Abstract
Mitochondria experience genomic and functional instability in diabetes, and mitochondrial dysfunction has a critical role in the development of diabetic retinopathy. Diabetes also alters expressions of many long noncoding RNAs (LncRNAs), the RNAs with >200 nucleotides and no open reading frame. LncRNAs are mainly encoded by the nuclear genome, but mtDNA also encodes three LncRNAs. Our goal was to investigate the effect of hyperglycemia on mtDNA-encoded LncRNA cytochrome B (LncCytB) in mtDNA stability in diabetic retinopathy. Retinal endothelial cells, transfected with LncCytB-overexpressing plasmids or siRNA, incubated in 5 mmol/L d-glucose (normal glucose [NG]) or 20 mmol/L d-glucose (high glucose [HG]) for 4 days, were analyzed for LncCytB expression by strand-specific PCR and its mitochondrial localization by RNA fluorescence in situ hybridization. Damage-sensitive mtDNA regions were examined by micrococcal nuclease (MNase) digestion sequencing and LncCytB occupancy at mtDNA by chromatin isolation by RNA purification. Protective nucleoids in mtDNA were analyzed by SYBR Green-MitoTracker Red staining and confirmed in isolated mitochondria by flow cytometry. Compared with NG, HG downregulated LncCytB by >50% but had no significant effect on the other mtDNA-encoded LncRNAs. mtDNA packaging was impaired, MNase sensitivity was increased, and LncCytB occupancy at mtDNA was decreased. While LncCytB overexpression ameliorated mtDNA damage and decrease in nucleoids and copy numbers, LncCytB-siRNA exacerbated damage and further reduced nucleoids. Retinal microvessels from streptozotocin-induced diabetic mice and human donors with diabetic retinopathy presented a similar decrease in LncCytB and mtDNA nucleoids. Thus, LncCytB has a major role in maintaining mitochondrial genomic stability, and its downregulation in the hyperglycemic milieu contributes to increased vulnerability of mtDNA to damage.
ARTICLE HIGHLIGHTS
In the pathogenesis of diabetic retinopathy, an mtDNA-encoded long noncoding RNA, long noncoding cytochrome B (LncCytB), is downregulated.
Downregulation of LncCytB results in the reduced number of protective nucleoids, and the mtDNA becomes vulnerable to the damage.
The damaged mtDNA compromises the electron transport chain system, increasing superoxide levels, and the vicious cycle of free radicals continues to self-propagate.
Preventing the downregulation of LncCytB in diabetes will interfere in the self-perpetuating vicious cycle of free radicals and should inhibit the development of diabetic retinopathy.
Introduction
Mitochondrial dysfunction plays a critical role in the development of diabetic retinopathy. Retinal mitochondrial structural and functional stability are impaired, and cytochrome C leaks into the cytosol, accelerating capillary cell apoptosis, a phenomenon that precedes the histopathology characteristic of this blinding disease (1–3). Mitochondria have their own DNA (mtDNA), which encodes genes for 13 proteins, and these proteins are crucial for the functioning of the electron transport chain (ETC). Mitochondrial DNA is damaged in diabetes, and the transcription of mtDNA-encoded ND1 and ND6 of complex I and cytochrome B (CYTB) of complex III are decreased (4–6). Complexes I and III of the ETC are the major sources of superoxide radicals within the mitochondrial matrix, and in diabetes, increased glucose oxidation by the tricarboxylic acid cycle pushes more electron donors into the ETC. When the increase in the voltage gradient across the mitochondrial membrane reaches a critical threshold, electron transfer inside complex III is blocked, resulting in more superoxide generation (7). In diabetic retinopathy, while complex III activity is inhibited, complex I remains unchanged (8).
Recent technical advances have documented that only one-fifth of the transcription across the human genome is associated with protein-coding genes, and noncoding RNAs (ncRNAs) are approximately four times more than the coding RNA sequences (9). Among these, long ncRNAs (LncRNAs) are the nonprotein coding sequence transcripts that contain >200 nucleotides but have no open reading frame for translation (10,11). LncRNAs can bind to DNA or RNA in a sequence-specific manner, act as sponges for miRNAs or as scaffolds to provide stability, or facilitate DNA binding activity of their associated transcription factors (12–14). Aberrant changes in their expression is implicated in several diseases, including many metabolic abnormalities associated with diabetic retinopathy (15–18). The majority of LncRNAs are encoded by nuclear DNA (nDNA), but 15% of the mtDNA also has ncRNAs, including three major LncRNAs, two for complex I (LncRNAs ND5 and ND6) and one for complex III (LncRNA CYTB [LncCytB]) (19,20), corresponding to the regions complementary to the mitochondrial ND5, ND6, and CYTB genes. The role of mtDNA-encoded LncRNAs in diseases, including diabetic retinopathy, is unclear.
mtDNA, unlike nDNA, lacks protective histones and is more vulnerable to damage, but it is packaged into protein-DNA complexes known as nucleoids. The organization of mtDNA in nucleoids greatly influences defects in mtDNA and its biogenesis; nucleoid damage and/or their aberrant organization is implicated in aging and many chronic diseases (21–23). LncRNAs can act as scaffolds to promote protein interactions and can control chromatin remodeling complexes, altering the nucleosome spacing (11,24). In hyperglycemia, mtDNA nucleoids are decreased, and increased mitochondrial translocation of nDNA-encoded LncRNAs, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), and nuclear-enriched abundant transcript 1 (NEAT1) is implicated with this decrease (25,26). Whether LncCytB, which itself is encoded by mtDNA, has any role in maintaining mtDNA stability in diabetes remains unclear.
Our goal was to investigate the role of LncCytB in mtDNA stability in diabetic retinopathy. Using human retinal endothelial cells (HRECs), genetically manipulated for LncCytB, the effect of high glucose (HG) on LncCytB and its role in mtDNA damage and stability (nucleoids) was determined. Results were confirmed in the retinal microvessels from streptozotocin-induced diabetic mice and from human donors with documented diabetic retinopathy.
Research Design and Methods
HRECs
HRECs (cat. no. ACBRI 181; Cell Systems, Kirkland, WA), were cultured in DMEM (cat. no. D5523; Sigma-Aldrich, St. Louis, MO) containing 12% heat-inactivated FBS, 15 μg/mL endothelial cell growth supplement, and 1% each of insulin, transferrin, selenium, GlutaMAX, and antibiotic/antimitotic. Cells from the fifth to eighth passage were incubated in 5 mmol/L d-glucose (normal glucose [NG]) or 20 mmol/L d-glucose (HG) for 96 h, a duration when mtDNA damage and capillary cell apoptosis can be detected (27,28). As an osmotic/metabolic control, each experiment also included HRECs in 20 mmol/L l-glucose (L-Gl), instead of HG (25,26). Cells were provided with fresh medium (NG, HG, or L-Gl) every 24 h.
LncCytB overexpression was performed in HRECs from the fifth to sixth passage by transfecting them with LncCytB construct clone in pCMV6-AC vector (cat. nos. CW308041 and PS100020, respectively; OriGene Technologies, Rockville, MD) using TurboFectin 8.0 transfection reagent (cat. no. TF81001; OriGene Technologies). For silencing, Antisense LNA GapmeR LncCytB-siRNAs (cat. no. 339511; QIAGEN, Valencia, CA) and Lipofectamine RNAiMAX (cat. no. 13778-030; Invitrogen, Carlsbad, CA) were used. Cells transfected with pCMV6-AC empty vector or scrambled control (SC) RNA were used as controls. Each experiment was performed with cells from the same batch/passage and repeated in three to four different preparations. LncCytB overexpression/silencing efficiency was evaluated by quantifying LncCytB expression.
Mice
Diabetes was induced in mice (20 g body weight) overexpressing manganese superoxide dismutase (encoded by Sod2 gene) (Tg) or wild-type (WT) mice by injecting streptozotocin (55 mg/kg body weight i.p.) for 4 consecutive days. Three days after the last injection, mice presenting with blood glucose >250 mg/dL were considered diabetic and were maintained as diabetic for 6 months. Age-matched nondiabetic Tg and WT mice were used as their respective controls. Each group had 8–10 mice, with similar numbers of males and females. Blood glucose values in Tg or WT diabetic mice were similar (350–500 mg/dL), and were three to four times higher than their nondiabetic controls (8). The treatment of the animals conformed to the Association for Research in Vision and Ophthalmology Resolution on the Use of Animals in Research, and the protocol was approved by the Wayne State University animal care and use committee.
Human Donors
Eye globes from donors, enucleated within 6–8 h of death, were obtained from Eversight Eye Bank (Ann Arbor, MI). The diabetic retinopathy group had seven donors (three male and four female, 55–75 years of age, and 16–35 years of diabetes), and the age-matched nondiabetic control group had six donors (three male and three female). The exclusion criteria included donors with any other ocular diseases, chronic diseases (e.g., Alzheimer disease, cancer, HIV), and drug use or smoking within the 3 years before death (16).
Retinal Microvessels
One mouse retina, or one-sixth to one-eighth of human retina, was incubated in 5–10 mL of deionized water for 60 min at 37°C in a shaking water bath. Retinal microvessels were isolated under a dissecting microscope (25,26).
Gene Expression
Gene transcripts were quantified using SYBR Green–based real-time quantitative PCR using β-actin (human) or 18S rRNA (mouse) as a housekeeping gene (25,26). The list of gene/species-specific primers is provided in Supplementary Table 1.
LncCytB expression was confirmed by strand-specific RT-PCR. Two micrograms RNA was mixed with 10 μmole/L each of the forward primers of LncCytB (antisense strand specific) and β-actin/18S rRNA (human/mouse) and incubated at 70°C for 5 min, followed by transfer on ice for secondary structure denaturation and primer annealing. The reverse transcriptase buffer with deoxynucleoside triphosphate (dNTP) and reverse transcriptase enzyme was added as per the manufacturer’s protocol (cat. no. 4368814; Applied Biosystems, Foster City, CA), and the reaction mixture was incubated at 42°C for 60 min, followed by 85°C for 5 min (29). The strand-specific cDNA was used as a template, and the forward and reverse primers for LncCytB and β-actin/18S rRNA were used to amplify the cDNA.
RNA Fluorescence In Situ Hybridization
Using asymmetric PCR amplification, a fluorescein-12-dUTP–incorporated probe was prepared, and the PCR products were gel purified. HRECs fixed in 4% paraformaldehyde were dehydrated with 70–100% ethanol and after air drying, incubated at 37°C for 3 h with the denatured probe in formamide-containing buffer (10% dextran sulfate, 10% formamide, and 4× saline-sodium citrate buffer [pH 7.0]). The cells were then washed with the hybridization buffer, followed by PBS, and mounted using DAPI-containing mounting medium (cat. no. H-1000; Vector Laboratories, Burlingame, CA). The images were captured by Zeiss microscope using a 20× objective. The hybridized probes were determined by visualizing signals of fluorescein-12-dUTP–incorporated probe, and the images were calibrated with the Zeiss Zen 2.6 pro inbuilt software package and modules. The arithmetic mean intensity (AMI) was determined by Zeiss colocalization software module (16,25). Mitochondrial localization of LncCytB was performed by RNA fluorescence in situ hybridization (FISH) immunofluorescence technique using CoxIV as a mitochondrial marker (1:500 dilution, cat. no. ab153709; Abcam, Cambridge, MA). Texas Red–labeled anti-rabbit antibody was used as complex IV secondary antibody (1:500 dilution, cat. no. TI-1000-1.5; Vector Laboratories).
Micrococcal Nuclease Digestion and Sequencing
Nucleoid association with proteins protect mtDNA from nuclease digestion (30). To investigate the effect of HG on the packaging/organization of mtDNA and identification of vulnerable regions, micrococcal nuclease (MNase) cleavage was performed. As described previously (16), mitochondria were incubated with 0.5 units of MNase at 25°C for 15 min in 100 μL of MNase digestion buffer. DNA was isolated from the extract using EZ Nucleosomal DNA Prep Kit (cat. no. D5220; Zymo Research, Irvine, CA), and after checking for the uniformity, 100 ng of DNA was processed for library preparation (31). The library was outsourced for Illumina next-generation sequencing by GENEWIZ (South Plainfield, NJ). The raw FASTQ sequencing files were uploaded on the https://www.usegalaxy.org server and aligned against the hg38 human genome data set using the Bowtie 2 module. The files in BigWig format were analyzed in the University of California, Santa Cruz, browser (https://genome.ucsc.edu/) (32).
Chromatin Isolation by RNA Purification Sequencing
Chromatin isolation by RNA purification (ChIRP) sequencing was performed to determine LncCytB occupancy in mtDNA using chromatin extract prepared from 1% formaldehyde cross-linked cells. Fluorescein-12-dUTP–incorporated LncCytB probe (100 pmol, cat. no. FERR0101; Thermo Fisher Scientific, Waltham, MA) was hybridized, and the RNA-chromatin complex was immunoprecipitated by antifluorescein antibody–bound A/G PLUS-Agarose beads (cat. no. 11426320001; Roche Diagnostics GmbH, Mannheim, Germany) (16,33). RNA immunoprecipitation-associated DNA (100 ng) was used for library preparation using NEBNext Ultra II DNA library preparation kit for Illumina and NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) (cat. nos. E7645S and E7335S, respectively; New England Biolabs, Ipswich, MA). The library was outsourced for Illumina next-generation sequencing to GENEWIZ. Relative LncCytB occupancy in the CYTB region (15–16 kb) was plotted using the log ratio of NG and HG.
mtDNA Damage and Copy Numbers
mtDNA damage was evaluated by extended length PCR, a technique based on the ability of the damaged DNA to avert progression of polymerase along the DNA template (4,34). Using semiquantitative PCR, long and short regions of mtDNA (8.8 kb and 223 bp for human and 10 kb and 116 bp for mice) were amplified, and the products were separated on 1.2–2.0% agarose gel. Relative amplification of the long to the short product, which is inversely proportional to the damage, was quantified. Mitochondrial copy numbers were assayed in the genomic DNA by quantifying the ratio of gene transcripts of mtDNA-encoded CYTB and nDNA-encoded β-actin (4,5,25).
Mitochondrial Nucleoids
Nucleoids were analyzed in live cells using a costaining technique with SYBR Green for DNA and MitoTracker Deep Red (MTR) for mitochondria. Cells on the coverslips were incubated with 200 nmol/L MTR (cat. no. M22426; Thermo Fisher Scientific) for 30 min at 37°C in a 5% CO2 incubator. After rinsing with PBS, cells were fixed in 100% methanol for 15 min, and permeabilized with 0.5% Triton X-100. Cells were rinsed with PBS and incubated in 1 mL 1× SYBR DNA Gel Stain (cat. no. S33102; Invitrogen) for 1 h at room temperature and after rinsing with PBS (twice for 5 min), mounted using VECTASHIELD antifade mounting medium. The images were acquired using a Zeiss microscope with a 40× objective. After subtracting the background fluorescence and masking the nuclear fluorescence by adjusting the threshold, the number of nucleoids/cell were counted using the automatic particle counting module of ImageJ software (National Institutes of Health, Bethesda, MD) (18,25).
Nucleoids were also quantified in the isolated mitochondria. Briefly, 10 μg mitochondria (HRECs or retina prepared by the centrifugation procedure [8]) was stained with MTR and 1× SYBR Green at 37°C for 30 min in the dark and resuspended in the flow buffer (25 mmol/L Tris HCl [pH 7.4], 250 mmol/L sucrose, 2 mmol/L EDTA, and 3% BSA). They were then scanned under FL1 488-nm and FL3 640-nm wavelength in a BD Accuri C6 Plus Flow Cytometer (BD Biosciences, San Jose, CA). Raw Flow Cytometry Standard files were analyzed using WinList 3D version 9 (Verity Software House, Topsham, ME) and FlowJo version 10.8.1 (BD Biosciences) software. Relative nucleoid content was estimated by plotting forward scatter area with forward scatter on the z-axis, SYBR Green on the x-axis, and MTR on the y-axis. Mitochondria without any staining with SYBR Green or MTR were used as controls.
Statistical Analysis
The statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA), and the results are presented as mean ± SD. Significance of variance was analyzed using one-way ANOVA, and P < 0.05 was considered statistically significant.
Data and Resource Availability
The data sets generated during the current study are available from the corresponding author upon reasonable request.
Results
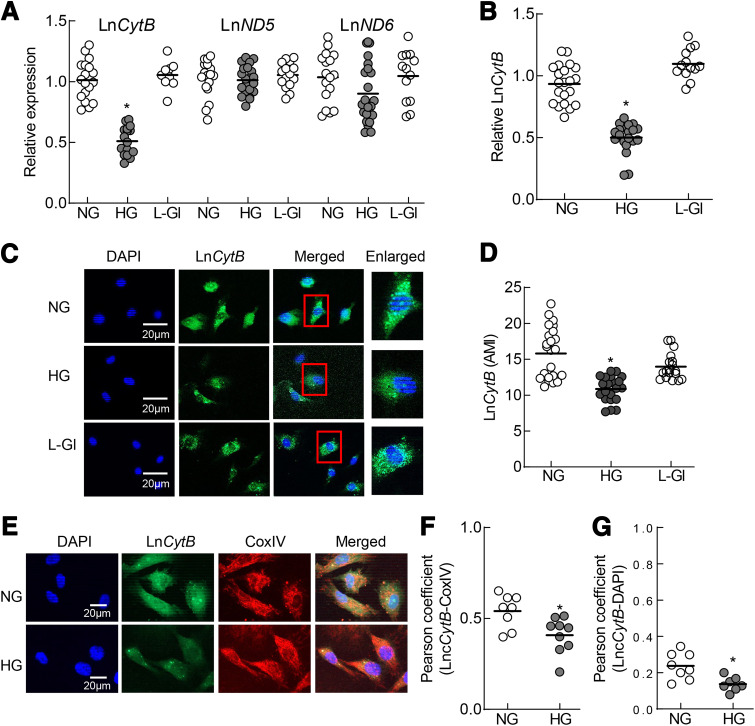
Compared with NG, HG downregulated LncCytB in HRECs by >50% (P < 0.05) and LncND6 by >10% (P > 0.05) and had no effect on LncND5 (Fig. 1A). Strand-specific PCR showed similar (>50%) downregulation of LncCytB in HG (Fig. 1B). Incubation of cells in L-Gl instead of HG had no effect on the expression of these LncRNAs. Downregulation of LncCytB was further confirmed by RNA FISH. Green fluorescence intensity was significantly lower in HG versus NG (Fig. 1C), which was also confirmed by AMI quantification (Fig. 1D). Mitochondrial levels of LncCytB were determined by RNA FISH immunofluorescence technique. Compared with NG, HG significantly decreased fluorescence intensity of LncCytB (green) in the mitochondria (Fig. 1E), and the Pearson correlation coefficient between LncCytB and CoxIV (red) showed a 40% decrease in HG versus NG (Fig. 1F). Some LncCytB staining was also observed in the nucleus, and as indicated by Pearson correlation coefficient between LncCytB and the nuclear marker DAPI (blue), it was also decreased by HG (Fig. 1G).
Figure 1.
Effect of HG on mtDNA-encoded LncRNAs. A: Transcripts of LncCytB, LncND5, and LncND6 were quantified by quantitative RT-PCR using β-actin as a housekeeping gene. B and C: LncCytB was quantified by strand-specific PCR using strand-specific cDNA as a template (B) and RNA FISH technique (C). D: AMI of LncCytB was determined using Zeiss software. E: Mitochondrial levels of LncCytB were determined by RNA FISH immunofluorescence technique using fluorescein-12-dUTP–labeled LncCytB probe (green) and Texas Red (red)–conjugated secondary antibody against mitochondrial marker CoxIV. F and G: Pearson correlation between LncCytB and CoxIV (F) and DAPI (G) was calculated using a random region of interest from ≥25 cells in each group. Graph data are mean ± SD obtained from three to four different cell preparations, with each measurement made in duplicate. *P < 0.05 vs. NG.
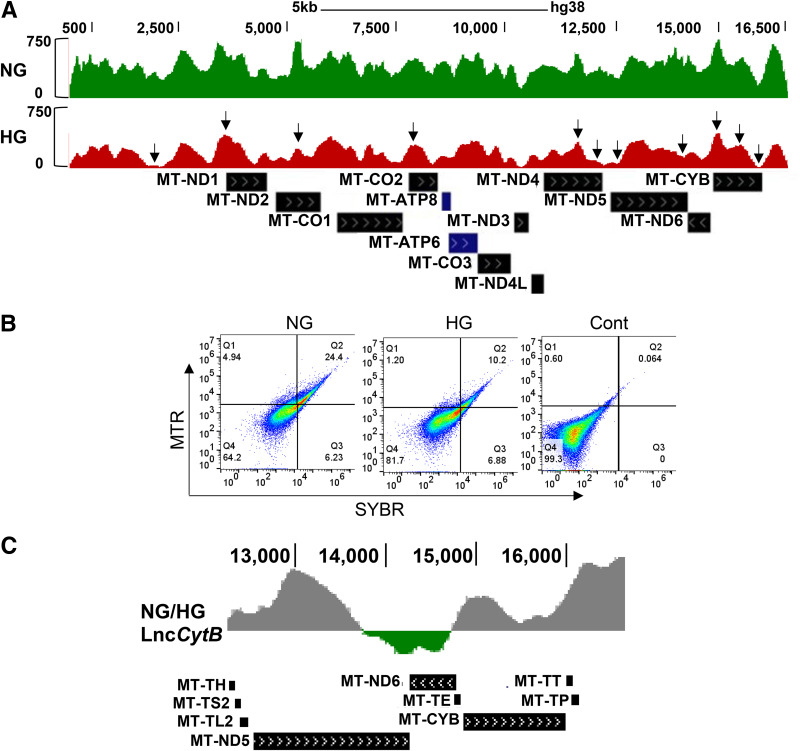
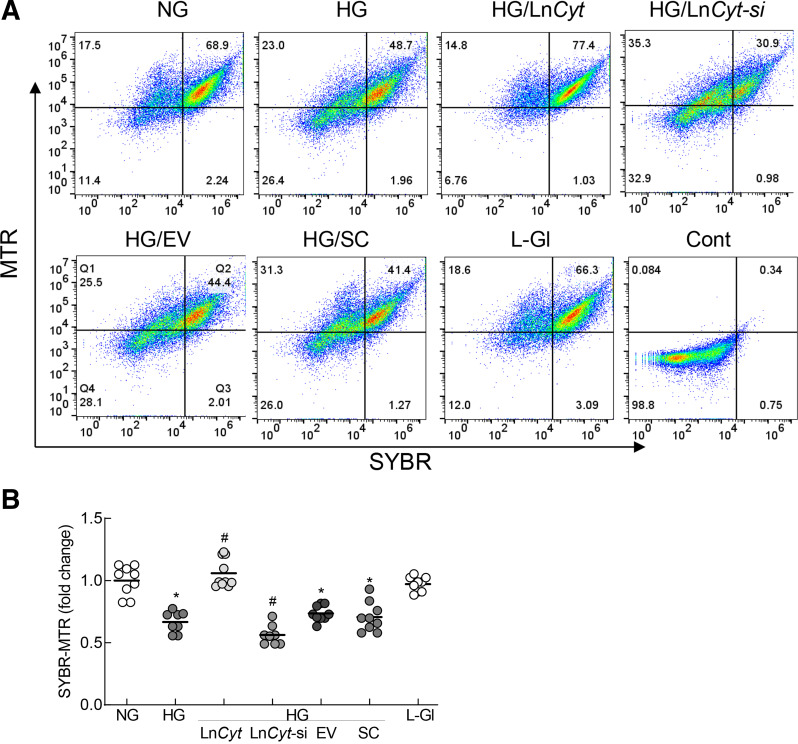
To characterize the regions of mtDNA prone to the damage, MNase sequencing was performed (35). Global cleavage of mtDNA was significantly increased in HG versus NG, and the peaks were either smaller or missing, suggesting increased disorganization (Fig. 2A). Packaging of mtDNA was further confirmed in the isolated mitochondria by dual dye–stained fluorescence flow cytometry, and Fig. 2B shows significantly decreased scattering of SYBR Green-MTR–stained cells in the second quadrant (Q2) in HG, suggesting decreased packaging. LncRNAs provide protection to the DNA by binding with proteins and DNA (14); therefore, to investigate the role of LncCytB in mtDNA damage/instability, the occupancy of LncCytB at mtDNA was determined by ChIRP sequencing. Compared with NG, HG significantly decreased LncCytB occupancy at mtDNA. Figure 2C shows the log ratio of LncCytB occupancy on mtDNA in NG and HG, especially in the CYTB region of the mtDNA.
Figure 2.
Effect of HG on mtDNA sensitivity to MNase digestion and occupancy of LncCytB on mtDNA. A: Nucleosome remodeling on mtDNA was assessed by MNase-assisted nucleosomal DNA sequencing, showing the nucleosomal DNA remodeling patterns in the mtDNA. B: Packaging of mtDNA was accessed in the isolated mitochondria by staining with MTR and SYBR Green and flow cytometry under FL1 488-nm and FL3 640-nm wavelength excitation. Raw Flow Cytometry Standard files were analyzed using WinList 3D version 9. C: Log ratio of LncCytB occupancy on mtDNA in NG and HG in the CYTB region of the mtDNA (13,000–16,000 bp), performed by ChIRP using fluorescein-12-dUTP–incorporated LncCytB followed by next-generation sequencing. Arrowheads indicate blunted or missing peaks. Cont, unstained mitochondria.
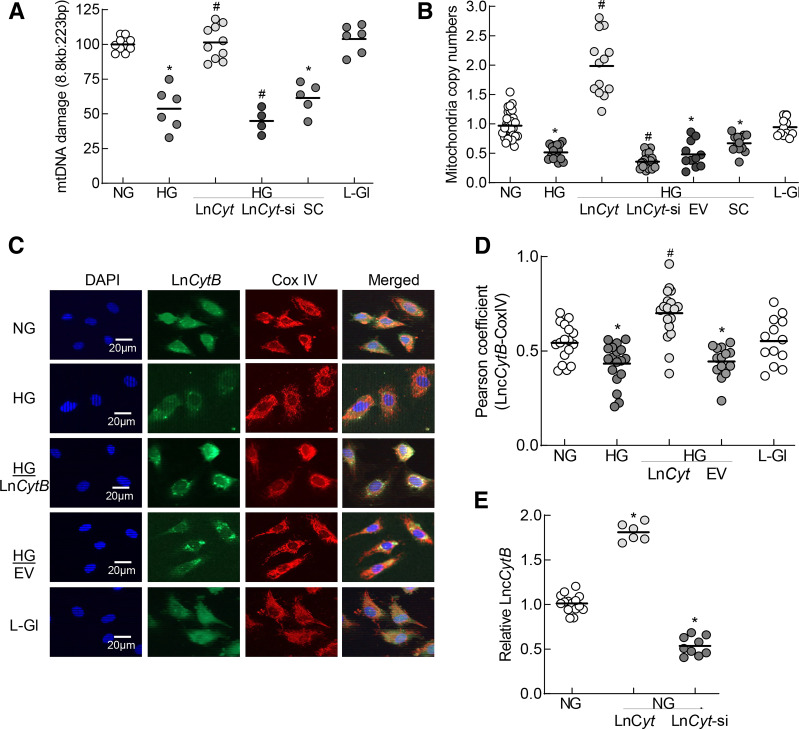
To investigate the role of LncCytB in mtDNA stability, the effect of regulation of LncCytB on mtDNA damage and copy numbers was determined. Compared with untransfected cells in HG, glucose-induced increase in mtDNA damage and decrease in their copy numbers were significantly ameliorated in LncCytB-overexpressing cells. The values from cells overexpressing LncCytB were significantly different compared with those from untransfected cells. However, LncCytB-siRNA exacerbated glucose-induced mtDNA damage and reduction in the copy numbers (P < 0.05 vs. HG). Values from cells transfected with empty vector or SC RNA or untransfected cells, incubated in HG, were not different from one another (Fig. 3A and B). LncCytB overexpression also significantly increased its mitochondrial expression, as evidenced by increased colocalization of LncCytB and CoxIV and their Pearson correlation coefficients. Cells in L-Gl had values similar to cells in NG (Fig. 3C and D). Figure 3E shows the transfection efficiencies with an approximately twofold increase and 60% decrease in LncCytB transcripts in cells transfected with LncCytB-overexpressing plasmids and LncCytB-siRNA, respectively.
Figure 3.
Effect of LncCytB regulation on mtDNA damage and copy numbers. A: mtDNA damage was quantified by extended length PCR using long mtDNA (8.8 kb) and short (223 bp) amplicons of the mtDNA; relative amplification of the long to the short product, which is inversely proportional to the damage, was quantified. B: Copy numbers of mtDNA were determined in the genomic DNA by quantifying the ratio of gene transcripts of mtDNA-encoded CYTB and nDNA-encoded β-actin. C: Mitochondrial localization of LncCytB was performed by RNA FISH using fluorescein-12-dUTP–labeled LncCytB probe (green) and Texas Red (red)–conjugated secondary antibody against CoxIV. D: Pearson correlation coefficient of CoxIV with LncCytB. E: Transfection efficiencies of LncCytB-overexpressing plasmids and siRNA in HRECs were determined by strand-specific PCR, using β-actin as a housekeeping gene. Each measurement was made in duplicate/triplicate in three to five different cell preparations, and the histogram data are mean ± SD. HG/LnCyt, HG/LnCyt-si, HG/EV, and HG/SC indicate cells transfected with LncCytB-overexpressing plasmids, LncCytB-siRNA, empty vector, and SC RNA, respectively, and incubated in HG. *P < 0.05 vs. NG; #P < 0.05 vs. HG.
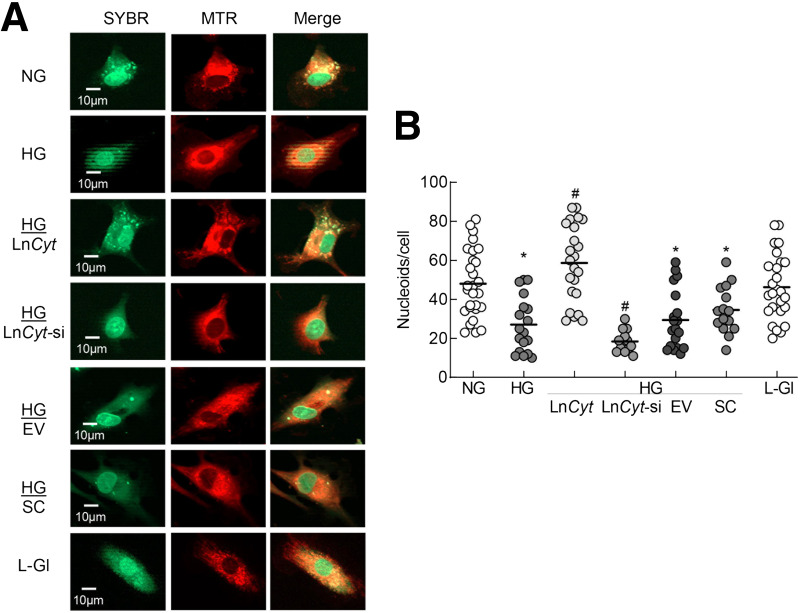
Nucleoids contain multiple DNA binding proteins, and LncRNAs can complex with DNA protein (21,23,24). To investigate the role of LncCytB in mtDNA stability, the effect of LncCytB regulation on mtDNA nucleoids was investigated. Compared with NG, as evidenced by decreased costaining of SYBR Green and MTR, HG significantly reduced nucleoid numbers. While overexpression of LncCytB prevented the decrease in nucleoids, LncCytB-siRNA further reduced their numbers. Nucleoids in untransfected cells or cells transfected with the empty vector or SC RNA, in HG, were not different from one another but were significantly lower compared with cells in NG or L-Gl (Fig. 4A and B).
Figure 4.
Regulation of LncCytB and nucleoids in mtDNA. A: Nucleoids in live cells were determined using SYBR Green for DNA and MTR for mitochondria. B: The number of nucleoids counted in five to eight images per group using ImageJ software. Measurements were made in duplicate/triplicate in three to four cell preparations, and the data are mean ± SD. HG/LnCyt, HG/LnCyt-si, HG/EV, and HG/SC indicate transfected cells with LncCytB-overexpressing plasmids, LncCytB-siRNA, empty vector, and SC RNA, respectively, in HG. *P < 0.05 vs. NG; #P < 0.05 vs. HG.
The role of LncCytB in nucleoids was further confirmed by flow cytometry. As shown in Fig. 5A and B, cells positive for SYBR Green and MTR, using the scattering values obtained in Q2, were significantly lower in HG versus NG or L-Gl. This was ameliorated by LncCytB overexpression but was further decreased by LncCytB-siRNA.
Figure 5.
Nucleoids in isolated mitochondria. A: Nucleoids in isolated mitochondria were analyzed by flow cytometry using MTR and SYBR Green dual staining. B: Relative SYBR Green-MTR–positive cells were analyzed using the scattering values obtained in Q2. HG/LnCyt, HG/LnCyt-si, HG/EV, and HG/SC indicate cells transfected with LncCytB-overexpressing plasmids, LncCytB-siRNA, empty vector, and SC RNA, respectively, and incubated in HG. *P < 0.05 vs. NG; #P < 0.05 vs. HG. Cont, unstained mitochondria.
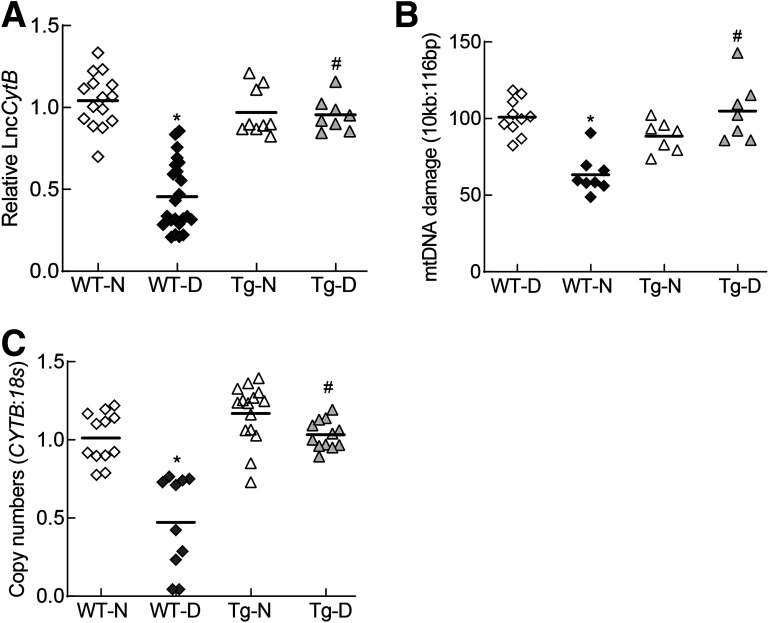
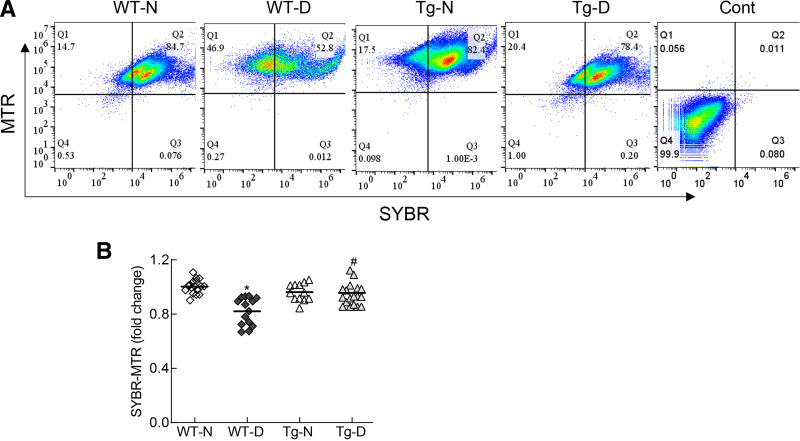
Consistent with the results from HRECs, strand-specific PCR showed a >50% decrease in the relative expression of LncCytB in the retinal microvasculature from WT diabetic mice (WT-D) compared with their age-matched WT normal mice (WT-N) (Fig. 6A). In the same diabetic mice, mtDNA damage was significantly increased, and copy numbers were decreased (Fig. 6B and C). Flow cytometry analysis performed in the isolated mitochondria showed a significantly reduced number of SYBR Green-MTR–positive cells in WT-D compared with WT-N (Fig. 7A and B). However, in the Tg diabetic mice (overexpressing Sod2) (Tg-D), relative LncCytB expression was similar to that in WT-N. Similarly, the increase in mtDNA damage and decrease in mtDNA copy numbers and nucleoids seen in WT-D were also protected in Tg-D. Tg normal mice (Tg-N) had similar LncCytB expression, mtDNA damage, copy numbers, and nucleoids as WT-N, and these values were not significantly different from those in Tg-D (Figs. 6 and 7).
Figure 6.
Effect of diabetes on LncCytB expression and mtDNA damage and copy numbers. A: LncCytB transcripts were quantified in the retinal microvessels by strand-specific PCR using 18S as a housekeeping gene. B: mtDNA damage was assessed by extended length PCR using primers for long (10 kb) and short (116 bp) amplicons of mtDNA. C: mtDNA copy numbers were determined in the genomic DNA by quantifying the ratio of gene transcripts of mtDNA-encoded CYTB and nDNA-encoded 18S. Data are mean ± SD obtained from five to eight mice in each experimental group. *P < 0.05 vs. WT-N; #P < 0.05 vs. WT-D.
Figure 7.
Effect of diabetes on nucleoids. A: Nucleoids were quantified in the isolated mitochondria by flow cytometry using dual staining with MTR and SYBR Green. B: SYBR Green-MTR–positive cells in Q2 were plotted. Data are mean ± SD obtained from six to eight mice in each experimental group (WT-N, WT-D, Tg-N, and Tg-D). *P < 0.05 vs. WT-N; #P < 0.05 vs. WT-D. Cont, unstained mitochondria.
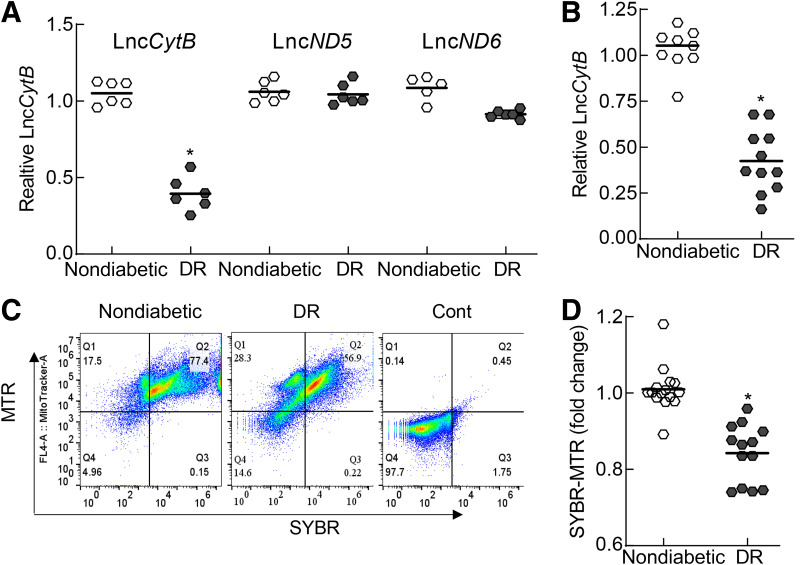
Consistent with the results from in vitro and in vivo models, retinal microvessels from donors with diabetic retinopathy had 50% less LncCytB compared with their age-matched donors without diabetes, but LncND5 and LncND6 were not significantly different in these two groups (Fig. 8A). Decrease in LncCytB was further confirmed by strand-specific PCR, showing a similar decrease in LncCytB in donors with diabetic retinopathy (Fig. 8B), and was accompanied by a significant reduction in the nucleoids, as evidenced by flow cytometry analysis showing a reduced number of SYBR Green-MTR–positive cells (Fig. 8C and D).
Figure 8.
Mitochondrial LncRNAs in the retinal microvessels from donors with documented diabetic retinopathy (DR) compared with age-matched donors without diabetes. A: Transcripts of LncCytB, LncND5, and LncND6 were quantified by RT-PCR using β-actin as a housekeeping gene. B: LncCytB transcripts were quantified by strand-specific PCR. C: FACS analysis was performed in the isolated mitochondria using SYBR Green-MTR dual staining. D: SYBR Green-MTR–positive cells in Q2 were plotted. Each measurement was made in duplicate with five to six donors per group. Data are mean ± SD, *P < 0.05 vs. age-matched donors without diabetes. Cont, unstained mitochondria.
Discussion
Diabetes impairs structural, functional, and genomic stability of the retinal mitochondria, and dysfunctional mitochondria play a central role in the development of diabetic retinopathy (1,2). Increased glucose-derived pyruvate oxidation by the tricarboxylic acid cycle in hyperglycemia elevates flux of electron donors into the ETC system, which increases the voltage gradient across the mitochondrial membrane, and blockage of electron transfer inside complex III results in increased superoxide generation (7). mtDNA is damaged in diabetes, and the activity of complex III is compromised, fueling a self-propagating vicious cycle of free radicals (2,4,5). The mitochondrial genome encodes for LncND5, LncND6, and LncCytB, and these LncRNAs form intermolecular duplexes with their functional mRNA counterparts (19,20). Our data show for the first time that while hyperglycemia has no significant effect on LncND5 and LncND6, LncCytB is downregulated and its occupancy at the mtDNA reduced. Packaging of the histone-free mtDNA is impaired, the number of protective nucleoids is decreased, the sensitivity of mtDNA to MNase digestion is increased, and LncCytB binding on the mtDNA is reduced. While overexpression of LncCytB prevents reduction in the protective nucleoids and copy numbers, LncCytB-siRNA exacerbates mtDNA damage and further reduces nucleoids and copy numbers. Consistent with in vitro results, retinal microvessels from diabetic mice and from human donors with documented diabetic retinopathy also showed a significant reduction in LncCytB expression and nucleoids and increased mtDNA damage. These results clearly suggest that LncCytB downregulation in the hyperglycemic milieu contributes to the increased vulnerability of mtDNA to damage, suggesting its major role in mitochondrial genomic stability.
The human genome has approximately four times more ncRNAs than coding RNA sequences, but the mitochondrial transcriptome has only ∼15% ncRNAs (36), which includes three LncRNAs. These mtDNA-encoded LncRNAs are the counterpart antisense transcripts of the mitochondrial ND5 and ND6 (complex I) and CYTB (complex III) mRNAs (19,20). Superoxide radicals are generated by both complex I and complex III of the ETC (37), but in the hyperglycemic milieu, as mentioned above, complex III becomes its major source (7,38). Our results show that although hyperglycemia has no significant effect on LncND5 and LncND6, expression of LncCytB is significantly decreased. In support, we have shown previously that while the activity of complex III is decreased in diabetes, complex I remains unchanged (8), strengthening the importance of LncCytB in diabetic retinopathy. The literature has shown that mtDNA-encoded LncRNAs can shuttle between the mitochondria and nucleus (39), and aberrant shuttling of LncCytB in the nucleus has been suggested to play a role in abnormal mitochondrial metabolism in the cancer cells (40). Here, we show that in the hyperglycemic milieu, LncCytB expression in the nucleus is also reduced, suggesting a possible effect of LncCytB on the nuclear genome in diabetic retinopathy.
LncRNAs can stabilize the protein-DNA complex and help to recruit the transcription factors onto the targeted genes (11,24,41). Decreased occupancy of LncCytB on mtDNA, including in the CYTB region, supports the role of LncCytB in maintaining mtDNA stability. Unlike nDNA, mtDNA is devoid of nucleosomes but is organized as protein-DNA complexes (21), and these nucleoids protect mtDNA from the damaging free radicals. Nucleoids can remodel in response to cellular demands by attaining a more open shape in respiring growth conditions and compressing in repressed conditions, and in compact structure, they are resistant to nuclease digestion (32,42). Here, we show that mtDNA digestion by MNase, an enzyme that preferentially cleaves the linker regions between nucleosomes and digests the free DNA ends toward the core nucleosome (43), produces blunted/missing peaks throughout the mtDNA, including in its CYTB region, suggesting a loosely packed-relaxed mtDNA. Nucleoids lack any delimiting membrane but are coated with proteins forming a macromolecular structure, and maintenance of nucleoid structure is associated with mitochondrial organization and function (25,44–46). Our results show that while glucose-induced increase in mtDNA damage and decrease in nucleoids are prevented by LncCytB overexpression, LncCytB-siRNA further worsens this, confirming a major role of LncCytB in maintaining mtDNA integrity. Furthermore, nucleoids also help in accurate propagation of the mitochondrial genome and are considered as the general center of mitochondrial biogenesis (21,23,47). The data presented here clearly show that regulation of LncCytB (overexpression or siRNA) affects mtDNA copy numbers, further strengthening the role of LncCytB in mtDNA biogenesis.
Results from the in vitro model are supported by our in vivo model. Compared with the normal mice, retinal microvessels from the diabetic mice had decreased LncCytB transcripts and nucleoids and high mtDNA damage and reduced copy numbers. However, overexpression of Sod2, in addition to ameliorating mtDNA damage, prevents decrease in LncCytB, nucleoids, and copy numbers. In support, mammalian nucleoids also contain Sod2, and Sod2 is considered as a fidelity protein in the mitochondrial nucleoids protecting nucleoid POLG from damage. Furthermore, a decrease in nucleoids can induce mitochondrial stress, and Sod2 helps to protect mtDNA from superoxide-induced oxidative damage (48–50). Sod2-overexpressing mice are also protected from a diabetes-induced decrease in mitochondrial functional and genomic alterations, including complex III activity, mtDNA damage, and copy numbers, and from developing diabetic retinopathy (5,8). Moreover, our results from donors with diabetic retinopathy also show a similar decrease in LncCytB and nucleoids but not in LncND5 and LncND6, further supporting the role of LncCytB in the development of diabetic retinopathy.
We recognize that LncCytB is an antisense LncRNA and that there is a possibility that LncCytB overexpression is preventing a glucose-induced decrease in CYTB expression-complex III activity, which could be helping in maintaining mitochondrial integrity. Moreover, our study is focused on the role of LncCytB in mtDNA stability. The role of other factors, including an impaired mtDNA repair mechanism and epigenetic modifications, in regulating mtDNA stability in diabetic retinopathy, however, cannot be ruled out.
In summary, LncCytB has a major role in the regulation of mtDNA stability in diabetic retinopathy. Its reduction in the hyperglycemic milieu contributes to decreased nucleoids and makes mtDNA more vulnerable to damage, resulting in a compromised ETC system and a continuing vicious cycle of free radical self-propagation. Thus, preventing LncCytB downregulation will protect mtDNA and the ETC system and will interfere in the self-perpetuating vicious cycle of free radicals; this should inhibit/arrest the development of diabetic retinopathy.
Article Information
Acknowledgments. The authors thank Gina Polsinelli, Wayne State University, for help with maintaining the animal colony.
Funding. The study was supported in part by National Eye Institute grants EY014370, EY017313, EY022230, and EY333516 and a grant from the Thomas Foundation to R.A.K. and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology, Wayne State University.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.K. and G.M. contributed to the experimental plan, researched data, interpreted data, and edited the manuscript. K.A. researched data and edited the manuscript. R.A.K. contributed to the experimental plan, data interpretation, literature search, and manuscript writing and editing. R.A.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.21760475.
J.K. and G.M. contributed equally to this work.
References
- 1. Kowluru RA, Mishra M. Oxidative stress, mitochondrial damage and diabetic retinopathy. Biochim Biophys Acta 2015;1852:2474–2483 [DOI] [PubMed] [Google Scholar]
- 2. Kowluru RA. Mitochondrial stability in diabetic retinopathy: lessons learned from epigenetics. Diabetes 2019;68:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizutani M, Kern TS, Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest 1996;97:2883–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Madsen-Bouterse SA, Mohammad G, Kanwar M, Kowluru RA. Role of mitochondrial DNA damage in the development of diabetic retinopathy, and the metabolic memory phenomenon associated with its progression. Antioxid Redox Signal 2010;13:797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santos JM, Tewari S, Goldberg AFX, Kowluru RA. Mitochondrial biogenesis and the development of diabetic retinopathy. Free Radic Biol Med 2011;51:1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tewari S, Santos JM, Kowluru RA. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxid Redox Signal 2012;17:492–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes 2005;54:1615–1625 [DOI] [PubMed] [Google Scholar]
- 8. Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci 2007;48:3805–3811 [DOI] [PubMed] [Google Scholar]
- 9. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet 2015;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018;172:393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics 2016;14:42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–1325 [DOI] [PubMed] [Google Scholar]
- 14. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv 2017;3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gordon AD, Biswas S, Feng B, Chakrabarti S. MALAT1: a regulator of inflammatory cytokines in diabetic complications. Endocrinol Diabetes Metab 2018;1:e00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Radhakrishnan R, Kowluru RA. Long noncoding RNA MALAT1 and regulation of the antioxidant defense system in diabetic retinopathy. Diabetes 2021;70:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Biswas S, Coyle A, Chen S, Gostimir M, Gonder J, Chakrabarti S. Expressions of serum lncRNAs in diabetic retinopathy - a potential diagnostic tool. Front Endocrinol (Lausanne) 2022;13:851967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowluru RA. Long noncoding RNAs and mitochondrial homeostasis in the development of diabetic retinopathy. Front Endocrinol (Lausanne) 2022;13:915031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rackham O, Shearwood A-MJ, Mercer TR, Davies SMK, Mattick JS, Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011;17:2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Paepe B, Lefever S, Mestdagh P. How long noncoding RNAs enforce their will on mitochondrial activity: regulation of mitochondrial respiration, reactive oxygen species production, apoptosis, and metabolic reprogramming in cancer. Curr Genet 2018;64:163–172 [DOI] [PubMed] [Google Scholar]
- 21. Kolesnikov AA. The mitochondrial genome. The nucleoid. Biochemistry (Mosc) 2016;81:1057–1065 [DOI] [PubMed] [Google Scholar]
- 22. Bogenhagen DF. Does mtDNA nucleoid organization impact aging? Exp Gerontol 2010;45:473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee SR, Han J. Mitochondrial nucleoid: shield and switch of the mitochondrial genome. Oxid Med Cell Longev 2017;2017:8060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem 2017;17:1750–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mohammad G, Kowluru RA. Nuclear genome-encoded long noncoding RNAs and mitochondrial damage in diabetic retinopathy. Cells 2021;10:3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohammad G, Kowluru RA. Mitochondrial dynamics in the metabolic memory of diabetic retinopathy. J Diabetes Res 2022;2022:3555889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santos JM, Tewari S, Lin JY, Kowluru RA. Interrelationship between activation of matrix metalloproteinases and mitochondrial dysfunction in the development of diabetic retinopathy. Biochem Biophys Res Commun 2013;438:760–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Invest Ophthalmol Vis Sci 2003;44:5327–5334 [DOI] [PubMed] [Google Scholar]
- 29. Strydom E, Pietersen G. Development of a strand-specific RT-PCR to detect the positive sense replicative strand of soybean blotchy mosaic virus. J Virol Methods 2018;259:39–44 [DOI] [PubMed] [Google Scholar]
- 30. Miyakawa I, Fujimura R, Kadowaki Y. Use of the nuc1 null mutant for analysis of yeast mitochondrial nucleoids. J Gen Appl Microbiol 2008;54:317–325 [DOI] [PubMed] [Google Scholar]
- 31. Chereji RV, Bryson TD, Henikoff S. Quantitative MNase-seq accurately maps nucleosome occupancy levels. Genome Biol 2019;20:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J Cell Sci 2008;121:1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp 2012. (61):3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang AL, Lukas TJ, Yuan M, Neufeld AH. Increased mitochondrial DNA damage and down-regulation of DNA repair enzymes in aged rodent retinal pigment epithelium and choroid. Mol Vis 2008;14:644–651 [PMC free article] [PubMed] [Google Scholar]
- 35. Muthurajan U, Mattiroli F, Bergeron S, et al. Chapter One - In Vitro Chromatin Assembly: Strategies and Quality Control. In Methods in Enzymology. Vol. 573. Marmorstein R, Ed. Cambridge, MA, Academic Press, 2016, pp. 3–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu X, Shan G. Mitochondria encoded non-coding RNAs in cell physiology. Front Cell Dev Biol 2021;9:713729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bleier L, Dröse S. Superoxide generation by complex III: from mechanistic rationales to functional consequences. Biochim Biophys Acta 2013;1827:1320–1331 [DOI] [PubMed] [Google Scholar]
- 38. Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res 2010;107:1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Y, Sun L, Wang RR, Hu JF, Cui J. The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: new players in an old arena. Crit Rev Oncol Hematol 2018;131:76–82 [DOI] [PubMed] [Google Scholar]
- 40. Zhao Y, Liu S, Zhou L, et al. Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells. Am J Cancer Res 2019;9:999–1008 [PMC free article] [PubMed] [Google Scholar]
- 41. Kukat C, Larsson NG. mtDNA makes a U-turn for the mitochondrial nucleoid. Trends Cell Biol 2013;23:457–463 [DOI] [PubMed] [Google Scholar]
- 42. Sumitani M, Kasashima K, Ohta E, Kang D, Endo H. Association of a novel mitochondrial protein M19 with mitochondrial nucleoids. J Biochem 2009;146:725–732 [DOI] [PubMed] [Google Scholar]
- 43. Davis IJ, Pattenden SG. Chapter 1-3: chromatin accessibility as a strategy to detect changes associated with development, disease, and exposure and susceptibility to chemical toxins. In Toxicoepigenetics. McCullough SD, Dolinoy DC, Eds. Cambridge, MA, Academic Press, 2019, pp. 85–103 [Google Scholar]
- 44. Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell 2003;14:1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ambro L, Pevala V, Bauer J, Kutejová E. The influence of ATP-dependent proteases on a variety of nucleoid-associated processes. J Struct Biol 2012;179:181–192 [DOI] [PubMed] [Google Scholar]
- 46. Feric M, Demarest TG, Tian J, Croteau DL, Bohr VA, Misteli T. Self-assembly of multi-component mitochondrial nucleoids via phase separation. EMBO J 2021;40:e107165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rebelo AP, Dillon LM, Moraes CT. Mitochondrial DNA transcription regulation and nucleoid organization. J Inherit Metab Dis 2011;34:941–951 [DOI] [PubMed] [Google Scholar]
- 48. Kienhöfer J, Häussler DJ, Ruckelshausen F, et al. Association of mitochondrial antioxidant enzymes with mitochondrial DNA as integral nucleoid constituents. FASEB J 2009;23:2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bakthavatchalu V, Dey S, Xu Y, et al. Manganese superoxide dismutase is a mitochondrial fidelity protein that protects Polγ against UV-induced inactivation. Oncogene 2012;31:2129–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bao D, Zhao J, Zhou X, et al. Mitochondrial fission-induced mtDNA stress promotes tumor-associated macrophage infiltration and HCC progression. Oncogene 2019;38:5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]