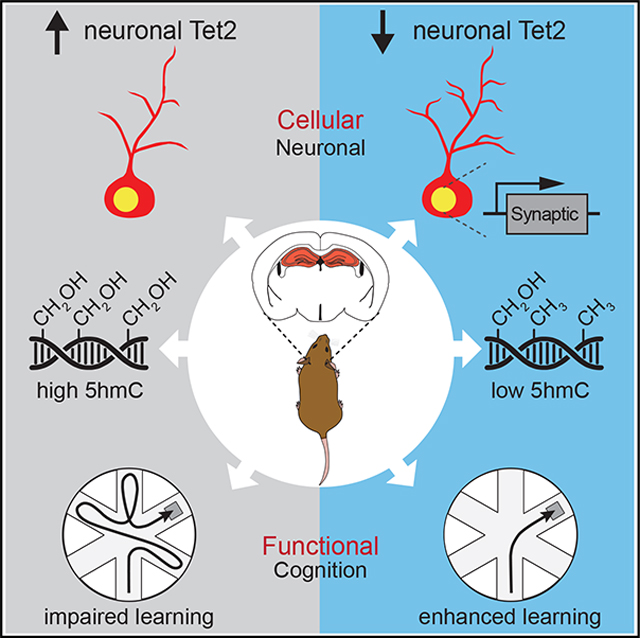
SUMMARY
DNA methylation has emerged as a critical modulator of neuronal plasticity and cognitive function. Notwithstanding, the role of enzymes that demethylate DNA remain to be fully explored. Here, we report that loss of ten-eleven translocation methylcytosine dioxygenase 2 (Tet2), which catalyzes oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), in adult neurons enhances cognitive function. In the adult mouse hippocampus, we detected an enrichment of Tet2 in neurons. Viral-mediated neuronal overexpression and RNA interference of Tet2 altered dendritic complexity and synaptic-plasticity-related gene expression in vitro. Overexpression of neuronal Tet2 in adult hippocampus, and loss of Tet2 in adult glutamatergic neurons, resulted in differential hydroxymethylation associated with genes involved in synaptic transmission. Functionally, overexpression of neuronal Tet2 impaired hippocampal-dependent memory, while loss of neuronal Tet2 enhanced memory. Ultimately, these data identify neuronal Tet2 as a molecular target to boost cognitive function.
In brief
Pratt et al. identify neuronal Tet2 as a molecular target to boost cognitive function. Tet2-mediated neuronal hydroxymethylation and transcriptional changes negatively regulate adult hippocampal synaptic processes. Increasing neuronal Tet2 impairs hippocampal-dependent memory, while loss of neuronal Tet2 enhances memory.
Graphical Abstract
INTRODUCTION
Modification of DNA by methylation is dynamically regulated in mature neurons to enable processes necessary for cognition (Jaenisch and Bird, 2003; Moore et al., 2013). DNA methylation changes occur in mature neurons at activity-dependent genes following learning paradigms, after synchronous activation by electroconvulsive stimulation, and in response to cognitive enhancement interventions such as exercise (Miller and Sweatt, 2007; Guo et al., 2011). Mice lacking the DNA methyltransferases DNMT1 and DNMT3a in mature forebrain excitatory neurons exhibit deficits in hippocampal-dependent learning and memory (Feng et al., 2010). Conversely, induction of DNMT3a in mature neurons improves memory in aged mice, indicating that increased methylation enhances engram stability (Gulmez Karaca et al., 2020). Much less is known about the functional consequences of demethylation in mature neurons.
TET1–3 iteratively oxidize 5-methyl-cytosine (5mC) to produce three stepwise intermediates, 5-hydroxymethyl-cytosine (5hmC), 5-formyl-cytosine (5fC), and 5-carboxyl-cytosine (5caC) that can be converted back to cytosine (Tahiliani et al., 2009; Ito et al., 2011). 5hmC is enriched in the brain, predominantly in neurons and on exon boundaries of active synaptic genes in humans and mice (Kriaucionis and Heintz, 2009; Khare et al., 2012). Correlative studies have indicated that 5hmC may be involved in neuronal function, with health interventions such as caloric restriction resulting in decreased 5hmC in the brain (Chouliaras et al., 2012), while patients with the neuropsychiatric disorder schizophrenia have increased 5hmC (Dong et al., 2012).
In the hippocampus, TET enzymes have emerged as critical regulators of cognitive function. Constitutive loss of Tet1 impairs memory extinction in young mice (Rudenko et al., 2013; Kaas et al., 2013), and knockdown of Tet3 in primary hippocampal neurons reduces glutamatergic synaptic scaling and plasticity (Yu et al., 2015). We and others have shown that age-related loss of Tet2 in adult neural stem cells promotes impairments in adult hippocampal neurogenesis and associated cognitive processes (Gontier et al., 2018; Li et al., 2017). However, the functional role of Tet2 in mature neurons remains unexplored.
While decreased TET expression has been implicated in cognitive impairments, here we report that loss of Tet2 in adult mature excitatory forebrain neurons enhances hippocampal-dependent memory. Using in vitro and in vivo viral-mediated neuronal overexpression and RNA interference approaches, neuron-specific conditional genetic knockout mouse models, RNA sequencing (RNA-seq), and antibody-based 5hmC immunoprecipitation combined with deep sequencing (hMeDIP-seq) approaches, we identified differentially expressed genes and hydroxymethylated regions in genes relating to synaptic processes. Increased neuronal Tet2 impaired hippocampal-dependent memory, while loss of neuronal Tet2 enhanced memory. These findings indicate that Tet2-mediated neuronal hydroxymethylation and transcriptional changes negatively regulate adult hippocampal synaptic and cognitive processes.
RESULTS
Tet2 mRNA expression is enriched in hippocampal neurons
Given neuronal enrichment of 5hmC in the brain (Szulwach et al., 2011; Globisch et al., 2010), we first characterized Tet2 mRNA expression in NeuN-positive neurons, GFAP-positive astrocytes, and IBA1-positive microglia by RNAScope in combination with immunohistochemical analysis of cell markers. We detected a significant enrichment of Tet2 in adult neurons compared with other cell types (Figure 1A).
Figure 1. Tet2 is enriched in neurons and regulates neuronal complexity.
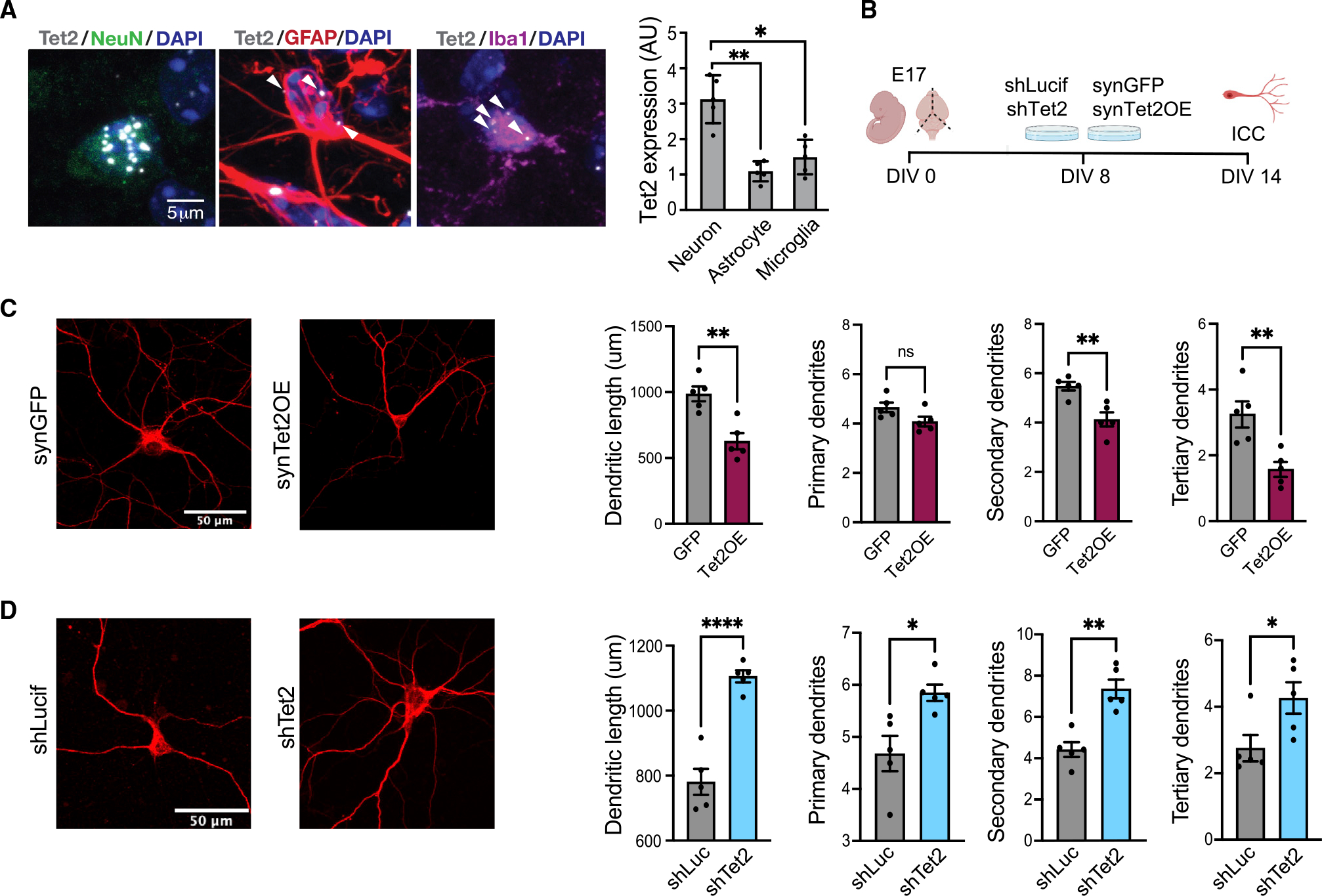
(A) Representative image and quantification of Tet2 RNA scope expression in hippocampal neurons, astrocytes and microglia of young (3 months old) adult mice (n = 5 mice with 20–40 cells per animal; one-way ANOVA; *p < 0.05, **p < 0.01).
(B) Schematic of experimental design. Primary mouse neurons were cultured for 8 days and subsequently infected with lentivirus encoding Tet2 (Tet2 OE) or GFP control sequences under the neuron-specific Synapsin1 promoter or encoding shRNA sequences targeting Tet2 (shTet2) or luciferase control (shCtrl).
(C) Representative images and quantification of dendritic length and number of primary, secondary, and tertiary dendrites for MAP2-postive neurons following viral-mediated Tet2 overexpression (n = 5 per group; t test; *p < 0.05, **p < 0.01).
(D) Representative images and quantification of dendritic length and number of primary, secondary, and tertiary dendrites for MAP2-postive neurons following viral-mediated Tet2 knockdown (n = 5 per group; t test; *p < 0.05, **p < 0.01, ****p < 0.0001).
Data are represented as mean ± SEM.
See also Figure S1.
Tet2 alters neuronal morphology and gene-expression profiles of synaptic-plasticity-related genes in vitro
We next investigated the role of neuronal Tet2 using an in vitro model. We used a cell-type-specific, viral-mediated overexpression approach in which mature primary mouse neurons were infected with lentiviral constructs encoding either Tet2 (Tet2 overexpression [OE]) or GFP control under the control of the neuron-specific Synapsin1 promoter (Figure 1B). Tet2 OE was confirmed by qPCR (Figure S1A). We assessed cellular changes by immunocytochemical analysis and detected a decrease in dendritic complexity of MAP2-positive neurons overexpressing Tet2 (Figure 1C). To complement these studies, mature neurons were infected with lentivirus encoding short hairpin RNA (shRNA) sequences targeting Tet2 (shTet2) or luciferase control (shLuc) (Figure 1B). Knockdown of Tet2 was confirmed by qPCR (Figure S1D). Decreased neuronal Tet2 expression resulted in an increase in dendritic complexity of MAP2-positive neurons (Figures 1D and S1F). No differences in neuronal cell density or toxicity were observed under OE or RNA interference conditions (Figures S1B, S1C, S1E, and S1F). These data indicate that Tet2 promotes morphological changes in mature neurons in vitro.
To gain insight into molecular changes elicited by viral-mediated OE and knockdown of Tet2, we characterized the neuronal transcriptome by RNA-seq (Figure 2A). We detected 4,057 differentially expressed genes following Tet2 OE and 3,115 differentially expressed genes following Tet2 abrogation compared with control conditions (Table S1). Gene Ontology analysis identified processes involved in synaptic vesicle recycling for Tet2 OE (Figure 2B) and synaptic signaling for Tet2 abrogation (Figure 2D). We surveyed expression of synaptic plasticity genes (Koopmans et al., 2019) and observed bidirectional changes in activity-dependent immediate-early genes that included activity-regulated cytoskeleton-associated protein (Arc), early growth response 1 (Egr1), neuronal pas domain protein 4 (Npas4), and junb proto-oncogene (JunB) following Tet2 OE (Figure 2C) and Tet2 abrogation (Figure 2E). In sum, we detected 552 genes that were bidirectionally changed by Tet2 OE and Tet2 abrogation (Figure 2F). Gene Ontology analysis of the bidirectionally changed gene set identified processes involved in neuronal projection development and regulation of synaptic vesicle recycling (Figures 2G and 2H). These data raise the possibility that neuronal Tet2 modulates synaptic signaling and plasticity.
Figure 2. Tet2 regulates synaptic-related gene expression in vitro.
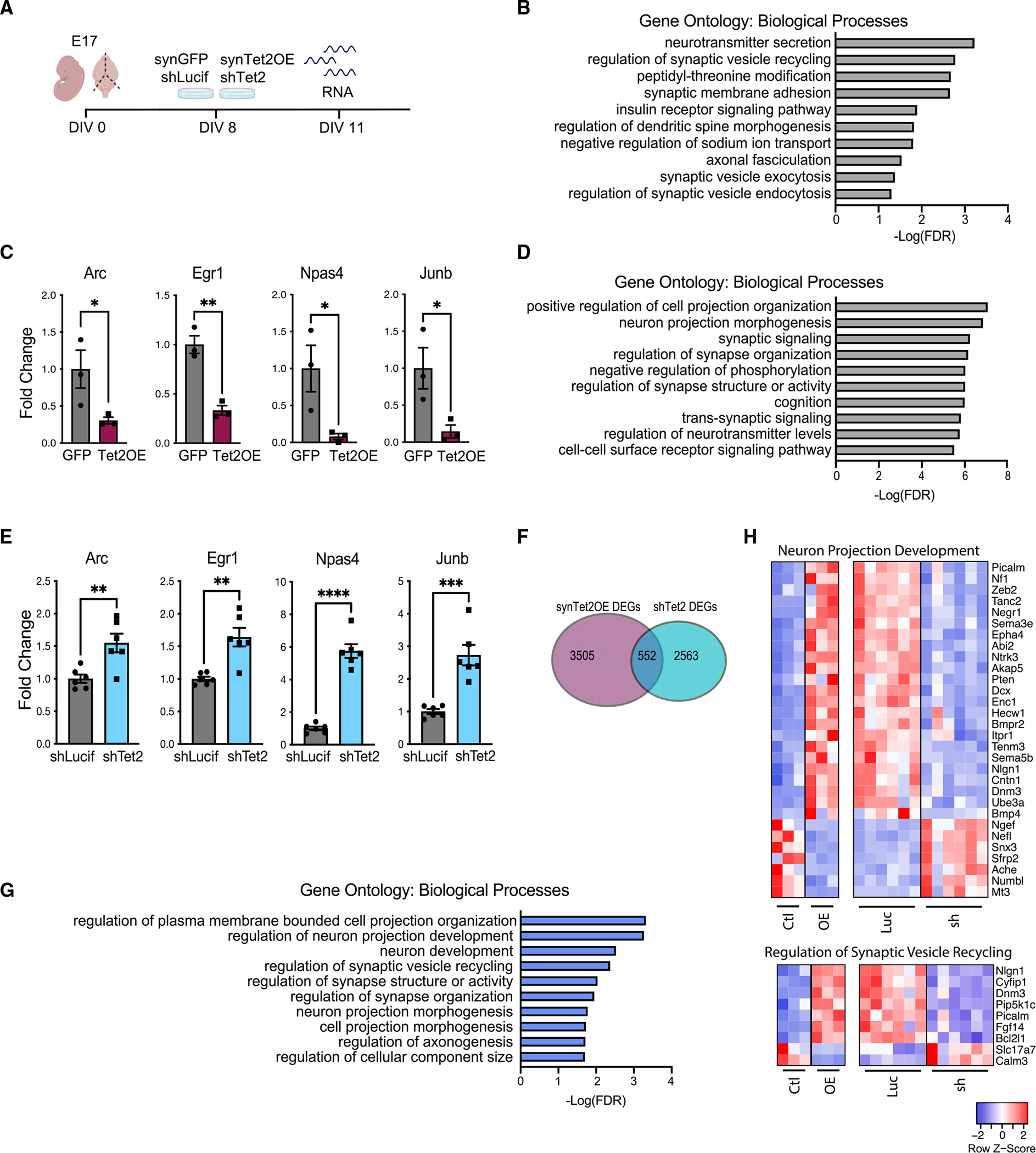
(A) Schematic of experimental design. Primary mouse neurons were cultured for 8 days and infected with lentivirus encoding Tet2 (Tet2 OE) or GFP control sequences under the neuron-specific Synapsin1 promoter or encoding shRNA sequences targeting Tet2 (shTet2) or luciferase control (shCtrl).
(B) Gene Ontology analysis of biological processes on differentially expressed genes between Tet2 OE and control.
(C) Fold change of representative genes from Tet2 OE RNA-seq dataset (n = 3 per group; adjusted p value with Bonferroni’s multiple testing hypothesis correction, *p < 0.05, **p < 0.01).
(D) Gene Ontology analysis of biological processes on differentially expressed genes between shTet2 and control.
(E) Fold change of representative genes from shTet2 RNA-seq dataset (n = 6 per group; adjusted p value with Bonferroni’s multiple testing hypothesis correction,**p < 0.01, ***p < 0.001, ****p < 0.0001).
(F) Venn diagram of differentially expressed genes from Tet2 OE and shTet2 RNA-seq datasets and overlap of bidirectional differentially expressed genes between conditions. Significance threshold set at q <0.05 with absolute fold change >0.5. Overlap p < 1.579 × 10−148 by hypergeometric test.
(G) Gene Ontology analysis of biological process on the 552 genes overlapping between the Tet2 OE and shTet2 RNA-seq datasets.
(H) Heatmaps of differentially expressed genes from the overlapping gene set in the regulation of neuron projection development (top) and regulation of synaptic vesicle recycling (bottom) biological process categories.
Data are represented as mean ±
See alsoTable S1.
Increased Tet2 in adult hippocampal neurons alters hydroxymethylation on genes related to synaptic transmission and synaptic signaling in vivo
To assess the effect of increasing neuronal Tet2 in vivo, young adult wild-type mice were given bilateral stereotaxic injections of high-titer virus encoding Tet2 or GFP under the control of the neuron-specific Synapsin1 promoter into their hippocampi (Figure 3A). NeuN-positive neuronal nuclei were isolated by fluorescence-activated cell sorting (FACS) from the hippocampi (Figures S2A and S2B). We used hMeDIP-seq to assess the influence of Tet2 OE on the DNA hydroxymethlyation landscape in vivo. OE of Tet2 in hippocampal neurons led to 270 hMeDIP peaks being gained, while 29 were lost (Figure 3B). Gained hMeDIP peaks were distributed across the genome (Figure 3C; Table S2) and were enriched in CpG islands, exons, neuronal proximal enhancers, and promoters (Figure 3D), suggesting a functional role in neurons. Performing Gene Ontology analysis of genes associated with the gained hMeDIP peaks (Figure 3E), we find enrichment of terms related to synaptic transmission, regulation of ion channel activity, and synaptic signaling (Figure 3F). Several of these genes have well-established roles in neuronal function and behavior, such as fibroblast growth factor 14 (Fgf14), Slit- and Trk-like protein (Slitrk2), and DLG-associated protein 2 (Dlgap2) (Figure 3G).
Figure 3. Increased neuronal Tet2 in the adult hippocampus alters hydroxymethylation on genes related to synaptic processes and impairs memory.
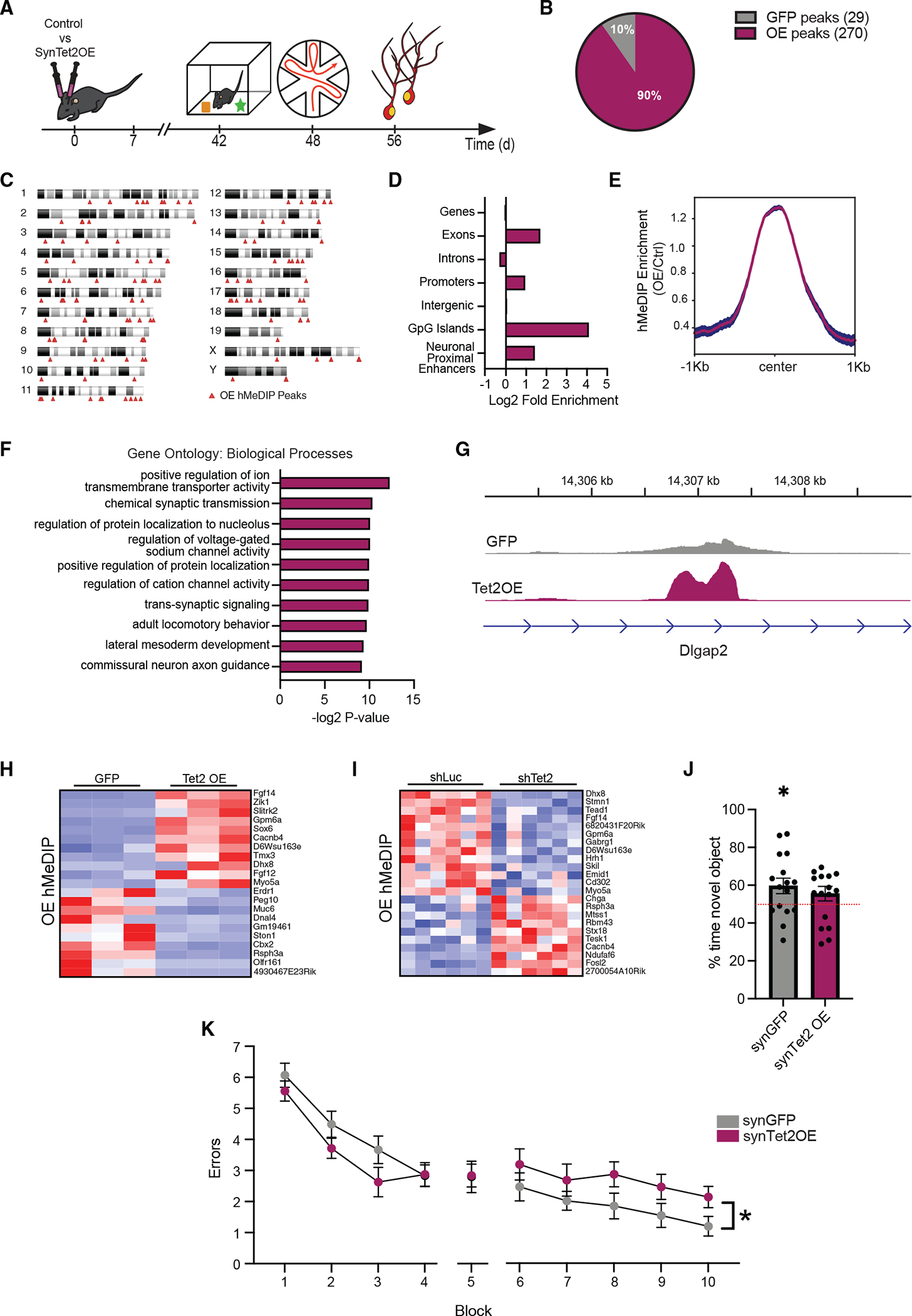
(A) Schematic of experimental paradigm. Adult (3–4 months old) wild-type mice were given bilateral stereotaxic injections of lentivirus (LV) encoding either Tet2 (Tet2 OE) or GFP control sequences driven by the neuron-specific Synapsin1 promoter into the hippocampus and subject to behavioral and molecular analysis 5 weeks later.
(B) Pie chart of 5hmC peaks gained (270 peaks) and lost (29 peaks) in Tet2 OE neurons compared with GFP control (q = 0.05).
(C) Ideogram of genomic location of hMeDIP peaks gained following neuronal Tet2 overexpression.
(D) Enrichment of gained hMeDIP peaks over genomic elements in Tet2 OE neurons.
(E) Metagene profile of gained hMeDIP peaks following neuronal Tet2 overexpression. Mean ± SEM.
(F) Gene Ontology analysis of biological processes on the 131 genes associated with the 270 gained hMeDIP peaks following neuronal Tet2 overexpression.
(G) Example gene track of the gained hMeDIP peak associated with Dlgap2 in Tet2 OE neurons.
(H) Heatmap of differentially expressed genes in the in vitro Tet2 OE RNA-seq dataset that have associated gained hMeDIP peaks in the in vivo Tet2 OE hMeDIP-seq dataset.
(I) Heatmap of differentially expressed genes in the in vitro shTet2 RNA-seq dataset that have associated gained hMeDIP peaks in the in vivo Tet2 OE hMeDIP-seq dataset.
(J) Quantification of percentage of interaction time during novel object recognition testing (n = 15–16 mice per group; t test, *p < 0.05).
(K) Quantification of the number of entry errors during radial arm water maze training and testing (n = 15–16 mice per group; repeated-measures ANOVA with Bonferroni post hoc correction, *p < 0.05).
Data are represented as mean ± SEM
To determine the effects of hydroxymethylation changes on gene expression in neurons, we compared our in vivo Tet2 OE hMeDIP-seq dataset with our in vitro OE and knockdown RNA-seq datasets (Figure 3H and 3I). We find that 21 genes that were significantly altered following Tet2 OE in vitro also gained hMeDIP peaks with Tet2 OE in vivo (Figure 3H), while 23 genes significantly changed following Tet2 knockdown in vitro had associated hMeDIP peaks in vivo (Figure 3I). Several of these genes are bidirectionally changed by Tet2 OE and abrogation and regulate neuronal function, such as Fgf14, glycoprotein M6A (Gpm6a), and Myosin 5a (Myo5a). These results suggest that Tet2-mediated hydroxymethylation modulates expression of genes important for neuronal function.
Increased Tet2 in adult neurons impairs hippocampal-dependent memory
To investigate the effect of increased neuronal Tet2 on cognitive processes, hippocampal-dependent learning and memory was assessed using novel object recognition (NOR) and radial arm water maze (RAWM) paradigms in young adult mice following viral-mediated neuronal Tet2 OE in the hippocampus (Figure 3A). During NOR testing, control mice were biased toward a novel object relative to a familiar object compared with mice overexpressing neuronal Tet2 (Figure 3J). In RAWM, all mice showed similar learning capacity during the training phase (Figure 3K). Increased neuronal Tet2 resulted in significantly more errors in locating the target platform during spatial memory testing compared with control mice (Figure 3K). As a control, we profiled general health using an open-field paradigm (Figure S2C) and observed no differences in overall activity, total distance traveled, or time spent in the center of the open field, indicative of normal motor and anxiety functions (Figure S2D). These behavioral data indicate that increased neuronal Tet2 impairs hippocampal-dependent recognition memory and spatial memory.
Loss of Tet2 in adult excitatory neurons alters hydroxymethylation on genes related to synaptic transmission and synapse structure in vivo
To investigate the effect of abrogating neuronal Tet2 in vivo, we generated Tet2flox/flox mice carrying an inducible CamK2a-Cre-ERT2 gene, in which Tet2 is excised specifically in adult glutamatergic forebrain neurons upon tamoxifen administration (Tet2 conditional knockout [cKO]) (Figure 4A). Young adult Tet2 cKO and littermate control (Tet2flox/flox) mice were administered tamoxifen, and molecular changes were assessed at 8 months of age. Decreased Tet2 expression was confirmed in neuronal nuclei isolated from Tet2 cKO by qPCR (Figure S3A). No differences in Tet1 and Tet3 expression were observed between genotypes (Figure S3B). We used hMeDIP-seq to assess how loss of neuronal Tet2 affects the DNA hydroxymethlyation landscape in vivo. We discovered 241 hMeDIP peaks with reduced signal when comparing KO with wild-type neurons, while there were 103 hMeDIP peaks with augmented signals (Figure 4B). The 241 hMeDIP peaks were distributed across the genome, with the most enrichment on chromosomes 9 and X (Figure 4C; Table S3), and overrepresentation at neuronal proximal enhancers (Figure 4D). These 241 hMeDIP peaks are associated with 204 unique genes (Figure 4E). Gene Ontology analysis determined an overrepresentation of processes relating to synaptic structure and synaptic transmission for the associated genes (Figure 4F), including neuregulin 3 (Nrg3), Fgf14, gamma-amino-butyric acid type a receptor of subunit gamma1 (Gabrg1), and glutamate ionotropic receptor kainite type subunit 2 (Grik2) (Figure 4G). Several genes involved in the identified neuronal processes (Figure 4F) also had hMeDIP peaks that were gained with Tet2 OE, such as smoothened (Smo), Fgf14, Gabrg1, and Slitrk2.
Figure 4. Loss of Tet2 in adult excitatory neurons alters hydroxymethylation on genes related to synaptic processes and enhances hippocampal-dependent memory.
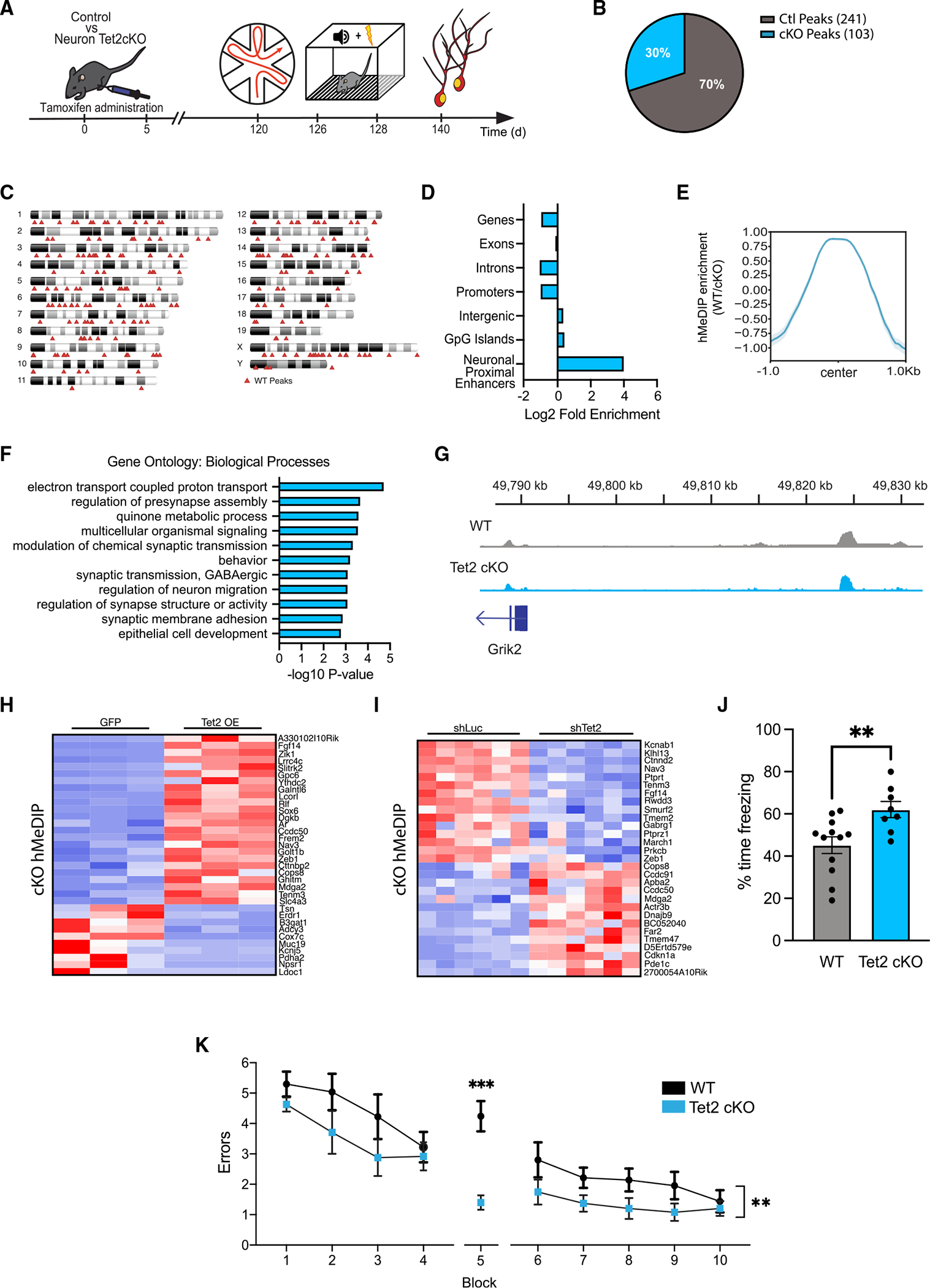
(A) Schematic of experimental paradigm. Adult (4 months old) CamK2aCre-ERT2;Tet2flox/flox excitatory neuron-specific knockout (Tet2 cKO) and littermate Tet2flox/flox control (wild-type [WT]) mice were administered tamoxifen and subjected to behavioral and molecular assays after 4 months.
(B) Pie chart of hMeDIP peaks gained (103 peaks) and lost (241 peaks) in Tet2 cKO neurons compared with control (q = 0.05).
(C) Ideogram of genomic location of hMeDIP peaks lost following loss of neuronal Tet2.
(D) Enrichment of lost hMeDIP peaks over genomic elements in Tet2 cKO neurons. (E) Metagene profile of lost hMeDIP peaks following loss of neuronal Tet2. Mean ± SEM.
(F) Gene Ontology analysis of biological processes on the 202 genes associated with the 241 lost hMeDIP peaks following loss of neuronal Tet2.
(G) Example gene track of the lost hMeDIP peak associated with Grik2 in Tet2 cKO neurons.
(H) Heatmap of differentially expressed genes in the in vitro Tet2 OE RNA-seq dataset that have associated lost hMeDIP peaks in the in vivo Tet2 cKO hMeDIP-seq dataset.
(I) Heatmap of differentially expressed genes in the in vitro shTet2 RNA-Seq dataset that have associated lost hMeDIP peaks in the in vivo Tet2 cKO hMeDIP-seq dataset.
(J) Quantification of the percentage of freezing 24 h after contextual fear conditioning training is shown (n = 8–12 mice per group; t test, **p < 0.01).
(K) Quantification of the number of entry errors during radial arm water maze training and testing (n = 8–12 mice per group; repeated-measures ANOVA with Bonferroni post hoc correction, **p < 0.01, ***p < 0.001).
Data are represented as mean ± SEM
To link changes in hydroxymethylation after loss of neuronal Tet2 to changes in gene expression, we integrated our in vivo Tet2 cKOhMeDIP-seq dataset with our in vitro OE and knockdown RNA-seq datasets (Figures 4H and 4I). hMeDIP peaks lost after deletion of neuronal Tet2 in vivo were associated with altered expression of 34 genes following Tet2 OE in vitro (Figures 4H) and 29 genes following Tet2 abrogation in vitro (Figure 4I). Several of these genes were bidirectionally regulated by opposite Tet2 manipulations, such as Fgf14, neuron navigator 3 (Nav3), and teneurin transmembrane protein 3 (Tenm3). These results indicate that loss of Tet2 reduces DNA hydroxymethylation at neuronally relevant genomic elements with concordant changes in gene expression. Our combined sequencing analysis suggests that Tet2 modulates functionally relevant gene-expression programs in neurons through directed hydroxymethylation changes.
Loss of Tet2 in adult excitatory neurons enhances hippocampal-dependent memory
Behavioral data indicate that increasing neuronal Tet2 in the adult hippocampus impairs memory; however, it remains an open question whether decreasing neuronal Tet2 can enhance cognitive function. Hippocampal-dependent learning and memory was assessed using fear-conditioning and RAWM paradigms in 8-month-old adult Tet2 cKO and littermate control mice (Figure 4A). During fear-conditioning training, mice exhibited no differences in baseline freezing time between genotypes (Figure S3B). Loss of neuronal Tet2 resulted in increased freezing time during contextual (Figure 4J), but not cued (Figure S3B), memory testing. In RAWM, all mice showed similar learning capacity during the training phase (Figure 4K). Loss of Tet2 in adult glutamatergic forebrain neurons resulted in significantly less errors in locating the target platform during spatial memory testing compared with control mice (Figure 4K). We observed no differences in general health using an open-field paradigm (Figures S3C–S3D). These data demonstrate that loss of Tet2 in adult excitatory neurons enhances hippocampal-dependent associative fear memory acquisition and spatial memory.
DISCUSSION
Cumulatively, our data indicate that neuronal Tet2 negatively regulates hippocampal-dependent memory. Our in vitro neuronal RNA-seq data in combination with our in vivo neuronal hMeDIP-seq data dissect the involvement of Tet2 in regulating synaptic processes at the DNA modification and transcriptional levels and implicate downstream synaptic plasticity gene targets as cell-autonomous regulators of neuronal function. We demonstrate that OE of neuronal Tet2 in the hippocampus impairs memory, while loss of neuronal Tet2 in excitatory forebrain neurons enhances hippocampal-dependent memory. Our data identify neuronal Tet2 as a molecular target to boost cognitive function in the adult brain.
Despite similar enzymatic activity, TET enzymes exhibit unique regulatory functions (Rasmussen and Helin, 2016). In the brain, Tet1 regulates neuronal activity and memory formation, with abrogation of Tet1 leading to aberrant long-term depression and impairments in memory extinction (Kaas et al., 2013; Rudenko et al., 2013). Tet3 regulates glutamatergic synaptic scaling in vitro, and knockdown of Tet3 impairs cocaine-induced memory consolidation (Yu et al., 2015; Liu et al., 2018). In contrast, we show that loss of neuronal Tet2 modulates synaptic gene expression and enhances hippocampal-dependent memory. These differences likely arise from unique genomic targeting of TET family members, particularly as Tet2 lacks a DNA-binding domain and is guided by binding partners.
We and others identified a role for adult neural stem cell Tet2 in promoting neurogenesis and enhancing cognition in older mice (Gontier et al., 2018; Li et al., 2017). Demethylation processes and Tet2 specifically are required for efficient cell-fate decisions and differentiation (Li et al., 2017), with age-related loss of Tet2 in adult neural stem cells inhibiting neurogenesis and impairing associated cognitive processes (Gontier et al., 2018). Our present work investigating the role of neuronal Tet2 in regulating hippocampal-dependent memory complements our previous findings in adult neural stem cells, highlighting the cell-type-specific and context-dependent roles of Tet2 in the adult and aging hippocampus.
Dysregulation of the neuroepigenome, particularly DNA methylation, is a hallmark of brain aging and age-related neurodegenerative disorders (Hwang et al., 2017). Our data support recent research delineating the role of Tet2 in promoting Alzheimer’s disease (Carrillo-Jimenez et al., 2019) and the neuroprotective role of Tet2 loss in Parkinson’s disease (Marshall et al., 2020). Cumulatively, this work posits neuronal Tet2 inhibition as an exciting therapeutic approach.
Limitations of the study
While confirmed at the mRNA level, OE or knockdown of Tet2 protein was difficult to ascertain using commercially available antibodies. Neuronal in vivo hMeDIP-seq data in combination with in vitro RNA-seq data begin to dissect the involvement of Tet2 in regulating synaptic processes at the transcriptional level. We note that this analysis postulated that 5hmC could be altered similarly in vitro and in vivo following neuronal Tet2 manipulation. Further hMeDIP-seq analysis of primary neurons is warranted to gain a full understanding of the convergent changes in neuronal 5hmC following Tet2 manipulation in vitro versus in vivo. Additionally, in vivo neuronal hMeDIP-seq and in vitro neuronal RNA-seq implicate downstream synaptic plasticity gene networks as regulators of neuronal function and cognitive processes. We anticipate that these data will prompt mechanistic investigations delineating the contribution of identified genes within the observed gene networks in mediating memory enhancements observed following loss of neuronal Tet2.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Saul Villeda (saul.villeda@ucsf.edu).
Materials availability
Plasmids generated in this study are available upon request.
Data and code availability
All RNA-Seq and hMeDIP-Seq data have been deposited in the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-NeuN, clone A60 | Millipore | Cat# MAB377; RRID:AB_2298772 |
| Rabbit polyclonal anti-IBA1 | FUJIFILM Wako Shibayagi | Cat# 016-26721; RRID:AB_2811160 |
| Guinea pig polyclonal anti-GFAP | Synaptic Systems | Cat# 173 004; RRID:AB_10641162 |
| Mouse monoclonal anti-MAP2 | Sigma Aldrich | Cat# M1406; RRID:AB_477171 |
| Rabbit polyclonal TurboGFP | Thermo Fisher | Cat# PA5-22688; RRID:AB_2540616 |
| Mouse monoclonal anti-NeuN-AlexaFluor488, clone A60 | Millipore | Cat# MAB377X; RRID:AB_2149209 |
| Rabbit polyclonal anti-5hmC | Active Motif | Cat# 39769; RRID:AB_10013602 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| TSA Plus Cyanine 3 | Akoya Biosciences | SKU NEL744001KT |
| Superscript II | Thermo Fisher | 18064-014 |
| KAPA HiFi Hotstart | Kapa Biosystems | KK2601 |
| Ampure XP Beads | Beckman Coulter | A63881 |
| Tamoxifen | Sigma Aldrich | T5648 |
| Poly-L-lysine hydrobromide | Sigma Aldrich | P6282 |
| Protein A magnetic beads | NEB | S1425S |
| Proteinase K | Sigma Aldrich | 03115879001 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAScope Multiplex Fluorescent Reagent Kit V2 | ACD Bio | 323100 |
| RNA-Protein Co-Detection Ancillary Kit | ACD Bio | 323180 |
| Papain dissociation system | Worthington | LK003153 |
| RNeasy Kit | Qiagen | 74104 |
| High-Capacity cDNA Reverse Transcription kit | Thermo Fisher | 4374966 |
| KAPA Sybr Fast qPCR Kit | Kapa Biosystems | KK4601 |
| Nextera Kit | Illumina | FC-131-1096 |
| qPCR Library Quantification Kit | Kapa Biosystems | KK4824 |
| End-It DNA End-Repair Kit | Lucigen | ER0720 |
| NEB Ultra DNA Library Kit | NEB | E7805S |
|
| ||
| Deposited data | ||
|
| ||
| RNA-Seq Data | This paper | GEO: GSE171295 |
| Tet2 OE hMeDIP-Seq | This paper | GEO: GSE198910 |
| Tet2 cKO hMeDIP-Seq | This paper | GEO: GSE199597 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: Tet2flox/flox (B6;129S-Tet2tm1.1Iaai/J) | The Jackson Laboratory | RRID:IMSR_JAX:017573 |
| Mouse: CamKIIa-CreERT2 (B6;129S6-Tg(Camk2a-cre/ERT2)1Aibs/J) | The Jackson Laboratory | RRID:IMSR_JAX:012362 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Tet2 RNAScope probe | ACD Bio | 511591 |
| shTet2-C targeting sequence: GAATGTAACCTTGATTGTTAT |
This paper | N/A |
| shTet2-D targeting sequence: ACCACACTCGATGCGGTATTTC |
This paper | N/A |
| shLuc targeting sequence: GCCATTCTATCCTCTAGAGGA |
This paper | N/A |
| Nextera Index Kit Set A | Illumina | FC-131-1002 |
| Smart-Seq2 poly-dT anchored oligonucleotide: AAGCAGTGGTATCAACGCAGAGTACT(30)VN |
Picelli et al. (2014) | N/A |
| TSO primer: AAGCAGTGGTATCAACGCAGAGT ACATrGrG + G |
Picelli et al. (2014) | N/A |
| Smart-Seq2 PCR Primer: AAGCAGTGGTATCA ACGCAGAGT |
Picelli et al. (2014) | N/A |
| NEBNext Multiplex Oligos for Illumina (Set 1) | NEB | E7335L |
| qPCR primer sequences | Gontier et al. (2018); This paper | Table S4 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pGreenPuro shRNA expression lentivector (CMV) | System Biosciences | SI505A-1 |
| pGreenPuro-shTet2-C | This paper | N/A |
| pGreenPuro-shTet2-D | This paper | N/A |
| pGreenPuro-shLuc | This paper | N/A |
| pENTR-D-TOPO | Thermo Fisher | K240020 |
| Synapsin-GFP lentivirus | N/A | |
| psPax2 | Addgene | Addgene plasmid # 12260; RRID:Addgene_12260 |
| pCMV-VSV-G | Addgene; Stewart et al. (2003) | Addgene plasmid # 8454; RRID:Addgene_8454 |
|
| ||
| Software and algorithms | ||
|
| ||
| STAR v2.7.3a | Dobin et al. (2013) | https://github.com/alexdobin/STAR |
| RSEM v1.3.1 | Li and Dewey, 2011 | https://github.com/deweylab/RSEM |
| R v4.0.2 | R Core Team, 2022 | https://www.r-project.org/ |
| DESeq2 v1.28.1 | Love et al. (2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Galaxy | Afgan et al. (2018) | www.usegalaxy.org |
| Bowtie 2 | Langmead and Salzberg, 2012 | https://github.com/BenLangmead/bowtie2 |
| MACS2 | Feng et al., 2012 | https://github.com/macs3-project/MACS |
| PANTHER | The Gene Ontology Consortium | http://geneontology.org |
| Graphpad Prism 6.0 | Graphpad Software | https://www.graphpad.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal models
The following mouse lines were used: C57BL/6J (The Jackson Laboratory; stock number: 000,664), Tet2 flox/flox (The Jackson Laboratory; stock number: 017573) mice, and CamKIIa-CreERT2 mice (The Jackson Laboratory; stock number: 012362). All mice used were on a C57BL/6 genetic background. Mice carrying a Tet2 flox/flox gene were crossed with mice carrying an inducible CamKIIa-Cre-ERT2 gene to obtain Tet2 flox/flox/CamKIIa-CreERT2+/0 mice. All studies were done in male mice. Studies were performed in mice starting at 3–4 months of age with Tet2 flox/flox/CamKIIa-CreERT2+/0 behavioral testing performed at 8 months of age. Animals were individually housed after stereotaxic injections or tamoxifen injections for the duration of behavioral testing. The numbers of mice used to result in statistically significant differences was calculated using standard power calculations with α = 0.05 and a power of 0.8. We used an online tool (http://www.stat.uiowa.edu/~rlenth/Power/index.html) to calculate power and sample size on the basis of experience with the respective tests, variability of the assays and inter-individual differences within groups. Mice were housed under specific pathogen-free conditions under a 12-h light-dark cycle and all animal handling and use was in accordance with institutional guidelines approved by the University of California San Francisco Institutional Animal Care and Use Committee (IACUC).
Primary neuron cultures
Primary mouse hippocampal and cortical neurons were dissociated into single-cell suspensions from E17 mouse embryo (C57Bl/6J) brains with a papain dissociation system (Worthington, Cat# LK003153). Neurons were seeded onto poly-L-lysine (Sigma Aldrich, Cat# P6282) coated plates [0.1% (wt/vol)] and grown in Neurobasal medium (Thermo Fisher, Cat# 21103049) supplemented with B-27 serum-free supplement (Thermo Fisher, Cat# 17504044), GlutaMAX (Thermo Fisher, Cat# 35050061) and penicillin–streptomycin (Thermo Fisher, Cat# 15140122) in a humidified incubator at 37°C, with 5% CO2. Half-media changes were performed every 4–5 days. Neurons were plated on 12 mm glass coverslips (Carolina Biological Supplies cat# 633009) in 24-well plates (100,000 cells/well) and in 12-well plates for molecular studies (500,000 cells/well). Neurons were infected with lentivirus on day 8 in vitro (DIV 8), and processed for further analyses on DIV11 for RNA analysis and DIV14 for immunohistochemical analysis. For immunocytochemistry, cells were fixed for 10 min with 4% paraformaldehyde, then washed and stained with MAP2 antibody (Sigma-Aldrich, Cat# M1406, RRID:AB_477171), turbo-GFP antibody (Thermo Fisher, Cat# PA5–22688, RRID:AB_2540616) to confirm lentiviral infection, and Hoechst 33342 (Thermo Fisher, Cat# H3570). Cells were lysed in Buffer RLT (Qiagen, Cat# 74004) for downstream RNA isolation.
METHOD DETAILS
RNAScope
In situ hybridization experiments and fluorescent immunostainings were performed using the RNAScope Multiplex Fluorescent Reagent Kit V2 and the RNA-Protein Co-Detection Ancillary Kit (ACD Bio, Cat# 323100 and 323180). The manufacturer recommended workflow for RNAScope with immunofluorescence co-detection was adapted for free-floating fixed-frozen sections. Briefly, cryopreserved 40 μm thick coronal brain sections were washed in PBS and pre-treated in 200ul hydrogen peroxide provided in the RNAScope Reagent kit. Antigen retrieval was performed in 1x co-detection target retrieval buffer for 10 min at 95°C. Sections were washed two times in PBS before incubation with the primary antibodies (NeuN (Millipore Cat# MAB377, RRID:AB_2298772), Iba1 (FUJIFILM Wako Shibayagi Cat# 016–26721, RRID:AB_2811160), GFAP (Synaptic Systems Cat# 173 004, RRID:AB_10641162)) in co-detection antibody diluent overnight at 4°C. The next day, the sections were washed in PBS (Thermo Fisher, Cat# 10010023), mounted in phosphate buffer on Superfrost plus slides and air dried for 1h at room temperature. Downstream RNAScope processing followed the manufacturer’s instructions. Briefly, sections were treated for 10 min with RNAScope Protease III, incubated for 2 h with the Tet2 RNAScope probe (ACD Bio, Cat# 511591) in a HybEZ oven set at 40°C. Following amplification steps, mouse Tet2 transcripts were detected using TSA Plus Cy3 reagents (Akoya Biosciences, Cat# SKU NEL744001KT). For immunohistochemical labeling with antibodies following the RNAScope assay, tissues were incubated with Alexa-Fluor-conjugated secondary antibodies in co-detection antibody diluent immediately after developing the HRP-TSA Plus signal. Sections were placed on a coverslip using Prolong Diamond and stored at 4°C. All slides were imaged within two weeks using a Zeiss LSM 800 confocal microscope with 20x and 63x objectives. For quantifications, stacks of 1μm thick slices were a acquired of the dentate gyrus in the hippocampus. Number of puncta per cell (neurons, microglia, astrocytes with clearly identifiable DAPI-positive nuclei) were counted and averaged for 20–40 cells from 3 sections per animal.
Viral plasmids and viruses
Lentiviruses (LVs) encoding shRNAs targeting endogenous mouse Tet2 were generated using the lentiviral pGreenPuro shRNA expression system (System Biosciences, Cat# SI505A-1) according to the manufacturer’s instructions. The following Tet2 targeted sequences were used (Tet2-SH-C, 5′-GAATGTAACCTTGATTGTTAT-3′; Tet2 SH-D, 5′-ACCACACTCGATGCGGTATTTC-3′). A virus encoding a shRNA targeting luciferase (SH-Luc, 5′- GCCATTCTATCCTCTAGAGGA-3′) was used as a control. The lentiviral murine Tet2 overexpression construct was generated in a two-step cloning process. First, the Tet2 coding sequence (CDS) and part of the 5′ and -3′ untranslated regions (UTRs) was amplified from adult mouse hippocampal cDNA and cloned into the pENTR-D-TOPO vector (Thermo Fischer, cat# K240020). After sequence validation, the CDS was further PCR amplified and restriction enzyme sites (NheI and AscI) were incorporated into the forward and reverse primers. Tet2 was then ligated into a Synapsin promoter-based lentiviral plasmid using traditional restriction enzyme-based cloning strategy. A Synapsin-GFP construct based on the same lentiviral plasmid backbone was used as a control. All plasmids were validated by Sanger sequencing prior to virus production. LV particles were generated as previously described (Lin et al., 2021) with viral titers between 1.2 × 109 to 1.5 × 109 viral particles per mL. Knockdown and overexpression of Tet2 was validated by qRT-PCR.
Lentiviruses production
293T cells were lipotransfected with 4:3:1 μg of lentiviral vector:psPax2:pCMV-VSVG (psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260); pCMV-VSV-G was a gift from Bob Weinberg (Addgene plasmid # 8454, RRID:Addgene_8454) (Stewart et al., 2003). After 48 h lentivirus-containing media was centrifuged for 5 min at 1000g and filtered through 45μm filter to remove cellular debris. Media underwent utracentrifugation (24000 RPM for 1.5 h) to concentrate virus. Viral pellets were gently resuspended in PBS. Lentiviral titers were between 1.2 × 109 to 1.5 × 109 viral particles per mL. Primary neuron cell cultures were infected at an ROI of 1, and viral solutions were diluted to 1.0 × 108 viral particles/mL prior to in vivo stereotaxic injection.
RNA extraction, cDNA synthesis and qPCR
Total RNA was isolated using phenol/chloroform extraction according to manufacturer’s protocols combined with columns from the RNeasy kit (QIAGEN, Cat# 74104). To quantify mRNA expression levels, equal amounts of cDNA were synthesized using the High-Capacity cDNA Reverse Transcription kit (ThermoFisher, Cat# 4374966) and mixed with the KAPA SYBR Fast mix (Roche, Cat# KK4601), and primers. GAPDH mRNA was amplified as an internal control. Quantitative RT-PCR was carried out in a CFX384 Real Time System (Bio-Rad).
Library generation and RNA sequencing
After RNA isolation, RNA-sequencing libraries were constructed using the Smart-Seq2 protocol from (Picelli et al., 2014), with modifications. Briefly, 8 ng of high-quality RNA was reverse transcribed using SuperScript II (Life Technologies, Cat# 18064–014) with a poly-dT anchored oligonucleotide primer, and a template switching oligonucleotide (TSO) primer that generated homotypic PCR primer binding sites. The cDNA underwent 10 rounds of PCR amplification using KAPA HiFi Hotstart (Kapa Biosystems, Cat# KK2601), followed by Ampure bead (Beckman Coulter, Cat# A63881) clean-up. The quality of the amplified cDNA was tested using Qubit nucleic acid quantitation (Life Technologies). 2ng of high-quality amplified cDNA was fragmented with the Tn5 transposase from the Illumina Nextera kit (Illumina, Cat# FC-131-1096) to a median size of ~500 bp. The fragmented libraries were amplified with indexed Nextera primers (Illumina, Cat# FC-131-1002) using 12 rounds of PCR. Final libraries were purified with AMPure beads and quantified on an Agilent Bioanalyzer. Libraries were pooled for sequencing on an Illumina NovaSeq with an SP flow cell (paired reads 2 × 150 bp).
RNA-sequencing analysis
Alignment of RNA-sequencing reads to the mouse mm10 transcriptome was performed using STAR v2.7.3a (Dobin et al., 2013) following ENCODE standard options, read counts were generated using RSEM v1.3.1 (Li and Dewey, 2011), and differential expression analysis was performed in R v4.0.2 using the DESeq2 v1.28.1 (Love et al., 2014) (detailed pipeline v2.1.2 and options available on https://github.com/emc2cube/Bioinformatics/). All data available under Gene Expression Omnibus accession no. GSE171295 (Token code: ehunomscxdedtkz). The derived differential gene expression lists underwent gene ontology analysis using Panther.
Stereotaxic injections
Sterotaxic injections were performed according to the protocol previously described (Lin et al., 2021). Animals were placed in a stereotaxic frame and anesthetized with 2% isoflurane (Patterson Veterinary) (2 L per min oxygen flow rate) delivered through an anesthesia nose cone. Fur around the incision area was trimmed and ophthalmic eye ointment was applied to the cornea to prevent desiccation during surgery. Viral solutions were injected bilaterally into the hippocampal CA1 and dentate gyrus regions using the following coordinates: (from bregma) anterior = −2 mm, lateral = 1.5 mm and (from skull surface) height = −1.7 and −2.1 mm. A 2 μL volume of viral solution was injected stereotaxically over 10 min (injection speed: 0.20 μL per min) using a 5-μL 26s-gauge Hamilton syringe into each locale. To limit reflux along the injection track, the needle was maintained in situ for 8 min, slowly pulled out half-way and kept in position for an additional 2 min. The skin was closed using silk suture and VetBond glue. Each mouse was injected subcutaneously with saline, enrofloxacin (Bayer) antibiotic, carprofen (Patterson), and buprenorphine (Butler Schein) as directed for pain, single-housed, and monitored during recovery.
Tamoxifen administration
All experimental genetic mouse models (Tet2 flox/flox control and Tet2 flox/flox/CamKIIa-CreERT2+/0 mutant mice) received tamoxifen. At 4 months mice were administered tamoxifen to induce Tet2 excision specifically in mature forebrain excitatory (adult glutamatergic) neurons termed Neuron Tet2 cKO. Tamoxifen (Sigma-Aldrich, Cat# T5648) was dissolved in sunflower seed oil/ethanol (10:1) at 30 mg/mL and administered intraperitoneally at 180 mg/kg body weight once per day for 5 days.
Neuronal nuclei isolation
Neuronal nuclei were isolated based on the protocol previously described with minor modifications (Krishnaswami et al., 2016). Briefly, flash-frozen dissected hippocampi were dounce homogenized (Wheaton, Cat# 357538) in 750 μL of Homogenization buffer (HB: 250 mM sucrose, 25mM KCl, 5 mM MgCl2, 10mM Tris pH = 8.0, 1 μM DTT, 0.1% v/v Triton X-100, 1x RNAse Inhibitor, 1x protease inhibitor, Nuclease-free H2O) with 12 strokes of the loose pestle and 20 strokes of the tight pestle. Samples were filtered through a 40 μm filter and centrifuged at 500 RCF for 6 min at 4°C. Samples were resuspended in 750 μL HB and spun at 500 RCF for 8 min at 4°C. Samples were incubated for 15 min on ice in staining buffer (0.5% wt/vol BSA in PBS). Conjugated mouse monoclonal anti-NeuN-AlexaFluor488 antibody (Millipore, Cat# MAB377X, RRID:AB_2149209) was added to the tube at a final dilution of 1:250. Samples were incubated on a tube rotator for 60 min at 4°C and then spun for 5 min at 500g at 4°C. Samples were washed 2x with 1000 μL staining buffer and Hoechst 44,432 was added to staining buffer at a final concentration of 0.01 μg/mL. Samples were then filtered through a 35μm FACS tube filter and sorted.
Fluorescence-activated cell sorting (FACS)
Nuclei were sorted on a BD FACSAria Fusion with a 70 μm nozzle and with a flow rate of 1–2.5. Nuclei were first gated by forward and side scatter, then gated for doublets with height and width. Nuclei that were both Hoechst+ and NeuN+ were sorted into Tri Reagent (Sigma Aldrich, T9424) for RNA analysis or PBS for DNA analysis.
hMeDIP-seq library preparation
For each library, 200ng of DNA was sonicated to 300–500 base pairs, and after end cleanup and A-tailing (End-It DNA End-Repair Kit, Lucigen, Cat# ER0720), NEBNext adapters were ligated onto the samples (NEB Ultra DNA library kit, NEB, Cat# E7805S). Samples were then divided into input and immunoprecipitation samples. DNA was denatured by heating at 95°C for 10 min, followed by plunging the samples on ice. Ice-cold 10x hMeDIP buffer (100mM NaPO4 pH 7.0, 1.4 mM NaCl, and 0.5% Triton X-100) was added to the immunoprecipitation sample to a final concentration of 1x. 0.2 μL of 5hmC DNA antibody (Active Motif, Cat# 39769, RRI-D:AB_10013602) was added to the samples. The samples were rotated overnight at 4°C. Protein A magnetic beads (NEB, Cat# S1425S) were added to the reaction, and rotated at 4°C for 2 h. The samples were collected on a magnetic rack, and the samples were washed three times (with 10 min incubations) with ice-cold 1x hMeDIP buffer. DNA was eluted from the beads by shaking the samples in lysis buffer (50mM Tris pH 8.0, 10mM EDTA, and 0.5% SDS) with proteinase K (100 μg/mL, Sigma Aldrich, Cat# 03115879001) at 55°C for 3 + hours. The DNA was purified by phenol/chloroform extraction followed by ethanol precipitation. After purification, the libraries were PCR amplified (KAPA HiFi, kk2602) for 14 cycles using paired-end indexed primers (NEBNext Multiplex Oligos (Set 1), NEB, Cat# E7335L). Libraries were pooled and purified with 1.8X Agencourt AMpure XP beads (Beckman Coulter, A63881). Individual libraries were quantified on the Agilent Bioanalyzer with the DNA sensitivity protocol and pooled for sequencing.
hMeDIP-seq analysis
hMeDIP-Seq libraries were sequenced on the Illumina HiSeq4000 to a average depth of over 50 million reads. Fastq files were uploaded to Galaxy (usegalaxy.org) for mapping using Bowtie 2 and peak finding with MACS2 (Afgan et al., 2018; Feng et al., 2012; Langmead and Salzberg, 2012). Libraries were mapped to the mm10 genome using Bowtie 2 with default parameters. MACS2 was used to determine peaks that were lost after overexpression of Tet2 or deletion of Tet2 with the following change to default parameters: minimum enrichment changed to 2-fold. Peaks were associated with genes if they overlapped or were the closest gene (by transcriptional start site) to the peak. The derived gene list underwent gene ontology analysis using Panther. All data available under Gene Expression Omnibus accession no. GSE198910 (Token code: uvmzaumuxneljsd) and GSE199597 (Token code: cvytgeeqjpybzwr).
Open field
Mice were placed in the center of an open 40 cm × 40 cm square chamber (Kinder Scientific) with no cues or stimuli and allowed to move freely for 10 min. Infrared photobeam breaks were recorded and movement metrics analyzed by MotorMonitor software (Kinder Scientific).
Novel object recognition
The novel object recognition task was preformed according to the protocol previously described (White et al., 2020). During the habituation phase (day 1), mice could freely explore an empty arena for 10 min. During the training phase (day 2), two identical objects were placed in the habituated arena, and mice could explore the objects for 5 min. For the testing phase (day 3), one object was replaced with a novel object, and mice could explore the objects for 5 min. Time spent exploring each object was quantified using the Smart Video Tracking Software (Panlab; Harvard Apparatus). Two different sets of objects are used. To control for any inherent object preference, half of the mice are exposed to object A as their novel object and half to object B. To control for any potential object-independent location preference, the location of the novel object relative to the trained object is also varied. The objects were chosen based on their ability to capture the animal’s interest, independent of genetic background or age. To determine percent time with novel object, we calculate (Time with novel object)/(Time with Trained Object + Time with Novel Object) * 100. In this preference index, 100% indicates full preference for the novel object, and 0% indicates full preference for the trained object. A mouse with a value of 50% would have spent equal time exploring both objects. Mice that did not explore both objects during the training phase were excluded from analysis.
Radial arm water maze
Spatial learning and memory was assessed using the radial arm water maze (RAWM) paradigm according to the protocol previously described (Alamed et al., 2006). In this task the goal arm location containing a platform remains constant throughout the training and testing phase, while the start arm is changed during each trial. Spatial cues are posted on all four walls of the RAWM area. Entry into an incorrect arm is scored as an error, and errors are averaged over training blocks (three consecutive trials). On day one, during the training phase, mice are trained for 15 trails, with trials alternating between a visible and hidden platform for blocks 1–4 and then switching to only a hidden platform in block 5. On day 2 during the testing phase, mice are tested for 15 trials with a hidden platform for blocks 6–10. Investigators were blinded to genotype and treatment when scoring.
Fear conditioning
In this task, mice learned to associate the environmental context (fear conditioning chamber) with an aversive stimulus (mild foot shock; unconditioned stimulus, US) enabling testing for hippocampal-dependent contextual fear conditioning. To also assess amygdala-dependent cued fear conditioning, the mild foot shock was paired with a light and tone cue (conditioned stimulus, CS). Conditioned fear was displayed as freezing behavior. Specific training parameters are as follows: tone duration is 30 s; level is 70 dB, 2 kHz; shock duration is 2 s; intensity is 0.6 mA. This intensity is not painful and can easily be tolerated but will generate an unpleasant feeling. On day one, each mouse was placed in a fear-conditioning chamber and allowed to explore for two minutes before delivery of a 30-s tone (70 dB) ending with a 2-s foot shock (0.6 mA). Two minutes later, a second CS-US pair was delivered. On day 2, each mouse was first placed in the fear-conditioning chamber containing the same exact context, but with no CS or foot shock. Freezing was analyzed for 2 min. One hour later, the mice were placed in a new context containing a different odor, cleaning solution, floor texture, chamber walls and shape. Animals were allowed to explore for 2 min before being re-exposed to the CS. Freezing was analyzed for 3 min using a FreezeScan video tracking system and software (Cleversys, Inc).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data and statistical analysis
Graphed data are expressed as mean ± SEM. Statistical analysis was performed with Prism 8.0 software (GraphPad Software). Unless otherwise noted, means between two groups were compared with two-tailed, unpaired Student’s t-test. Comparisons of means from multiple groups with each other or against one control group were analyzed with one-way ANOVA and Tukey’s post-hoc test. All histology and behavior experiments conducted were done in a randomized and blinded fashion. For each experiment, the overall size of the experimental groups corresponded to distinct animals or cultures. Unique samples were not measured repeatedly within the same characterization of a given cohort.
Supplementary Material
Highlights.
Tet2 is enriched in neurons in the adult mouse hippocampus
Tet2 negatively regulates neuronal complexity and synaptic gene expression in vitro
Adult hippocampal neuronal Tet2 alters hydroxymethylation on synaptic gene networks
Neuronal Tet2 negatively regulates memory in the adult hippocampus
ACKNOWLEDGMENTS
Work was funded by a Frontiers in Medical Research fellowship (K.J.B.P.), an NIH Ruth L. Krischstein NRSA fellowship (J.M.S.; F32-AG055292), an Irene Diamond Fund AFAR award (G.G.), the Simons Foundation (S.A.V.), and the NIA (RF1 AG062357, R01 AG077770). We acknowledge the UCSF Parnassus Flow Core (RRID: SCR_018206) and support by DRC Center Grant NIH P30 DK063720 for assistance with flow cytometry. We thank the UCSF Center for Advanced Technology for assistance with RNA-seq. Some computing was performed on the Sherlock cluster. We thank Stanford University and the Stanford Research Computing Center for providing computational resources and support.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111612.
REFERENCES
- Afgan E, Jalili V, Goonasekera N, Taylor J, and Goecks J (2018). Federated Galaxy: biomedical computing at the frontier. In 2018 IEEE 11th International Conference on Cloud Computing (CLOUD). 10.1109/cloud.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamed J, Wilcock DM, Diamond DM, Gordon MN, and Morgan D (2006). Two-day radial-arm water maze learning and memory task; robust resolution of amyloid-related memory deficits in transgenic mice. Nat. Protoc. 1, 1671–1679. 10.1038/nprot.2006.275. [DOI] [PubMed] [Google Scholar]
- Carrillo-Jimenez A, Deniz Ö, Niklison-Chirou MV, Ruiz R, Bezerra-Salomão K, Stratoulias V, Amouroux R, Yip PK, Vilalta A, Cheray M, et al. (2019). TET2 regulates the neuroinflammatory response in microglia. Cell Rep. 29, 697–713.e8. 10.1016/j.celrep.2019.09.013. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DLA, Kenis G, Keitel S, Hof PR, van Os J, Steinbusch HWM, Schmitz C, and Rutten BPF (2012). Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr. Alzheimer Res. 9, 536–544. 10.2174/156720512800618035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Gavin DP, Chen Y, and Davis J (2012). Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl. Psychiatry 2, e159. 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, and Liu XS (2012). Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740. 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, and Fan G (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430. 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, and Carell T (2010). Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5, e15367. 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontier G, Iyer M, Shea JM, Bieri G, Wheatley EG, Ramalho-Santos M, and Villeda SA (2018). Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981. 10.1016/j.celrep.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulmez Karaca K, Kupke J, Brito DVC, Zeuch B, Thome C, Weichenhan D, Lutsik P, Plass C, and Oliveira AMM (2020). Neuronal ensemble-specific DNA methylation strengthens engram stability. Nat. Commun. 11, 639. 10.1038/s41467-020-14498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. (2011). Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351. 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J-Y, Aromolaran KA, and Zukin RS (2017). The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 18, 347–361. 10.1038/nrn.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, and Zhang Y (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, and Bird A (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G-L, King JR, Song H, and Sweatt JD (2013). TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093. 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, et al. (2012). 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 19, 1037–1043. 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans F, van Nierop P, Andres-Alonso M, Byrnes A, Cijsouw T, Coba MP, Cornelisse LN, Farrell RJ, Goldschmidt HL, Howrigan DP, et al. (2019). SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234.e4. 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, and Heintz N (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, et al. (2016). Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 11, 499–524. 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12 (323). 10.1186/1471-2105-12-323.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yao B, Chen L, Kang Y, Li Y, Cheng Y, Li L, Lin L, Wang Z, Wang M, et al. (2017). Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun. 8, 15903. 10.1038/ncomms15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Bieri G, Gontier G, Müller S, Smith LK, Snethlage CE, White CW, Maybury-Lewis SY, and Villeda SA (2021). MHC class I H2-Kb negatively regulates neural progenitor cell proliferation by inhibiting FGFR signaling. PLoS Biol. 19, e3001311. 10.1371/journal.pbio.3001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sun X, Wang Z, Le Q, Liu P, Jiang C, Wang F, and Ma L (2018). Retrieval-induced upregulation of Tet3 in pyramidal neurons of the dorsal Hippocampus mediates cocaine-associated memory reconsolidation. Int. J. Neuropsychopharmacol. 21, 255–266. 10.1093/ijnp/pyx099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall LL, Killinger BA, Ensink E, Li P, Li KX, Cui W, Lubben N, Weiland M, Wang X, Gordevicius J, et al. (2020). Epigenomic analysis of Parkinson’s disease neurons identifies Tet2 loss as neuroprotective. Nat. Neurosci. 23, 1203–1214. 10.1038/s41593-020-0690-y. [DOI] [PubMed] [Google Scholar]
- Miller CA, and Sweatt JD (2007). Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, and Fan G (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- R Core Team (2022). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing; ). https://www.R-project.org/. [Google Scholar]
- Rasmussen KD, and Helin K (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, and Tsai L-H (2013). Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122. 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song C-X, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. (2011). 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616. 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, and Rao A (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CW, Fan X, Maynard JC, Wheatley EG, Bieri G, Couthouis J, Burlingame AL, and Villeda SA (2020). Age-related loss of neural stem cell O-GlcNAc promotes a glial fate switch through STAT3 activation. Proc. Natl. Acad. Sci. USA 117, 22214–22224. 10.1073/pnas.2007439117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Su Y, Shin J, Zhong C, Guo JU, Weng Y-L, Gao F, Geschwind DH, Coppola G, Ming G.l., and Song H (2015). Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat. Neurosci. 18, 836–843. 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-Seq and hMeDIP-Seq data have been deposited in the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-NeuN, clone A60 | Millipore | Cat# MAB377; RRID:AB_2298772 |
| Rabbit polyclonal anti-IBA1 | FUJIFILM Wako Shibayagi | Cat# 016-26721; RRID:AB_2811160 |
| Guinea pig polyclonal anti-GFAP | Synaptic Systems | Cat# 173 004; RRID:AB_10641162 |
| Mouse monoclonal anti-MAP2 | Sigma Aldrich | Cat# M1406; RRID:AB_477171 |
| Rabbit polyclonal TurboGFP | Thermo Fisher | Cat# PA5-22688; RRID:AB_2540616 |
| Mouse monoclonal anti-NeuN-AlexaFluor488, clone A60 | Millipore | Cat# MAB377X; RRID:AB_2149209 |
| Rabbit polyclonal anti-5hmC | Active Motif | Cat# 39769; RRID:AB_10013602 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| TSA Plus Cyanine 3 | Akoya Biosciences | SKU NEL744001KT |
| Superscript II | Thermo Fisher | 18064-014 |
| KAPA HiFi Hotstart | Kapa Biosystems | KK2601 |
| Ampure XP Beads | Beckman Coulter | A63881 |
| Tamoxifen | Sigma Aldrich | T5648 |
| Poly-L-lysine hydrobromide | Sigma Aldrich | P6282 |
| Protein A magnetic beads | NEB | S1425S |
| Proteinase K | Sigma Aldrich | 03115879001 |
|
| ||
| Critical commercial assays | ||
|
| ||
| RNAScope Multiplex Fluorescent Reagent Kit V2 | ACD Bio | 323100 |
| RNA-Protein Co-Detection Ancillary Kit | ACD Bio | 323180 |
| Papain dissociation system | Worthington | LK003153 |
| RNeasy Kit | Qiagen | 74104 |
| High-Capacity cDNA Reverse Transcription kit | Thermo Fisher | 4374966 |
| KAPA Sybr Fast qPCR Kit | Kapa Biosystems | KK4601 |
| Nextera Kit | Illumina | FC-131-1096 |
| qPCR Library Quantification Kit | Kapa Biosystems | KK4824 |
| End-It DNA End-Repair Kit | Lucigen | ER0720 |
| NEB Ultra DNA Library Kit | NEB | E7805S |
|
| ||
| Deposited data | ||
|
| ||
| RNA-Seq Data | This paper | GEO: GSE171295 |
| Tet2 OE hMeDIP-Seq | This paper | GEO: GSE198910 |
| Tet2 cKO hMeDIP-Seq | This paper | GEO: GSE199597 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: Tet2flox/flox (B6;129S-Tet2tm1.1Iaai/J) | The Jackson Laboratory | RRID:IMSR_JAX:017573 |
| Mouse: CamKIIa-CreERT2 (B6;129S6-Tg(Camk2a-cre/ERT2)1Aibs/J) | The Jackson Laboratory | RRID:IMSR_JAX:012362 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Tet2 RNAScope probe | ACD Bio | 511591 |
| shTet2-C targeting sequence: GAATGTAACCTTGATTGTTAT |
This paper | N/A |
| shTet2-D targeting sequence: ACCACACTCGATGCGGTATTTC |
This paper | N/A |
| shLuc targeting sequence: GCCATTCTATCCTCTAGAGGA |
This paper | N/A |
| Nextera Index Kit Set A | Illumina | FC-131-1002 |
| Smart-Seq2 poly-dT anchored oligonucleotide: AAGCAGTGGTATCAACGCAGAGTACT(30)VN |
Picelli et al. (2014) | N/A |
| TSO primer: AAGCAGTGGTATCAACGCAGAGT ACATrGrG + G |
Picelli et al. (2014) | N/A |
| Smart-Seq2 PCR Primer: AAGCAGTGGTATCA ACGCAGAGT |
Picelli et al. (2014) | N/A |
| NEBNext Multiplex Oligos for Illumina (Set 1) | NEB | E7335L |
| qPCR primer sequences | Gontier et al. (2018); This paper | Table S4 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pGreenPuro shRNA expression lentivector (CMV) | System Biosciences | SI505A-1 |
| pGreenPuro-shTet2-C | This paper | N/A |
| pGreenPuro-shTet2-D | This paper | N/A |
| pGreenPuro-shLuc | This paper | N/A |
| pENTR-D-TOPO | Thermo Fisher | K240020 |
| Synapsin-GFP lentivirus | N/A | |
| psPax2 | Addgene | Addgene plasmid # 12260; RRID:Addgene_12260 |
| pCMV-VSV-G | Addgene; Stewart et al. (2003) | Addgene plasmid # 8454; RRID:Addgene_8454 |
|
| ||
| Software and algorithms | ||
|
| ||
| STAR v2.7.3a | Dobin et al. (2013) | https://github.com/alexdobin/STAR |
| RSEM v1.3.1 | Li and Dewey, 2011 | https://github.com/deweylab/RSEM |
| R v4.0.2 | R Core Team, 2022 | https://www.r-project.org/ |
| DESeq2 v1.28.1 | Love et al. (2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Galaxy | Afgan et al. (2018) | www.usegalaxy.org |
| Bowtie 2 | Langmead and Salzberg, 2012 | https://github.com/BenLangmead/bowtie2 |
| MACS2 | Feng et al., 2012 | https://github.com/macs3-project/MACS |
| PANTHER | The Gene Ontology Consortium | http://geneontology.org |
| Graphpad Prism 6.0 | Graphpad Software | https://www.graphpad.com |


