treadmilling alone cannot drive septum constriction without a functioning PGS machinery. While treadmilling of FtsZ filaments has been discussed extensively as driving force of septum constriction (17, 34–38), several studies have shown that FtsZ GTPase and therefore treadmilling function become nonessential for cell division after assembly of the septal peptidoglycan synthase complex (7, 20, 25, 39) while still contributing to the velocity of septum constriction (25). Concomitantly, a two-phase septum constriction model has been proposed for S. aureus (7). A first phase of constriction driven by FtsZ treadmilling leads to septal recruitment of the lipid II flippase MurJ. Lipid II translocation in turn recruits PBP2 to the septum, which then drives a second constriction phase via its peptidoglycan synthase activity. In line with this, our observation of a complete stop of septum closure by glycopeptide antibiotics provides clear evidence that PGS is the essential driving force of septum constriction. Furthermore, our results expand the abovementioned model as they show that PGS is essential not only in the second constriction phase but also throughout the entire process of cell division. This corroborates recent findings that FtsZ treadmilling is essential in the first phase of cell division for CWBM recruitment but plays a secondary role in septum constriction (25). The essential SEDS-PBP pair FtsW/PBP1 (27) may drive septum constriction in the first phase, which is in line with recent findings that TPase activity of PBP1 is essential for cell division in S. aureus (28).
Septum constriction was also massively disturbed by the beta-lactam antibiotic oxacillin, consolidating the importance of PGS for cell division. In contrast to the glycopeptides, the effects of oxacillin depended on the cell division stage. Treatment led to a complete constriction stop only in cells in an early stage of cell division, while it slowed down the speed of septum constriction in late-stage cells. The more intricate impact of oxacillin toward Z-ring dynamics offers a new perspective on the role of PBP2 for S. aureus cell division. It was previously shown that the TPase function of PBP2 plays an essential role in the protein’s localization (14). This was concluded from the observation that blocking the TPase active site with oxacillin results in loss of septal PBP2 signal (14). However, it was not differentiated whether oxacillin delocalizes PBP2 from the septum or prevents accurate recruitment to the septum (14). Our results from live-cell microscopy of PBP2 localization dynamics under antibiotic treatment unequivocally demonstrated that oxacillin inhibits recruitment of PBP2 while having no qualitative effect on PBP2 already at the septum at the time of exposure. Therefore, the TPase site is crucial for PBP2 recruitment to the septum, but septal localization seems to be secured via a different mechanism, e.g., protein-protein interactions (14, 27, 28, 40).
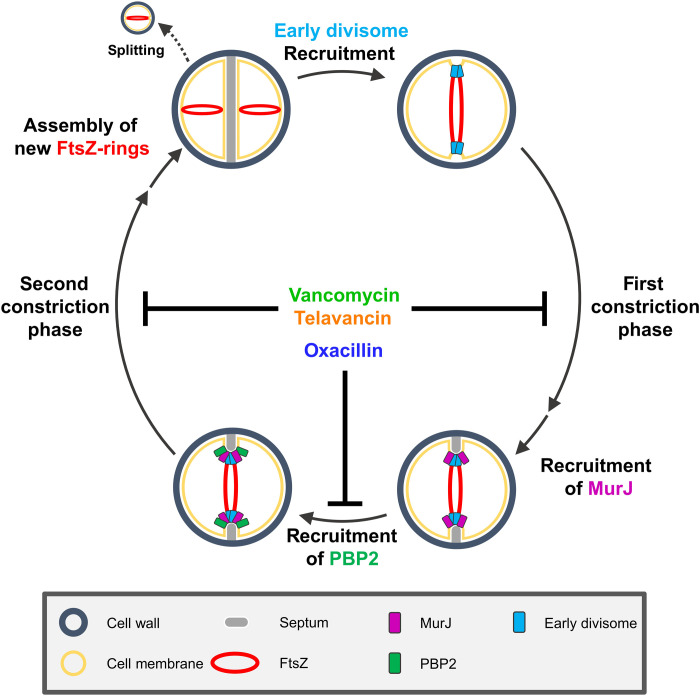
In conclusion, we propose a model of how PGS-targeting antibiotics affect cell division (Fig. 7). Glycopeptides inhibit TGase and TPase reactions of early divisome proteins [e.g., PBP1/FtsW (7, 27)] by substrate blocking, effectively stopping the first constriction phase. In contrast, oxacillin inhibits septal recruitment of PBP2 by TPase active site binding. It thereby prevents cells from transitioning to the second phase, where septum closure is driven by PBP2 activity. Similar to the first phase, glycopeptides completely stop the second constriction phase by substrate blocking, preventing both PGS reactions catalyzed by PBP2. Oxacillin considerably slows down the second constriction phase by TPase inhibition, indicating that TGase and TPase reactions may both contribute to septum constriction. Our model identifies inhibition of cell division as key cellular effect of antibiotics targeting cell wall biosynthesis.
Fig. 7. Proposed model for cell cycle inhibition by the CWBM-targeting antibiotics vancomycin, telavancin, and oxacillin.
MATERIALS AND METHODS
Strains and plasmids
All strains, plasmids, and primers used in this study are listed in table S1. pCQ11-mNeonGreen was constructed by restriction (Nhe I and Asc I) digest substitution of gfp in pCQ11–green fluorescent protein (GFP) with mNeongreen amplified from pLOM-S-mNeongreen-EC18153 (J. Hibberd, Addgene plasmid #137075; http://n2t.net/addgene:137075; RRID:Addgene_137075) using primers mNeon-for and mNeon-rev. For the construction of pCQ11-MurJ-mNeonGreen, murJ was amplified from S. aureus RN4220 genomic DNA using primers MurJ-for and MurJ-rev, and the resulting fragment was cloned into pCQ11-mNeonGreen using Nhe I and Sma I. pCQ11-MurJ-mNeonGreen was transformed into electrocompetent S. aureus RN4220 as previously described (41) using Escherichia coli StellarTM (Takara Bio) as shuttle strain.
pCQ11-PBP4–yellow fluorescent protein (YFP) was constructed by amplifying pbpD from genomic DNA using primers PBP4_NheI-FW and PBP4_SpeI-RV. The yfp gene was amplified using primers YFP_SpeI_pCQ11_Ct-FW and YFP_AscI_pCQ11_Ct-RV. The two polymerase chain reaction products were ligated and cloned into pCQ11 using Nhe I and Asc I. The strain producing a RodA-GFP fusion protein from the native rodA promotor was constructed as previously described for FtsW-GFP (42) with the exception that primers RodA-c-v pMAD-5′ and RodA-c-v pMAD-3′ were used to amplify rodA, and RodA-c-h pMAD-5 and RodA-c-h pMAD-3′ were used to amplify the downstream fragment from genomic S. aureus DNA.
Growth conditions
Müller-Hinton Broth (MHB; Oxoid) prepared according to Clinical and Laboratory Standards Institute (CLSI) standards was used for all experiments throughout this study, including preparation of agar plates for strain storage (see table S1 for selective antibiotics used) and killing kinetics. For cultivation, overnight precultures (see table S1 for selective antibiotics used) were diluted 1:100 in identical medium. Cultures without need for induction of protein production were grown at 37°C under constant shaking until they reached optical density at 600 nm (OD600) = 0.5, if not stated otherwise. If induction was necessary, 0.1 mM isopropyl-β-d-thiogalactopyranosid (IPTG; Thermo Fisher Scientific) was added, and cultures were grown for 4 hours at 37°C under constant shaking and adjusted to OD600 = 0.5 with identical medium before further treatment.
Antibiotic concentrations
Antibiotic concentrations used for treatment were vancomycin (8 μg/ml; Hikma), telavancin (4 μg/ml; Sanofi), oxacillin (2 μg/ml; Sigma-Aldrich), and nisin (16 μg/ml; Sigma-Aldrich), if not stated otherwise.
Wide-field microscopy
For wide-field microscopy, strains were grown as stated above to an OD600 = 0.5, followed by addition of the compound of interest and further incubation for 25 min at 37°C. Cells were transferred to a microscopy slide coated with 1% agarose for microscopy.
In case of S. aureus RN4220 pCQ11-FtsZ-SNAP, FtsZ-SNAP was labeled with 0.2 μM SNAP-Cell TMR-Star (New England Biolabs) during the last 20 min of main culture incubation and washed three times in medium before addition of antibiotics. For microscopic covisualization of FtsZ-SNAP and DiBAC4(3) (Biotium), 10 μM DiBAC4(3) was added for the last 15 min of main culture incubation before addition of antibiotics. For cell membrane visualization, 20 nM Nile Red in 1% dimethyl sulfoxide (DMSO) was applied on top of the agarose-mounted cells.
Wide-field fluorescence microscopy of SNAP-Cell TMR-Star, FtsW-GFPmut2, PBP2-GFP, MurJ-mNeonGreen, and DiBAC4(3) was performed on a Carl Zeiss AxioObserver Z1 equipped with a HXP 120 C lamp, an αPlan-APOCHROMAT 100×/1.46 oil immersion objective, and an AxioCam MRm camera. Wide-field fluorescence microscopy of RodA-GFP, PBP4-YFP, and Nile Red was performed on a Carl Zeiss AxioObserver Z1 equipped with a X-Cite Xylis LED Lamp, an αPlan-APOCHROMAT 100×/1.46 oil immersion objective, and a Teledyne Photometrics Prime BSI express camera. Visualization of FtsW-GFPmut2, PBP2-GFP, MurJ-mNeonGreen, RodA-GFP, PBP4-YFP, and DiBAC4(3) was achieved using Carl Zeiss filter set 38 (450- to 490-nm excitation, 495-nm beam splitter, and 500- to550-nm emission). Visualization of SNAP-Cell TMR-Star and Nile Red was achieved using Carl Zeiss filter set 43 (538- to 562-nm excitation, 570-nm beam splitter, and 570- to 640-nm emission).
Live-cell microscopy using HiLo illumination
For HiLo live-cell microscopy (43), strains were grown as stated above to an OD600 = 0.5 followed by centrifugation and resuspension in 1× phosphate-buffered saline (PBS). A coverslip (1.5 H, 24 mm by 24 mm) was prepared with 60 μl of a human BD Fc Block (BD Biosciences) diluted 1:5 in 1× PBS for 5 min, washed with 1× PBS, and covered with 60 μl of washed culture for 5 min. The coverslip was washed with 1× PBS and 60 μl of 3% bovine serum albumin in 1× PBS. After 5 min at room temperature, the solution was removed from the coverslip, which was then placed into the preheated stage-top incubator with a special insert on the microscope. The lower surface of the coverslip was in contact with the immersion oil on the preheated objective. The sample volume of the custom incubation chamber was filled with MHB plus erythromycin (10 μg/ml) and 0.1 mM IPTG. Z-stacks with 0.25-μm z-steps were recorded at 10-min intervals over 90 min upon HiLo illumination. Immediately after acquisition of the third stack (corresponding to t = 20 min), the sample volume was exchanged for MHB plus erythromycin (10 μg/ml) and 0.1 mM IPTG supplemented with the compound of interest. Then, the experiment was continued. S. aureus RN4220 pCQ11-ftsZ-mCherry was used for live-cell imaging of FtsZ-rings, and S. aureus RN4220 RNpPBP2-31 was used for live-cell imaging of PBP2.
HiLo live-cell wide-field microscopy was performed on a Nikon Eclipse Ti equipped with a CFI Apo TIRF 100×/1.49 oil immersion objective (Nikon), a Prime BSI (Photometrics), and an external phase contrast (Nikon) with 100× PH4. Fluorescence was excited using a 488-nm laser (Cobalt 06-MLP 488 nm, 120 mW) and a 561-nm laser (Cobalt 06-DPL 561 nm, 100 mW) using a quadband filter set (ET 405/488/561/640-nm Laser Quad Band Set by Chroma). Live-cell images were deconvoluted with synthetic point spread functions (PSFs) using the ImageJ plugin deconvolutionlab2 (44, 45). PSFs were generated with the ImageJ plugin PSFGenerator (46) for the corresponding emission wavelengths.
Confocal Airyscan superresolution microscopy
For confocal Airyscan superresolution microscopy, strains were grown as stated above to an OD600 = 0.5, followed by addition of the compound of interest and further incubation for 25 min at 37°C. Then, cells were transferred to a coverslip coated with 1% agarose. In the case of S. aureus RN4220 pCQ11-FtsZ-SNAP, FtsZ-SNAP was labeled with 0.2 μM SNAP-Cell 505-Star (New England Biolabs) during the last 20 min of main culture incubation and washed three times in medium before addition of antibiotics. Cell membrane was labeled with 1:100 CellBrite Fix 640 during the last 20 min of main culture incubation and washed three times in medium before addition of antibiotics. Confocal Airyscan microscopy was performed using a Carl Zeiss LSM 880 equipped with a Plan-Apochromat 63×/1.4 oil immersion objective upon excitation with 488 nm for SNAP-Cell 505-Star and 640 nm for CellBrite Fix 640.
DiSC3(5) membrane depolarization quantification
Cultures were grown as stated above until they reached OD600 = 0.3. DiSC3(5) (Biomol) was added to a final concentration of 1 μM, and DMSO concentration was adjusted to 1%. Two hundred microliters of the culture was transferred into flat-bottom black polystyrene 96-well plates (Greiner). DiSC3(5) fluorescence was measured using a Tecan Spark 10M equipped with a monochromator every minute until the signal remained stable for 5 min, indicating maximal uptake and auto-quenching of the dye. Antibiotic was added, and DiSC3(5) fluorescence was measured for at least 30 min. Valinomycin (5 μM; Sigma-Aldrich) plus 300 mM KCl were used as positive control (47).
Minimal inhibitory concentrations
Determination of minimal inhibitory concentrations (MICs) was performed according to CLSI Standards (48). Briefly, 100 μl of a 1:2 serial dilution of the respective antibiotic was prepared in a sterile 96-well plate with u-shaped wells. One hundred microliters of a main culture grown as stated above to OD600 = 0.3 and diluted 1:100 in identical medium was added to each well. Plates were sealed and incubated at 37°C. The MIC was determined as the well with lowest antibiotic concentration, where no visible growth was observable after 20 hours of incubation. MICs were confirmed in a total of three biologically independent experiments. To evaluate MICs with an increased inoculum, the identical procedure was applied to cultures grown to OD600 = 1 and diluted 1:50.
Killing kinetics
Cultures were grown as stated above to OD600 = 0.5 and diluted 1:100 in identical medium. One hundred microliters of culture was transferred into a sterile 96-well plate with u-shaped wells. A 1:10 dilution series in identical medium was prepared, and 50 μl was streaked on half area of agar plates. Antibiotic was added to the culture, and the plate was sealed. For the untreated control, no antibiotic was added before sealing. After 1, 2, 4, and 24 hours of antibiotic addition, dilution and streaking were repeated. Agar plates were incubated for 24 hours after culture streaking at 37°C. Colony-forming units (CFU) were counted for each dilution. CFU per milliliter were calculated for each dilution with total CFU between 20 and 300. Final CFU per milliliter of each sample was determined as the mean of all dilutions, where CFU per milliliter were calculated.
Data analysis
Generation of CAPs
Single-cell images were extracted from the raw microscopy images using the cell recognition and segmentation algorithm of the ImageJ Plug-In MicrobeJ (49). Single-cell images were stacked using the Fiji Images to Stack function with bicubic interpolation to scale (smallest). Stacked single-cell images were convolved using the Fiji ImageJ Convolve Process with the convolution Kernel {-1 -1 -1 -1 -1; -1 -1 -1 -1 -1; -1 -1 24 -1 -1; -1 -1 -1 -1 -1; -1 -1 -1 -1 -1} and projected using the Fiji average projection function. An overview of the process is provided in fig. S1.
Septal axis plots and medial/lateral ratios
Septal axis plots were measured on CAPs using the Fiji Line Plot Profile Function with a line width of 3. Values of each plot were adjusted relative to the medial maximum value of the plot, which was determined within a 3–pixel (px) range around the median px of the measurement. Lateral maxima were determined from the relative numbers as the highest values outside the range used for determination of the medial maximum. Two lateral maxima were determined per plot, one for the px number smaller and one for the px number higher than the medial maximum, yielding a left and a right lateral maximum. The two lateral maximum values were used to calculate a mean lateral maximum value. Medial/lateral ratios were calculated as the quotient of the mean lateral maximum and the medial maximum.
Abundance of septal signal
The abundance of septal signal was determined using the single-cell images extracted for CAPs. Each set of images was manually sorted for the visual appearance of a distinctive septal signal after individual contrast adjustments for each image using the Fiji enhance contrast function with 0.3% saturated px and normalization. A distinctive septal signal was defined as a visible septum morphology in any stage of septum progression with any orientation to the visual axis. If a potentially septal signal orthogonal to the visual axis could not be distinguished from an unspecific membrane signal, then cells were handled as not showing a septal signal.
Cell cycle phase distribution
The distribution of cell cycle phases was determined using single-cell images of cells stained with Nile Red. Cells were manually assessed and classified according to a previously described three-phase model of staphylococcal cell membrane morphology throughout the cell cycle (29).
Z-ring diameter
Z-ring diameters were measured in confocal Airyscan superresolution microscopy images along the longest traversing axis of each septum using the Fiji Line Plot Profile Function with a line width of 3. Z-ring diameters were determined as the distance between the left and right fluorescence maximum of each plot with a lower threshold of 300 nm.
Live-cell measurements of Z-ring diameter
The progression of Z-ring diameters over time was analyzed using deconvoluted HiLo live-cell microscopy data. To determine the behavior of Z-ring size over the course of one cell cycle in S. aureus, about 30 cells at the start of a cell division cycle (Z-ring assembly predivision of the parent cell) were chosen randomly at t = 0 min. Assembly state was defined via visibility of a newly formed Z-ring before parent cell division. To analyze Z-ring dynamics during treatment, cells were chosen randomly at t = 0 min until about 30 cells per category (assembly state, early-, mid-, and late-stage cell division states) were found. Early-, mid-, and late-stage cell division states of cells were determined via the measured Z-ring diameter at t = 0 min (early: ≥780 nm; mid: 585 to 715 nm; late: ≤ 520 nm; increments corresponding to px size). For each individual cell, the Z-ring diameter was measured for every 10 min from t = −20 min to t = 70 min with px-size accuracy (65 nm). The Z-ring diameter was defined as described above in the analysis of confocal Airyscan superresolution microscopy, with a lower threshold of 325 nm. To determine the distribution of division stage categories at t = 0 min, cells were chosen randomly and categorized as described above. Z-ring diameter was denoted 0 nm if the Z-ring diameter was below the lower threshold and cell division was observed in the phase contrast channel. Septum closure was achieved if the Z-ring diameter was decreased to 0 nm according to this definition. The time needed for septum closure was measured from t = 0 min or from the time where a septal Z-ring was imaged first. Time of active septum constriction was defined as the interval between the first time point with a decrease in Z-ring size compared to the time point before and septum closure. Active septum progression was defined as ≥130-nm decrease of Z-ring size compared to t = 0 min.
Live-cell PBP2-GFP localization
The localization of PBP2-gfp over time was analyzed using HiLo live-cell microscopy data. For each replicate, a total of 30 cells were chosen randomly at t = −20 min and qualitatively assessed for septal PBP2 localization every 10 min from t = −20 min to t = 70 min. If cells divided, then a daughter cell was chosen randomly for analysis of the remaining time points. Fluxes between each time point were classified into four categories: (i) cell division: Septal PBP2 becomes non-septal in the daughter cell; (ii) PBP2 recruitment: PBP2 localization changes from nonseptal to septal; (iii) PBP2 stays nonseptal; and (iv) PBP2 stays septal. A theoretical fifth category where PBP2 localization becomes nonseptal without division was not needed as no cells behaved this way in our analysis.
Statistical analysis
Statistical analysis of data was performed using GraphPad Prism 8. Statistical significance was determined against the respective untreated control in unpaired two-tailed Student’s t tests with a 95% confidence interval. All experiments were repeated three times with biologically independent replicates. Statistical analysis was always performed on either the population mean values of the independent replicates or the CAP data. The numbers of cells used for statistical analysis of each dataset are listed in table S2. All graphs show the mean of all three population mean values with error bars denoting the SD. Statistical significance was denoted as not significant (ns); *P = 0.05 to 0.01; **P = 0.01 to 0.001; ***P = 0.001 to 0.0001; ****P ≤ 0.0001. Individual cell values or population mean values were plotted either as superplots or in another appropriate way to increase data intelligibility whenever applicable (50).
Software
Image Analysis was performed using Fiji (ImageJ) version 2.0.0-rc-69/1.52p; Java 1.8.0_172 (64 bit) (51) and the ImageJ Plug-In MicrobeJ version 5.13l (20)–beta (49). Statistical analysis was performed using GraphPad Prism 8.0.2 (263).
Acknowledgments
We thank G. Marke for constructing S. aureus RN4220 pCQ11-MurJ-mNeonGreen. We thank H.-G. Sahl for critically reading the manuscript. We thank J.-P. Siebrasse for providing superb technical support with regard to the microscopic equipment. We thank the Quantitative Biology Center (QBiC) at the University of Tübingen for support in storage and management of microscopy data.
Funding: This work was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Project-ID 398967434-TRR261 (T.S., U.K., and F.G.), supported by the DZIF (German Center for Infection Research).
Author contributions: Conception and design of the work: J.-S.P., D.B., T.S., U.K., and F.G. Data acquisition: J.-S.P. and D.B. Data analysis and interpretation: J.-S.P., D.B., U.K., and F.G. Manuscript drafting and edition: J.-S.P., D.B., T.S., U.K., and F.G. All authors approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S6
Tables S1 to S2
References
REFERENCES AND NOTES
- 1.M. G. Pinho, M. Kjos, J.-W. Veening, How to get (a)round: Mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 11, 601–614 (2013). [DOI] [PubMed] [Google Scholar]
- 2.D.-J. Scheffers, M. G. Pinho, Bacterial cell wall synthesis: New insights from localization studies. Microbiol. Mol. Biol. Rev. 69, 585–607 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.F. A. Rubino, A. Mollo, S. Kumar, E. K. Butler, N. Ruiz, S. Walker, D. E. Kahne, Detection of transport intermediates in the peptidoglycan flippase MurJ identifies residues essential for conformational cycling. J. Am. Chem. Soc. 142, 5482–5486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.F. A. Rubino, S. Kumar, N. Ruiz, S. Walker, D. E. Kahne, Membrane potential is required for MurJ function. J. Am. Chem. Soc. 140, 4481–4484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. F. F. Pereira, A. O. Henriques, M. G. Pinho, H. de Lencastre, A. Tomasz, Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189, 3525–3531 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.M. G. Pinho, S. R. Filipe, H. de Lencastre, A. Tomasz, Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183, 6525–6531 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. M. Monteiro, A. R. Pereira, N. T. Reichmann, B. M. Saraiva, P. B. Fernandes, H. Veiga, A. C. Tavares, M. Santos, M. T. Ferreira, V. Macário, M. S. VanNieuwenhze, S. R. Filipe, M. G. Pinho, Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.T. Schneider, H.-G. Sahl, An oldie but a goodie - Cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 300, 161–169 (2010). [DOI] [PubMed] [Google Scholar]
- 9.J. M. Ghuysen, Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 2, 372–380 (1994). [DOI] [PubMed] [Google Scholar]
- 10.F. Grein, T. Schneider, H.-G. Sahl, Docking on lipid II-A widespread mechanism for potent bactericidal activities of antibiotic peptides. J. Mol. Biol. 431, 3520–3530 (2019). [DOI] [PubMed] [Google Scholar]
- 11.P. E. Reynolds, Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8, 943–950 (1989). [DOI] [PubMed] [Google Scholar]
- 12.B. Salamaga, L. Kong, L. Pasquina-Lemonche, L. Lafage, M. von zur Und Zur Muhlen, J. F. Gibson, D. Grybchuk, A. K. Tooke, V. Panchal, E. J. Culp, E. Tatham, M. E. O’Kane, T. E. Catley, S. A. Renshaw, G. D. Wright, P. Plevka, P. A. Bullough, A. Han, J. K. Hobbs, S. J. Foster, Demonstration of the role of cell wall homeostasis in Staphylococcus aureus growth and the action of bactericidal antibiotics. Proc. Natl. Acad. Sci. U.S.A. 118, e2106022118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. A. Kohanski, D. J. Dwyer, J. J. Collins, How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 8, 423–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. G. Pinho, J. Errington, Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55, 799–807 (2005). [DOI] [PubMed] [Google Scholar]
- 15.X. Yang, R. McQuillen, Z. Lyu, P. Phillips-Mason, A. de La Cruz, J. W. McCausland, H. Liang, K. E. DeMeester, C. C. Santiago, C. L. Grimes, P. de Boer, J. Xiao, A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 6, 584–593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. J. F. Egan, W. Vollmer, The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 1277, 8–28 (2013). [DOI] [PubMed] [Google Scholar]
- 17.R. McQuillen, J. Xiao, Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-Ring. Annu. Rev. Biophys. 49, 309–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. J. Perez, Y. Cesbron, S. L. Shaw, J. Bazan Villicana, H.-C. T. Tsui, M. J. Boersma, Z. A. Ye, Y. Tovpeko, C. Dekker, S. Holden, M. E. Winkler, Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 116, 3211–3220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.V. A. Lund, K. Wacnik, R. D. Turner, B. E. Cotterell, C. G. Walther, S. J. Fenn, F. Grein, A. J. Wollman, M. C. Leake, N. Olivier, A. Cadby, S. Mesnage, S. Jones, S. J. Foster, Molecular coordination of Staphylococcus aureus cell division. eLife 7, e32057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.X. Yang, Z. Lyu, A. Miguel, R. McQuillen, K. C. Huang, J. Xiao, GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. W. Bisson-Filho, Y.-P. Hsu, G. R. Squyres, E. Kuru, F. Wu, C. Jukes, Y. Sun, C. Dekker, S. Holden, M. S. VanNieuwenhze, Y. V. Brun, E. C. Garner, Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Pazos, K. Peters, M. Casanova, P. Palacios, M. VanNieuwenhze, E. Breukink, M. Vicente, W. Vollmer, Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat. Commun. 9, 5090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N. Baranova, P. Radler, V. M. Hernández-Rocamora, C. Alfonso, M. López-Pelegrín, G. Rivas, W. Vollmer, M. Loose, Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nat. Microbiol. 5, 407–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.G. R. Squyres, M. J. Holmes, S. R. Barger, B. R. Pennycook, J. Ryan, V. T. Yan, E. C. Garner, Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol. 6, 553–562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K. D. Whitley, C. Jukes, N. Tregidgo, E. Karinou, P. Almada, Y. Cesbron, R. Henriques, C. Dekker, S. Holden, FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat. Commun. 12, 2448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. F. F. Pereira, A. O. Henriques, M. G. Pinho, H. de Lencastre, A. Tomasz, Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol. Microbiol. 72, 895–904 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.N. T. Reichmann, A. C. Tavares, B. M. Saraiva, A. Jousselin, P. Reed, A. R. Pereira, J. M. Monteiro, R. G. Sobral, M. S. VanNieuwenhze, F. Fernandes, M. G. Pinho, SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat. Microbiol. 4, 1368–1377 (2019). [DOI] [PubMed] [Google Scholar]
- 28.K. Wacnik, V. A. Rao, X. Chen, L. Lafage, M. Pazos, S. Booth, W. Vollmer, J. K. Hobbs, R. J. Lewis, S. J. Foster, Penicillin-binding protein 1 (PBP1) of Staphylococcus aureus has multiple essential functions in cell division. MBio 13, e0066922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. M. Monteiro, P. B. Fernandes, F. Vaz, A. R. Pereira, A. C. Tavares, M. T. Ferreira, P. M. Pereira, H. Veiga, E. Kuru, M. S. VanNieuwenhze, Y. V. Brun, S. R. Filipe, M. G. Pinho, Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 6, 8055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B. M. Saraiva, M. Sorg, A. R. Pereira, M. J. Ferreira, L. C. Caulat, N. T. Reichmann, M. G. Pinho, Reassessment of the distinctive geometry of Staphylococcus aureus cell division. Nat. Commun. 11, 4097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.G. K. Best, N. H. Best, A. V. Koval, Evidence for participation of autolysins in bactericidal action of oxacillin on Staphylococcus aureus. Antimicrob. Agents Chemother. 6, 825–830 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.G. Sakoulas, G. M. Eliopoulos, V. G. Fowler Jr., R. C. Moellering Jr., R. P. Novick, N. Lucindo, M. R. Yeaman, A. S. Bayer, Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49, 2687–2692 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. A. Kohanski, D. J. Dwyer, B. Hayete, C. A. Lawrence, J. J. Collins, A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- 34.M. Osawa, H. P. Erickson, Liposome division by a simple bacterial division machinery. Proc. Natl. Acad. Sci. U.S.A. 110, 11000–11004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.P. Szwedziak, Q. Wang, T. A. M. Bharat, M. Tsim, J. Löwe, Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3, e04601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.H. P. Erickson, D. E. Anderson, M. Osawa, FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. T. Nguyen, C. M. Oikonomou, G. J. Jensen, Simulations of Proposed Mechanisms of FtsZ-Driven Cell Constriction. J. Bacteriol. 203, 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.L. T. Nguyen, C. M. Oikonomou, H. J. Ding, M. Kaplan, Q. Yao, Y.-W. Chang, M. Beeby, G. J. Jensen, Simulations suggest a constrictive force is required for Gram-negative bacterial cell division. Nat. Commun. 10, 1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. Coltharp, J. Buss, T. M. Plumer, J. Xiao, Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 113, E1044–E1053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.V. R. Steele, A. L. Bottomley, J. Garcia-Lara, J. Kasturiarachchi, S. J. Foster, Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Mol. Microbiol. 80, 542–555 (2011). [DOI] [PubMed] [Google Scholar]
- 41.S. Schenk, R. A. Laddaga, Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94, 133–138 (1992). [DOI] [PubMed] [Google Scholar]
- 42.F. Grein, A. Müller, K. M. Scherer, X. Liu, K. C. Ludwig, A. Klöckner, M. Strach, H.-G. Sahl, U. Kubitscheck, T. Schneider, Ca2+-daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 11, 1455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Tokunaga, N. Imamoto, K. Sakata-Sogawa, Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 44.D. Sage, L. Donati, F. Soulez, D. Fortun, G. Schmit, A. Seitz, R. Guiet, C. Vonesch, M. Unser, DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 115, 28–41 (2017). [DOI] [PubMed] [Google Scholar]
- 45.C. A. Schneider, W. S. Rasband, K. W. Eliceiri, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Kirshner, F. Aguet, D. Sage, M. Unser, 3-D PSF fitting for fluorescence microscopy: Implementation and localization application. J. Microsc. 249, 13–25 (2013). [DOI] [PubMed] [Google Scholar]
- 47.J. D. Te Winkel, D. A. Gray, K. H. Seistrup, L. W. Hamoen, H. Strahl, Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front. Cell Dev. Biol. 4, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.I. Wiegand, K. Hilpert, R. E. W. Hancock, Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- 49.A. Ducret, E. M. Quardokus, Y. V. Brun, MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S. J. Lord, K. B. Velle, R. D. Mullins, L. K. Fritz-Laylin, SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.P. P. Navarro, A. Vettiger, V. Y. Ananda, P. M. Llopis, C. Allolio, T. G. Bernhardt, L. H. Chao, Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol. 7, 1621–1634 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. S. Lunde, S. R. Hartouni, J. W. Janc, M. Mammen, P. P. Humphrey, B. M. Benton, Telavancin disrupts the functional integrity of the bacterial membrane through targeted interaction with the cell wall precursor lipid II. Antimicrob. Agents Chemother. 53, 3375–3383 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.H. Strahl, L. W. Hamoen, Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U.S.A. 107, 12281–12286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. B. Tol, D. Morales Angeles, D.-J. Scheffers, In vivo cluster formation of nisin and lipid II is correlated with membrane depolarization. Antimicrob. Agents Chemother. 59, 3683–3686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.B. N. Kreiswirth, S. Löfdahl, M. J. Betley, M. O’Reilly, P. M. Schlievert, M. S. Bergdoll, R. P. Novick, The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 (1983). [DOI] [PubMed] [Google Scholar]
- 57.S. Herbert, A.-K. Ziebandt, K. Ohlsen, T. Schäfer, M. Hecker, D. Albrecht, R. Novick, F. Götz, Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infec. Immun. 78, 2877–2889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S6
Tables S1 to S2
References