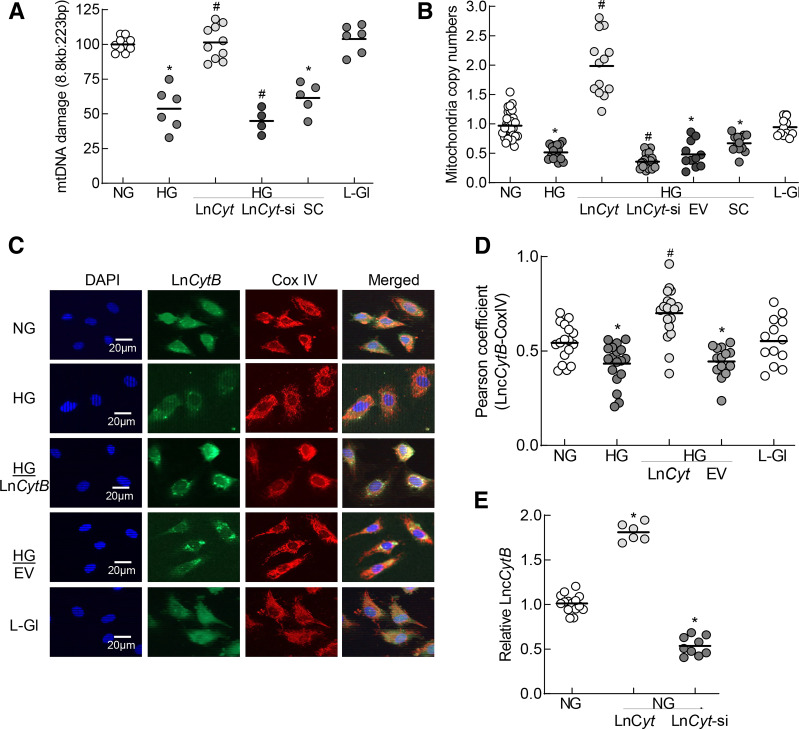
Figure 3.
Effect of LncCytB regulation on mtDNA damage and copy numbers. A: mtDNA damage was quantified by extended length PCR using long mtDNA (8.8 kb) and short (223 bp) amplicons of the mtDNA; relative amplification of the long to the short product, which is inversely proportional to the damage, was quantified. B: Copy numbers of mtDNA were determined in the genomic DNA by quantifying the ratio of gene transcripts of mtDNA-encoded CYTB and nDNA-encoded β-actin. C: Mitochondrial localization of LncCytB was performed by RNA FISH using fluorescein-12-dUTP–labeled LncCytB probe (green) and Texas Red (red)–conjugated secondary antibody against CoxIV. D: Pearson correlation coefficient of CoxIV with LncCytB. E: Transfection efficiencies of LncCytB-overexpressing plasmids and siRNA in HRECs were determined by strand-specific PCR, using β-actin as a housekeeping gene. Each measurement was made in duplicate/triplicate in three to five different cell preparations, and the histogram data are mean ± SD. HG/LnCyt, HG/LnCyt-si, HG/EV, and HG/SC indicate cells transfected with LncCytB-overexpressing plasmids, LncCytB-siRNA, empty vector, and SC RNA, respectively, and incubated in HG. *P < 0.05 vs. NG; #P < 0.05 vs. HG.