Abstract
Hepatic stellate cells (HSCs) are liver-resident cells best known for their role in vitamin A storage under physiological conditions. Upon liver injury, HSCs activate into myofibroblast-like cells, a key process in the onset of liver fibrosis. Lipids play an important role during HSC activation. Here, we provide a comprehensive characterization of the lipidomes of primary rat HSCs during 17 days of activation in vitro. For lipidomic data interpretation, we expanded our previously described Lipid Ontology (LION) and associated web application (LION/Web) with the LION-PCA heatmap module, which generates heatmaps of the most typical LION-signatures in lipidomic datasets. Furthermore, we used LION to perform pathway analysis to determine the significant metabolic conversions in lipid pathways. Together, we identify two distinct stages of HSC activation. In the first stage, we observe a decrease of saturated phosphatidylcholine, sphingomyelin, and phosphatidic acid and an increase in phosphatidylserine and polyunsaturated bis(monoacylglycero)phosphate (BMP), a lipid class typically localized at endosomes and lysosomes. In the second activation stage, BMPs, hexosylceramides, and ether-linked phosphatidylcholines are elevated, resembling a lysosomal lipid storage disease profile. The presence of isomeric structures of BMP in HSCs was confirmed ex vivo in MS-imaging datasets of steatosed liver sections. Finally, treatment with pharmaceuticals targeting the lysosomal integrity led to cell death in primary HSCs but not in HeLa cells. In summary, our combined data suggest that lysosomes play a critical role during a two-stage activation process of HSCs.
Keywords: hepatic stellate cell, phospholipid turnover, polyunsaturated fatty acid, lysosome, bioinformatics, lipidomics
Hepatic stellate cells (HSCs) are nonparenchymal liver cells that store the majority of vitamin A in vertebrates, thus acting as a retinoid reservoir. Most vitamin A present in these cells is stored as all-trans retinyl esters (REs) in lipid droplets (LDs) (1, 2). LDs are ubiquitous spherical organelles that store a variety of neutral lipids including triacylglycerols (TGs), cholesteryl esters (3, 4), REs (5), and acylceramides (6). Important functions of LDs include serving as reservoirs for energy and acting as building blocks for membranes or signaling molecules. The neutral lipids are protected against lipases by a monolayer of phospholipids and associated proteins (4). In quiescent HSCs, LDs have a relatively large diameter, most likely a reflection of their function—the long-time storage of vitamin A (1, 2, 7). Upon liver injury, HSCs transdifferentiate into cells with a myofibroblast-like appearance. This activation process is accompanied by deposition of collagen and loss of retinoids-laden LDs, thereby depleting the retinoid stores in HSCs. HSC activation in vivo is one of the first steps leading to hepatic fibrosis, a condition associated with increased stiffness and reduced liver function. Ignored and untreated liver fibrosis can develop into cirrhosis and may eventually result in the need for liver transplantation (1, 2). The molecular mechanisms driving HSC activation are, however, poorly understood.
Lipids have been implicated in HSC activation, as intracellular cholesterol (8, 9) as well as eicosanoids (10), ceramides, and endocannabinoids (11) increase during the activation process. Remodeling of neutral lipids, present in LDs, has also been implicated in HSC activation. In previous work, we provided evidence for the existence of two distinct LD populations during the activation process of HSCs (7, 12, 13). The first population, comprising large LDs enriched in vitamin A, is formed via a seipin-independent pathway (14) and is degraded during activation via the autophagy pathway (15, 16) and lysosomal lipase (17). During activation, a second LD population emerges, characterized by small LDs containing TGs with polyunsaturated fatty acids (PUFAs). In this population, neutral lipids are dynamically recycled by two opposing activities: synthesis by diacylglycerol O-acyltransferase 1 (DGAT1) and degradation by adipose triglyceride lipase (7, 13).
The appearance of this PUFA-enriched TG-pool is puzzling and its fate is currently not fully understood. It has been proposed that PUFA-TGs may provide building blocks for phospholipid synthesis—PUFAs are typically present at the sn-2 positions—to accommodate the membrane synthesis required for the proliferation and drastic morphological transformation during HSC activation (12). In addition, PUFA-TGs might serve as a reservoir, possibly via phospholipid synthesis, for the synthesis of bioactive lipids such as eicosanoids and endocannabinoids (10, 11).
To investigate the involvement and dynamics of lipids in the activation of HSCs, we performed (phospho)lipidomics on rat HSCs during the activation process. We report the comprehensive characterization of the lipidome of activating rat HSCs during the first 17 days and describe a remarkable reorganization of phospholipid composition. Using two new analysis modules in LION/web (18), we were able to associate the appearance of PUFAs with their incorporation in bis(monoacylglycero)phosphates (BMPs), a lipid class predominantly found in late endosomes and lysosomes. We propose an alternative hypothesis to explain the reported PUFA-TG accumulation in activating HSCs.
Results
A two-step reorganization of HSC lipidomes during activation
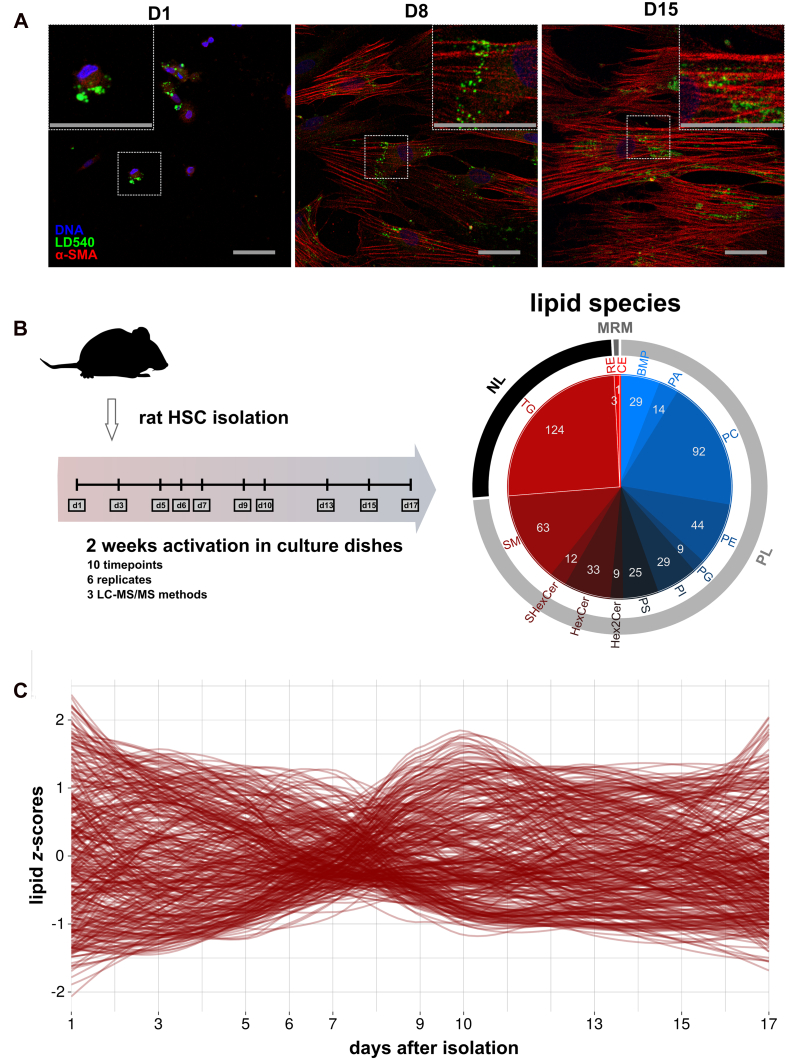
Primary HSCs from male rats were cultured at 37 °C in cell-culture medium. Under these conditions, HSCs undergo spontaneous activation (1, 2). We fixed the cells at three timepoints during this activation process (at days 1, 8, and 15) and stained the cells to examine typical markers of activation: loss of large LDs and increased levels of alpha-smooth muscle actin (α-SMA) (Fig. 1A). In line with previous observations (19), we observed increased levels of α-SMA between days 1 and 8, accompanied by a shift from large perinuclear LDs on day 1 to smaller peripheral LDs. Changes in LD dynamics and morphology have been quantified previously (13, 17, 19).
Figure 1.
Lipidomic profiling of in vitro activating rHSCs.A, confocal microscopy images of rHSCs cultured for 1, 8, or 15 days, stained by Hoechst (DNA, blue), LD540 (lipid droplets, green), and antibodies against α-SMA (HSC activation marker, red). Scale bars indicate 40 μm. B, graphical representation of the setup of the lipidomics experiments. Primary rHSCs were cultured between day 1 and day 17 and harvested at 10 timepoints. Three LC-MS/MS methods were used to measure lipid profiles: 'NL' (black in the surrounding circle), 'PL' (gray), and 'MRM' (dark gray). Values inside the pie represent the number of species within the respective lipid class that was identified. The 14 lipid classes are color-coded from blue to red: BMP (bis-monoacylglycero-phosphate), PA (phosphatidic acid), PC (phosphatidylcholine), PE (phosphatidylethanolamine), PG (phosphatidylglycerol), PI (phosphatidylinositol), PS (phosphatidylserine), Hex2Cer (dihexosylceramide), HexCer (hexosylceramide), SHexCer (sulfatide), SM (sphingomyelin), TG (triacylglycerols), RE (retinyl esters), and CE (cholesteryl esters). C, dynamics of all annotated lipids obtained by ‘PL’ over time. Lipids are scaled to z-scores and loess-smoothing was applied. α-SMA, α-smooth muscle actin; HSC, hepatic stellate cell; BMP, bis(monoacylglycero)phosphate; MRM, multiple reaction monitoring; rHSC, rat HSC.
To study changes in the HSC lipidome during activation in detail, we expanded our experimental setup to 10 timepoints (six replicates each) between 1 and 17 days after isolation (Fig. 1B). After lipid extraction, we performed lipidomics by combining three LC-MS/MS setups: Hydrophilic interaction liquid chromatography/ESI for phospholipid and sphingolipid analysis (“PL”), reverse phase/atmospheric pressure chemical ionization (APCI) in full scan-mode for neutral lipid analysis (“NL”), and in multiple reaction monitoring-mode for the specific detection of retinoids (see Experimental procedures for details). In total, we measured and annotated 487 lipid species covering 14 lipid classes (Fig. 1B). The complexity and dynamics of the obtained data are illustrated in Figure 1C.
Using this approach, we confirmed and expanded our previously reported changes in the profiles of neutral lipids; (1) REs were lost during HSC activation as determined by targeted lipidomics. At day 1, retinyl palmitate was the most abundant RE species, followed by retinyl stearate and retinyl oleate (Fig. S1A). After 3 days, the majority of these retinoids were lost; (2) the abundance of cholesterol esters showed a strong increase in the first 5 days, followed by a moderate decrease until day 9 (Fig. S1A); (3) the TG species-profile shifts dramatically within a week (Fig. S1B). To further illustrate this, we expressed all TG-species between day 1 to 7 as a percentage of the total TG and performed principal component analysis (PCA) on the obtained compositions (Fig. S1, C and D). As reported previously (11, 19), the TG-composition shifts from species with a lower to a higher degree of unsaturation (Fig. S1D, left) and from shorter to longer fatty acyl (FA) chain lengths (Fig. S1D, right) during activation. This shift is most pronounced in principal component 1 (hereafter PrC-1, to avoid confusion with the abbreviation for phosphatidylcholine (PC)), as indicated by the color-coded loading plots (Fig. S1D). PrC-1 predominantly separates the early timepoints day 1 to 3 from day 5 to 7 (Fig. S1C).
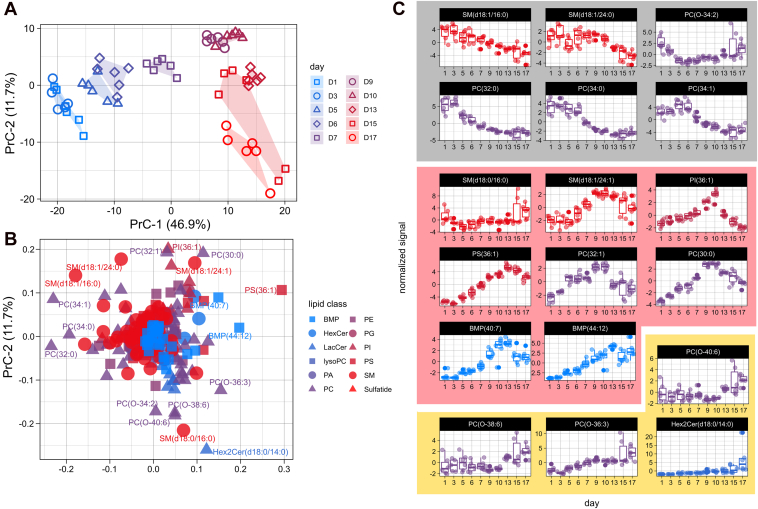
To investigate whether PUFA accumulation also occurs in membrane lipids, we performed PCA on the phospholipid and sphingolipid profiles (Fig. 2, A and B) The PCA score plot (Fig. 2A) showed that the lipidomes of activating HSCs changed gradually over time, with the most apparent differences occurring between days 1 and 9 (PrC-1 explains 46.9% of the total variance). Smaller changes were observed after these timepoints, as shown by a turn in the direction of the sample groups over time into the PrC-2 dimension (11.7% explained variance). These observations indicate a two-step change of the HSC lipidome during activation.
Figure 2.
Phospho/sphingolipid dynamics of in vitro activating rHSCs.A–C, principal component analysis of phospho and sphingolipids between day 1 and 17, visualized as score (A) and loading (B) plots for PrC-1 and 2. A, PCA score plot with samples shape- and color-coded for timepoints. Polygons with identical fill colors were added to indicate the outer regions with all samples of the sample groups. B, corresponding PCA loading plot with shapes and colors indicating lipid class (see legend Fig. 1B for lipid abbreviation). C, boxplots of 18 lipid species with the longest distance from the origin as a function of activation time. Boxplots are grouped into three categories: decreasing (gray), early emerging (light red), and late emerging (light yellow) lipids. Signals are normalized and scaled as z-scores. PCA, principal component analysis; rHSC, rat HSC.
To identify which lipid species were mainly responsible for this two-step shift, we examined the corresponding PCA loading plot (Fig. 2B). A scattered appearance of lipid species was observed around the origin. The first stage of activation, predominantly associated with PrC-1, was characterized by a decrease of PCs and sphingomyelins with saturated or monounsaturated fatty acids, as indicated by their presence on the left side of the plot (Figs. 2B and S1E). This decrease was accompanied by increased levels of lipids with a considerably higher level of unsaturation and with longer fatty acyl chain lengths, as shown by the fatty acid color-coded loading plots (Fig. S1E). The second stage of activation, predominantly associated with PrC-2, was mainly driven by changes in ether-linked PCs and dihexosylceramide (Hex2Cer) after day 9 (Figs. 2B and S1E). This two-stage activation process of HSCs is illustrated by the individual dynamics of 18 lipid species with the strongest contribution to PrC-1 and PrC-2 (Fig. 2C). Here, three groups can be distinguished: lipids with high levels at early timepoints (gray block, associated with HSC quiescence), early emerging lipids (red block, drivers of activation stage 1), and late emerging lipids (yellow block, stage 2) (Fig. 2C). The two-step activation, as observed by PCA, was confirmed by three alternative methods for dimensionality reduction: multidimensional scaling (MDS), t-distributed stochastic neighbor embedding (t-SNE), and uniform manifold approximation and projection (UMAP) (Fig. S2). In all three projections, samples from day 1 to 7 separate well from samples after day 9. Our combined results suggest a two-stage activation with different lipids involved in each activation step.
Multivariate analysis using LION-associations reveals increased BMP metabolism
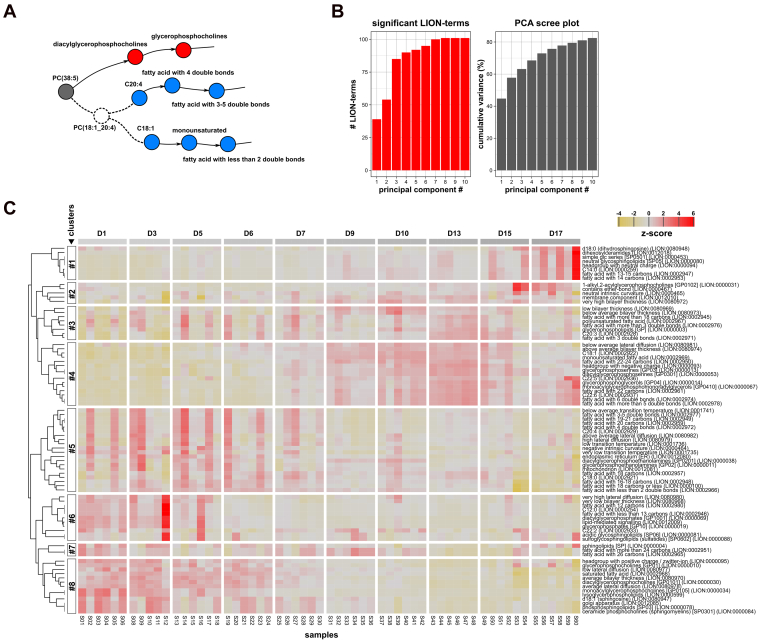
The PCA used in the previous section highlights individual features that are characteristic of lipidome changes during HSC activation. The obtained principal components, however, do not represent intrinsic biological meaning. Therefore, we developed an alternative dimension reduction method making use of Lipid Ontology (LION) (18), an ontology that associates lipid species with chemical, biophysical, and biological information. This method, named the LION-PCA heatmap algorithm, is an adaptation of GO-PCA (20) which performs similar tasks for RNA-seq, microarray, and proteomics data.
LION-PCA was implemented as a module of the web-application LION/web (https://heatmap.lipidontology.com/) and visualizes the most characteristic LION-signatures of a given lipidomic dataset in a heatmap (see Experimental procedures). To validate the newly incorporated LION-PCA module, we used a previously described public dataset containing the lipidomes of CHO-k1 cells incubated with either control, palmitic acid (C16:0), linoleic acid (C18:2), or arachidonic acid (C20:4) (18). LION-PCA heatmap was able to reduce the lipidomes containing 360 lipids into 60 LION-signatures. As anticipated, reported signatures included 'C16:0', 'C18:2', and 'C20:4', which were all grouped into different clusters associated with the respective FA-incubation conditions (Fig. S3).
Next, we used LION-PCA heatmap to explore characteristic lipid signatures during HSC activation (Fig. 3). To make optimal use of the potential of LION, we linked each lipid annotated with the sum of its fatty acids to its most likely associated fatty acids. For example, the most likely fatty acid composition of PC(34:1) in mammalian cells is 16:0_18:1 (12, 21). Hence, this species is dynamically linked to the fatty acids C16:0 and C18:1 (see Experimental procedures for more specific details). This approach allows LION to take fatty acid unsaturation and chain length into account (Fig. 3A). Based on the resulting dataset, the algorithm selected 79 LION-signatures of interest, covering lipid classification, chemical properties, biophysical associations, and predominant subcellular localizations. LION/web was set to group LION-signatures with similar dynamics into eight clusters. In line with the PCA-analysis, three major patterns were visible: clusters with high LION-signature at early timepoints (clusters 5–8, associated with quiescent HSCs), clusters with early emerging LION-signature signals (clusters 3–4, drivers of HSC activation stage 1), and clusters with late emerging LION-signature signals (clusters 1–2, drivers of stage 2) (Fig. 3C).
Figure 3.
Multivariate analysis of phospho/sphingolipid dynamics of in vitro–activating rHSCs.A, graphical outline of the rationale behind FA-composition prediction, with colored nodes representing LION-terms and lines representing LION-relations. By making use of predicted FA-compositions (indicated with dashed lines), LION-terms describing fatty acid building blocks will be taken into account during analysis. B and C, LION-PCA heatmap analysis of rHSC-activation lipidomics data. B, bar graphs showing the number of significant LION-terms (left) or the cumulative variance explained (right), per set number of principal components. C, heatmap generated by the LION-PCA heatmap module in LION/web with the number of principal components set to 4 (see panel B). Heatmap colors (from yellow to red) indicate the mean z-score for a given LION-signature per sample. The number of lipids present in each LION-signature is shown in Table S1. LION-signatures are grouped into eight clusters by hierarchical clustering. Names and IDs of the selected terms are displayed on the right. Samples are sorted by sample group (from days 1 to 17), indicated by the blocks on top of the graph (from light to dark gray). LION, Lipid Ontology; PCA, principal component analysis; rHSC, rat HSC.
Inspection of the heatmap revealed a remarkable decrease in the LION-signatures 'lysoglycerophospholipids' (four lipids) and 'glycerophosphates' (11 lipids) during the first activation stage. These lipid categories both play prominent roles in intracellular signaling. In addition, sphingomyelins (30 lipids) showed a notable drop during the course of the experiment (Fig. 3C). Furthermore, a shift was observed from ‘saturated fatty acids’ (157 lipids) shortly after isolation, to the increased abundance of ‘monounsaturated fatty acids’ (52 lipids) and ‘fatty acid with more than five double bonds' (34 lipids) at later timepoints. This increase was mainly driven by lipids associated with 'C22:5' (nine lipids) and 'C22:6' (30 lipids), both present in cluster 4 (Fig. 3C). The shift in fatty acid unsaturation is in line with the initial observations in Figure 2C and seems to be a major driver of the first activation stage.
Interestingly, the increased level of unsaturation in cluster 4 was accompanied by the rise of 'glycerophosphoserines' and 'glycerophosphoglycerols' (36 lipids), the latter predominantly consisting of 'monoacylglycerophospho-monoradylglycerols' (28 lipids) (Fig. 3C). This lipid class, also known as BMP or lysobisphosphatidic acid, is mainly found in late endosomes and lysosomes (21, 22) and increased levels have been reported in lysosomal storage disorders (LSDs) (21, 22, 23). Other lipids that are often reported in LSD are glycosphingolipids. Remarkably, our analysis also reports the accumulation of 'neutral glycosphingolipids' (23 lipids) and related terms at very late timepoints, present in cluster 1 and associated with the second stage of activation (Fig. 3C). Combined, these findings suggest a phenotypic similarity between activated HSCs with LSD-affected cells.
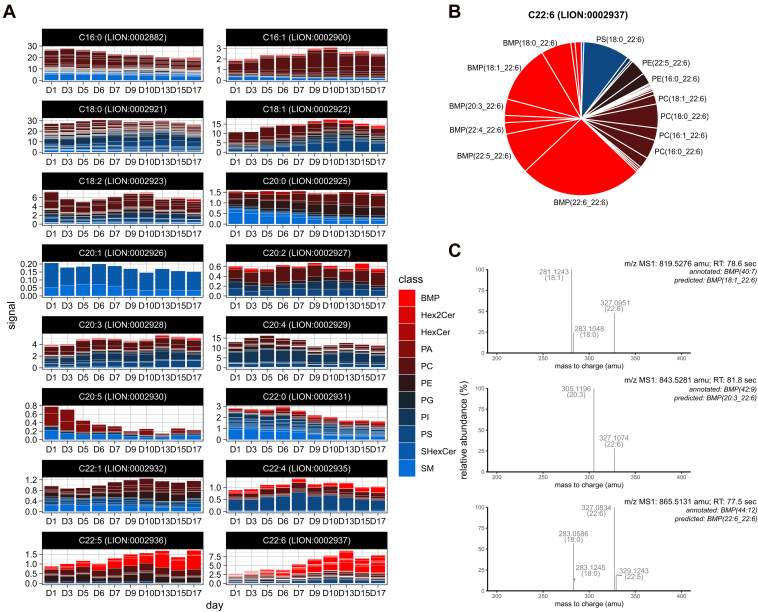
The synchronized rise of BMPs and highly unsaturated lipids prompted us to investigate whether these effects were interconnected. To this end, we rearranged the dataset by fatty acid associations, facilitated by the LION framework. For every fatty acid LION-term, we visualized lipid class abundance over time (Fig. 4A). In line with the observations in Figure 3C, a drop of lipids with saturated fatty acids (C16:0, C22:0) was observed (sum of the stacked bar graphs). This drop could not be attributed to specific lipid classes (fractions of individual stacked bars remained present). By contrast, the increase of lipids associated with C22:5 and C22:6 was predominantly driven by BMPs (Fig. 4A, bottom panels). This class is responsible for as much as 62% of the total signal of C22:6-lipids on day 17 (Fig. 4B). The increase of C22:5 and C22:6 during activation is based on the FA-composition prediction of lipids, as described in the previous section. To validate the predicted acyl composition of the increased BMP-species, we obtained high-resolution MS/MS spectra of these respective species (Fig. 4C). Indeed, the MS/MS spectra revealed the presence of C22:5 or C22:6 FAs. Combined, activation of HSCs is accompanied by an increase of highly unsaturated BMP lipids, followed by glycosphingolipid accumulation.
Figure 4.
Lipid class distribution of 16 fatty acid LION-terms.A, bar graphs of the total signal of lipids present in the indicated fatty acid LION-terms, averaged per timepoint. Bar graphs are stacked per lipid species, fills are color-coded by lipid class (red to blue, see Fig. 1B for lipid class abbreviation). Of note, a given species can be part of multiple LION-terms, e.g., PC(16:0_18:1) is associated with both C16:0 (LION:0002882) and C18:1 (LION:0002922). B, pie chart showing the relative abundance of lipid species associated with C22:6 (LION:0002937) at day 17, annotated with the corresponding FA-predicted lipid name. Identical fill colors as panel A are used. C, MS-spectra of data-dependent MS2-fragmentation, performed by a high-resolution Orbitrap mass spectrometer, to validate the predicted presence of C22:6 fatty acid in three abundant BMPs. In all three panels, [M-H]− fragments, indicative for C22:6 (theoretical m/z: 327.233), are observed. BMP, bis(monoacylglycero)phosphate; LION, Lipid Ontology.
Lipid metabolism pathway analysis during HSC activation
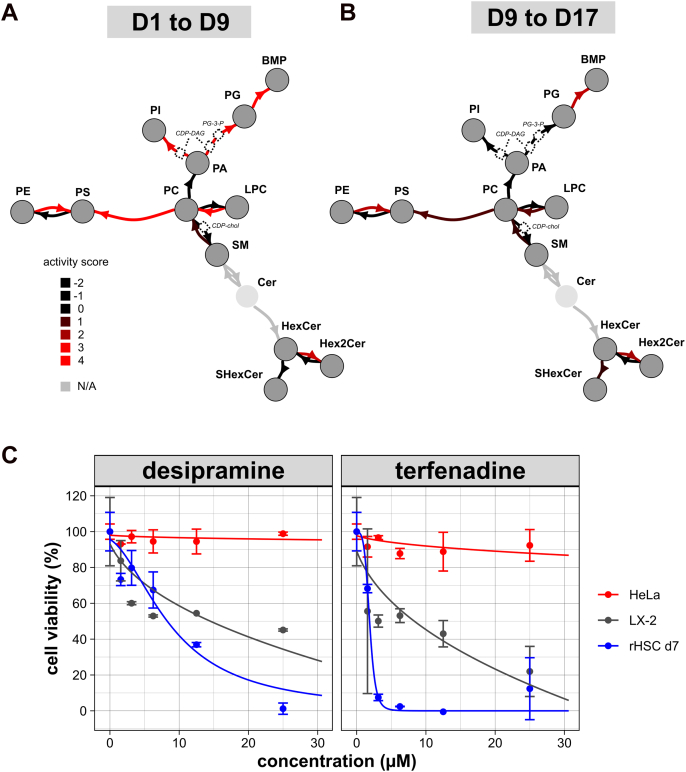
Next, we built upon a recently described method (24, 25) to list possible lipid pathways that are most characteristic for the two identified HSC-activation stages. The pathway analysis method was combined with the high level of annotation of lipids by LION (see Experimental procedures under ‘LION pathway analysis’). We compared the HSC lipidomes between day 1 to 9 (stage 1, Fig. 5A) and day 9 to 17 (stage 2, Fig. 5B). Pathway conversions were scored by the algorithm and visualized in the network as color-coded edges (Fig. 5, A and B). Aggregate activity scores—indicating a series of reaction paths—are shown in Table 1.
Figure 5.
Pathway analysis of phospho/sphingolipid metabolism in activating rHSCs. Graphs are showing active lipid pathways of rat HSCs between days 1 and 9 (A) or days 9 and 17 (B), as generated by the pathway analysis module in LION/web. Dark gray nodes indicate lipid classes, represented by LION-terms (see Table S2 for details). Light gray nodes represent lipids not present in the dataset, dashed boxes pathway intermediates. The directed edges indicate conversions; colors are scaled to their respective activity scores (black to red). Activity scores that could not be calculated due to missing data are depicted in gray. The values of individual activity scores, as well as aggregate activity scores of paths with more than one reaction, are shown in Table 1. C, effect of increasing concentrations of desipramine or terfenadine on cell viability of HeLa cells, LX-2 cells, and HSCs (after 7 days of culture) after 24 h of incubation. HSC, hepatic stellate cell; LION, Lipid Ontology; rHSC, rat HSC.
Table 1.
Z-scores of pathway analysis of phospholipid dynamics of in vitro activating rHSCs
| Phospholipid metabolism pathway | D1–D9 | D9–D17 |
|---|---|---|
| LPC > PC > PS | 7.021∗∗∗ | 1.498ns |
| PC > PS | 5.705∗∗∗ | 0.974ns |
| PE > PS | 4.166∗∗∗ | 2.226∗ |
| PA > PG > BMP | 4.356∗∗∗ | 1.628ns |
| SM > PC > PS | 4.575∗∗∗ | 1.128ns |
| PG > BMP | 3.225∗∗∗ | 2.339∗∗ |
| LPC > PC | 4.224∗∗∗ | 1.143ns |
| LPC > PC > PA > PG > BMP | 3.288∗∗∗ | 1.327ns |
| HexCer > Hex2Cer | 2.476∗∗ | 1.995∗ |
| LPC > PC > PS > PE | 3.648∗∗∗ | 0.022ns |
| PA > PG | 2.935∗∗ | −0.037ns |
| LPC > PC > SM | 2.341∗∗ | 0.475ns |
| SM > PC > PA > PG > BMP | 1.559ns | 1.066ns |
| PC > PA > PG > BMP | 1.358ns | 0.872ns |
| LPC > PC > PA > PG | 1.935∗ | 0.182ns |
| PA > PI | 2.876∗∗ | −0.83ns |
| LPC > PC > PA > PI | 1.901∗ | −0.276ns |
| SM > PC | 0.765ns | 0.621ns |
| SM > PC > PS > PE | 1.651∗ | −0.28ns |
| PC > PS > PE | 1.48ns | −0.782ns |
| LPC > PC > PA | 0.294ns | 0.249ns |
| SM > PC > PA > PG | −0.062ns | −0.12ns |
| SM > PC > PA > PI | −0.096ns | −0.578ns |
| PC > PA > PG | −0.617ns | −0.586ns |
| PC > SM | −0.914ns | −0.471ns |
| PC > PA > PI | −0.659ns | −1.147ns |
| HexCer > SHexCer | −2.851ns | 0.623ns |
| SM > PC > PA | −2.152ns | −0.121ns |
| SM > PC > LPC | −1.634ns | −0.909ns |
| PC > PA | −3.808ns | −0.792ns |
| PC > LPC | −3.077ns | −1.907ns |
| PS > PE | −3.611ns | −2.081ns |
| Hex2Cer > HexCer > SHexCer | −3.963ns | −2.087ns |
| Hex2Cer > HexCer | −2.754ns | −3.574ns |
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant, rows ordered by average score.
Inspection of the pathway scores in the first stage of activation (day 1–9) reveals a complex remodeling of the major phospholipids (phosphatidylcholine [PC], phosphatidylethanolamine [PE], phosphatidylserine [PS], phosphatidylinositol [PI]), with PS as the most prominent endpoint (Fig. 5A and Table 1). Furthermore, and in line with our lipidomic analysis (Figs. 3C and 4C), the analysis reveals increased BMP synthesis (phosphatidic acid (PA) > phosphatidylglycerol (PG) > BMP as the most significant path)). In the second stage of activation (day 9–17), overall lipid remodeling is slowed down, as indicated by lower pathway scores (Fig. 5A and Table 1). At this stage, the most significant conversions are PG > BMP, PC > PS, and HexCer > Hex2Cer, which are all present at the extreme ends of the pathway graph. Together, this pathway analysis shows no evident characteristic lipid pathways in the first stage but reveals lipidome regulation towards PS and lipids associated with endosomes and lysosomes during the second stage of activation.
Lysosomal targeting of activating HSCs
The profound appearance of lipids associated with lysosomes indicates a role of lysosomes in the activation of HSCs. Using our HSC model system, we tested two pharmaceuticals reported to cause lysosome destabilization, namely desipramine (26) and terfenadine (27). Similar to the autophagy inhibitors, these drugs resulted in dramatic cell death in HSCs (Fig. 5C). By contrast, the more generic cell line HeLa was viable within the same concentration range. LX-2 cells, a human-derived cell line originating from HSCs, were also sensitive to the pharmaceuticals, albeit to a lesser extent (Fig. 5C). Together, this observation underlines the importance of lysosomes for cell survival of HSCs and HSC-like cells.
Structural isomers of polyunsaturated BMPs are present at vitamin A-enriched foci of diseased human livers
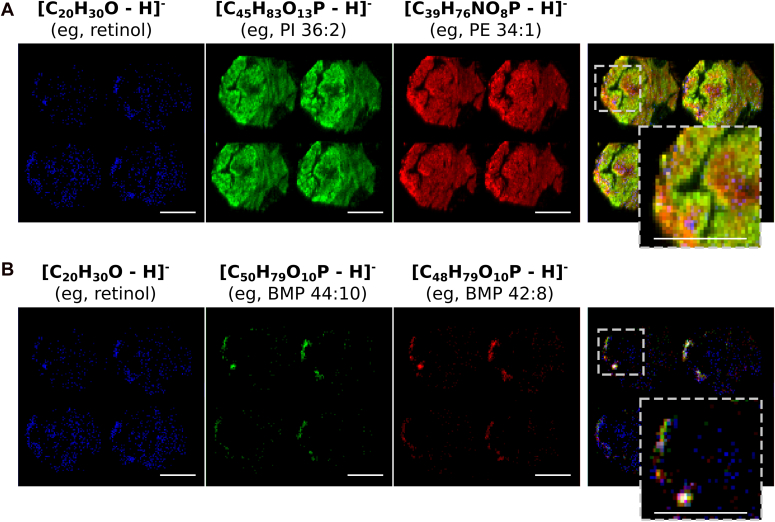
To examine the physiological relevance of our findings, we analyzed spatial metabolomics data of human diseased liver from the public database METASPACE (28, 29). As these datasets do not contain histological information, we used vitamin A (retinol) localization to determine HSC loci in the sections. Next, we listed all vitamin A colocalizing metabolites (Table S3) as a proxy for their presence in HSCs. Metabolite annotation in spatial metabolomics datasets is usually more ambiguous as compared to LC-MS/MS bulk lipidomics due to the lack of chromatographic separation of metabolites prior to MS-analysis, as well as the use of MS-1 measurements without subsequent fragmentation and analysis by tandem-MS (30). Nevertheless, the high-resolution MS technology used here can distinguish isobaric species and can identify molecular formulas. Hence, all metabolites are annotated with a number of structural isomers (molecules with the exact same elemental composition). Interestingly, from the 10 obtained vitamin A colocalizing metabolites (Table S3), five were annotated with the structural isomers PG or BMP. Moreover, these lipids were generally highly unsaturated, a feature that is usually more frequently observed for BMPs than PGs. Next, we visualized vitamin A with two abundant non colocalizing lipids (PI 36:2 and PE 34:1) (Fig. 6A) or two colocalizing lipids (BMP 44:10 and 42:8) (Fig. 6B) to show their spatial distribution in the liver sections. Vitamin A was enriched in several spots (Fig. 6, A and B) of about 10 to 20 μm in diameter, in line with the expected dimensions of HSCs in liver. The colocalizing PG/BMP lipids showed very similar patterns, indicative of their presence in HSCs. In contrast, the non colocalizing lipids were not spatially enriched and present in the full liver section (Fig. 6A). Combined, these results demonstrate the putative enrichment of polyunsaturated BMP-species in the HSCs of diseased human livers.
Figure 6.
Spatial metabolomics of steatosed human liver. Public dataset (METASPACE ID: ‘2017-05-11_13h41m04s’) containing liver sections of steatosed human liver, containing 389 annotated metabolites. Scale bars indicate 100 μm. A, spatial localization of [C20H30O-H]− (e.g., retinol) with two noncolocalizing metabolites: [C45H83O13P-H]− (e.g., PI 36:2, green) and [C39H76NO8P-H]− (e.g., PE 34:1, red). B, spatial localization of [C20H30O-H]− (e.g., retinol) with two colocalizing metabolites: [C50H79O10P-H]− (e.g., BMP 44:10, green) and [C48H79O10P-H]− (e.g., BMP 42:8, red). Please note that panels A and B are from the same tissue section. The left ion-images [C20H30O-H]− (e.g., retinol) are identical and serve as molecule localization reference. BMP, bis(monoacylglycero)phosphate.
Discussion
In the past decade, a number of studies showed that HSCs undergo a remarkable reprogramming during activation, both reflected by the proteome (31, 32, 33) and transcriptome (34, 35, 36). Although some targeted lipidomics approaches have been reported (7, 10, 11, 17, 19), a comprehensive time-course study of the lipidomes in these cells was lacking.
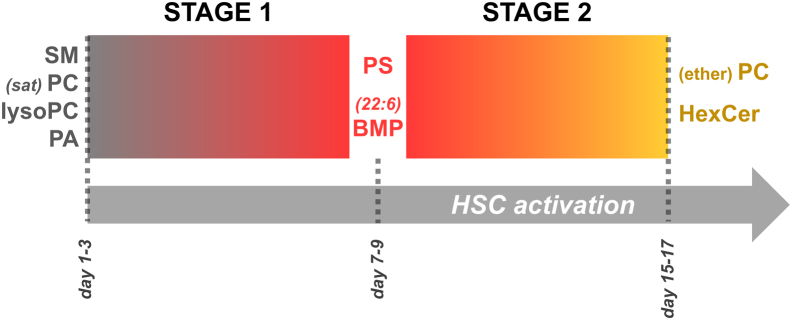
To our knowledge, we present here the most detailed lipidomics study of activating HSCs to date (Fig. 1), which provides novel insights into the differentiation dynamics in the activation process of HSCs. Inspection of the dataset by PCA revealed two stages of lipidome adaptation during activation (an overview is shown in Figure 7). Two-stage activation of HSC has been proposed earlier (37, 38, 39), referred to as ‘initiation’ followed by ‘perpetuation’. The first ‘initiation’ stage is initiated by extracellular signals and characterized by induction of growth factor receptors, differentiation into a fibrogenic phenotype, and changes in growth factor signaling. Hallmarks of the second ‘perpetuation’ stage are increased cell proliferation, scar formation, contractility, matrix degradation, and proinflammatory signaling. More recently, single cell transcriptomic analysis of hepatic stellate cells isolated from fibrotic revealed different populations (40, 41). How our in vitro observations are reflections of these in vivo features remain to be elucidated.
Figure 7.
Graphical overview of lipid dynamics during the two activation processes in HSCs. The main lipid (sub)classes are indicated for the three timepoints (d1–3, d7–9, d15–17) that separate the two activation stages (stage 1 as gradient from gray to red, stage 2 from red to orange) of rHSCs. See Figure 1 for lipid class abbreviations; sat indicates lipids containing saturated fatty acids; 22:6 indicates lipids containing docosahexaenoic acid. HSC activation is indicated from left to right. HSC, hepatic stellate cell.
To interpret our complex lipidomic data, we expanded LION/web, our recently reported web-tool to perform LION enrichment analysis of lipidomic data (18). We strongly improved the overall performance of LION/web by increasing the lipid coverage in LION, by the introduction of ‘smart matching’ to increase lipid mapping, and by the inclusion of fatty acid composition prediction of phospholipids (see Experimental procedures). In addition and most notably, we incorporated the new web-tool module ‘PCA-LION heatmap’. The module offers an analysis approach that was not yet available for lipidomics. Using lipid subsets provided by the LION framework, ‘PCA-LION heatmap’ selects and visualizes the signatures that are most characteristic for the dataset. This approach offers an attractive way to quickly browse through meaningful trends in the dataset, observations that may be obscured at first sight. The approach contrasts to other often used multivariate analyses in lipidomics such as PCA and partial least squares discriminant analysis (42), which focus on individual features. Our method is well-suited to process and evaluate all available information at once, including biophysical properties and predominant subcellular localization. When we used the ‘PCA-LION heatmap’ module to investigate the dynamics of the HSC dataset, we added fatty acid composition prediction to enhance the matching to LION. Accordingly, besides major trends on class level, the module also reported the putative increase of the fatty acids C22:5 and C22:6 (Fig. 3). The increased levels in these PUFAs, whose identities were validated by MS/MS-fragmentation, appeared to be found predominantly in the lipids of class BMP (Fig. 4).
It is important to stress that the ‘PCA-LION heatmap’ module primarily aims to provide clues about a lipidomic dataset by using a visualization approach and should not be used as a means to test hypotheses. Also, the quality of the output depends on the underlying database, LION. For many well-established lipids, LION contains numerous data points. For less common ones, however, characteristics such as cellular localization or biophysical properties may be absent and therefore are not taken in consideration. We advise all users of the web-tool to routinely consult the logs about what individual lipid species are matched to LION-terms. Furthermore, datasets that are submitted to the web-tool must comply with a few minimal requirements that are similar to the requirements described for the LION enrichment analysis web-tool (18): data should be normalized, samples at least in triplicate and lipid names should be in line with the LIPIDMAPS nomenclature.
We also made use of LION in the pathway analysis, an approach proposed by Nguyen et al. (24, 25). By making use of existing pathway information, they established a method to calculate activity scores for all potential biochemical reactions between the provided lipids. We merged this pathway analysis method with the LION ontology by using LION-definitions to define pathway nodes (Table S2). Comparison between the two HSC-activation stages using LION pathway analysis revealed primarily differences in overall lipid metabolism activity, with actively remodeling HSCs in the first stage and with somewhat decelerated activities in the second stage. Thus, in the pathway analysis, the two-stage activation appeared to be less pronounced than the heatmap analysis. This difference might be explained by the focus on lipid classes and lack of description of fatty acids in the pathway module. Nevertheless, also LION pathway analysis highlighted BMP synthesis as one of the major emerging pathways during HSC activation (Fig. 5). In support of increased BMP levels during HSC activation, we used two drugs, desipramine and terfenadine, that belong to the group of cationic amphiphilic drugs (CADs). CADs accumulate in acidic lysosomes by binding to BMP, thereby inhibiting the activity of lipases and affecting lysosomal membrane integrity (43). These CADs have also been used in cancer cells to induce cancer-specific cytotoxicity in vitro and in vivo (27).
BMP, also referred to as lysobisphosphatidic acid, is present in low amounts (1–3% of phospholipid pool) in mammalian cells and is enriched in the cellular compartments’ late endosomes and lysosomes (21, 22). Lysosomes are reported in several studies to play a central role during HSC activation. For example, the degradation of REs, a hallmark of HSC activation, is primarily autophagy-dependent (16, 39). In this catabolic pathway, lysosomes fuse with autophagosomes to form autophagolysosomes, the compartment responsible for cargo breakdown. In addition, we reported that lysosomal-resident lysosomal acid lipase is the predominant activity to catalyze RE-degradation (17). Interestingly, the in vitro activity of lysosomal acid lipase has been shown to be dependent on the presence of BMP (44). Lysosomes have also been suggested to have a critical role in cholesterol accumulation during HSC activation. In a catch-22 scenario, an overload of cholesterol is undermining the performance of lysosomes, leading to impaired toll-like receptor 4 breakdown. As a result, elevated levels of this receptor culminated in increased sensitivity to TGFβ-induced activation and interfered with a feedback mechanism mediated by sterol regulatory element-binding protein 2 (SREBP2) to maintain cellular cholesterol homeostasis (8, 9), initiating a repetitive chain of effects. Lastly, a link between PS (which increased during the first activation stage) and intracellular cholesterol transport after endosomal uptake was recently reported (45). The combined accumulation of cholesterol (8, 9), PS, BMPs (first activation stage), and glycosphingolipids (second activation stage) in HSCs (Figs. 3B, 5B and 7) is reminiscent of lipid profiles observed in LSD. These metabolic disorders are characterized by lysosomal dysfunction, resulting in the progressive build-up of substrates, such as complex glycolipids and other lipids (46). In this respect, it is interesting to note that researchers showed a decrease of HSC activation by overexpression of Niemann-Pick Type C2, a protein that is defective in the LSD Niemann-Pick C2 disease (47).
BMPs were found to be enriched in PUFAs, which was most pronounced for docosapentaenoic acid (C22:5) and docosahexaenoic acid (C22:6, DHA). The enrichment of DHA in BMPs has been reported earlier for other cell types and organs (48) and might be associated with the function of BMPs. It was speculated that the presence of DHA in BMPs serves as an antioxidant in lysosomes. In vitro experiments with liposomes containing cholesterol and BMP(22:6/22:6) showed decreased oxysterol-formation compared to liposomes containing cholesterol and BMP(18:1/18:1) (48). Interestingly, retinoids are known to be extremely sensitive to oxidation. In the light of its high abundance in HSCs and described routing to lysosomes, DHA-containing BMPs might serve an important role in preventing undesired retinoid oxidation.
Our newly presented data thus suggest an alternative explanation for the increase in PUFAs in TGs during activation: to provide building blocks for lysosomal antioxidants in the form of DHA-containing BMPs. Due to the lack of full structural elucidation of our TG-species, it is not possible to identify the presence of DHA in HSC TG-pool (Fig. S2). Nevertheless, any omega-3 fatty acid—e.g., α-linolenic acid (C18:3), eicosapentaenoic acid (C20:5)—can be converted to DHA, and TGs with more than three double bonds were observed abundantly (Fig. S2) (10, 19). In contrast, omega-3 fatty acids cannot be synthesized by de novo synthesis from monounsaturated fatty acids. This inability might explain the cell’s urge to store PUFAs whenever they are available to create storage buffers for future use.
In summary, our comprehensive lipidomics data of activating rat HSCs suggest a two-stage activation of HSCs that is guided by an increase in PUFAs and BMPs. We anticipate that this study provides novel clues for the elucidation of the molecular mechanisms involved in HSC activation as well as new leads for the identification of therapeutic targets to treat liver inflammation.
Experimental procedures
Hepatic stellate cell isolation and cell culture
HSCs were isolated from a 10 to 12 week-old male rat as described previously (49). Animals were handled according to governmental and international animal experimentation guidelines and laws. Experiments were approved by the Animal Experimentation Committee (Dierexperimentencommissie; DEC) of Utrecht University (DEC number 2013.III.02.016). After isolation, cells were protected from light and cultured on coverslips in 24-well plates for microscopy (Nunc) or in 6-well dishes (Nunc) for lipidomics. Cells were grown in Dulbecco Modified Eagle Medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all obtained from Gibco, Invitrogen GmbH) in 37 °C/5% CO2 incubators. Two experiments were performed with one rat each. The first experiment was selected as a representative study and is used in the presented figures. After isolation, the viability of the cells was routinely tested (>90% viability) using the Cell Counting Kit-8 (Sigma-Aldrich) according to the instructions (96-well plate protocol) provided by the manufacturer. Yields were about 16 million HSCs per rat, which were all divided over 60 dishes (thus, 266k cells per dish). Cell numbers per dish remained approximately constant over the course of the experiment, although the cell culture surface area covered by the cells increased as HSCs expand during activation.
Confocal microscopy
To visualize LDs and α-SMA, cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences Hatfield). Subsequently, cells were stained with DAPI and LD540 (kindly donated by Dr C. Thiele) (50). Staining for immunofluorescence was performed with anti-α SMA (Thermo Fisher Scientific), followed by goat-anti-mouse-alexa647 or donkey-a-rabbit-alexa647 (Life Technologies). Cells were mounted with FluorSave (Calbiochem) and subsequently imaged with a Leica TCS SPE Laser Scanning Spectral Confocal Microscope using preset settings for the representative dyes.
Lipidomics by LC-MS/MS
After cell culturing, lipids were extracted as described previously (51), dried under nitrogen, and dissolved in 100 μl chloroform/methanol (1:1). For the analysis of phospholipids and sphingolipids, extracts were injected on a HILIC column (2.6 μm HILIC 100 Å, 50 × 4.6 mm, Phenomenex) using an Infinity II 1290 UPLC (Agilent). Using a constant flow rate of 1 ml min−1, a gradient was generated (time in min, %B): (0, 0), (1, 50), (3, 50), (3.1, 100), (4, 100), using solvent A (acetonitrile/acetone, 9:1, v/v) and B (acetonitrile/H2O, 7:3, v/v, with 10 mM ammonium formate). Both solvents contained 0.1% formic acid. Subsequently, the column effluent was connected to a heated electrospray ionization source of an LTQ XL mass spectrometer (Thermo Fisher Scientific). The LTQ mass spectrometer was operated in both positive and negative ionization mode. Source and capillary temperatures were set to 450 °C and 400 °C, respectively and the ionization voltage to 2.5 kV. Full scan spectra were collected in the range from 450 to 1150 amu at a scan speed of 3 scans/s. For the measurement of neutral lipids (sterols, TGs, steryl esters), extracts were injected on a Kinetex/HALO C8 column (2.6 μm, 150 × 3.00 mm; Phenomenex) using a gradient of methanol:H2O (5:5 v/v, solvent A) and methanol:isopropanol (8:2 v/v, solvent B), which was generated by an Infinity II 1290 UPLC (Agilent). A constant flow rate of 600 μl min−1 (0 min; 100% A, 2 min; 0% A, 8 min; 0% A, 8.5 min; 100% A, 10 min; 100% A). Lipids were measured using APCI in positive mode coupled to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). To measure retinoids, extracts were injected and separated using a 250 × 3.0 mm Synergi 4u Max-RP 80A column (4 μm particle size, Phenomenex), a flow rate of 350 μl min−1, with the following gradient: (solvent A; acetonitrile:water (95:5), solvent B; acetone:chloroform (85:15), 0 min; 90% A, 5 min; 40% A, 17 min; 0% A, 19 min; 90% A, 25 min; 90% A), generated by a Flexar UHPLC system (PerkinElmer). The column effluent was connected to a triple quadrupole mass spectrometry (API 4000 QTRAP, MDS Sciex/Applied Biosystems) equipped with an APCI ionization source (set to 500 °C). Multiple reaction monitoring in positive ion mode was used to detect RE species with settings and m/z transitions as described previously (7). Raw datafiles were converted to mzXML files by msConvert (part of ProteoWizard v3.0.913) (52) and processed with the R package xcms v2.99.3 (53), as described (18, 54). Lipid amounts per sample were expressed as signal relative to the sum of the signals.
Dimensionality reduction of lipidomics data
Dimensionality reduction of lipidomic datasets was performed by PCA, MDS, t-SNE, and UMAP. First, datasets were normalized by expressing lipid signals as percentage of the sum per sample and subsequently z-scaled (mean = 0, SD = 1). PCA and MDS (euclidean distance, 2 dimensions) were performed by R base-functions. t-SNE (R package ‘Rtsne’, 2 dimensions, perplexity 18) and UMAP (R package ‘umap’) were performed using default settings.
Increased lipid coverage of LION
In comparison to the previous version of LION/web (18), LION now also includes lipids with odd-numbered fatty acids, as well as LION-terms representing odd-numbered fatty acids building blocks, starting from fatty acid C13:0 up to C25:7. When applicable, the newly added lipids were linked to biophysical properties as described (18).
Increased lipid mapping to LION by ‘smart matching’
A significant expansion in comparison to the previous version of LION/web (18) is the implementation of a new strategy—called 'smart matching'—to map lipid identifiers to LION. Previously, input lipids were matched to an association table containing lipids and corresponding LION IDs. Unmatched lipids were not used for further analysis. With 'smart matching', LION/web proceeds with a second strategy using dynamic associations in case direct matching was not successful for a given lipid. In this strategy, lipids are systematically disentangled into their building blocks and more general structures by text-mining. Subsequently, each lipid species is dynamically associated with these building blocks or simplified structures if they are represented by a LION-term. For example, TG(17:0/17:0/18:0) is not present in LION. With 'smart matching' switched on, this lipid will be dissected into their fatty acids (C17:0 and C18:0, represented by LION:0022231 and LION:0002921, respectively) and its more general structure, TG(52:0) (LION:0000759). As these features are represented by LION-nodes, TG(17:0/17:0/18:0) will be dynamically linked to these terms. In case, also TG(52:0) is not available in LION, the web-tool selects LION-terms higher in the hierarchy, here TGs (LION:0000622). This strategy strongly increases identifier mapping to LION and yields equal statistical output as when the LION-term TG(17:0/17:0/18:0) would have been present in the ontology.
Another expansion of LION/web is the addition of unique integers to all input lipids. Thus, when two identical lipids are submitted in LION/web, they are now processed as distinct features. The reason is that in lipidomics practice, most lipid names represent a collection of unique structures. For example, even PC(18:1/20:5) is not unique due to the undefined positions (Δ1, 2,…) and nature (cis/trans) of the double bonds in C18:1 and C20:5. Hence, the assumption that identical names represent identical structures cannot be made. Previously, duplicated lipid names were neglected.
Fatty acid composition prediction of phospholipids
The prediction of the fatty acid composition of lipid species is now incorporated in LION/web as an optional setting (‘predict fatty acid associations’) in the settings panel. Fatty acid compositions of glycerolipids(phospho)lipids annotated with the sum of its individual fatty acids were estimated based on multinomial distributions of reported fatty acid compositions with k = 2, 3, or 4 fatty acids (Extended Data Table 1 in (55)). To avoid an erroneous increase in the number of lipids n and statistical power in the dataset, the most abundant fatty acid combination per lipid was selected and used in the analysis. Summed lipids with 2 (PC, PS, PE, PI, PG, BMP, DG), 3 (TG), and 4 fatty acids (CL) were included in the prediction. For example, PC(32:0) can represent PC(16:0_16:0), PC(14:0_18:0), etc. From these subspecies, probabilities P(14:0 = 0, 16:0 = 1, 16:0 = 1, 18:0 = 0, … = 0) and P(14:0 = 1, 16:0 = 0, 16:0 = 0, 18:0 = 1, … = 0) were calculated and compared. For every species, the fatty acid composition with the highest probability was chosen. Cross-validation using a dataset containing PL(FA_FA) identifications (56) showed a good agreement of identified versus predicted fatty acids (Fig. S4).
LION-PCA heatmap analysis
LION-PCA is an adaptation of GO-PCA (20), a method designed for RNAseq datasets. LION-PCA processes lipidomics datasets as follows: the sum-normalized samples are scaled as sample z-scores (mean = 0, sd = 1). Then, PCA is performed and from a selected number of PrCs, the loadings are used to rank lipid features. For all selected PrCs, LION-term enrichment is assessed by using the ranking-mode using two-sided KS-tests as described (18). Significantly enriched LION-terms in any of the selected PrCs are selected. From these LION-terms, associated lipid feature z-scores are averaged per sample and visualized in a heatmap using R package ‘pheatmap’ v1.0.12. Using R package ‘shiny’ v.1.3.2 and the R-code for the user interface of LION/web (available via https://github.com/martijnmolenaar/LION-web), the described LION-PCA heatmap analysis pipeline was cast into a new module of LION/web, with similar feel and workflow as the enrichment analysis module, hosted on shinyserver.io and accessible via http://heatmap.lipidontology.com.
LION pathway analysis
Lipid pathway analysis was performed as described by Nguyen et al. (24, 25), with a number of adaptations. Briefly, for major lipid pathways, we constructed a simplified directional network of biochemical reactions, consisting of triplets of substrate, reaction direction, and product. Definitions of the biochemical reactions were obtained from the KEGG-database (57). The substrates and products of the reactions are presented by LION-terms. For example, conversion from lysoPC to PC was represented by the terms LION:0000030 (diacylglycerophosphocholines) and LION:0000034 (monoacylglycerophosphocholines). In our implementation, only the shortest paths between all the pathway’s metabolites are evaluated. Paths or individual reactions with z-scores higher than 1.645, corresponding with p < 0.05, are considered to be statistically significant, z-scores > 2.33: very significant (p < 0.01), and z-scores > 3.09: extremely significant (p < 0.001).
Software and R packages
‘shiny’ v1.3.2; ‘shinyBS’ v0.61; ‘shinyWidgets’ v0.4.8; ‘shinyTree’ v0.2.7; ‘pheatmap’ v1.0.12; ‘igraph’ v1.2.4.1; ‘ggplotify’ v0.0.4; ‘ggplot2’ v3.2.1; ‘ggthemes’ v4.2.0; ‘RSQLite’ v2.1.2; ‘RColorBrewer’ v1.1.2; ‘cowplot’ v1.0.0; ‘formattable’ v0.2.0.1; ‘rhandsontable’ v0.3.7; ‘reshape2’ v1.4.3; ‘Rtsne’ v0.15; ‘umap’ v0.2.5.0.
Data availability
All data are contained within the article and supporting information.
Supporting information
This article contains supporting information (18, 56).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Jos Brouwers for help during the initial stages of this project and Jeroen Jansen for expert technical assistance with the lipidomic experiments.
Author contributions
M. R. M., A. B. V., M. H., and J. B. H. conceptualization; M. R. M. and J. B. H. methodology; M. R. M. software; M. R. M. validation; M. R. M. and M. W. H. formal analysis; M. R. M. and M. W. H. investigation; M. R. M. and J. B. H. writing–original draft; A. B. V., M. H., and J. B. H. writing–review and editing; J. B. H. supervision.
Reviewed by members of the JBC Editorial Board. Edited by Dennis Voelker
Supporting information
References
- 1.Blaner W.S., O'Byrne S.M., Wongsiriroj N., Kluwe J., D'Ambrosio D.M., Jiang H., et al. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim. Biophys. Acta. 2009;1791:467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiam A.R., Beller M. The why, when and how of lipid droplet diversity. J. Cell Sci. 2017;130:315–324. doi: 10.1242/jcs.192021. [DOI] [PubMed] [Google Scholar]
- 4.Walther T.C., Farese R.V. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Byrne S.M., Blaner W.S. Retinol and retinyl esters: biochemistry and physiology: thematic review series: fat-soluble vitamins: vitamin A. J. Lipid Res. 2013;54:1731–1743. doi: 10.1194/jlr.R037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senkal C.E., Salama M.F., Snider A.J., Allopenna J.J., Rana N.A., Koller A., et al. Ceramide is metabolized to acylceramide and stored in lipid droplets. Cell Metab. 2017;25:686–697. doi: 10.1016/j.cmet.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajat M., Molenaar M.R., Brouwers J.F.H.M., Vaandrager A.B., Houweling M., Helms J.B. Hepatic stellate cells retain the capacity to synthesize retinyl esters and to store neutral lipids in small lipid droplets in the absence of LRAT. Biochim. Biophys. Acta. 2017;1862:176–187. doi: 10.1016/j.bbalip.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Teratani T., Tomita K., Suzuki T., Oshikawa T., Yokoyama H., Shimamura K., et al. A high-cholesterol diet exacerbates liver fibrosis in mice via accumulation of free cholesterol in hepatic stellate cells. Gastroenterology. 2012;142:152–164.e10. doi: 10.1053/j.gastro.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 9.Tomita K., Teratani T., Suzuki T., Shimizu M., Sato H., Narimatsu K., et al. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology. 2014;59:154–169. doi: 10.1002/hep.26604. [DOI] [PubMed] [Google Scholar]
- 10.Tuohetahuntila M., Spee B., Kruitwagen H.S., Wubbolts R., Brouwers J.F., van de Lest C.H., et al. Role of long-chain acyl-CoA synthetase 4 in formation of polyunsaturated lipid species in hepatic stellate cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2015;1851:220–230. doi: 10.1016/j.bbalip.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Shmarakov I.O., Jiang H., Liu J., Fernandez E.J., Blaner W.S. Hepatic stellate cell activation: a source for bioactive lipids. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:629–642. doi: 10.1016/j.bbalip.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molenaar M.R., Vaandrager A.B., Helms J.B. Some lipid droplets are more equal than others: different metabolic lipid droplet pools in hepatic stellate cells. Lipid Insights. 2017;10 doi: 10.1177/1178635317747281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuohetahuntila M., Molenaar M.R., Spee B., Brouwers J.F.H.M., Houweling M., Vaandrager A.B., et al. ATGL and DGAT1 are involved in the turnover of newly synthesized triacylglycerols in hepatic stellate cells. J. Lipid Res. 2016;57:1162–1174. doi: 10.1194/jlr.M066415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molenaar M.R., Yadav K.K., Toulmay A., Wassenaar T.A., Mari M.C., Caillon L., et al. Retinyl esters form lipid droplets independently of triacylglycerol and seipin. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernández-Gea V., Ghiassi-Nejad Z., Rozenfeld R., Gordon R., Fiel M.I., Yue Z., et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thoen L.F.R., Guimarães E.L.M., Dollé L., Mannaerts I., Najimi M., Sokal E., et al. A role for autophagy during hepatic stellate cell activation. J. Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Tuohetahuntila M., Molenaar M.R., Spee B., Brouwers J.F.H.M., Wubbolts R., Houweling M., et al. Lysosome-mediated degradation of a distinct pool of lipid droplets during hepatic stellate cell activation. J. Biol. Chem. 2017;292:12436–12448. doi: 10.1074/jbc.M117.778472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molenaar M.R., Jeucken A., Wassenaar T.A., Van De Lest C.H.A., Brouwers J.F., Helms J.B. LION/web: a web-based ontology enrichment tool for lipidomic data analysis. Gigascience. 2019;8 doi: 10.1093/gigascience/giz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Testerink N., Ajat M., Houweling M., Brouwers J.F., Pully V.V., van Manen H.J., et al. Replacement of retinyl esters by polyunsaturated triacylglycerol species in lipid droplets of hepatic stellate cells during activation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner F. GO-PCA: an unsupervised method to explore gene expression data using prior knowledge. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi T., Stang E., Fang K.S., De Moerloose P., Parton R.G., Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi T., Beuchat M.H., Lindsay M., Frias S., Palmiter R.D., Sakuraba H., et al. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1999;1:113–118. doi: 10.1038/10084. [DOI] [PubMed] [Google Scholar]
- 23.Moreau D., Vacca F., Vossio S., Scott C., Colaco A., Paz Montoya J., et al. Drug-induced increase in lysobisphosphatidic acid reduces the cholesterol overload in Niemann–Pick Type C cells and mice. EMBO Rep. 2019;20 doi: 10.15252/embr.201847055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen A., Rudge S.A., Zhang Q., Wakelam M.J.O. Using lipidomics analysis to determine signalling and metabolic changes in cells. Curr. Opin. Biotechnol. 2017;43:96–103. doi: 10.1016/j.copbio.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen A., Guedán A., Mousnier A., Swieboda D., Zhang Q., Horkai D., et al. Host lipidome analysis during rhinovirus replication in HBECs identifies potential therapeutic targets. J. Lipid Res. 2018;59:1671–1684. doi: 10.1194/jlr.M085910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kölzer M., Werth N., Sandhoff K. Interactions of acid sphingomyelinase and lipid bilayers in the presence of the tricyclic antidepressant desipramine. FEBS Lett. 2004;559:96–98. doi: 10.1016/S0014-5793(04)00033-X. [DOI] [PubMed] [Google Scholar]
- 27.Petersen N.H.T., Olsen O.D., Groth-Pedersen L., Ellegaard A.M., Bilgin M., Redmer S., et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24:379–393. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Alexandrov T., Ovchinnikova K., Palmer A., Kovalev V., Tarasov A., Stuart L., et al. METASPACE: a community-populated knowledge base of spatial metabolomes in health and disease. bioRxiv. 2019 doi: 10.1101/539478. [preprint] [DOI] [Google Scholar]
- 29.Palmer A., Phapale P., Chernyavsky I., Lavigne R., Fay D., Tarasov A., et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods. 2016;14:57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- 30.Alexandrov T. Spatial metabolomics and imaging mass spectrometry in the age of artificial intelligence. Annu. Rev. Biomed. Data Sci. 2020;3:61–87. doi: 10.1146/annurev-biodatasci-011420-031537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji J., Yu F., Ji Q., Li Z., Wang K., Zhang J., et al. Comparative proteomic analysis of rat hepatic stellate cell activation: a comprehensive view and suppressed immune response. Hepatology. 2012;56:332–349. doi: 10.1002/hep.25650. [DOI] [PubMed] [Google Scholar]
- 32.Paulo J.A., Kadiyala V., Banks P.A., Conwell D.L., Steen H. Mass spectrometry-based quantitative proteomic profiling of human pancreatic and hepatic stellate cell lines. Genomics Proteomics Bioinformatics. 2013;11:105–113. doi: 10.1016/j.gpb.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Chen F., Fan X., Lin C., Hao Y., Wei H., et al. Quantitative proteomic analysis on activated hepatic stellate cells reversion reveal STAT1 as a key regulator between liver fibrosis and recovery. Sci. Rep. 2017;7:44910. doi: 10.1038/srep44910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krenkel O., Hundertmark J., Ritz T., Weiskirchen R., Tacke F. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells. 2019;8:503. doi: 10.3390/cells8050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcher AB, Bendixen SM, Terkelsen MK, Hohmann SS, Hansen MH, Larsen BD, et al. Transcriptional regulation of hepatic stellate cell activation in NASH. Sci Rep. 2019;9:2324. doi: 10.1038/s41598-019-39112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman S.L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 38.Haaker M.W., Vaandrager A.B., Helms J.B. Retinoids in health and disease: a role for hepatic stellate cells in affecting retinoid levels. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865:158674. doi: 10.1016/j.bbalip.2020.158674. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 40.Rosenthal S.B., Liu X., Ganguly S., Dhar D., Pasillas M.P., Ricciardelli E., et al. Heterogeneity of HSCs in a mouse model of NASH. Hepatology. 2021;74:667–685. doi: 10.1002/hep.31743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W., He H., Wang T., Su N., Zhang F., Jiang K., et al. Single-cell transcriptomic analysis reveals a hepatic stellate cell-activation roadmap and myofibroblast origin during liver fibrosis in mice. Hepatology. 2021;74:2774–2790. doi: 10.1002/hep.31987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia J., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolter T., Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 44.Makino A., Ishii K., Murate M., Hayakawa T., Suzuki Y., Suzuki M., et al. D-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol alters cellular cholesterol homeostasis by modulating the endosome lipid domains. Biochemistry. 2006;45:4530–4541. doi: 10.1021/bi052104y. [DOI] [PubMed] [Google Scholar]
- 45.Trinh M.N., Brown M.S., Goldstein J.L., Han J., Vale G., McDonald J.G., et al. Last step in the path of LDL cholesterol from lysosome to plasma membrane to ER is governed by phosphatidylserine. Proc. Natl. Acad. Sci. U. S. A. 2020;117:18521–18529. doi: 10.1073/pnas.2010682117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platt F.M., D'Azzo A., Davidson B.L., Neufeld E.F., Tifft C.J. Lysosomal storage diseases. Nat. Rev. Dis. Primers. 2018;4:27. doi: 10.1038/s41572-018-0025-4. [DOI] [PubMed] [Google Scholar]
- 47.Twu Y.-C., Lee T.-S., Lin Y.-L., Hsu S.-M., Wang Y.-H., Liao C.-Y., et al. Niemann-Pick Type C2 protein mediates hepatic stellate cells activation by regulating free cholesterol accumulation. Int. J. Mol. Sci. 2016;17:1122. doi: 10.3390/ijms17071122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouvier J., Zemski Berry K.A., Hullin-Matsuda F., Makino A., Michaud S., Geloën A., et al. Selective decrease of bis(monoacylglycero)phosphate content in macrophages by high supplementation with docosahexaenoic acid. J. Lipid Res. 2009;50:243–255. doi: 10.1194/jlr.M800300-JLR200. [DOI] [PubMed] [Google Scholar]
- 49.Riccalton-Banks L., Bhandari R., Fry J., Shakesheff K.M. A simple method for the simultaneous isolation of stellate cells and hepatocytes from rat liver tissue. Mol. Cell. Biochem. 2003;248:97–102. doi: 10.1023/a:1024184826728. [DOI] [PubMed] [Google Scholar]
- 50.Spandl J., White D.J., Peychl J., Thiele C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 2009;10:1579–1584. doi: 10.1111/j.1600-0854.2009.00980.x. [DOI] [PubMed] [Google Scholar]
- 51.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 52.Adusumilli R., Mallick P. Data conversion with ProteoWizard msConvert. Methods Mol. Biol. 2017;1550:339–368. doi: 10.1007/978-1-4939-6747-6_23. [DOI] [PubMed] [Google Scholar]
- 53.Smith C.A., Want E.J., O'Maille G., Abagyan R., Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 54.Jeucken A., Brouwers J.F. High-throughput screening of lipidomic adaptations in cultured cells. Biomolecules. 2019;9:42. doi: 10.3390/biom9020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma C., Kesarwala A.H., Eggert T., Medina-Echeverz J., Kleiner D.E., Jin P., et al. NAFLD causes selective CD4+ T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531:253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofmann S., Krajewski M., Scherer C., Scholz V., Mordhorst V., Truschow P., et al. Complex lipid metabolic remodeling is required for efficient hepatitis C virus replication. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:1041–1056. doi: 10.1016/j.bbalip.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information.