Abstract
Background
Recently many long-term prospective studies have involved serial collection and storage of blood or tissue specimens. This has spurred nested case-control studies that involve testing some specimens for various markers that might predict cancer. Until now there has been little guidance in statistical design and analysis of these studies.
Methods
To develop statistical guidelines, we considered the purpose, the types of biases, and the opportunities for extracting additional information.
Results
The following guidelines:
(1) For the clearest interpretation, statistics should be based on false and true positive rates – not odds ratios or relative risks
(2) To avoid overdiagnosis bias, cases should be diagnosed as a result of symptoms rather than on screening.
(3) To minimize selection bias, the spectrum of control conditions should be the same in study and target screening populations.
(4) To extract additional information, criteria for a positive test should be based on combinations of individual markers and changes in marker levels over time.
(5) To avoid overfitting, the criteria for a positive marker combination developed in a training sample should be evaluated in a random test sample from the same study and, if possible, a validation sample from another study.
(6) To identify biomarkers with true and false positive rates similar to mammography, the training, test, and validation samples should each include at least 110 randomly selected subjects without cancer and 70 subjects with cancer.
Conclusion
These guidelines ensure good practice in the design and analysis of nested case-control studies of early detection biomarkers.
Background
Most current methods of cancer early detection, such as mammography or cervical cytology, are based on anatomic changes in tissues or morphologic changes in cells. Recently, various molecular markers, such as protein or genetic changes have been proposed for cancer early detection [1-4]. This has spurred many investigators with long-term cohort studies to serially collect and store blood or tissue specimens. The aim is to later perform a nested-case control study, where specimens from subjects with a particular type of cancer (cases) and specimens from a random sample of subjects without the cancer (controls) are tested for various molecular markers. Sometime this sort of study is called a retrospective longitudinal study [6] although retrospective longitudinal data could arise in other ways, as well. Unlike cross-sectional study designs, the markers are measured on specimens collected well before the onset of clinical disease in cases. This avoids the potential confounding effect of the target disease on the marker.
For example, in the ATBC (alpha-tocopherol, beta-carotene) [7] and CARET [8] studies, subjects were randomized to placebo or drug to in a long-term study to determine the effect of the drug on lung cancer mortality. During the course of the trial serum was serially collected and stored in a biorepository. In a subsequent nested case-control study, stored serum samples from all cases of prostate cancer and a random sample of controls were tested for prostate-specific antigen (PSA).
Importantly the nested case-control study of early detection biomarkers may be distinct from the original long-term study from which serum were collected. It is designed to answer a different question, it typically studies subjects with a different disease, and it often ignores the intervention in the original long-term study.
Methods
We had three considerations in formulating appropriate guidelines. First we wanted to link the analysis to the goal of study, namely, to help decide on further study of the biomarker as a trigger for early intervention. Second we wanted to minimize possible biases in the selection of cases and the controls and in the investigation of many markers. Third, we wanted to extract as much information as possible relevant to the evaluation.
Results
We offer the following guidelines for the design and analysis of nested case-control studies of early detection cancer biomarkers.
1. For the clearest interpretation, statistics for binary markers should be based on true and false positive rates or predictive values based on the true prevalence – not odds ratios, relative risks, or predictive values based on the prevalence in the study
A promising marker should have a high degree of accuracy in discriminating between subjects who are likely to get cancer from those who are not. For a binary marker, which is either positive or negative, the basic measures of this type of accuracy are the true positive and false positive rates. Consider the hypothetical data in the Table 1 from a nested case-control study of early detection biomarkers. The true positive rate (TPR), or the test sensitivity, is the probability the marker is positive given cancer. The false positive rate (FPR), or 1 – specificity, is the probability the marker is positive given no cancer. In Table 1, TPR is estimated by 80/100 = .80, and FPR is estimated by 10/1000 = 01. For a perfect test, TPR = 1 and the FPR = 0. There is always a tradeoff between the TPR and FPR, so it is meaningless to assess one without assessing the other. For example, one could make the TPR equal to 1 simply by classifying every subject as positive for cancer. But this would be a poor classification rule because the FPR would also be 1.
Table 1.
Hypothetical data for a binary marker from a nested case-control study
| Marker - | Marker + | Total | |
| No cancer | 990 | 10 | 1000 |
| Cancer | 20 | 80 | 100 |
True positive rate (TPR) = 80/100 = .80 = sensitivity False positive rate (FPR) = 10/1000 = .01 = 1 – specificity
Guidelines for FPR and TPR
Because the underlying prevalence of cancer is so low in average risk populations, for acceptable cancer screening of asymptomatic people, the FPR should be very small [9]. As a starting point we recommend basing guidelines for FPR and TPR on the FPR and TPR for mammography, which, as discussed below, is FPR=.01 and TPR=.80. A mammogram is analogous to a biomarker test for cancer but there is extra information from various studies that makes it useful for setting guidelines.
One reason for using FPR and TPR from mammography as a standard is that, unlike biomarker measurements in nested case-control studies, in mammography studies there is a biopsy at the time of a positive test. This biopsy is the gold standard for determining cancer status in subjects with positive mammograms and is used for computing TPR in a way not possible with biomarkers in nested case-control studies. The TPR for mammography is the probability of a positive biopsy as a direct result of mammography in women with cancer and is estimated via mathematical models or data collected after following subjects not biopsied. As discussed in Baker and Pinsky [10], estimates ranged from .74 to 1.00 with .80 a conservative value.
Importantly the estimated TPR for mammography is not likely to be affected by overdiagnosis, which means that some screening-detected cancers would never have caused medical problems during the patient's life [11]. This would make the biomarker appear more promising than actually the case. Results from the HIP screening trial of mammography and clinical self-examination [12] suggest that if there were overdiagnosis with mammography, it would be relatively small. At the time of the last breast screening in the HIP trial, there were more cancers in the group randomized to screening than in the controls. But with further follow-up the number of cancers in the control group eventually equaled the number in the screened group, which would not have occurred if there were substantial overdiagnosis.
A second reason for using FPR and TPR from mammography is that, based on various randomized trials with cancer mortality endpoint, mammography is generally considered an acceptable screening modality. The implication is that a similar FPR and TPR for a biomarker would lead to an acceptable screening modality. For a particular biomarker, these target values of FPR and TPR from mammography may need modification depending on various factors. One factor is the invasiveness of a follow-up procedure to investigate a positive test (e.g. needle biopsy of the prostate to investigate an abnormal PSA versus laparotomy to investigate an abnormal CA125). The more invasive the follow-up procedure, the lower the FPR must be to gain acceptance in practice. A second factor is additional work-up prior to a biopsy. If a positive biomarker is unlikely to trigger additional diagnostic work-ups prior to biopsy, a higher FPR might be acceptable.
One caveat when using FPR from mammography is to be careful as to its definition. The restricted definition is the probability of a positive biopsy as a direct result of mammography in women without cancer. The less restricted definition is the probability of a suspicious mammogram warranting additional diagnostic follow-up of any type in women without cancer. Typically nested-case control studies of early detection biomarkers do not provide information on additional diagnostic follow-up. Therefore they cannot be used to estimate a less restricted FPR involving diagnostic follow-up. However, because nested case-control biomarker studies provide data on cancer diagnosis, they can be used to estimate a more restricted FPR based on unnecessary biopsies. Therefore the target FPR is based on the more restricted definition of FPR in mammography. For mammography the more restricted FPR is estimated by the fraction of women screened by mammography who received a biopsy in which no cancer was detected. As discussed in Baker and Pinsky [10], estimates of FPR from three studies ranged from .005 to .013 with a middle value of around .010.
Inappropriateness of odds ratio and relative risk
When evaluating binary early detection markers, it is inappropriate to report an odds ratio or relative risk, as is common in epidemiology or clinical trials. Because an odds ratio or relative risk is a single number, it does not capture the tradeoff between correctly classifying cancer and incorrectly classifying non-cancers. Also the odds ratio or relative risk can lead to an overoptimistic impression of the performance of an early detection test if the interpretation is based on experience in epidemiology or clinical trials. In the latter settings an odds ratio of 3 is often considered large. Much larger odds ratio are needed from early detection tests for useful application in the screening setting [13]. For example, for the target values of FPR = .01 and TPR = .80, the odds ratio equals (TPR × (1-FPR)/ ((1-TPR) × FPR) = 396, as in Table 1.
Appropriate computation of predictive values
It is sometimes useful to use the FPR and TPR to compute the predictive value negative (PVN), the probability of no disease if the marker is negative, and the predictive value positive (PVP), the probability of disease if the marker is positive. For cancer screening, it is the PVP that is most important to the physician in clinical decision-making. Because the likelihood of any individual cancer type in an asymptomatic person is nearly always very low, a negative early detection test usually adds little information to the clinical impression. The computation of the PVN and PVP depend on the prevalence of cancer as well as on the FPR and the TPR, as shown below,
PVP = (TPR × prevalence) / (TPR × prevalence + FPR (1-prevalence))
PVN = (1-FPR) × (1-prevalence) / ((1-TPR) × prevalence + (1-FPR) (1-prevalence))
When the prevalence is small, as with cancer screening, a small absolute change in FPR, but not TPR, can have a large impact on the PVP. It is erroneous to directly compute the prevalence from data in the nested case-control study, as in Table 1, because the ratio of cases to controls in the study is not the same as in the population. For example suppose FPR = .01, TPR = .80, and prevalence = .003. We obtain,
PVP = = (.8 × .003) /(.8 × .003 + .01 .997)= .19
PVN = (.99 × .997) / (.2 × .003 + .99 × .997) = .999
If we had incorrectly substituted the apparent prevalence in Table 1 of 100/1100 = .091, we would have incorrectly computed PVP = .89 and PVN = .980. For this reason, calculation and reporting of PVP and PVN using only data from a nested case-control study is not useful or appropriate.
Extension to ordered categories via ROC curves
Many markers for the early detection of cancer can be reported as ordered categories. Some markers, such as sputum cytology, inherently involve ordered categories, such as no evidence of cancer, slight atypia, moderate atypia, severe atypia, and frank cancer. Other markers, such as PSA, involve a continuous measure for which higher values indicate a greater probability of cancer. Dividing these continuous measures into ranges (either based on predetermined values or percentiles) gives ordered categories.
With ordered categories, the statistics should still be based on FPR and TPR. Each ordered category is a possible cutpoint, where values equal to or higher than the cutpoint are called positive and values lower than the cutpoint are called negative. For each cutpoint, one can compute FPR and TPR (Table 2) and generate a receiver-operating characteristic (ROC) curve, which is a plot of TPR versus FPR for the various pairs [14]. (See Figure 1). The higher and farther left the points on the ROC curve the better the test performance. As mentioned previously, acceptable cancer screening requires very small false positive rates. Therefore, for evaluating cancer biomarkers, we are only interested in the leftmost sliver of the ROC curve in Figure 1.
Table 2.
Hypothetical Data for an Ordered Marker From a Nested Case-Control Design
| 1 | 2 | 3 | 4 | 5 | ||
| No cancer | 960 | 20 | 10 | 8 | 2 | 1000 |
| Cancer | 0 | 10 | 10 | 20 | 60 | 100 |
For cutpoint 4, the true positive rate is (20+60)/100 = .80 For cutpoint 4, the false positive rate is (8 + 2)/1000 = .01
Figure 1.
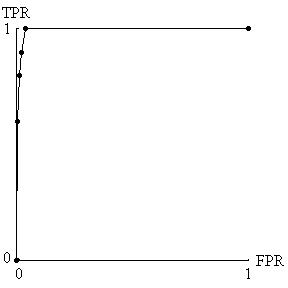
ROC curve derived from hypothetical data in Table 2. Because acceptable cancer screening requires a very small false positive rate (FPR) and a reasonable true positive rate (TPR), only the four points farthest to the left are relevant for biomarker evaluation.
2. To avoid overdiagnosis bias, cases should be diagnosed as a result of symptoms rather than on screening
For the TPR in the biomarker study to reflect the true TPR, cases should be diagnosed as a result of symptoms rather than on screening. For example in the study of PSA in the ATBC trial [7], cases were subjects diagnosed with prostate cancer as a result of symptoms. If the prostate cancer cases were detected as the result of screening, say with ultrasound, the TPR could be artificially elevated if there were overdiagnosis, as previously discussed.
3. To minimized selection bias, the spectrum of control conditions should be the same in study and target screening populations
For the FPR in the nested case-control study to reflect the true FPR in the target population, the spectrum of control conditions should be the same as in the target population. By control conditions, we mean characteristics of the population, such as the presence of other diseases or certain known risk factors that could elevate the false positive rate.
The spectrum of conditions could differ considerably if the retrospective biomarker study were embedded in a randomized trial with strict eligibility requirements. For example, consider a biomarker for the early detection of lung cancer where the data comes from a biorepository arising from a randomized trial of healthy subjects. It would be inappropriate to apply the results to a population with a high prevalence of chronic obstructive lung disease, bronchitis, or viral pnuemonitis because these conditions could increase the number of positive readings in subjects without lung cancer. Because FPR is very small for screening to be acceptable, this spectrum bias could have important consequences in a clinical application.
It would not always be possible to identify all relevant control conditions, but to the extent possible, the control conditions should be similar in both populations.
4. To extract additional information, criteria for a positive test should be based on combinations of individual markers and changes in marker levels over time
Data from multiple markers present an opportunity to extract potentially valuable information not available by analyzing markers separately. Consider the hypothetical data in Table 3. The left side corresponds to subjects without cancer, and the right corresponds to subjects with cancer. In our idealized circumstance, markers A and B are independent for subjects without cancer and are perfectly correlated for subjects with cancer. The indicated region for A = 2 or A = 3 and B = 2 or B = 3 designates a positive test that has FPR = .04 and the TPR = .80. Suppose that marker A and marker B were evaluated separately. The indicated region corresponding to A only, namely A = 2 or A = 3 and the indicated region corresponding to B only, namely B = 2 and B = 3, each designates a positive test that has FPR=. 20 and TPR=. 80. Thus, in this particular example, the combination of markers leads to a much better test than the separate markers, with a smaller FPR (.04 versus .20) for the same TPR of .80. One could create a similar example with any number of marker levels. For an ideal test in which FPR = 0 and TPR = 1, the region would encompass all subjects with cancer but no subjects without cancer.
Table 3.
Hypothetical data for two ordered markers, A and B
| Subjects without cancer (N = 1000) | Subjects with cancer (N = 100) | |||||||
| A = 1 | A = 2 | A = 3 | B only | A = 1 | A = 2 | A = 3 | B only | |
| B = 1 | 640 | 80 | 80 | 800 | 20 | 0 | 0 | 20 |
| B = 2 | 80 | 10a | 10a | 100e | 0 | 20b | 0b | 20f |
| B = 3 | 80 | 10a | 10a | 100e | 0 | 0b | 60b | 60f |
| A only | 800 | 100c | 100c | 20 | 20d | 60d | ||
For A and B combined; aFalse positive rate (FPR) of indicated region (10+10+10+10)/1000=. 04; bTrue positive rate (TPR) of indicated region = (20 +0+0+60)/100=. 80 For A only; cFalse positive rate (FPR) of indicated region =(100+100)/1000=. 20; dTrue positive rate (TPR) of indicated region = (20+60)/100=. 80 For B only; eFalse positive rate (FPR) of indicated region =(100+100)/1000= .20; fTrue positive rate (TPR) of indicated region = (20+60)/100=.80
A real application of how marker combinations provide extra information comes from a recent study by Mok et al [15] on CA125 and Prostasin as markers for ovarian cancer. Although the data are not from a nested case-control study and have not been validated in subsequent studies, they are useful for illustration. Based on Figure 5 of Mok et al [15] with approximate values for the outpoints, we investigated regions with FPR=.02. The combination for a positive test of CA 125 greater than or equal to 40 U/ml and Prostasin greater than or equal to 5 μg/ml had FPR = .02 and TPR = .92. Evaluating CA 125 alone, the criterion for a positive test of CA 125 greater than 80 U/ml had FPR=.02 and TPR =.78. Evaluating Prostasin alone, the criterion for a positive test of Prostasin greater than 15 μg/ml had FPR=. 02 and TPR=. 32. Thus, in this real example, the combination of markers led to a better test than the separate markers, with a larger TPR (.92 versus .78 or .32) for the same FPR of .02.
To more generally compare performance of a combination of markers with a single marker, we need to compare ROC curves. Creating an ROC curve from a combination of markers is different from creating an ROC curve from a single marker. With a combination of markers, the number of possible regions, as in Table 3 a, for calling a marker positive is extremely large. Some choices of regions correspond to AND rules, for example A>1 and B>1, as in Table 3 a. Some choices of regions correspond to OR rules, for example A>2 or B>2. Other choices are also possible, but for biological reasons, one would usually require all regions to be contiguous. A plot of the FPR and TPR for each region would lead to a cloud of points, rather than the smooth curve in Figure 1. To create the best ROC curve, one should select those points that are highest and farthest to the left, which is generally all that would need to be presented and only for small false positive rates. For complicated situations, Baker [16] proposed an algorithm to select the regions creating the best ROC curve without the need to enumerate all the regions. Mathematically, in any sample of data, the best ROC curve for a combination of markers must be as good or better than the ROC curve for any of the markers evaluated separately. The reason is that the set of possible regions for calling a combination of markers positive includes as a special case the regions for calling any single marker positive.
Alternative approaches that do not directly optimize the ROC curve include linear logistic regression or linear discriminant analysis [17], which choose regions based on linear combinations of the markers, and neural networks [18], which choose regions in a very complicated nonlinear manner. Due to the potential for overfitting (to follow), it is not possible to make a blanket statement as to which approach for choosing regions is best. If one takes the set of regions for calling a combination positive that gives a good ROC curve in a random sample of data, it may give a poor ROC curve in another random sample of the data, simply due to selecting chance patterns in the first sample. As discussed in the section on overfitting, this motivates splitting the data into two random samples, training and test, and using the regions from the training sample to compute the definitive ROC curve based on data in the test sample.
Changes in marker values over time also provide potentially valuable information not available when examining markers at a single time point. With marker measurements at two different times per subject (and approximately the same interval between times), a common summary measure is the slope. If investigators believe that both slope and baseline level predict cancer, the combination can be evaluated using the previously discussed methods for evaluating multiple markers, namely, treating baseline level and slope as two "separate" markers. With measurements at more than two times per subject, investigators may identify a more complicated feature, such as whether or not there is a sudden increase in marker levels [19].
When biomarker measurements occur at regular time intervals (and allowing different numbers of measurements for each subject), one can estimate TPR and FPR by using a first order Markov chain in reverse time, as described by Baker and Tockman [20] for the analysis of precancerous lesions for lung cancer.
5. To avoid overfitting, the criteria for a positive marker combination developed in a training sample should be evaluated in a random test sample from the same study and, if possible, a validation sample from another study.
With a single marker and a large number of subjects, there is usually no concern with overfitting. However with many combinations of markers, overfitting could invalidate results. Overfitting is often associated with step-wise regression models [21] but it can occur in other situations as well. Overfitting of a larger number of markers to a relatively small number of subjects produces a model that is overly sensitive to chance fluctuations in the data. As a simple example, overfitting occurs when a sports announcer reports that a baseball player had a very high batting average against left-handed pitchers in ballpark X over the past month. This average is not very reliable for future predictions because the particular set of factors, left-handed pitchers and ballpark X, were selected to give a high average. In reality the high average is more likely the result of chance factors that coincided with left handed pitchers at ballpark X during that particular month. One way to adjust for overfitting is to apply the factors in another sample not used for initial reporting. The first sample is known as the training sample, and the second sample is known as the test sample. For example, the prediction of the average against left handed pitchers in the ballpark X could be tested on data from a different month. Similarly, a standard statistical approach to adjust for overfitting is to randomly split the data into a training and test samples. This is called the split-sample approach. Promising marker combinations are identified in the training sample, but more reliable FPR and TPR measurements are made in the test sample because it involves different data. Baker [16] used the split-sample approach to evaluate the performance of four markers for prostate cancer.
More sophisticated adjustments for overfitting include cross-validation and bootstrapping. Suppose the interest lies in a single statistic, such as the area under the part of the ROC curve corresponding to a small FPR. In a typical cross-validation analysis (i) the data are randomly split into ten samples each with 10% of the data, (ii) one of the 10% samples is deleted, (iii) the statistic is computed by applying the classification rule recalculated from the remaining 90% of the sample to the deleted 10% sample and (iv) an average is taken of the statistic over all ten 10% samples [22]. Although one could use a different fraction than 10%, ROC computation it is not possible with the most extreme version of cross-validation that involves leaving out one subject at a time. With bootstrapping, (i) the entire data set is resampled with replacement many times, (ii) on each resampling the statistics is computed, and (iii) the statistics from each resampling are combined in a special way [22]. In a recent study comparing these adjustments with other types of statistics in a different setting, Steyerberg et al [22] found that the split-sample approach tended to underestimate performance, cross-validation performed poorly on some statistics that were not normally distributed, and bootstrapping performed best overall. For our purposes of estimating an entire ROC curve rather than a summary statistic, more research is needed for cross-validation and bootstrapping, as it is not clear how best to combine ROC curves over different samples.
Regardless of the method used to adjust for overfitting in forming a classification rule, to obtain the most reliable FPR and TPR measurements, the classification rule should ideally be evaluated in a validation sample from a different study, as in Baker [16].
6. To identify biomarkers with true and false positive rates similar to mammography, the training, test, and validation samples should each include at least 110 randomly selected subjects without cancer and 70 subjects with cancer (as based on FPR and TPR for mammography)
The sample size is based on the need to determine if the biomarker is sufficiently promising for investigation as a trigger for early intervention in a future trial. As discussed previously, based on considerations from mammography, our target values are FPR = .01 and TPR= .80. In most situations, we think it would be of interest to specify a 95% confidence interval for TPR of (.70, .90). Using a normal approximation the target standard error is approximately .05. Setting the standard error of TPR, TPR × (1-TPR) / (square root of n), equal to .05 and solving for the sample size n, we obtain n= 64, which we round up to 70. In addition we think that in most situations the largest reasonable value of FPR would be .03 which is 3 times the number of false positives as with mammography screening. Because FPR is so small, we do not use a normal approximation. We specify a sample size of n = 110, so that under the binomial distribution with FPR=.01, the upper 2.5% bound equals .03 × 110. Strictly, these sample sizes apply only after a single criterion for a positive test has been identified. For a training sample, one might consider larger sample sizes.
Conclusion
A major advantage of nested case-control studies for early detection biomarkers is that they can be done quickly if serum from a long-term study has been stored in a biorepository. Importantly the retrospective aspect does not compromise the validity. There are none of the usual problems with retrospective studies such as recall bias. Thus we anticipate that in the coming years, there will be many reports in literature from studies of this design.
These guidelines should greatly help investigators design and analyze nested case-control studies for early detection biomarkers and help readers of the literature to interpret them. It bears emphasis that these studies do not prove clinical efficacy of the markers. Rather, they suggest which markers or marker combinations are the most promising candidates for further study as a trigger for early intervention in definitive trials with cancer-mortality endpoints.
Competing interests
None declared
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgement
The authors thank David Ransohoff for his helpful comments.
Contributor Information
Stuart G Baker, Email: sb16i@nih.gov.
Barnett S Kramer, Email: bk76p@nih.gov.
Sudhir Srivastava, Email: ss1a@nih.gov.
References
- Xu Y, Shen ZZ, Wiper DW, Wu MZ, Morton RE, Elson P, Kennedy AW, Belinson J, Markman M, Casey G. Lysophosphatidic acid as a potential biomarker for ovarian and other gynecologic cancers. JAMA. 1998;280:719–723. doi: 10.1001/jama.280.8.719. [DOI] [PubMed] [Google Scholar]
- Signorello LB, Brismar K, Bergstrom R, Andersson SO, Wolk A, Trichopoulos D, Adami HO. Insulin-like growth factor-binding protein-1 and prostate cancer. Journal of the National Cancer Institute. 1999;91:1965. doi: 10.1093/jnci/91.22.1965. [DOI] [PubMed] [Google Scholar]
- You WC, Blot WJ, Zhang L, Kneller RW, Li JY, Jin ML, Chang YS, Zeng XR, Zhao L, Fraumeni JF, Xu GW, Samloff MI. Serum pepsinogens in relation to precancerous gastric lesions in a population at high risk for gastric cancer. Cancer Epidemiology, Biomarkers, and Prevention. 1993;2:113–117. [PubMed] [Google Scholar]
- Nam RK, Diamandis EP, Toi A, Trachtenberg J, Magklara A, Scorilas A, Papnastasiou PA, Jewett MAS, Narod SA. Serum human glandular kallikrein-2 protease levels predict the presence of prostate cancer among men with elevated prostate specific antigen. Journal of Clinical Oncology. 2000;18:1036–1042. doi: 10.1200/JCO.2000.18.5.1036. [DOI] [PubMed] [Google Scholar]
- Langholz B. Case-control study, nested. In: Armitage P, Colton T, editor. In The Encyclopedia of Biostatistics, Chichester: John Wiley & Sons; 1998. pp. 514–519. [Google Scholar]
- Pepe MS, Etzioni R, Feng S, Potter JD, Thompson ML, Thornquist M, Winget M, Yasui Y. Phases of biomarker development for early detection of cancer. Journal of the National Cancer Institute. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- The ATBC Cancer Prevention Study Group The Alpha-Tocopherol, Beta-Carotene Lung Cancer Prevention Study: Design, methods, participant characteristics, and compliance. Annals of Epidemiology. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Omenn GS, Goodman GE, Thornquist MD, Balmes J, Cullen MR, Glass A, Keogh JP, Meyskens FL, Jr, Valanis B, Williams JH, Barhart S, Cherniack MG, Brodkin CA, Hammar S. Risk factors for lung cancer and for intervention effects in CARET, the Beta-Carotene and Retinol Efficacy Trial. Journal of the National Cancer Institute. 1996;88:1550–9. doi: 10.1093/jnci/88.21.1550. [DOI] [PubMed] [Google Scholar]
- Lillienfeld AM. Some limitations and problems of screening for cancer. Cancer. 1974;35:1720–1724. doi: 10.1002/1097-0142(197406)33:6+<1720::aid-cncr2820330705>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Baker SG, Pinsky PF. A proposed design and analysis for comparing digital and analog mammography: special receiver-operating characteristic methods for cancer screening. Journal of the American Statistical Association. 2001;96:421–428. doi: 10.1198/016214501753168136. [DOI] [Google Scholar]
- Kramer BS, Brown ML, Prorok PC, Potosky AL, Gohagan JK. Prostate-cancer screening-what we know and what we need to know. Annals of Internal Medicine. 1993;119:914–923. doi: 10.7326/0003-4819-119-9-199311010-00009. [DOI] [PubMed] [Google Scholar]
- Shaprio S, Venet W, Strax P, Venet L. Periodic Screening for Breast Cancer, The Health Insurance Plan Project and Its Sequelae, 1963–1986, Baltimore, Johns Hopkins University Press.
- Emir B, Wieand S, Su JQ, Cha S. Analysis of repeated markers used to predict progression of cancer. Statistics in Medicine. 1998;17:2563–78. doi: 10.1002/(SICI)1097-0258(19981130)17:22<2563::AID-SIM952>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, Berkowitz RS, Cramer DW. Prostasin, a potential serum marker for ovarian cancer: Identification through microarray technology. Journal of the National Cancer Institute. 2001;93:1458–64. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- Baker SG. Identifying combinations of cancer markers for further study as triggers of early intervention. Biometrics. 2000;56:1082–1087. doi: 10.1111/j.0006-341x.2000.01082.x. [DOI] [PubMed] [Google Scholar]
- Gail MH, Muenz L, McIntire KR, Radovich B, Braunstein G, Brown PR, Deftos L, Dnistrian A, Dunsmore M, Elashoff R, Geller N, Go VLW, Hirji K, Klauber MR, Pee D, Petroni G, Schartz M, Wolfsen AR. Multiple markers for lung cancer diagnosis: validation of models for advanced lung cancer. Journal of the National Cancer Institute. 1986;76:805–826. [PubMed] [Google Scholar]
- Hastie T. Neural networks. In: Armitage P, Colton T, editor. In The Encyclopedia of Biostatistics, Chichester: John Wiley & Sons; 1998. pp. 2986–2989. [Google Scholar]
- Morell CH, Pearson JD, Carter HB, Brant LJ. Estimating unknown transition times using a piecewise nonlinear mixed-effects model in men with prostate cancer. Journal of the American Statistical Association. 1995;90:45–53. [Google Scholar]
- Baker SG, Tockman MS. Evaluating serial observations of precancerous lesions for further study as a trigger for early intervention. Statistics in Medicine. [DOI] [PubMed]
- Harrell FE, Lee KL, Califf RM, Pryor DB, Rosati RA. Modelling strategies for improved prognostic prediction. Statistics in Medicine. 1984;3:143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- Steyerberg EW, Harrell FE, Jr, Borsboom GJJM, Eijkemans MJC, Vergouwe Y, Habbema JDF. Internal validation of predictive models; Efficiency of some procedures for logistic regression. Journal of Clinical Epidemiology. 2001;54:774–781. doi: 10.1016/S0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]


