Abstract
Homologous recombinational repair of DNA double-strand breaks and crosslinks in human cells is likely to require Rad51 and the five Rad51 paralogs (XRCC2, XRCC3, Rad51B/Rad51L1, Rad51C/Rad51L2 and Rad51D/Rad51L3), as has been shown in chicken and rodent cells. Previously, we reported on the interactions among these proteins using baculovirus and two- and three-hybrid yeast systems. To test for interactions involving XRCC3 and Rad51C, stable human cell lines have been isolated that express (His)6-tagged versions of XRCC3 or Rad51C. Ni2+-binding experiments demonstrate that XRCC3 and Rad51C interact in human cells. In addition, we find that Rad51C, but not XRCC3, interacts directly or indirectly with Rad51B, Rad51D and XRCC2. These results argue that there are at least two complexes of Rad51 paralogs in human cells (Rad51C–XRCC3 and Rad51B–Rad51C–Rad51D–XRCC2), both containing Rad51C. Moreover, Rad51 is not found in these complexes. X-ray treatment did not alter either the level of any Rad51 paralog or the observed interactions between paralogs. However, the endogenous level of Rad51C is moderately elevated in the XRCC3-overexpressing cell line, suggesting that dimerization between these proteins might help stabilize Rad51C.
INTRODUCTION
The eukaryotic Rad51 protein is related to the prokaryotic RecA protein, and is the key protein facilitating both mitotic and meiotic homologous recombination (1). In addition to Rad51 and the closely related meiotic DMC1 protein, there are five Rad51-related proteins (or paralogs) in human cells: XRCC2 (2–4), XRCC3 (2,5,6), Rad51B/Rad51L1 (7–9), Rad51C/Rad51L2 (10) and Rad51D/Rad51L3 (9,11,12). These Rad51 paralogs share 20–30% sequence identity with Rad51 and with each other, and probably arose by gene duplication, followed by the development of new functions (for review see 13–15). The Rad51 paralogs were first implicated in homologous recombinational repair (HRR) on the basis of their sequence similarity to Rad51. In addition, there is now extensive evidence for an important role in HRR from analyses with mutations in hamster and chicken DT40 cell lines (4,6,16–18). To date, the precise functions of the set of five Rad51 paralogs in vertebrate cells have not been determined.
In Saccharomyces cerevisiae there are only two Rad51 paralogs (Rad55 and Rad57). These proteins form a heterodimer that stimulates Rad51-mediated strand-exchange activity by facilitating Rad51’s displacement of RPA from single-stranded DNA (19). As there are several similarities between the Rad51 paralogs in yeast and in vertebrate cells, it is reasonable to expect that some or all of the mammalian Rad51 paralogs might perform an analogous function. As with the yeast paralogs, each of the human Rad51 paralogs interacts with other paralogs, but not with itself (20). Several studies argue that the vertebrate Rad51 paralogs may function as Rad51 accessory factors (17,18,21–23). Several of the human Rad51 paralogs have been purified, with the goal of determining their function(s). The Rad51D protein has both single-strand DNA binding and DNA-stimulated ATPase activities, and interacts with XRCC2 in vivo (21). The purified XRCC3/Rad51C heterodimer also binds to single-stranded DNA and forms networks of protein and DNA as seen by electron microscopy (23). This heterodimer was also reported to have homologous pairing activity (24).
Here we report studies of the physical interactions of the Rad51 paralogs in human cells, as a follow-up to our findings in the yeast two-hybrid and baculovirus systems (20). Protein extracts of human lymphoblastoid cell lines expressing tagged versions of XRCC3 and Rad51C were used to pull down these recombinant proteins and to identify interactions with other Rad51 paralogs. We also examined whether DNA damage affected the protein level and/or pattern of interactions of any of the Rad51 paralogs. Our results support the simultaneous existence of at least two complexes of Rad51 paralogs in human cells, both containing Rad51C, but only one containing XRCC3.
MATERIALS AND METHODS
Cell culture and DNA transfections
TK6 and WTK1 are human lymphoblastoid cell lines derived from the same progenitor cell line (25). Cells were grown at 37°C in suspension cultures in a humidified 5% CO2 atmosphere in RPMI 1640 medium supplemented with 10% heat-inactivated horse serum, 100 U/ml penicillin and 100 µg/ml streptomycin (Life Technologies). Cells were maintained in exponential growth at densities from 1 to 10 × 105 cells/ml.
The (His)6-HA-tagged XRCC3 expression plasmid (pDS158) is a derivative of the pcDNA3 vector (Invitrogen) and has been described previously (2). XRCC3 is expressed from a CMV promoter and contains a C-terminal HA-tag flanked downstream by a (His)6-tag. The expression plasmid for hRad51C (pDS266) was constructed similarly and contains the hRAD51C ORF that was ligated at the C-terminus to an HA-tag plus a (His)6-tag and subcloned into the pcDNA3 vector (Invitrogen). Stable cell lines containing either pDS158 (TK6 cells) or pDS266 (WTK1 cells) were generated by electroporation (625 V/cm, 950 µF) using the Gene Pulser II apparatus (Bio-Rad). One million cells were transfected according to the methods of van den Hoff et al. (26) in 400 µl cytomix buffer using 10 µg of plasmid. Forty hours post-transfection, cells were seeded at 1000 cells/well in 96-well plates in standard growth medium supplemented with 0.5 or 1.0 mg/ml geneticin (Life Technologies) for TK6 cells and WTK1 cells, respectively. Plates were fed every 3–4 days with medium supplemented with the appropriate concentration of geneticin. Geneticin-resistant clones were isolated after 14 days of growth and checked for expression of the recombinant protein by western blot analysis.
To measure cell killing, cells were exposed to graded doses of X-rays in T-25 flasks at cell densities of 8–10 × 105/ml using a 160 kVp Pantak industrial X-ray generator with 1 mm Cu and 1 mm Al filtration and a dose rate of 1 Gy/min. Other cells were treated with mitomycin C (MMC; Sigma) at concentrations from 0.062 to 0.5 µg/ml for 4 h at 37°C. Aliquots of treated and untreated cultures were seeded into 96-well plates at 1–20 cells/well to determine the surviving fraction, as calculated according to standard methods (27).
Native cellular protein extracts
Native (i.e. non-denatured) protein extracts were prepared according to ‘General Guidelines for Purification of 6× His-tagged Proteins’ (Qiagen). Cells were spun down for 5 min at 1000 g, washed with cold PBS and re-pelleted. Approximately 3 × 107 cells were re-suspended in 750 µl native extract lysis buffer [300 mM NaCl, 50 mM NaH2PO4, 1% Triton X-100, 10 mM imidazole, 10 mM β-mercaptoethanol, 2 mM Pefabloc serine protease inhibitor (Roche), pH 8.0; Qiagen] and incubated on a shaker for 10 min at 4°C. The lysate was centrifuged at 10 000 g for 10 min at 4°C to pellet cellular debris and DNA. The supernatant was aspirated and protein was quantified using the modified Lowry protein DC assay according to the manufacturer’s directions (Bio-Rad).
Ni2+ pull-down experiments
Ni2+–nitrilotriacetate (NTA) magneto-capture was used to isolate (His)6-tagged recombinant proteins. Native protein extract containing 1–1.5 mg of protein was diluted in 1 ml of native lysis buffer containing 10 mM imidazole, 10 mM β-mercaptoethanol, 2 mM Pefabloc and 50 µl magnetic beads (Qiagen). The suspension was incubated on a nutator for 2 h at 4°C. The beads were magnetically separated (magnetic separator, Dynal) for 5 min at 4°C, the supernatant was discarded and the beads were washed three times for 10 min in 1 ml native lysis buffer containing 20 mM imidazole at 4°C. Bound protein complexes were eluted in 50 µl lysis buffer containing 250 mM imidazole. Generally, 5–20 µl aliquots were subjected to SDS–PAGE and western blotting (see below).
Some of the Ni2+ pull-down experiments were carried out on extracts from cells treated first with X-rays or MMC. For X-ray treatment, the cells were exposed to 8 Gy of X-rays in T-75 flasks. For MMC treatment, 5 × 107 cells/time point were treated for 4 h at 37°C with three concentrations of MMC (0.125, 0.25 and 0.5 µg/ml). Following the treatment, cells were resuspended in fresh medium for either 0, 4 or 8 h at 37°C, washed in fresh medium and converted to extracts as described above.
DNase I digestion and ethidium bromide treatment
In order to ensure that DNA was not responsible for any of the observed protein associations, cellular protein extracts were subjected to digestion with DNase I prior to the pull-down reactions. Approximately 1500 µg of native protein was incubated with 500 U of DNase I (Roche) for 30 min at 37°C in a total volume of 1 ml of native lysis buffer. Alternatively, the pull-down reactions and each of the subsequent wash steps were carried out in native lysis buffer in the presence of 100 µg/ml ethidium bromide (28). Analyses of protein complexes were carried out by western blotting as described below.
Immunoprecipitation
Standard methods were tried for immunoprecipitation of Rad51C, but we found that it non-specifically bound to the beads in the absence of anti-Rad51C antibody (data not shown). We therefore used the following procedure, which avoided this problem. Immunoprecipitation of endogenous Rad51C protein was carried out using MagnaBind goat anti-mouse IgG magnetic beads (Pierce Chemical). A 200 µl aliquot of bead suspension was added to 1 ml native protein lysis buffer containing either ∼0.5 µg of monoclonal mouse anti-Rad51C antibody or ∼0.5 µg of control antibody (mouse IgG, Santa Cruz Biotech.). The mixture was incubated on a nutator at 4°C for 1 h. The beads were magnetically separated at 4°C for 5 min, washed twice in 1 ml lysis buffer to remove unbound antibody and resuspended in 0.25 ml lysis buffer containing ∼1 mg of native cellular protein extract and 2 mM Pefabloc (Roche); the mixture was incubated at 4°C for 1 h. The beads were magnetically separated for 5 min, and the supernatant was aspirated and discarded. The beads were washed three times for 15 min each in 1 ml lysis buffer. Immune complexes were eluted in 50 µl IgG elution buffer (Pierce Chemical).
Western analysis
The NuPAGE Bis–Tris electrophoresis system (Invitrogen) was used for polyacrylamide gel electrophoresis under reducing conditions according to the manufacturer’s instructions. Gels (10 and 12%) were used with NuPAGE reagents. Gels were run at 200 V for ∼70 min (10% gels) or ∼2.5 h (12% gels). Proteins were transferred to PVDF membranes (Millipore) and probed with specific antibodies. Following incubation with a primary antibody, detection was carried out using horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Santa Cruz Biotech.) or goat anti-rabbit secondary antibody (Jackson ImmunoResearch; 0.4 mg/ml). Filters were incubated with Supersignal WestPico substrate kit (Pierce Chemical) or ECL-Plus (Amersham Pharmacia Biotech.), followed by exposure with Hyperfilm ECL (Amersham Pharmacia Biotech.). Signal intensities were quantified using the ChemiImager 4400 Low Light Imaging System (Alpha Innotech Corporation) and AlphaEase software (version 5.0).
Antibodies against the human Rad51 paralogs
For detection of Rad51C we used either polyclonal rabbit anti-Rad51C antibody (1:4000; kindly provided by P. Sung) or monoclonal mouse anti-Rad51C (1:3000; isolated in J. Albala’s laboratory). For detection of Rad51B we used polyclonal rabbit anti-Rad51B antibodies 1918 or 1919 (1:600; isolated in J. Albala’s laboratory). For detection of XRCC3 we used two different polyclonal rabbit anti-XRCC3 antibodies, one isolated against the whole protein (1:20 000; kindly provided by P. Sung) and one isolated against the C-terminal part of XRCC3 (1:6000; Novus Biologicals). For detection of Rad51D we used polyclonal rabbit anti-Rad51D antibody (1:1000; Novus Biologicals), and for detection of XRCC2 we used polyclonal rabbit anti-XRCC2 antibody (1:2000; kindly provided by P. Sung). Note that we found no evidence that any of the antibodies against specific Rad51 paralogs cross-reacted against any of the other Rad51 paralogs in our experiments, and each of the anti-Rad51 paralog antibodies used recognized a protein of the expected molecular weight.
To control for loading variation during western analyses, we normalized to the protein signals of either β-tubulin or the transcription factor QM. We used rabbit polyclonal anti-β-tubulin antibody (1:1000, H-235; Santa Cruz Biotech.) or rabbit polyclonal anti-QM antibody (1:3000, SC-798; Santa Cruz Biotech.). For the detection of hRad51 we used polyclonal rabbit anti-hRad51 antibody (1:3000, PC-130; Calbiochem) or monoclonal mouse anti-hRad51 antibody (1:1000, NB100-148; Novus Biologicals).
RESULTS
Isolation of human lymphoid cell lines overexpressing tagged XRCC3 and Rad51C
TK6 and WTK1 human cell lines overexpressing tagged XRCC3 and Rad51C, respectively, were isolated in order to characterize their in vivo protein interactions and to study the phenotypic effect of their overexpression. These (His)6-tagged proteins were used for interaction studies because their characterization gave much cleaner results than immunoprecipitation of the native proteins using polyclonal antibodies and because a monoclonal antibody against XRCC3 is not yet available. TK6 cells are human B-lymphoblasts derived from WIL-2 cells that express wild-type P53 (also referred to as Tp53) (29–31). TK6 cells were transfected with pDS158 to ectopically express XRCC3-HA-(His)6 [hence referred to as just XRCC3-(His)6] using the CMV promoter. The plasmid pDS158 has already been shown to complement an xrcc3 mutation in a CHO cell line (2). Several TK6 clones were isolated that overexpressed XRCC3-(His)6 as determined by western blot. The stable transfectant having the highest level of XRCC3-(His)6 expression was named TK6-XRCC3. The level of XRCC3-(His)6 protein in TK6-XRCC3 cells was determined to be ∼10-fold higher than that of native XRCC3 protein (Fig. 1A). We intended to generate TK6 cells overexpressing a similarly tagged recombinant Rad51C protein, but were unsuccessful in several attempts. We speculate that this result reflects either a cytotoxic or cytostatic function of elevated levels of Rad51C that is particularly evident in cells expressing wild-type P53. We were able to isolate stable Rad51C overexpressing clones from WTK1 cells, a sister cell line of TK6 that expresses homozygous mutant P53 (29–31). Western analysis indicated that the clone (WTK1-Rad51C) that appeared to express the highest level of recombinant Rad51C protein expressed Rad51C-(His)6 at only ∼35–50% of the level of the native Rad51C (Fig. 1B).
Figure 1.
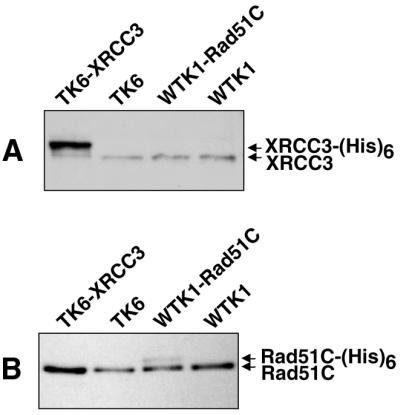
Expression levels of recombinant proteins in stable transfectants of human cell lines. Western blot analysis of 25 µg of protein extracted from each cell line was used to monitor the protein expression levels of recombinant XRCC3-(His)6 in TK6-XRCC3 cells and Rad51C-(His)6 in WTK1-Rad51C cells. (A) Native and recombinant XRCC3 was detected using polyclonal anti-XRCC3 antibody (P. Sung). (B) Native and recombinant Rad51C were detected using polyclonal anti-Rad51C antibody (P. Sung).
Constitutive and X-ray-induced protein levels of Rad51 paralogs in human lymphoid cells
Before characterizing the patterns of interactions among the Rad51 paralogs and whether DNA damage affected these patterns, we wanted to make sure that each paralog was detectable in our cell lines and determine if DNA damage altered their protein levels. Relative protein expression levels for each of the Rad51 paralogs and for hRad51 were determined, before and after X-ray treatment, in the four human cell lines we used in this study (WTK1, WTK1-Rad51C, TK6 and TK6-XRCC3) (Fig. 2). Densitometry of signal intensities was corrected for loading, using the immunoblot signals for either β-tubulin or transcription factor QM. As each antibody used has a different strength of interaction, it is not possible to determine the actual amount of each paralog, nor make comparisons between the amounts of the different paralogs. The levels of the native Rad51 paralogs and of hRad51 do not differ between TK6 and WTK1 cells. The relative native protein levels of the different paralogs and of hRad51 did not vary between WTK1 and WTK1-Rad51C, or between TK6 and TK6-XRCC3, with one notable exception: TK6-XRCC3 cells appear to contain higher levels of the native Rad51C protein (∼1.5-fold) than TK6 cells (Figs 1B and 2B). Different antibodies against XRCC3 were used in the experiments depicted in Figures 1 and 2. One polyclonal antibody (Novus, Fig. 2) was made against a C-terminal peptide of XRCC3. While it recognizes the native protein well, it does not recognize the C-terminally tagged recombinant XRCC3 protein nearly as well as does the polyclonal antibody (from P. Sung; Fig. 1) made against the complete XRCC3 protein. Therefore, Figure 1 most accurately depicts the true level of XRCC3 overexpression.
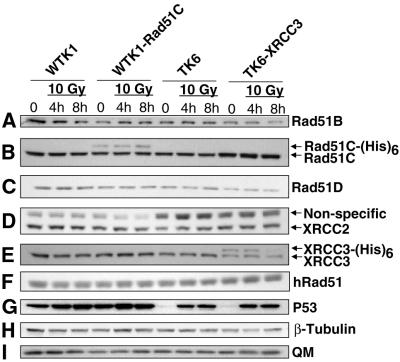
Figure 2.
Constitutive and X-ray-induced levels of the Rad51 paralogs in different human cell lines. Western blot analysis using different antibodies was used to determine the constitutive level of the Rad51 paralogs and of hRad51, and the level of these proteins 4 and 8 h after treatment with 10 Gy X-rays. P53 was used as a control for the X-ray treatment (see text), and either β-tubulin or transcription factor QM was used as loading standards for each western blot (only one representative blot for each is shown). Unlike Figure 1, the detection of XRCC3 here was done using an antibody from Novus that only weakly recognizes the recombinant XRCC3-(His)6 protein (see text).
In order to test whether any of the Rad51 paralogs were induced or suppressed by DNA damage, the relative amount of each paralog was measured in whole-cell extracts made 4 and 8 h post-irradiation with 10 Gy X-rays. To ensure that the X-irradiation stimulated the expression of a well-characterized damage-response protein, we monitored P53. For TK6 cells we detected an increase in P53 4 and 8 h post-irradiation, but no significant increase in the WTK1 background, as reported previously (30) (Fig. 2G). In contrast, there were no significant X-ray-induced changes in the expression of any of the native Rad51 paralogs. Minor differences seen within any single experiment may be attributed to slight differences in the amount of protein loaded onto the gels and/or in the amounts of the protein that were transferred onto the membrane, and were not reproducible. Thus, we conclude that X-irradiation failed to alter the level of any paralog. This experiment does not rule out small differences, for instance if they were much less than 2-fold. We did observe in two western blots that TK6-XRCC3 cells exposed to 10 Gy X-rays showed a decrease in the level of recombinant XRCC3 8 h post-irradiation; the reason for this decrease is unknown.
Effect of recombinant Rad51C and XRCC3 on DNA damage-induced cell death
The cytotoxic response to X-rays was determined for two independently isolated WTK1-Rad51C cell lines (clones 1–3 and clone 8). These results were compared with the parental WTK1 cell line and to two control cell lines that were stably transfected with the pcDNA3 vector that contains the empty neo-cassette. No differences were observed in the cytotoxic response to X-rays between these cell lines, suggesting that the modest increase in the expression of Rad51C was not of physiologic significance (data not shown). To test this further, we exposed WTK1-Rad51C cells and appropriate control cells to the inter-strand cross-linking agent MMC and also found no significant differences in survival. We also investigated the cytotoxic response of TK6-XRCC3 to MMC and found that the toxicity was very similar in TK6 cells and TK6-XRCC3 cells, but as this experiment was performed only once, this result is preliminary. Taken together, the ectopic expression of recombinant XRCC3 or Rad51C in TK6 or WTK1 cells does not appear to confer a change in the sensitivities to IR or MMC for the levels of overexpression that were achievable.
Protein associations of hRad51 paralogs in human cell lines expressing tagged proteins
To test for the interaction of XRCC3 and Rad51C in human cells, we used TK6-XRCC3 and WTK1-Rad51C cells. Each of the tagged proteins was pulled down from native cell extracts using Ni2+–NTA beads, and associated proteins were identified by western blotting. The experiments revealed the association of XRCC3 and Rad51C in extracts of both cell lines (Fig. 3A and B). In addition, we detected Rad51B, Rad51D and XRCC2 in Ni2+ pull downs in WTK1-Rad51C extracts, but these proteins were not detected in Ni2+ pull downs in TK6-XRCC3 extracts (Fig. 3C–E). The failure to detect proteins other than Rad51C in the XRCC3 pull down cannot be attributed to differences in the native abundance of these proteins. Instead, it is likely that two complexes of these proteins are present in undamaged cells: one containing Rad51C and XRCC3, and another containing Rad51C, Rad51B, Rad51D and XRCC2. As pulling down Rad51C-(His)6 did not pull down native Rad51C (Fig. 3A), and pulling down XRCC3-(His)6 did not pull down native XRCC3 (Fig. 3B), this experiment also demonstrates that neither Rad51C nor XRCC3 interacts with itself either directly or indirectly. Rad51 was not detected over background in either pull down experiment (Fig. 3F). Additional Ni2+ pull-down experiments were carried out in the presence of DNase I or ethidium bromide to test whether the observed interactions required DNA (see Materials and Methods); these treatments did not alter the observed interactions, demonstrating that these associations do not require the presence of DNA (data not shown).
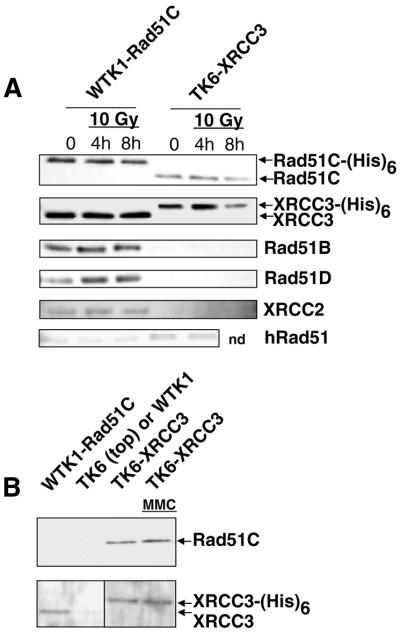
Figure 3.
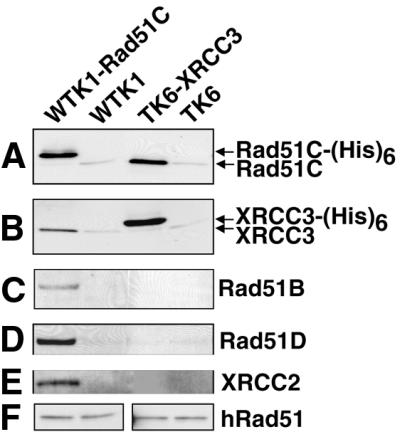
Interactions of XRCC3 and Rad51C, and of Rad51C with Rad51B, Rad51D and XRCC2 in human cells. Following Ni2+ pull down of recombinant Rad51C-(His)6 and XRCC3-(His)6 from untreated cells, western blot analysis was used to determine which of the Rad51 paralogs interacted constitutively. Rad51C-(His)6 is directly pulled down in the first lane, and XRCC3-(His)6 is directly pulled down in the third lane. WTK1 and TK6 (second and fourth lanes) are negative controls. (A) Western blot using antibody against Rad51C (from P. Sung). (B) Western blot using antibody against XRCC3 (P. Sung). (C–E) Western blots using antibodies against Rad51B (J. Albala), Rad51D (Novus) and XRCC2 (P. Sung), respectively, showing that these proteins are associated with Rad51C-(His)6, but not with XRCC3-(His)6. (F) Western blots using antibody against Rad51 (Calbiochem). A small fraction of native Rad51C, XRCC3 and Rad51 were non-specifically bound to the Ni2+ beads (A, B and F).
To test if DNA damage changed the protein interactions observed in untreated cells, the WTK1-Rad51C and TK6-XRCC3 cell lines were treated with 10 Gy X-rays. No difference in the pattern of protein interactions was observed following X-ray treatment (Fig. 4A). MMC treatment of TK6-XRCC3 cells did not modulate the interaction between recombinant XRCC3 and native Rad51C (Fig. 4B).
Figure 4.
Interactions between Rad51 paralogs in Ni2+ pull-down experiments are unaffected by DNA damage. (A) WTK1-Rad51C and TK6-XRCC3 cells were treated with 10 Gy X-ray and protein extracts (same as used in Fig. 2) were used in Ni2+ pull-down experiments. In the first three lanes, Rad51C-(His)6 is pulled down, and in the last three lanes, XRCC3-(His)6 is pulled down. (B) Treatment of TK6-XRCC3 with MMC (0.5 µg/ml for 4 h) does not affect the interaction of Rad51C with XRCC3-(His)6. In the first lane, Rad51C-(His)6 is pulled down, and in the last two lanes, XRCC3-(His)6 is pulled down. TK6 was the negative control in the top panel and WTK1 was used as a negative control in the bottom panel (second lane in each case). Please note that the upper part of (B) was probed with a monoclonal antibody against Rad51C, and this antibody does not recognize the recombinant form of Rad51C-(His)6. This is very probably due to the interference of the His6-tag with the C-terminally directed epitope of this antibody.
Co-immunoprecipitation of untagged Rad51C and the other Rad51 paralogs from TK6 cells
As we were not able to isolate a TK6 cell line stably overexpressing Rad51C, Ni2+ pull-down experiments with Rad51C were done in the WTK1-derived cells (see above). To assess whether Rad51C showed the same pattern of interactions in both cell lines, native Rad51C was immunoprecipitated from TK6 cells, and western analysis used to determine which other proteins were associated with Rad51C. Rad51C was found to interact with each of the other Rad51 paralogs, but not with Rad51 itself (Fig. 5), in agreement with the results of the Ni2+ pull-down experiments using extracts of WTK1-Rad51C cells.
Figure 5.
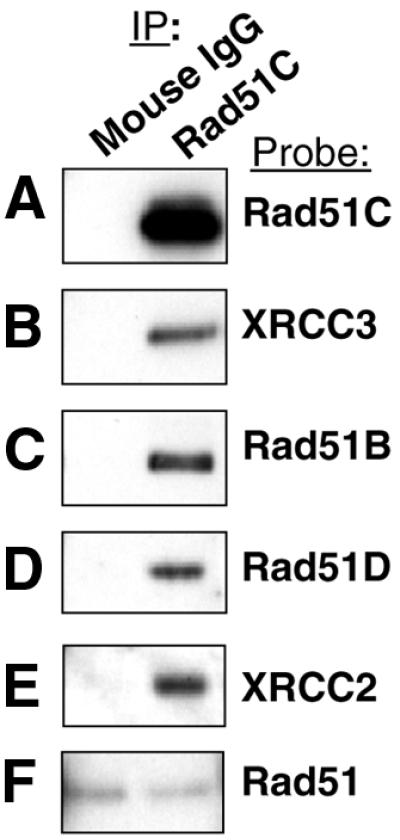
Immunoprecipitation of native Rad51C from TK6 cells results in co-immunoprecipitation of the other four Rad51 paralogs, but not Rad51. A monoclonal Rad51C antibody was used for immunoprecipitation (right hand lanes), and normal mouse IgG was used as a negative control (left hand lanes). (A–F) Polyclonal antibodies specific for each Rad51 paralog and for Rad51 were used in western detection.
DISCUSSION
The five Rad51 paralogs are each essential for the normal functioning of HRR. In chicken cells these proteins are involved in the repair of double-strand breaks (17,18), and the three mouse homologs that have been tested (Rad51B, Rad51D and XRCC2) are required for development (32–34). In order to characterize these proteins in human cells, two closely related human lymphoblast cell lines (TK6 and WTK1) were used. TK6 cells are wild-type with regard to P53, while WTK1 cells constitutively expresses only a mutant form of P53 (Met 237 to Ile) and are impaired in undergoing apoptosis after ionizing radiation (29–31). WTK1 cells manifest higher levels of homologous recombination and inter-homolog mitotic recombination, presumably due to the unusual P53 mutation (35–37). In the present study, human cell lines overexpressing two of the Rad51 paralogs (XRCC3 and Rad51C) were engineered. It is unclear why no stable transfectants of TK6 cells overexpressing Rad51C could be isolated, but one possibility is that overexpression of this protein is deleterious to cells containing wild-type P53.
Previously, it was reported that overexpression of the human Rad51B protein in CHO cells caused a G1 delay in untreated cells and an increased sensitivity to UV due to increased apoptosis (38). However, we found no effect of Rad51C or XRCC3 overexpression on MMC or X-ray sensitivity. Their results might be explained by heterologous overexpression of a human protein in hamster cells, whereas our experiments were performed with human cells overexpressing human proteins. Our yeast two-hybrid experiments showed differences in the abilities of human and mouse Rad51D proteins to interact with human Rad51C, suggesting that heterologous expression of some paralogs might not have the same effect as homologous expression (20).
Our experiments show that radiation exposure does not appear to significantly change the level of any of the Rad51 paralogs, suggesting that none of the proteins are induced by double-strand breaks. It has been reported that RAD51B is transcriptionally induced by X-rays (7), but we saw no evidence for induction of the Rad51B protein after exposure to X-rays. When comparing the TK6 parental and TK6-XRCC3 cell lines both with and without DNA damage, the level of native Rad51C protein is reproducibly higher (∼1.5-fold) in TK6-XRCC3 cells, but the levels of Rad51B, Rad51D and XRCC2 are normal (Fig. 2). Our favored explanation for this observation is that the higher level of the XRCC3 protein in TK6-XRCC3 cells helps stabilize specifically the Rad51C protein. This interpretation is consistent with our observation that XRCC3 is bound to Rad51C but is not associated with any of the other Rad51 paralogs (discussed below).
Previous work, from both our group and others, has shown that in the yeast two-hybrid and baculovirus systems each human Rad51 paralog has the ability to interact with one or more of the others. The previous reports on interactions among human Rad51 paralogs in human cells showed that in HeLa cells Rad51D interacts with XRCC2 (21), and XRCC3 interacts with Rad51C (23). Besides confirming that both of these interactions also occur in human lymphoblastoid cells (Figs 3 and 5), we demonstrate that other specific interactions observed in yeast or insect cells are actually occurring amongst the native proteins in human cells. We found that Rad51C interacts directly or indirectly with Rad51B, Rad51D and XRCC2. Conversely and somewhat surprisingly, we did not observe any direct or indirect interactions between XRCC3 and Rad51B, Rad51D or XRCC2. It seems extremely unlikely that this lack of interaction is an artifact due to the tagged nature of the XRCC3 protein, as this same tagged protein has been found to complement the xrcc3 mutation in IRS1-SF cells (2). X-irradiation did not change any of the observed interactions.
Previously, we suggested that all of the paralogs might interact simultaneously in a single complex (20). This speculation was based on results obtained using human proteins in yeast two- and three-hybrid experiments and in baculovirus experiments. We have presented evidence that in both unirradiated and X-irradiated human cells, the paralogs do not appear to form one large complex. Instead, there seem to be at least two complexes, both of which contain Rad51C: an XRCC3–Rad51C complex and a complex that probably contains Rad51B–Rad51C–Rad51D–XRCC2 (Fig. 6). Our Rad51C experiments involve direct isolation of Rad51C, by either Ni2+ pull down or by immunoprecipitation, and western analysis to detect the presence of other Rad51 paralogs that might be present in the Rad51C protein complexes. Using these methods, we cannot determine if there was a single complex containing Rad51B–Rad51C–Rad51D–XRCC2 or, instead, two different complexes: a Rad51B–Rad51C heterodimer and a Rad51C–Rad51D–XRCC2 heterotrimer (Fig. 6A). Our experiments cannot distinguish between direct and indirect interactions. Therefore, the presence of the Rad51C–Rad51D–XRCC2 heterotrimer is inferred based on the evidence that Rad51C is associated with both Rad51D and XRCC2 (Fig. 3), as well as results from both the yeast two- and three-hybrid and baculovirus systems indicating that Rad51C only interacts with XRCC2 when Rad51D is present (20). We therefore assume either that XRCC2 and Rad51C interact indirectly in human cells through their mutual interactions with Rad51D, or that they interact directly, but that Rad51D is required for this interaction, possibly by inducing a conformational change in either XRCC2 or Rad51C.
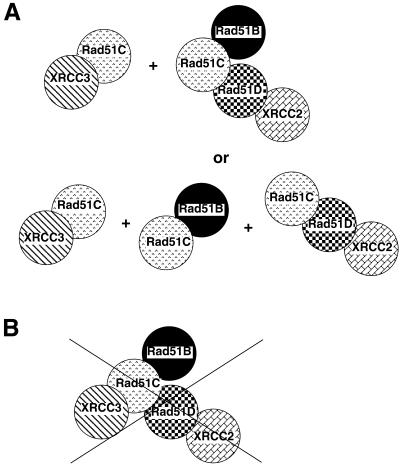
Figure 6.
Summary of interactions observed between Rad51 paralogs in human cells. (A) Evidence from this study suggests that human cells contain two different complexes of Rad51 paralogs (upper row), but our data could also be explained by three different complexes (lower row) or a mixture of these complexes. (B) One large complex containing all of the Rad51 paralogs seems unlikely to exist in human cells because XRCC3 does not appear to associate with Rad51B, Rad51D or XRCC2.
Three independent studies have found evidence that a heterotetramer containing Rad51B–Rad51C–Rad51D–XRCC2 exists in some human cells lines (39–41). These studies have also independently found that the only Rad51 paralog with which XRCC3 interacts is Rad51C, and that Rad51C can interact with each of the other paralogs. Our results agree with these observations, suggesting that the Rad51 paralogs form similar protein complexes in various human cell lines. Although our results suggest a heterotrimer and/or heterotetramer, it is very possible that individual dimers, such as XRCC2/Rad51D and Rad51B/Rad51C, may be present simultaneously in human cells.
Although our current results from human cells are consistent with most of our previous yeast two- and three-hybrid and baculovirus experiments, there are a few cases in which the current results diverge. One example is the apparent lack of interaction between either XRCC3 or Rad51C with Rad51 in our human cell lines, even though Rad51 is easily detected in both TK6 and WTK1 cells (Fig. 2). In the yeast two-hybrid system, Rad51 interacts strongly with XRCC3 and weakly with Rad51C, and the weak interaction of Rad51 with Rad51C is enhanced by simultaneous overexpression of XRCC3 (20). We also reported that both Rad51C and XRCC3 interact with Rad51 when the proteins are expressed in insect cells. In addition, the interaction of XRCC3 and Rad51 was seen in HeLa cells overexpressing recombinant XRCC3 (2). The lack of an interaction in the current experiments might relate to the cell lines used, or might be due to these interactions being either very weak or transient. A similar conundrum exists in yeast for Rad51 and one of its paralogs, Rad55. While Rad55 interacts strongly with Rad51 in the yeast two-hybrid system (42,43), the two proteins interact very weakly in biochemical experiments (19).
Evidence presented here shows that in human cells XRCC3 and Rad51B are not in the same complex, although each can interact independently with Rad51C. This finding is in contradistinction to our yeast three-hybrid data, as the direct interaction between XRCC3 and Rad51C was enhanced by expression of Rad51B (20). This difference may be reconciled by hypothesizing that in the yeast three-hybrid system the interaction between Rad51B and Rad51C changes the conformation of Rad51C, such that it can now more easily bind to XRCC3 in a type of exchange reaction. Alternatively, a temporary interaction between Rad51B and Rad51C might stabilize Rad51C, enabling more of this protein to interact independently with XRCC3. A third possibility is that the three-hybrid result was an artifact. In an unpublished yeast three-hybrid experiment, we also asked whether overexpression of Rad51C could act to bridge the interaction of XRCC3 and Rad51B, and we found that it could not. This result is consistent with our inability to see an indirect interaction between XRCC3 and Rad51B in human cells. Another difference from previous experiments involves evidence from the yeast three-hybrid system that Rad51B can interact with two molecules of Rad51C (20). We see no evidence for this in these human cells; when we pull down recombinant Rad51C, we also pull down native Rad51B, but do not detect native Rad51C [i.e. we do not observe the Rad51C-(His)6/Rad51B/Rad51C heterotrimer]. This argues that the Rad51C/Rad51B/Rad51C heterotrimer is unlikely to exist in these human cell lines.
In summary, we identified at least two protein complexes, each containing Rad51 paralogs. An explicit demonstration of the functional importance of the Rad51 paralogs in human cells in recombinational repair awaits the development of human cell mutants in which one or more of the paralogs is either knocked out or functionally deficient. Evidence already exists that the human Rad51 paralogs can restore recombinational repair proficiency when the corresponding native protein has been lost in rodent and chicken cell mutants (2,3,5,17,18). Our finding of at least two protein complexes, both containing Rad51C, suggests that there might be a type of hand-off or exchange reaction occurring between these complexes during recombinational repair in human cells.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Patrick Sung for generously sending us antibodies, Stacey Gauny for assisting with cell culture and the survival curves, and Jens Hain for suggesting the use of magnetic anti-IgG beads for immunoprecipitation experiments. We would also like to thank Kristi Miller, J.-Y. Masson, S. C. West and Nan Liu for sharing their results prior to publication. This work was supported by NIH grant GM30990 (to D.S.), NIH grant CA73966 (to A.K.), the NASA NSCORT in Radiation Health and NASA grant T-964W (to A.K.), all administered under DOE contract no. DE-AC03-76SF00098 to LBNL. J.A. was supported by NIH grant CA81019 and the California Breast Cancer Research Program 5KB-0123, and work by L.H.T. was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under contract no. W-7405-ENG-48.
REFERENCES
- 1.Bianco P.R., Tracy,R.B. and Kowalczykowski,S.C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci., 3, D570–D603. [DOI] [PubMed] [Google Scholar]
- 2.Liu N., Lamerdin,J.E., Tebbs,R.S., Schild,D., Tucker,J.D., Shen,M.R., Brookman,K.W., Siciliano,M.J., Walter,C.A., Fan,W., Narayana,L.S., Zhou,Z.Q., Adamson,A.W., Sorensen,K.J., Chen,D.J., Jones,N.J. and Thompson,L.H. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell., 1, 783–793. [DOI] [PubMed] [Google Scholar]
- 3.Cartwright R., Tambini,C.E., Simpson,P.J. and Thacker,J. (1998) The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res., 26, 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- 5.Tebbs R.S., Zhao,Y., Tucker,J.D., Scheerer,J.B., Siciliano,M.J., Hwang,M., Liu,N., Legerski,R.J. and Thompson,L.H. (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA, 92, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce A.J., Johnson,R.D., Thompson,L.H. and Jasin,M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev., 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice M.C., Smith,S.T., Bullrich,F., Havre,P. and Kmiec,E.B. (1997) Isolation of human and mouse genes based on homology to REC2, a recombinational repair gene from the fungus Ustilago maydis. Proc. Natl Acad. Sci. USA, 94, 7417–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albala J.S., Thelen,M.P., Prange,C., Fan,W., Christensen,M., Thompson,L.H. and Lennon,G.G. (1997) Identification of a novel human RAD51 homolog, RAD51B [Erratum, Genomics (1998) 51, 480]. Genomics, 46, 476–479. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright R., Dunn,A.M., Simpson,P.J., Tambini,C.E. and Thacker,J. (1998) Isolation of novel human and mouse genes of the recA/RAD51 recombination-repair gene family. Nucleic Acids Res., 26, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dosanjh M.K., Collins,D.W., Fan,W., Lennon,G.G., Albala,J.S., Shen,Z. and Schild,D. (1998) Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res., 26, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pittman D.L., Weinberg,L.R. and Schimenti,J.C. (1998) Identification, characterization and genetic mapping of Rad51d, a new mouse and human RAD51/RecA-related gene. Genomics, 49, 103–111. [DOI] [PubMed] [Google Scholar]
- 12.Kawabata M. and Saeki,K. (1998) Sequence analysis and expression of a novel mouse homolog of Escherichia coli recA gene. Biochim. Biophys. Acta, 1398, 353–358. [DOI] [PubMed] [Google Scholar]
- 13.Thompson L.H. and Schild,D. (1999) The contribution of homologous recombination in preserving genome integrity in mammalian cells. Biochimie, 81, 87–105. [DOI] [PubMed] [Google Scholar]
- 14.Thacker J. (1999) A surfeit of RAD51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- 15.Thompson L.H. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- 16.Brenneman M.A., Weiss,A.E., Nickoloff,J.A. and Chen,D.J. (2000) XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat. Res., 459, 89–97. [DOI] [PubMed] [Google Scholar]
- 17.Takata M., Sasaki,M.S., Sonoda,E., Fukushima,T., Morrison,C., Albala,J.S., Swagemakers,S.M., Kanaar,R., Thompson,L.H. and Takeda,S. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol., 20, 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takata M., Sasaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 20.Schild D., Lio,Y., Collins,D.W., Tsomondo,T. and Chen,D.J. (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem., 275, 16443–16449. [DOI] [PubMed] [Google Scholar]
- 21.Braybrooke J.P., Spink,K.G., Thacker,J. and Hickson,I.D. (2000) The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem., 275, 29100–29106. [DOI] [PubMed] [Google Scholar]
- 22.O’Regan P., Wilson,C., Townsend,S. and Thacker,J. (2001) XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem., 276, 22148–22153. [DOI] [PubMed] [Google Scholar]
- 23.Masson J.Y., Stasiak,A.Z., Stasiak,A., Benson,F.E. and West,S.C. (2001) Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl Acad. Sci. USA, 98, 8440–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurumizaka H., Ikawa,S., Nakada,M., Eda,K., Kagawa,W., Takata,M., Takeda,S., Yokoyama,S. and Shibata,T. (2001) Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc. Natl Acad. Sci. USA, 98, 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy J.A., Virolainen,M. and Defendi,V. (1968) Human lymphoblastoid lines from lymph node and spleen. Cancer, 22, 517–524. [DOI] [PubMed] [Google Scholar]
- 26.van den Hoff M.J., Moorman,A.F. and Lamers,W.H. (1992) Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res., 20, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furth E.E., Thilly,W.G., Penman,B.W., Liber,H.L. and Rand,W.M. (1981) Quantitative assay for mutation in diploid human lymphoblasts using microtiter plates. Anal. Biochem., 110, 1–8. [DOI] [PubMed] [Google Scholar]
- 28.Lai J.S. and Herr,W. (1992) Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl Acad. Sci. USA, 89, 6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia F., Wang,X., Wang,Y.H., Tsang,N.M., Yandell,D.W., Kelsey,K.T. and Liber,H.L. (1995) Altered p53 status correlates with differences in sensitivity to radiation-induced mutation and apoptosis in two closely related human lymphoblast lines. Cancer Res., 55, 12–15. [PubMed] [Google Scholar]
- 30.Little J.B., Nagasawa,H., Keng,P.C., Yu,Y. and Li,C.Y. (1995) Absence of radiation-induced G1 arrest in two closely related human lymphoblast cell lines that differ in p53 status. J. Biol. Chem., 270, 11033–11036. [DOI] [PubMed] [Google Scholar]
- 31.Zhen W., Denault,C.M., Loviscek,K., Walter,S., Geng,L. and Vaughan,A.T. (1995) The relative radiosensitivity of TK6 and WI-L2-NS lymphoblastoid cells derived from a common source is primarily determined by their p53 mutational status. Mutat. Res., 346, 85–92. [DOI] [PubMed] [Google Scholar]
- 32.Shu Z., Smith,S., Wang,L., Rice,M.C. and Kmiec,E.B. (1999) Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(–/–) background. Mol. Cell. Biol., 19, 8686–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pittman D.L. and Schimenti,J.C. (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis, 26, 167–173. [DOI] [PubMed] [Google Scholar]
- 34.Deans B., Griffin,C.S., Maconochie,M. and Thacker,J. (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J., 19, 6675–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amundson S.A. and Chen,D.J. (1996) Inverse dose-rate effect for mutation induction by gamma-rays in human lymphoblasts. Int. J. Radiat. Biol., 69, 555–563. [DOI] [PubMed] [Google Scholar]
- 36.Xia S.J., Shammas,M.A. and Shmookler Reis,R.J. (1997) Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol. Cell. Biol., 17, 7151–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiese C., Gauny,S.S., Liu,W.C., Cherbonnel-Lasserre,C.L. and Kronenberg,A. (2001) Different mechanisms of radiation-induced loss of heterozygosity in two human lymphoid cell lines from a single donor. Cancer Res., 61, 1129–1137. [PubMed] [Google Scholar]
- 38.Havre P.A., Rice,M.C., Noe,M. and Kmiec,E.B. (1998) The human REC2/RAD51B gene acts as a DNA damage sensor by inducing G1 delay and hypersensitivity to ultraviolet irradiation. Cancer Res., 58, 4733–4739. [PubMed] [Google Scholar]
- 39.Liu N., Schild,D., Thelen,M.P. and Thompson,L.H. (2002) Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res., 30, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller K.A., Yoshikawa,D.M., McConnell,I.R., Clark,R., Schild,D. and Albala,J.S. (2002) RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 41.Masson J.Y., Tarsounas,M.C., Stasiak,A.Z., Stasiak,A., Shah,R., McIlwraith,M.J., Benson,F.E. and West,S.C. (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev., 15, 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson R.D. and Symington,L.S. (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55 and Rad57. Mol. Cell. Biol., 15, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hays S.L., Firmenich,A.A. and Berg,P. (1995) Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55 and Rad57 proteins. Proc. Natl Acad. Sci. USA, 92, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]