Abstract
Background
Respiratory disease is a major cause of morbidity and mortality; however, surveillance for circulating respiratory viruses is passive and biased. Wastewater-based epidemiology has been used to understand SARS-CoV-2, influenza A, and respiratory syncytial virus (RSV) infection rates at a community level but has not been used to investigate other respiratory viruses. We aimed to use wastewater-based epidemiology to understand community viral respiratory infection occurrence.
Methods
A retrospective wastewater-based epidemiology surveillance study was carried out at a large wastewater treatment plant located in California, USA. Using droplet digital RT-PCR, we measured RNA concentrations of influenza A and influenza B viruses, RSV A and RSV B, parainfluenza (1–4) viruses, rhinovirus, seasonal coronaviruses, and metapneumovirus in wastewater solids three times per week for 17 months (216 samples) between Feb 1, 2021, and June 21, 2022. Novel probe-based RT-PCR assays for non-influenza viral targets were developed and validated. We compared viral RNA concentrations to positivity rates for viral infections from clinical specimens submitted to California Sentinel Clinical Laboratories (sentinel laboratories) to assess concordance between the two datasets.
Findings
We detected RNA from all tested viruses in wastewater solids. Human rhinovirus (median concentration 4300 [0–9500] copies per gram dry weight) and seasonal human coronaviruses (35 000 [17 000–56 000]) were found at the highest concentrations. Concentrations of viral RNA correlated significantly and positively with positivity rates of associated viral diseases from sentinel laboratories (tau 0·32–0·57, p<0·0009); the only exceptions were influenza B and RSV A, which were rarely detected in wastewater solids. Measurements from wastewater indicated coronavirus OC43 dominated the seasonal human coronavirus infections whereas parainfluenza 3 dominated among parainfluenza infections during the study period. Concentrations of all tested viral RNA decreased noticeably after the omicron BA.1 surge suggesting a connection between changes in human behaviour during the surge and transmission of all respiratory viruses.
Interpretation
Wastewater-based epidemiology can be used to obtain information on circulation of respiratory viruses at a localised, community level without the need to test many individuals because a single sample of wastewater represents the entire contributing community. Results from wastewater can be available within 24 h of sample collection, generating real time information to inform public health responses, clinical decision making, and individual behaviour modifications.
Funding
CDC Foundation.
Introduction
Acute respiratory illnesses are defined as symptomatic infections of the lower or upper respiratory tract and can also result in systemic symptoms and secondary infections. They are consistently the top causes of illness and mortality globally in children younger than 5 years, after the neonatal period, and represent a large burden of infectious diseases.1 Viral acute respiratory illnesses are most commonly caused by SARS-CoV-2, seasonal coronavirus, parainfluenza, metapneumovirus, influenza, respiratory syncytial virus, rhinovirus, adenoviruses, bocaviruses, and non-rhinovirus enteroviruses.2 In the USA, viral acute respiratory illnesses cause nearly 400 000 child hospitalisations at a cost of $1 billion annually.3
Respiratory disease surveillance in the USA is passive and draws from many sources, often relying on institutions to identify specific diseases through keywords in clinical records and death certificates, and testing of clinical specimens. The passive respiratory disease surveillance system is voluntary and biased towards identifying infections in individuals with comorbidities or for whom symptoms are severe and patients require clinical care.4 Since many acute respiratory illnesses are self-limiting, diagnostic testing of individuals with respiratory illness is uncommon. A lack of robust, unbiased data for disease occurrence and the resulting lack of knowledge on occurrence and trends in circulating respiratory diseases limits public health responses, institutional awareness that can guide clinical decision making regarding testing and treatment, and efforts to understand disease epidemiology.
Research in context.
Evidence before this study
Wastewater is a composite biological sample from the contributing community containing various human excretions. Wastewater-based epidemiology uses measured concentrations of pathogen targets in wastewater to understand occurrence and trends in infectious diseases. It has been applied in the past to understand circulation of poliovirus, as well as enteric viruses and bacteria. During the COVID-19 pandemic, wastewater-based epidemiology has been extensively used to understand the occurrence of COVID-19; to our knowledge, this work represents the first time wastewater-based epidemiology has been used to understand respiratory disease occurrence. On Aug 1, 2022, we searched PubMed and Google Scholar, for papers published since database inception, using the terms: wastewater AND (influenza OR metapneumovirus OR rhinovirus OR parainfluenza OR respiratory syncytial virus OR RSV OR “coronavirus OC43” OR “coronavirus HKU-1” OR “coronavirus NL63” OR “coronavirus 229E”); study language was not restricted to English and studies had to relate to wastewater concentrations of these viruses to respiratory disease occurrence. We found that the extension of wastewater-based epidemiology to other respiratory viruses was limited to two studies. One of those studies showed that concentrations of a respiratory syncytial virus (RSV) genomic RNA target in wastewater solids at two wastewater treatment plants correlated to a measure of RSV community infections, whereas the other showed that concentrations of an influenza A genomic RNA target in wastewater solids on two university campuses correlated well with active surveillance data of influenza A incidence in students. There was no other study that presented concentrations of other respiratory viruses in wastewater.
Added value of this study
This study fills the knowledge gaps identified in the literature review by providing measurements of concentrations of various respiratory viruses in wastewater solids including human rhinovirus, parainfluenza, metapneumovirus, influenza A and influenza B, RSV A and RSV B, and seasonal coronaviruses, and showing that their concentrations are associated with traditional measures of disease occurrence in the community. The work illustrates the use of wastewater-based epidemiology for understanding the occurrence of respiratory viral diseases beyond COVID-19.
Implications of all the available evidence
Viral respiratory infections cause substantial morbidity and mortality. In the USA, surveillance for respiratory infections is passive and voluntary, biased to only testing individuals with severe illness, subject to reporting delays of weeks, and temporally and spatially coarse. Wastewater measurements can be available within 24 h of sample collection and are inclusive of the entire contributing community, including those who are asymptomatic or unwilling or unable to seek clinical care, therefore allowing a rapid assessment of the presence of infections and infection trends. Such data can inform local clinical decision making including the prescription of drugs and testing, availability of community testing resources, and vaccination or behaviour-change campaigns; and can be communicated to the public so the community, particularly vulnerable individuals, can make informed decisions about behaviours that limit their risk. Clinical testing remains important for individualised care particularly for patients with severe disease.
Before the COVID-19 pandemic, viral respiratory infections largely circulated seasonally with predictable dynamics.2 However, seasonal occurrence of respiratory viruses has changed substantially during the pandemic,5 presumably due to both imposed and voluntary behaviour changes, such as masking and stay-at-home orders.6 If and when respiratory disease dynamics will return to those observed pre-pandemic is uncertain. More active approaches to disease surveillance are needed to better understand and respond to atypical respiratory disease dynamics.
Wastewater-based epidemiology uses information from wastewater to gain insights into infectious disease occurrence in contributing communities. Wastewater represents a composite biological sample containing bodily excretions including urine, faeces, sputum, and mucus. Excretions of individuals who are infected can contain markers of infectious disease (including live and dead organisms, proteins, and nucleic acids) and methods have been developed to detect and quantify these markers in the wastewater matrix. Wastewater-based epidemiology does not require that individuals who are infected receive medical care or testing or even have symptoms to be represented in the community-level data source. Wastewater-based epidemiology, however, cannot discern information about individual disease severity.
Wastewater-based epidemiology has been shown to be useful for understanding community circulation of poliovirus,7 salmonella,8 hepatitis A,9 and enteroviruses,10 but it has not been widely used for disease surveillance outside of polio in endemic regions. The application of wastewater-based epidemiology to respiratory diseases was prompted by the COVID-19 pandemic, during which concentrations of SARS-CoV-2 RNA in wastewater were shown to correlate to incident COVID-19.11 Thereafter, Hughes and colleagues12 and Wolfe and colleagues13 showed that respiratory syncytial virus (RSV) and influenza A RNA concentrations, respectively, in wastewater solids were associated with disease occurrence in associated sewer sheds.
In this study, we aimed to test the use of wastewater-based epidemiology for a range of viral respiratory diseases.
Methods
Study design
For this surveillance study, we developed and validated novel hydrolysis probe-based RT-PCR assays that target respiratory viral genomes and then applied the assays to wastewater solids collected three times per week at a wastewater treatment plant over 17 months during the COVID-19 pandemic. We measured concentrations of respiratory syncytial virus (RSV) A and RSV B, influenza A (IAV) and influenza B (IBV), human metapneumovirus (HMPV), human parainfluenza (HPIV; 1–4), seasonal human coronaviruses (HCoVs 229E, OC43, NL63, and HKU-1), and human rhinovirus (HRV) RNA. We chose to focus on these viruses because they are common viral aetiologies of respiratory infections.14 In addition, passive respiratory disease surveillance occurs in the region where the sampling was conducted, allowing us to compare wastewater to clinical data; concordance between the two would lend credence to the use of wastewater for disease surveillance. This study was reviewed by the State of California Health and Human Services Agency Committee for the Protection of Human Subjects and determined to be exempt from ethical oversight.
RT-PCR assays
We used published hydrolysis probe-based RT-PCR assays for IAV and IBV.15 We designed 13 novel RT-PCR internal hydrolysis probe assays for HRV (targeting HRV A, B, and C together); HPIV 1, 2, 3, 4A, and 4B; HCoVs 229E, NL63, OC43, and HKU-1; HMPV; and RSV A and B. Custom assays were needed because available commercial assays are proprietary. To develop the assays, viral genome sequences were downloaded from the National Center for Biotechnology Information (NCBI) between February and June, 2022, and aligned to identify conserved regions. Assays were developed in silico using Primer3Plus. Parameters used in assay development (eg, sequence length and GC content) are provided in the appendix (p 6). Primers and probes were screened for specificity in silico, and in vitro against virus panels, intact respiratory viruses, synthetic viral genomic RNA, or cDNA sequences (appendix pp 2–3, 7–9).
Sample collection
Samples were obtained at San José-Santa Clara Regional Wastewater Facility, a wastewater treatment plant that serves 1 500 000 residents of Santa Clara County (CA, USA; appendix p 12).11
Samples were collected daily for a COVID-19 wastewater-based epidemiology effort,11 and a subset of those samples (ie, three samples per week and 216 samples in total) were used in this study. The samples were chosen to span a 17-month period (from Feb 1, 2021, to June 21, 2022) that included implementation and easing of indoor mask mandates, changes in COVID-19 vaccine availability, public health promotion of hand hygiene and mask wearing, and periods of both high and low COVID-19 incidence.
Procedures
50 mL of settled solids were collected using sterile technique in clean bottles from the primary clarifier. 24-h composite samples were collected by combining grab samples from the sludge line every 6 h.11 Samples were stored at 4°C, transported to the laboratory, and processed within 6 h. Solids were then dewatered11 and frozen at –80°C for 4–60 weeks. Before further processing, samples were thawed overnight at 4°C, and then RNA was obtained from the dewatered solids following previously published protocols.11 RNA was obtained from ten replicate sample aliquots. Each replicate RNA extract from each sample was processed immediately to measure viral RNA concentrations using droplet digital RT-PCR using assays for IAV, IBV, HMPV, total HPIV (ie, HPIV 1, HPIV 2, HPIV 3, HPIV 4A, and HPIV 4B), total HCoV (ie, HCoV 229E, HCoV NL63, HCoV OC43, and HCoV HKU-1), RSV A, RSV B, HRV, and pepper mild mottle virus (PMMoV). PMMoV is highly abundant in wastewater globally16 and is used as an internal recovery and faecal strength control.17 18 samples (approximately one per month) were selected to measure each HCoV and HPIV individually (appendix p 10). Each 96-well PCR plate of wastewater samples included PCR positive (appendix pp 7–8) and negative controls, and extraction negative controls. For a sample to be recorded as positive, it was required to have at least three positive droplets. Additional details are available in the appendix (pp 2–3).
Concentrations of RNA targets were converted to concentrations per dry weight of solids in units of copies per gram dry weight using dimensional analysis. Standard deviations were calculated using QuantaSoft software (version 1.0.596; Bio-Rad, Hercules, CA, USA). The lowest detectable concentration (limit of quantification) corresponds to three positive droplets, which is equivalent to between about 500–1000 copies per gram; the range in values is a result of the range in the equivalent mass of dry solids added to the wells.
We obtained SARS-CoV-2 and PMMoV gene concentrations in solids at the plant from a regional monitoring programme.11 The measurements were used in a supplementary manner to provide insight into the progress of the COVID-19 pandemic, and to assess the effect of sample storage on measurements (appendix pp 3–4).
California Sentinel Clinical Laboratories (hereafter referred to as sentinel laboratories) test clinical specimens for IAV and IBV, RSV, HPIV 1–4, HCoV (229E, NL63, OC43, and HKU-1), HMPV, and HRV. Sentinel laboratories do not differentiate HRV from other enteroviruses. Specimens tested by the laboratories come from across the state (with a population of 40 million people) and represent inpatient or outpatient samples of people receiving medical care. Positivity rates were calculated using data from all state sentinel laboratories and are reported by Morbidity and Mortality Weekly Report week. Positivity rates were also aggregated at the county-level for comparison (appendix p 4). Incidence and prevalence rates for these diseases are not available; and inferring them from the positivity rate data is not possible because of the inherent bias in the populations seeking and receiving testing.
For context, state-aggregated and county-aggregated daily COVID-19 positivity rates were calculated using data from all laboratories (not just sentinel laboratories) that provide PCR testing of patient specimens.
Statistical analysis
Kendall's tau was used to test the null hypotheses that clinical specimen positivity rates were not associated with viral concentrations in wastewater solids, and that viral RNA concentrations were not associated with each other. Kendall's tau was used because variables were not normally distributed. Because positivity data are aggregated by Morbidity and Mortality Weekly Report week, we used median wastewater measurements from the same Morbidity and Mortality Weekly Report week to match clinical and wastewater data. 53 Kendall's tests of association were carried out so a p value of 0·0009 was used to identify tau values significantly different from 0 based on Bonferroni's correction. Statistical analyses were done using RStudio (version 1.4.1106).
Role of the funding source
The funder of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Results are reported as suggested in the Environmental Microbiology Minimal Information guidelines (appendix pp 4, 13).18 Extraction and PCR negative and positive controls performed as expected (ie, negative and positive, respectively) with the exception of a positive control that did not amplify for a HCoV run including samples collected before Feb 26, 2021; HCoV results before this date were therefore omitted. The median PMMoV concentration across the samples was 1·6 × 109 copies per gram, similar to measurements previously reported for the plant.11 PMMoV concentrations were stable across samples (IQR 1·3 × 109–2·0 × 109 copies per gram), suggesting consistent faecal strength and RNA extraction efficiency. Additionally, comparisons of PMMoV concentrations to those measured in the same samples with no storage indicates sample storage and freeze–thaw cycles had a limited effect on PMMoV RNA concentrations, and we assume that the effect was similarly minimal for the other RNA targets (appendix pp 3–4).
In-silico analysis indicated no cross reactivity of the novel RT-PCR probe-based assays (table 1 ) with sequences deposited in NCBI. The novel assays for HPIV, HMPV, HRV, and the HCoV were tested in vitro against non-target viral gRNA as well as target gRNA (appendix pp 7–9). No cross reactivity was observed (appendix p 15).
Table 1.
Forward and reverse primer and probe sequences for the detection of the respiratory viral RNA used in this study
| Forward primer sequence | Reverse primer sequence | Probe sequence | |
|---|---|---|---|
| Human rhinovirus (204 bp) | GCCYGCGTGGCKGCC | GAAACACGGACACCCAAAG | TCCTCCGGCCCCTGAATG |
| Human parainfluenza 1 (199 bp) | AAGTTCAGTACAAAGCGGGA | GCTCARTAGGGGTTCTCCTA | AGCAAAGCAGAGATCTCACACA |
| Human parainfluenza 2 (141 bp) | AATACAACAGGGCARTGGG | GATAAAATAGCGTGAGGACTGC | TCCTGTATATGGTGGTCTCATAAATGG |
| Human parainfluenza 3 (153 bp) | TGTGGTGACCAACAGATCAA | CCCTCCAAAGAATCGTCCTG | TCCAATGAAAACACTGATCCCAGA |
| Human parainfluenza 4A (170 bp) | TGAACGGTTGCATTCAGG | TTGACTGGTTGCACCTAATTCT | CTGGCAATCTCAACATAGACCATG |
| Human parainfluenza 4B (156 bp) | GGAGAACTTTGAAACCACCTCTAA | TCTCCTTTAACTACCCTATCTTTGC | ACCCCCATAAGGCAAGAAGC |
| Human coronavirus 229E (92 bp) | GGATGACATCATGAAGGCAG | TACCCGTTTTCGCTGACTTT | TTCCTGAGGCTTGTCAAAACCT |
| Human coronavirus NL63 (107 bp) | GAAGCGTGTTCCTACCAGAG | TGGCATCAACACCATTCTGA | CAGTGCTTTGGTCCTCGTGA |
| Human coronavirus OC43 (181 bp) | GTCTTTTACTCCTGGTAAGCAATC | GGGTACAACATTCCCTCCTG | CCGATCAGTCCGACCAGTTTAG |
| Human coronavirus HKU-1 (99 bp) | CCTGGTACGATTTTGCCTCA | ATTGGGTCCACGTGATTGAG | AGGCTCAGGAAGGTCTGCTT |
| Human metapneumovirus (109 bp) | ACTTTATTGGAGAAGGAGCAGG | GGGTAATGRTGATCAAGRTCA | AYTGGATGGCMAGAACAGCA |
| Influenza A virus (106 bp) | CAAGACCAATCYTGTCACCTCTGAC | GCATTYTGGACAAAVCGTCTACG | TGCAGTCCTCGCTCACTGGGCACG |
| Influenza B virus (103 bp) | TCCTCAAYTCACTCTTCGAGCG | CGGTGCTCTTGACCAAATTGG | CCAATTCGAGCAGCTGAAACTGCGGTG |
| Respiratory syncytial virus A (159 bp) | AGAGGTGGCAGTAGAGTTGA | CTCCACAACTTGTTCCATTTCTG | ATGGTGCAGGGCAAGTGATG |
| Respiratory syncytial virus B (90 bp) | TGACACTCCCAATTATGATGTGC | CCTGTGAATTTATGATTTGCATCTTCAG | ACACCTAAACAAACTATGTGGTATGC |
Additional details of the target region of the genomes are in the appendix (pp 7–8). Primers and probes were purchased from Integrated DNA Technologies (Coralville, IA, USA). All probes contained fluorescent molecules and quenchers (5′ FAM and or HEX/ZEN/3′ IBFQ); FAM, 6-fluorescein amidite; HEX, hexachloro-fluorescein; ZEN, a proprietary internal quencher from Intrgrated DNA Technologies (Coralville, IA, USA); and IBFQ, Iowa Black FQ. The appendix (p 2) indicates whether hexachloro-fluorescein or 6-fluorescein amidite molecules were used. The size of the product generated by the primers is provided in brackets in the first column. All assays run at an annealing temperature of 59°C (appendix pp 2–3).
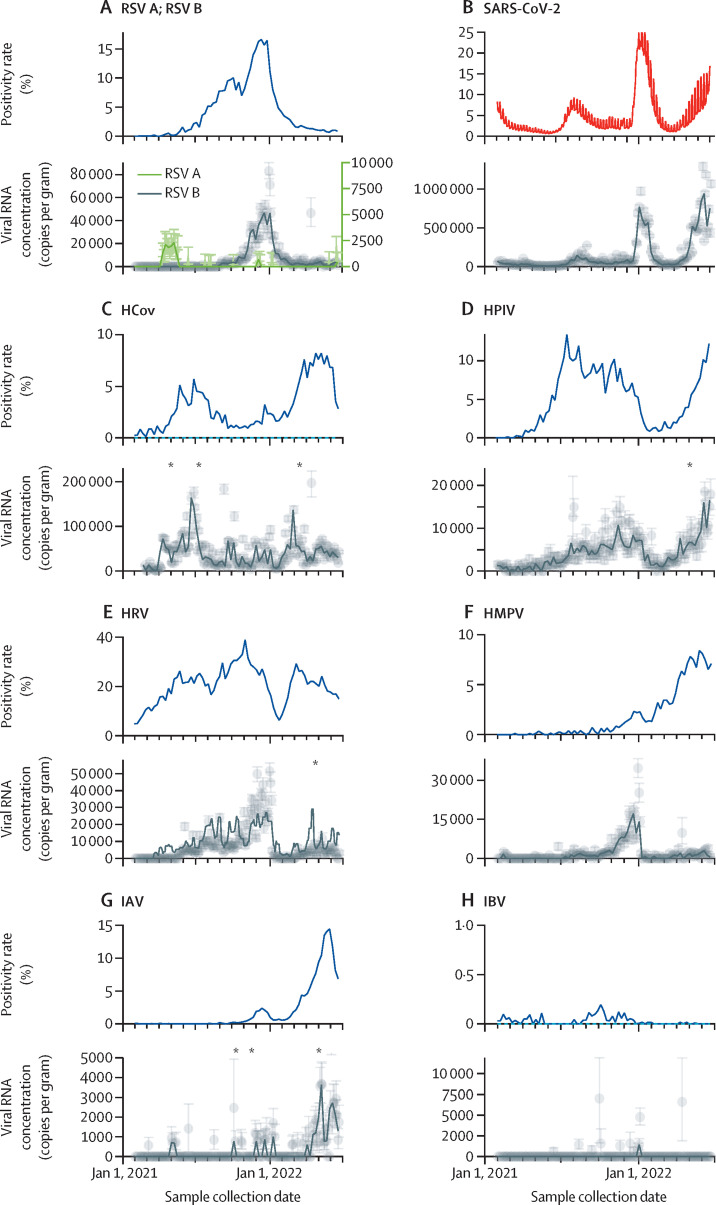
RNA from all viruses were detected in wastewater samples (figure 1 ). Concentrations varied from a median of 0 to more than a million copies per gram. In order of the highest to lowest, median (IQR) concentrations in units of copies per gram were 35 000 (17 000–56 000) for HCoV, 4300 (0–9500) for HRV, 3500 (1400–6300) for HPIV, 1700 (0–5400) for RSV B, 760 (0–2000) for HMPV, 0 (0–660) for IAV, 0 (0–0) for IBV, and 0 (0–0) for RSV A, for which 0 represents non-detects (table 2 ). Although median IAV, IBV, and RSV A were non-detects, viral RNA from IAV was detected in 60 (28%) of 216 samples, as was IBV in 12 (6%) of 216 samples and RSV A in 30 (14%) of 216 samples. For context, median concentration for SARS-CoV-2 was 48 000 (25 000–130 000) copies per gram. In general, concentrations were lowest at the beginning of the study (February, 2021), and increased until January, 2022, at which time RNA concentrations for all viruses showed a steep drop. After the steep drop, viral RNA concentrations began to increase until the end of the time series with the exception of RSV B for which we did not observe a rebound. HCoV concentrations rebounded first, when all other viral RNA concentrations, including those of SARS-CoV-2, were still relatively low (figure 2 ).
Figure 1.
State-aggregated positivity rate and viral RNA in wastewater solids
Grey symbols represent measurements; error bars are SD. Black lines represent Morbidity and Mortality Weekly Report weekly median wastewater measurements. Asterisks indicate dates for which a point is off scale. (A) RSV positivity rates include RSV A and RSV B. (B) SARS-CoV-2 wastewater results and state-aggregated positivity rate are from all laboratories in the state (not just sentinel laboratories) and are shown in red to distinguish from other clinical data. (C) HCoV is the sum of all four seasonal HCoVs (ie, OC43, HKU-1, 229E, and NL63); three measurements were located off scale (1·1 × 106 copies per gram on April 9, 2021, 3·6 × 105 on June 24, 2021, and 5·1 × 105 on March 3, 2022). (D) HPIV is the sum of HPIVs (1–4); one measurement was located off scale (3·6 × 104 copies per gram on April 14, 2022). (E) HRV includes HRV A, B, and C; one measurement was located off scale (7·6 × 104 copies per gram on April 14, 2022). (G) Three IAV measurements were located off scale (2·1 × 104 copies per gram on Sept 30, 2021, 7·5 × 104 on Oct 31, 2021, and 1·5 × 104 on April 14, 2022). RSV=respiratory syncytial virus. HCoV=human coronaviruses. HPIV=human parainfluenza viruses. HRV=human rhinovirus. HMPV=human metapneumovirus. IAV=influenza A virus. IBV=influenza B virus.
Table 2.
Summary statistics of measurements of viral RNA in wastewater solids
| Median viral RNA concentration (IQR) | Samples for which the viral RNA was not detected | Number of measurements made | |
|---|---|---|---|
| Human parainfluenza virus | 3500 (1400–6300) | 21 | 216 |
| Human coronaviruses | 35 000 (17 000–56 000) | 0 | 207 |
| Human metapneumovirus | 760 (0–2000) | 95 | 216 |
| Human rhinovirus | 4300 (0–9500) | 61 | 216 |
| Respiratory syncytial virus A | 0 (0–0) | 186 | 216 |
| Respiratory syncytial virus B | 1700 (0–5400) | 89 | 216 |
| Influenza A virus | 0 (0–660) | 156 | 216 |
| Influenza B virus | 0 (0–0) | 204 | 216 |
| SARS-CoV-2 | 48 000 (25 000–130 000) | 0 | 216 |
Viral RNA concentration is reported in copies per gram dry weight of wastewater solids. Human coronaviruses is the sum of all four non-severe acute respiratory syndrome human coronaviruses (OC43, HKU-1, 229E, and NL63). Human parainfluenza virus is the sum of human parainfluenza viruses 1–4. Human rhinovirus is the sum of human rhinovirus A, B, and C. SARS-CoV-2 data are included for context.
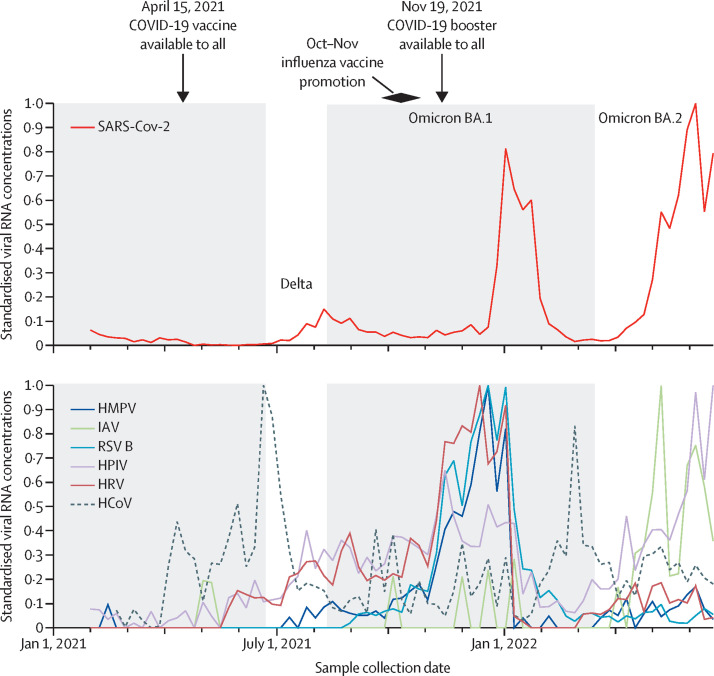
Figure 2.
Standardised concentrations of viral RNA
Standardised concentrations of viral RNA for all viruses except for RSV A and influenza B virus, which were rarely detected, to allow for clear visualisation of concordance in the reduction of virus concentrations after the omicron BA.1 surge, and the rise of HCoV concentrations shortly thereafter. The grey shaded backgrounds indicate periods of time that indoor masking mandates were in effect locally. The red SARS-CoV-2 line is labelled with the dominating circulating variant at the time. Standardised concentrations were calculated using the following formula: Cst(t) = (C(t) – Cmin)/(Cmax– Cmin), where Cst(t) is the standardised concentration at a given week t, C(t) is the Morbidity and Mortality Weekly Report weekly median concentration shown in figure 1 at week t, Cmin is the minimum concentration, and Cmax is the maximum concentration. HMPV=human metapneumovirus. IAV=influenza A virus. RSV=respiratory syncytial virus. HPIV=human parainfluenza viruses. HRV=human rhinovirus. HCoV=human coronavirus.
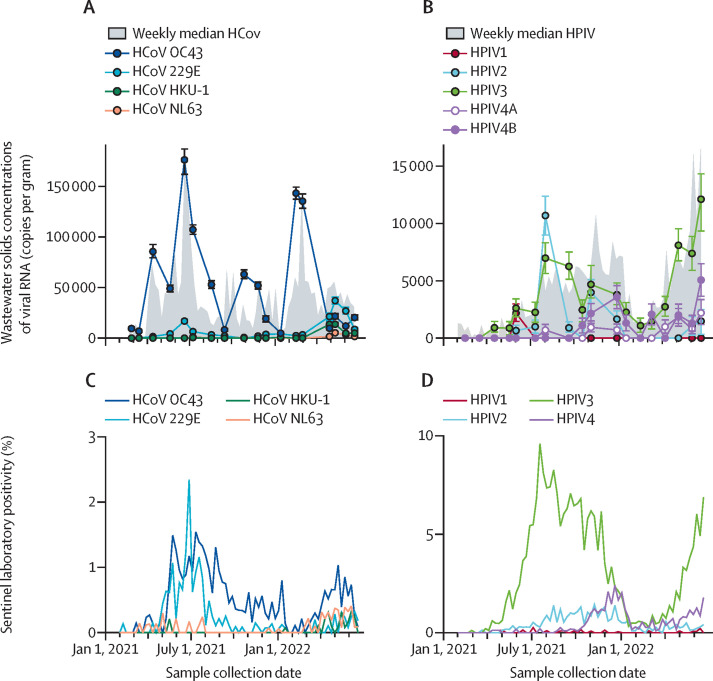
To assess which HCoV were circulating during the study, we measured concentrations of OC43, HKU-1, 229E, and NL63 RNA in 18 samples (figure 3 ). Most of the total HCoV RNA was OC43 RNA in 15 of 18 samples. For the three remaining samples collected in April, May, and early June, 2022, most of the total HCoV RNA was 229E. HKU-1 RNA and NL63 RNA were each detected in six samples (concentrations ranging from 600 to 14 000 copies per gram for HKU-1 RNA and 500 and 4000 copies per gram for NL63 RNA) and never represented the majority of HCoV RNA.
Figure 3.
Concentrations of viral RNA in wastewater solids and comparison to sentinel laboratory percent positivity data
(A) Concentration of total HCoV RNA (weekly medians from figure 1 shown as grey filled areas) and individual HCoVs. (B) Concentrations of total concentration of HPIV RNA (weekly medians from figure 1 shown as grey filled areas) and individual HPIVs. (C) State-aggregated percent positivity from sentinel laboratories for tests for individual HCoVs. (D) State-aggregated percent positivity from sentinel laboratories for tests for individual HPIVs. Percent positivity for unknown HCoV and unknown HPIV from the sentinel laboratories surveillance system are not shown in panels C and D. Error bars on individual HCoV and HPIV are SDs and in some cases are smaller than the symbol. HCoV=human coronavirus. HPIV=human parainfluenza virus.
Similarly, we measured RNA concentrations of each HPIV (1, 2, 3, 4A, and 4B) in 18 samples. RNA from all five HPIVs were present; HPIV 3 dominated in most samples, although HPIV 2 and HPIV 4B each dominated in one of the 18 tested samples. HPIV 1 and HPIV 4A contributed the least to total HPIV (figure 3).
We tested whether state-aggregated positivity rates for respiratory viral infections were associated with the wastewater concentrations of viral RNA. Sentinel laboratories tested a median (IQR) of 5918 (2321–6874) clinical specimens per week for influenza (IAV and IAB), 2419 (1171–6065) for RSV (RSV A and RSV B), 1084 (848–1363) for HPIV, HMPV, and HRV, and 931 (746–1165) for HCoV. The associations between wastewater RNA concentrations and positivity rates were significant and positive for all viruses except for IBV. Kendall's tau between positivity rates and wastewater concentration were 0·47 (p<10–7) for HRV, 0·45 (p<10–5) for IAV, 0·52 (p<10–9) for HPIV, 0·57 (p<10–10) for RSV (using RSV B wastewater concentrations as RSV A was mostly non-detect), 0·32 (p=0·0002) for HMPV, 0·32 (p=0·0001) for HCoV, and –0·010 (p=0·92) for IBV. Wastewater concentrations and positivity rates aggregated across viruses were positively correlated (tau=0·43, p <10–15; appendix p 16). Results using county-aggregated positivity rates, when available, are similar to those using state-aggregated rates (appendix pp 4, 17). All the tests of association were repeated with wastewater respiratory virus RNA concentrations normalised by PMMoV RNA concentrations and the results were unchanged.
State-aggregated positivity rates for individual HCoV infections are similar to observations of the relative occurrence of their RNA in wastewater solids (figure 3). OC43 had the highest positivity relative to other HCoVs, in agreement with the wastewater measurements. Wastewater indicates that 229E dominated at the end of the study (spring 2022), but this finding is not reflected in the state-aggregated clinical data. Positivity rates for individual HPIV suggest HPIV 3 dominated HPIV infections throughout the study period; HPIV 2 and 4 also circulated with lower positivity rates. HPIV 1 was rarely detected in clinical specimens. This finding agrees with the wastewater data that indicated HPIV 3 dominated with HPIV 2 and HPIV 4B also present at relatively high levels, and HPIV 1 was rarely detected (figure 3). Wastewater concentrations and positivity rates aggregated across individual HCoV and HPIV were positively correlated (tau 0·49 for HCoV and 0·66 and HPIV, both p<10–6; appendix p 18).
Viral RNA concentrations were associated with each other, as well as with SARS-CoV-2 N gene concentrations available from a regional monitoring programme at the plant (appendix p 11). HPIV, HRV, HMPV, RSV B, and SARS-CoV-2 RNA were significantly correlated with each other (p<0·0009) with tau from 0·16 (SARS-CoV-2 and HRV) to 0·58 (HRV and HMPV). Given the low occurrence of IAV, IBV, and RSV A RNA, the fact that they were not consistently correlated with other viral RNA concentrations is unsurprising. HCoV RNA concentrations were not correlated to concentrations of other viral targets.
Discussion
We detected RNA from all tested respiratory viruses in wastewater solids, including HRV, HPIV, HMPV, influenza, RSV, and seasonal coronaviruses. Additionally, we detected four seasonal HCoVs (OC43, 229E, NL63, and HKU-1), and five HPIVs (1, 2, 3, 4A, and 4B). Non-influenza respiratory viral RNA targets were measured using novel assays; these assays were found to be specific to the intended viral genomic targets when tested against a range of viruses. Previous studies have documented SARS-CoV-2,11, 19 HRV,10, 20 RSV,12 and influenza13 RNA in wastewater, but to our knowledge, no study to date has showed concentrations of other respiratory viruses.
RSV A RNA was rarely detected in wastewater, yet RSV B RNA concentrations were more than 10 000 copies per gram at times, and RSV B wastewater trends mirrored RSV clinical surveillance data. This finding suggests limited local cocirculation of the two RSV groups. Changes in dominant RSV strains have been documented to occur on continental21 and community scales22 with resultant changes in population immunity.23
The association between viral RNA in wastewater solids and the clinical specimen positivity rates for respective respiratory viruses, available through passive disease surveillance, suggests that wastewater data are representative of respiratory disease occurrence in the community. The available clinical data were aggregated across the entire state, which could obfuscate local disease occurrence patterns. For example, deviation in HMPV wastewater and state-aggregated positivity rate trends suggest localised patterns in HMPV infections. Additionally, state-aggregated HCoV case positivity suggests OC43 infections dominated HCoV infections, yet wastewater measurements suggest 229E dominated locally in the spring of 2022. Similarly, based on wastewater measurements, HPIV 2 and HPIV 4 could have contributed to more HPIV infections locally than on average across the state.
The analysis between wastewater data and clinical data is limited by the use of passive surveillance data and no local data matching the sewershed on respiratory disease occurrence. The state-aggregated clinical data used serves as a proxy for sewershed clinical data; it is a valid proxy if state-wide disease occurrence trends mimic those at the local scale. Limited county-aggregated sentinel laboratory positivity rates generally match state-aggregated positivity rates (appendix p 17) and associations between county-aggregated clinical data (as available) and wastewater are consistent with results obtained with state-aggregated clinical data (appendix pp 4–5). Clinical surveillance data are also biased towards those receiving medical care and not all specimens are tested for all viruses, making estimating disease occurrence from positivity rate data difficult. Positivity rates might not be associated with disease occurrence in the same manner for all viruses.
There are additional uncertainties associated with understanding disease occurrence using wastewater testing. Little information is available on concentrations of viral RNA excreted into wastewater from individuals who are infected, despite evidence showing that shedding does occur for viruses included in this study.24 Given the current state of knowledge, knowing how inputs of viral RNA to the wastewater system differ between individuals infected with different viruses and with different disease severities is difficult. Wastewater cannot be used to infer specifics about the severity of illness, or characteristics of infected subpopulations. Wastewater captures contributions from anyone using sewers in the sewershed, including transient populations.
Despite these limitations and uncertainties, the association between viral RNA in wastewater solids and case positivity holds when data from the measured viruses are combined, suggesting that the relative concentrations of viruses in wastewater solids are related to their different rates of disease occurrence. HCoV and HRV concentrations were highest during the study period, suggesting seasonal HCoV and HRV infections were most common. Conversely, IBV and RSV A concentrations were lowest, suggesting limited circulation of those viruses. Future work is needed to document time varying patterns of respiratory viral RNA inputs from individuals who are infected to the wastewater system to enable modelling of the number of individuals who are infected within a sewershed from wastewater viral RNA concentrations,25 and to enable modelling of epidemiological parameters such as the reproductive number from wastewater data.26
Viral RNA concentrations in wastewater solids, including those of SARS-CoV-2, generally followed similar trends over time, with the exception of HCoV. Evident in the trends is a notable drop-off in concentrations after the omicron BA.1 surge in January, 2022. The drop-off might suggest that disease mitigation measures practiced by the community in response to the surge (eg, isolation due to illness, working remotely, and wearing masks) reduced the spread of all respiratory viruses.27 Local indoor mask mandates ended shortly after the omicron surge (March 2, 2022), and this change was followed by an increase in concentrations of respiratory viruses in wastewater (figure 2). HCoVs, specifically OC43, were the first viruses to reappear in wastewater after the omicron surge. HCoVs are highly transmissible with R0 values higher, on average, than influenza and HRV,28 but lower than RSV and similar to HPIV; there are no R0 data for HMPV.14 Infection with omicron also possibly reduced susceptibility to other respiratory viruses and contributed to the decrease in all viral activity; antibody cross immunity has been suggested to control temporal patterns of some respiratory viruses.29, 30
Data generated from wastewater can be available within 24 h of sample collection, and samples represent the entire contributing population—even those with asymptomatic infections—thereby overcoming various biases and the inherent reporting delays of the passive clinical surveillance system. Results suggest that wastewater surveillance for multiple respiratory viruses that commonly circulate can provide information on a community scale on causes of acute respiratory illnesses. Additional studies from diverse geographical locations are needed to further confirm these results. The acute respiratory illnesses caused by the viruses included in this study have differing levels of likely severity and associated prevention measures, treatments, and complications. Therefore, more information about circulating viruses, obtained from wastewater, can inform local clinical, public health, and individual decision making. Medical doctors can use information on circulating viruses to aid in differential diagnosis and making decisions about patient specimen testing that could influence the use of therapeutics. Hospital directors can use the information to inform hospital staffing, and the stocking of therapeutics and prevention supplies in anticipation of needs when outbreaks begin. Wastewater data can be used to guide the provision of community testing resources and targeting of vaccination or behavioural change campaigns by public health departments. Finally, wastewater data can also be communicated to the public, similar to a weather report, so the community and particularly individuals who are considered to be vulnerable can make informed decisions about behaviours that could put them at risk.
Data sharing
Wastewater data are immediately available publicly at the Stanford Digital Repository. State-aggregated and county-aggregated sentinel laboratory surveillance data are available upon request from California Department of Health (InfluenzaSurveillance@cdph.ca.gov). COVID-19 disease occurrence data are available publicly via data from all laboratories (not just sentinel laboratories) that provide PCR testing of patient specimens.
Declaration of interests
BH, DD, VC-H, AB, and BJW are employees of Verily Life Sciences. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This work is supported by funding from the CDC Foundation. We acknowledge Payal Sarkar at the wastewater treatment plant for overseeing sample collection. We acknowledge Erin Murray, Monica Sun, Tomás Leon, and Alexander Yu at the California Department of Public Health for their assistance. This study was done on the ancestral and unceded lands of the Muwekma Ohlone people. We pay our respects to them and their Elders, past and present, and are grateful for the opportunity to live and work here.
Contributors
ABB contributed to the conceptualisation of the study, the resources, data curation, writing of the original draft, reviewing and editing of subsequent drafts, visualisation, supervision, and funding acquisition. BH and DD contributed to the investigation, data curation, and reviewing and editing of manuscript drafts. VC-H and AB contributed to the investigation and reviewing and editing of manuscript drafts. MKW contributed to the conceptualisation of the study and the reviewing and editing of manuscript drafts. BJW contributed to the conceptualisation of the study, reviewing and editing of manuscript drafts, and supervision. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication. ABB and BH verified the underlaying data of the study.
Supplementary Material
References
- 1.Wang H, Bhutta ZA, Coates MM, et al. Global, regional, national, and selected subnational levels of stillbirths, neonatal, infant, and under-5 mortality, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1725–1774. doi: 10.1016/S0140-6736(16)31575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moriyama M, Hugentobler WJ, Iwasaki A. Seasonality of respiratory viral infections. Annu Rev Virol. 2020;7:83–101. doi: 10.1146/annurev-virology-012420-022445. [DOI] [PubMed] [Google Scholar]
- 3.Henrickson KJ. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J. 2004;23(suppl 1):S6–10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 4.Killerby ME, Biggs HM, Haynes A, et al. Human coronavirus circulation in the United States 2014–2017. J Clin Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuitunen I, Artama M, Haapanen M, Renko M. Respiratory virus circulation in children after relaxation of COVID-19 restrictions in fall 2021—a nationwide register study in Finland. J Med Virol. 2022;94:4528–4532. doi: 10.1002/jmv.27857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu P, Xu M, Lu L, et al. The changing pattern of common respiratory and enteric viruses among outpatient children in Shanghai, China: two years of the COVID-19 pandemic. J Med Virol. 2022;94:4696–4703. doi: 10.1002/jmv.27896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouwer AF, Eisenberg JNS, Pomeroy CD, et al. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc Natl Acad Sci USA. 2018;115:E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diemert S, Yan T. Clinically unreported salmonellosis outbreak detected via comparative genomic analysis of municipal wastewater Salmonella isolates. Appl Environ Microbiol. 2019;85:e00139–e00219. doi: 10.1128/AEM.00139-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCall C, Wu H, O'Brien E, Xagoraraki I. Assessment of enteric viruses during a hepatitis outbreak in Detroit MI using wastewater surveillance and metagenomic analysis. J Appl Microbiol. 2021;131:1539–1554. doi: 10.1111/jam.15027. [DOI] [PubMed] [Google Scholar]
- 10.Brinkman NE, Fout GS, Keely SP. Retrospective surveillance of wastewater to examine seasonal dynamics of enterovirus infections. MSphere. 2017;2:e00099–e00117. doi: 10.1128/mSphere.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe MK, Topol A, Knudson A, et al. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in Northern California shows strong association with COVID-19 incidence. mSystems. 2021;6 doi: 10.1128/mSystems.00829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes B, Duong D, White BJ, et al. Respiratory syncytial virus (RSV) RNA in wastewater settled solids reflects RSV clinical positivity rates. Environ Sci Technol Lett. 2022;9:173–178. [Google Scholar]
- 13.Wolfe MK, Duong D, Bakker KM, et al. Wastewater-based detection of two influenza outbreaks. Environ Sci Technol Lett. 2022;9:687–692. [Google Scholar]
- 14.Leung NHL. Transmissibility and transmission of respiratory viruses. Nat Rev Microbiol. 2021;19:528–545. doi: 10.1038/s41579-021-00535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.US Center for Disease Control and Prevention CDC's influenza SARS-CoV-2 multiplex assay. 2022. https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex-primer-probes.html
- 16.Symonds EM, Nguyen KH, Harwood VJ, Breitbart M. Pepper mild mottle virus: a plant pathogen with a greater purpose in (waste)water treatment development and public health management. Water Res. 2018;144:1–12. doi: 10.1016/j.watres.2018.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClary-Gutierrez JS, Aanderud ZT, Al-Faliti M, et al. Standardizing data reporting in the research community to enhance the utility of open data for SARS-CoV-2 wastewater surveillance. Environ Sci Water Res Technol. 2021;9:1545–1551. doi: 10.1039/d1ew00235j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borchardt MA, Boehm AB, Salit M, Spencer SK, Wigginton KR, Noble RT. The Environmental Microbiology Minimum Information (EMMI) Guidelines: qPCR and dPCR quality and reporting for environmental microbiology. Environ Sci Technol. 2021;55:10210–10223. doi: 10.1021/acs.est.1c01767. [DOI] [PubMed] [Google Scholar]
- 19.Boehm AB, Hughes B, Wolfe MK, White BJ, Duong D, Chan-Herur V. Regional replacement of SARS-CoV-2 variant omicron BA.1 with BA.2 as observed through wastewater surveillance. Environ Sci Technol Lett. 2022;9:575–580. doi: 10.1021/acs.estlett.2c00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blomqvist S, Savolainen-Kopra C, Paananen A, Hovi T, Roivainen M. Molecular characterization of human rhinovirus field strains isolated during surveillance of enteroviruses. J Gen Virol. 2009;90:1371–1381. doi: 10.1099/vir.0.008508-0. [DOI] [PubMed] [Google Scholar]
- 21.Cantú-Flores K, Rivera-Alfaro G, Muñoz-Escalante JC, Noyola DE. Global distribution of respiratory syncytial virus A and B infections: a systematic review. Pathog Glob Health. 2022;116:398–409. doi: 10.1080/20477724.2022.2038053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peret TCT, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79:2221–2229. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- 23.Melero JA, Moore ML. In: Challenges and opportunities for respiratory syncytial virus vaccines. Anderson LJ, Graham BS, editors. Springer; Berlin, Heidelberg: 2013. Influence of respiratory syncytial virus strain differences on pathogenesis and immunity; pp. 59–82. Berlin Heidelberg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branche AR, Walsh EE, Formica MA, Falsey AR. Detection of respiratory viruses in sputum from adults by use of automated multiplex PCR. J Clin Microbiol. 2014;52:3590–3596. doi: 10.1128/JCM.01523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soller J, Jennings W, Schoen M, et al. Modeling infection from SARS-CoV-2 wastewater concentrations: promise, limitations, and future directions. J Water Health. 2022;20:1197–1211. doi: 10.2166/wh.2022.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huisman JS, Scire J, Caduff L, et al. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. Environ Health Perspect. 2022;130 doi: 10.1289/EHP10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Liang M, Gao L, et al. Face masks to prevent transmission of COVID-19: a systematic review and meta-analysis. Am J Infect Control. 2021;49:900–906. doi: 10.1016/j.ajic.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer JA, Shutt DP, Moser SK, et al. Distinguishing viruses responsible for influenza-like illness. J Theor Biol. 2022;545 doi: 10.1016/j.jtbi.2022.111145. [DOI] [PubMed] [Google Scholar]
- 29.Nickbakhsh S, Mair C, Matthews L, et al. Virus–virus interactions impact the population dynamics of influenza and the common cold. Proc Natl Acad Sci USA. 2019;116:27142–27150. doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya S, Gesteland PH, Korgenski K, Bjørnstad ON, Adler FR. Cross-immunity between strains explains the dynamical pattern of paramyxoviruses. Proc Natl Acad Sci USA. 2015;112:13396–13400. doi: 10.1073/pnas.1516698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Wastewater data are immediately available publicly at the Stanford Digital Repository. State-aggregated and county-aggregated sentinel laboratory surveillance data are available upon request from California Department of Health (InfluenzaSurveillance@cdph.ca.gov). COVID-19 disease occurrence data are available publicly via data from all laboratories (not just sentinel laboratories) that provide PCR testing of patient specimens.