Abstract
Our studies in chronic binge alcohol (CBA) treated simian immunodeficiency virus (SIV)-infected macaques and in people living with HIV (PLWH) show significant alterations in metabolic homeostasis. CBA promotes a profibrotic phenotype in adipose tissue and skeletal muscle (SKM) and decreases adipose-derived stem cell and myoblast differentiation, making adipose and SKM potential drivers in metabolic dysregulation. Furthermore, we have shown that the differential expression of microRNAs (miRs) in SKM contributes to impaired myoblast differentiation potential. Beyond modulation of intracellular responses, miRs can be transported in extracellular vesicles (EVs) to mediate numerous cellular responses through intercellular and interorgan communication. This study tested the hypothesis that CBA alters concentration and miR cargo of EVs derived from adipocytes and myotubes isolated from SIV-infected male macaques. Fourteen male rhesus macaques received either (CBA, 2.5g/kg/day) or sucrose (VEH) for 14.5 months. Three months following the initiation of CBA/VEH, all animals were infected with SIVmac251 and 2.5 months later initiated on antiretroviral therapy. SKM and adipose tissue samples were collected at the study endpoint (blood alcohol concentration= 0 mM). EVs were isolated by ultracentrifugation of myotube and adipocyte cell culture supernatant. Nanoparticle tracking revealed no differences in concentration or size of particles between VEH and CBA groups. Adipocyte-derived EVs from CBA animals showed decreased miR-let-7a expression (p=0.03). Myotube-derived EVs from CBA animals had decreased miR-16 (p=0.04) and increased miR-133a and miR-133b (both p=0.04) expression. These results indicate that CBA administration differentially regulates EV miR content but does not alter number of EVs from adipocytes or myotubes. Future studies are warranted to determine the functional relevance of CBA-altered EV miR cargo and their role in intercellular and interorgan communication and metabolic dysregulation.
Keywords: Chronic binge alcohol, microRNAs, extracellular vesicles, adipose, skeletal muscle
INTRODUCTION
Advances in antiretroviral therapy have led to a near-normal life expectancy of people living with HIV (PLWH). This extended life expectancy has highlighted earlier occurrence of comorbid conditions such as insulin resistance and diabetes in this population (1–4). Additionally, at-risk alcohol use in PLWH is twice as high as in the general population (5), and at-risk alcohol use is associated with altered glucose/insulin dynamics in PLWH (6, 7). However, the mechanisms responsible for altered glucose/insulin dynamics with alcohol use in HIV have yet to be elucidated.
Adipose tissue and skeletal muscle (SKM) are the major contributors to fat mass and lean mass, respectively. These tissues participate in intercellular and interorgan crosstalk through the secretion of soluble factors and extracellular vesicles (EVs) (8–10) that contribute to metabolic homeostasis. Compared to uninfected individuals, PLWH gain fat mass and lose lean mass despite viral suppression (11). Furthermore, we have shown that chronic binge alcohol (CBA) dysregulates functions of adipose tissue and skeletal muscle in a simian immunodeficiency virus (SIV)-infected macaque model. In the SIV-infected macaque model used in the current study, we have previously demonstrated that CBA dysregulates whole body glucose/insulin dynamics as measured by disposition index and acute insulin response to glucose (12). Additionally, CBA decreases adipose-derived stem cell and myoblast differentiation (13, 14) and increases collagen deposition in adipose tissue and skeletal muscle (13, 15) further demonstrating the adverse effects of CBA on these peripheral metabolically active organs.
While CBA causes adipose tissue and SKM dysregulation, the exact mechanisms of CBA-mediated effects on glucose/insulin dynamics and metabolic tissue dysregulation are still unknown. CBA differentially regulates SKM and circulating miRs in SIV-infected macaques (16), and decreased SKM miR-206 expression mechanistically contributes to impaired myoblast differentiation (14). Additionally, clinical studies suggest that there are differentially expressed circulating miRs in people with at-risk alcohol use (17, 18) and type II diabetes (19). Additionally, in vitro studies demonstrate that adipose and skeletal muscle-derived miRs have functional implications on metabolic organs such as the liver and pancreas (20, 21). Because miRs are differentially expressed with CBA in SIV-infected macaques (16), and CBA impairs whole-body glucose/insulin dynamics in this model (12), we postulate that differential miR expression may contribute to metabolic dyshomeostasis (14). Moreover, their release within EVs could potentially impact distant cell pathophysiological responses. EVs are nanometer-sized, lipid bilayer-bound vesicles that are released from virtually all cell types and protect bioactive cargo, including miRs, while being transported in circulation. Exosomes are the smallest form of EVs ranging in size from 30–150 nm and are packaged into multivesicular bodies prior to release from the host cell. Once released into circulation, EVs can target a recipient cell to deliver the bioactive cargo and affect recipient cell function (22). Alcohol increases EV release from hepatocytes by CYP2E1-mediated ethanol metabolism in vitro (23), and total circulating EVs are increased in alcohol-administered mice and in people with alcohol-induced hepatitis (24). Alcohol also alters the EV miR cargo in hepatocytes (25), monocytes (26), and neural stem cells (27); however, whether alcohol alters adipocyte or myotube-derived EV cargo is unknown.
The aim of this study was to determine the impact of CBA on concentration and miR cargo of EVs released from cultured adipocytes and myotubes isolated from SIV-infected macaques. Selected miRs measured were chosen based on their published roles in metabolic tissue dysregulation (14, 21, 28–34) and alcohol-related pathologies (35, 36). We hypothesized that CBA would increase EV release and alter metabolic miR EV cargo. Elucidating the effects of chronic alcohol on EV profile would provide a foundation for future studies to investigate the functional effects of alcohol-induced EV alterations on intercellular and interorgan communication and metabolic regulation in the context of SIV infection.
METHODS
Animal Study Design
The animal study design was approved the Institutional Animal Care and Use Committee at both Louisiana State University Health Sciences Center (New Orleans, LA) and Tulane National Primate Research Center (Covington, LA). The study adhered to the National Institutes of Health guidelines for the care and use of animals in research. Details of the experimental design were previously published (12, 37, 38). Briefly, rhesus macaques (Macaca mulatta) ages 4–6 years were administered CBA (13–14g of ethanol per kg of body mass per week) or VEH through a gastric catheter. After 3 months of CBA or VEH administration, macaques were rectally infected with simian immunodeficiency virus (SIVmac251), and 2.5 months later, all macaques used for the current study were treated with antiretroviral therapy (ART; 20mg/kg of tenofovir and 30mg/kg of emtricitabine; provided by Gilead Sciences, Inc.) for 9 months. Macaques continued to receive CBA or VEH throughout the duration of the study (14.5 months, Figure 1). Macaques were fasted and humanely euthanized 24 hours after their last alcohol administration (blood alcohol level = 0mM). Adipose tissue and skeletal muscle tissue were collected for downstream analysis.
Figure 1.
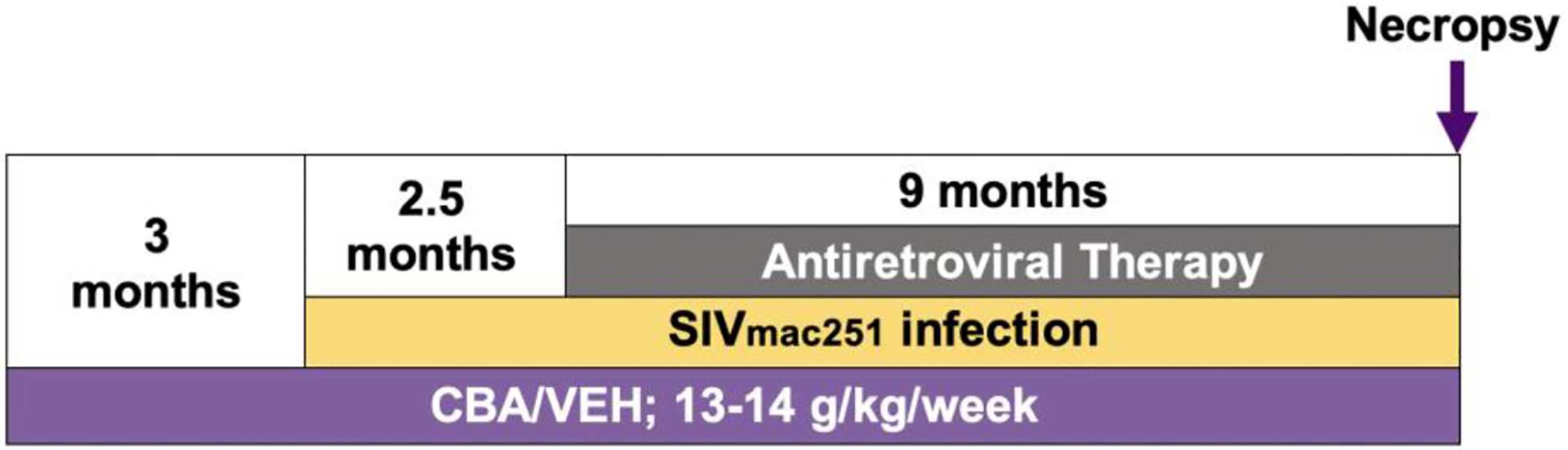
Schematic of nonhuman primate study timeline.
Cell Cultures
Primary adipose-derived stem cells from omental adipose tissue (OmAT) and myoblasts from quadriceps femoris muscle were isolated as previously described (13, 14, 39, 40). Briefly, rhesus macques OmAT (~50g) was washed, minced, and enzymatically digested in collagenase buffer [0.25% collagenase type I (Sigma-Aldrich, St. Louis, MO), 2% penicillin streptomycin (P/S), 0.25ug/ml Fungizone in HBSS] by incubating at 37°C, 5% CO2 for 30–60min. The tissue homogenate was filtered through a sterile gauze, the filtrate was centrifuged at 180xg for 7min, and the pellet was resuspended in red blood cell lysis buffer for 10 minutes (Qiagen, Hilden, Germany). The pellet was washed by centrifugation at 500xg for 5min, passed through a 70μm cell strainer, and plated in a 10cm tissue culture dish (passage 0).
Primary myoblasts were isolated from ~25mg of skeletal muscle tissue and enzymatically digested with 0.25% trypsin-EDTA diluted 1:4 in Ham’s F-12 during two 1h treatments. Cells were then plated for 5h for fibroblast separation. Nonadhered cells were collected and cultured in Ham’s F-12 medium with 10% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA) and 2.5ng/ml human fibroblast growth factor (R & D Systems, Minneapolis, MN) in 10cm dishes (passage 0). Primary adipose-derived stem cells (ADSCs) and myoblasts from passage 4 were used for the current study.
Differentiation to Adipocytes and Myotubes
ADSCs were plated at a density of 350,000 cells/10cm plate. Three 10cm plates of cells for each primary cell line were proliferated in stromal media (α-MEM media with 10% FBS, 1% L-glutamine, 2% P/S, and 0.25μg/ml Fungizone) until 90%−100% confluency. The cells were rinsed with PBS and differentiated in 10ml of adipogenic differentiation medium (0.5ug/ml dexamethasone, 0.5mM isobutylmethylxanthine, and 50μM indomethacin in stromal media). The media were replaced every 3 days. At day 12 of differentiation, cells were washed with PBS twice and serum-free differentiation media added. Serum-free media was used to avoid introducing EVs from serum to the culture. After 48h, culture supernatant was collected for EV isolation.
Myoblasts were plated at a density of 350,000 cells/10cm plate. Three 10cm plates of cells for each primary cell line were proliferated in Ham’s F-12 medium with 10% FBS, 2% L-glutamine, 1% P/S, 0.25μg/ml fungizone, and 2.5ng/ml human fibroblast growth factor. Once cells reached 70–80% confluency, cells were washed and medium was changed to differentiation medium [Ham’s F-12 medium with 2% horse serum (Lonza, Allendale, NJ), 2% L-glutamine, 1% P/S, 0.25ug/ml fungizone]. At day 3 of differentiation, cells were washed with PBS twice and serum-free differentiation media added. After 48h, culture supernatant was collected for EV isolation.
EV Isolation from Cell Culture
Cell culture supernatant from adipocyte or myotube cultures was collected from three 10cm plates (3–5 million cells) for each primary cell line and placed in 50ml conical tubes. Supernatant was centrifuged at 2,000xg for 10min to remove any cells or debris. The supernatant was transferred to a new 50ml conical tube and centrifuged at 15,500xg for 30min at 4°C to remove large particles including apoptotic bodies. The supernatant was filtered through a 0.22μm syringe filter (Whatman PVDF filter, Maidstone, UK) into polycarbonate tubes (Beckman Coulter, Brea, CA) and ultracentrifuged at 100,000xg for 1.5h at 4°C (Ti-70 rotor, Optima L-80XP, Beckman Coulter, Brea, CA). Supernatant was discarded leaving 1ml in the tube. The pellet was pipet mixed and transferred to 1.5ml ultracentrifuge tubes (Beckman Coulter) and ultracentrifuged at 100,000xg for 70min at 4°C (Optima TLX, Beckman Coulter). The pellet was resuspended in 100μl supernatant and mixed by pipetting.
EV isolates from adipocyte and myotube cultures were divided as follows: 10μl for nanoparticle tracking analysis, 30μl for protein isolation, 60μl for RNA isolation.
Electron Microscopy
Representative samples of adipocyte and myotube EV isolates were imaged using transmission electron microscopy (TEM) at the LSU Shared Instrumentation Facility (Baton Rouge, LA). Briefly, 3μl of sample was added to a 300-mesh carbon filmed TEM grid (EMS #CF300-CU) and dried at room temperature. The samples were stained with 2% uranyl acetate for 2min, inserted into a JEM-1400 transmission electron microscope (120 kV, JEOL Ltd., Akishima, Tokyo, Japan), and imaged with a Gatan digital camera (Gatan, Inc., Pleasanton, CA).
Western Blot for EV Markers
EV isolates (30μl) were homogenized in Tissue Protein Extraction Reagent (Thermo Fisher Scientific) buffer with added HALT protease inhibitor cocktail (ThermoFisher Scientific, Waltham, MA). Macaque whole skeletal muscle tissue was homogenized the same way to be used as a negative control for CD63 expression and positive control for calnexin expression. Concentration of protein samples was determined using a bicinchoninic acid assay (Pierce BCA Assay, Thermo Fisher Scientific). Protein (equal volumes of EV protein and skeletal muscle protein equal to average μg of EV protein) was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Bio-Rad Mini-PROTEAN TGX Precast Gels) and transferred overnight to polyvinylidene fluoride membranes (0.20μm pore size, EMD Millipore, Billerica, MA) for immunoblot analysis. Anti-CD63 1:250 (sc-5275, SantaCruz Biotechnology, Santa Cruz, CA) or anti-calnexin 1:1000 #2679, (Cell Signaling Technology, Danvers, MA) primary antibodies in 5% bovine serum albumin in Tris-Buffered Saline with 0.1% Tween (TBST) was incubated with membranes overnight. Anti-mouse or anti-rabbit HRP conjugated secondary antibodies (Cell Signaling) in 5% milk were incubated with membranes for 1h. Membranes were washed with 1X TBST three times for 5min between each step. Membranes were incubated for 5min with chemiluminescent substrate (Immobilon Forte Western HRP substrate) (EMD Millipore), and Blue Lite UHC films (Uniscience Corp, Miami, FL) were utilized to capture images.
Nanoparticle Tracking for EV Concentration and Size Distribution
Particle concentration and size distribution was measured using nanoparticle tracking analysis (NTA) with the Nanosight (NS300; Malvern, Worcestershire, UK). NTA uses Brownian motion to calculate particle size. Prior to running any samples, air, water, and then air again was used to flush the tubing leading to the chamber where the video was captured. Water used for cleaning was filtered through a 0.02μm syringe filter (Whatman PVDF filter). Samples were loaded into the chamber at a syringe pump speed of 1000μl/min until the fluid filled the chamber. Standard measurements of 60s each at a speed of 35μl/min were perfomed in triplicate for each sample. Prior to diluting the samples, EV isolates were fixed with zinc formalin (Sigma-Aldrich, St. Louis, MO) at a 1:1 ratio. Dilutions of each sample were adjusted so that more than 10 particles were seen per frame without any particles visually overlapping (dilution factor ~150). Between samples, the tubing was flushed with air, water, and then air again. Total particle concentration and size distribution were calculated by the NTA software (Malvern Panalytical, Malvern, UK).
EV miRNA Expression
Total RNA was isolated from 60μl of EV isolate using the miRNeasy mini kit (Qiagen) according to manufacturer’s instructions. cDNA was synthesized from 10ng of RNA and a master mix comprised of 0.15μl 100mM dNTP, 1U multiscribe RT enzyme, 1.5μl 10x RT buffer, 3.8U RNAse inhibitor, 1.16μl nuclease-free water, and 6μl of 5x RT primer pool. The RT primer pool consisted of forward and reverse primers of four miRs of interest including the housekeeping U6 (Applied Biosystems, Waltham, MA). Primer information can be found in Table 1. The reaction was carried out on a MyCyclerTM (Bio-Rad Laboratories, Hercules, CA) at 16°C for 30min, 42°C for 30min, and at 85°C for 5min. miRs were quantified using TaqMan Universal PCR Master Mix II (ThermoFisher, Waltham, MA). Each reaction was 20μl and composed of 10μl master mix, 1ul of the TaqMan Small RNA Assay, 7.7μl nuclease-free water, and 1.3μl of cDNA. The qPCR reaction was performed on a CFX96 real-time PCR system (Bio-Rad Laboratories, Hercules, CA) at 50 °C for 2min, 95°C for 10min, 95°C for 15sec, 60°C for 1min, 95°C with 0.5°C deceases until 60°C and repeated forty-four times. Each reaction was performed in duplicate. No template controls, no reverse transcriptase controls, and cell culture media-only samples were run for each primer as control measures. As a normalization control for RNA loading, small nucleolar RNA U6 was amplified in duplicate wells on the same multiwell plate. qPCR data were analyzed using the 2(-ΔΔCT) method.
Table 1.
miR primer sequences. Mature miRs were measured.
| Assay Name | Assay Number | Mature miR Sequence |
|---|---|---|
| hsa-let-7a-3p | 2307 | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-miR-9-5p | 583 | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-16-5p | 391 | UAGCAGCACGUAAAUAUUGGCG |
| hsa-miR-17-5p | 393 | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-26a-5p | 405 | UUCAAGUAAUCCAGGAUAGGCU |
| hsa-miR-103a-3p | 439 | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-125b-5p | 449 | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-142-5p | 2248 | CAUAAAGUAGAAAGCACUACU |
| hsa-miR-146a-5p | 468 | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-181a-5p | 480 | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-221-3p | 524 | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-720 | 2895 | UCUCGCUGGGGCCUCCA |
| hsa-miR-133a-3p | 2246 | UUUGGUCCCCUUCAACCAGCUG |
| hsa-miR-133b | 2247 | UUUGGUCCCCUUCAACCAGCUA |
| hsa-miR-144-3p | 197375_mat | UACAGUAUAGAUGAUGUACU |
| hsa-miR-203a-3p | 507 | GUGAAAUGUUUAGGACCACUAG |
| hsa-miR-206 | 510 | UGGAAUGUAAGGAAGUGUGUGG |
Because our previous work in PLWH and SIV-infected macaques suggests that alcohol or CBA has effects on metabolic homeostasis, miRs were selected based on their role in metabolic syndrome (28, 29, 21, 30–32) and pathologies associated with alcohol use such as liver fibrosis (35, 36) and muscle atrophy (14, 33, 34). miRs measured in adipocyte-derived EVs included miR-let-7a, miR-9, miR-16, miR-17, miR-26, miR-103, miR-125, miR-142, miR-146a, miR-181, miR-221, and miR-720. miRs measured in myotube-derived EVs included miR-16, miR-133a, miR-133b, miR-142, miR-144, miR-146a, miR-203, miR-206, and miR-720.
Statistical Analysis
Because the data did not fit assumptions of parametric statistics, the non-parametric Mann-Whitney U test was used to determine differences in EV content and miR expression between VEH and CBA groups. Hedges’ g was used to determine the effect size, and g ≥ 0.8 was considered large (41). GraphPad Prism 9 (San Diego, CA) was used to perform statistical analyses. The alpha level of significance was set at 0.05. Data are shown as mean ± standard error of the mean (SEM) unless otherwise indicated. Each data point on graphs represents a different animal.
RESULTS
Confirmation of EV Isolation
Following EV isolation from conditioned cell culture media, the isolates were imaged by TEM to ensure the presence of EVs. Figure 2 displays representative images of EVs isolated from adipocyte (Fig. 2a) and myotube (Fig. 2b) culture supernatant. Representative Western blots for expression of EV marker CD63 and negative control marker calnexin are shown in Figure 3.
Figure 2.
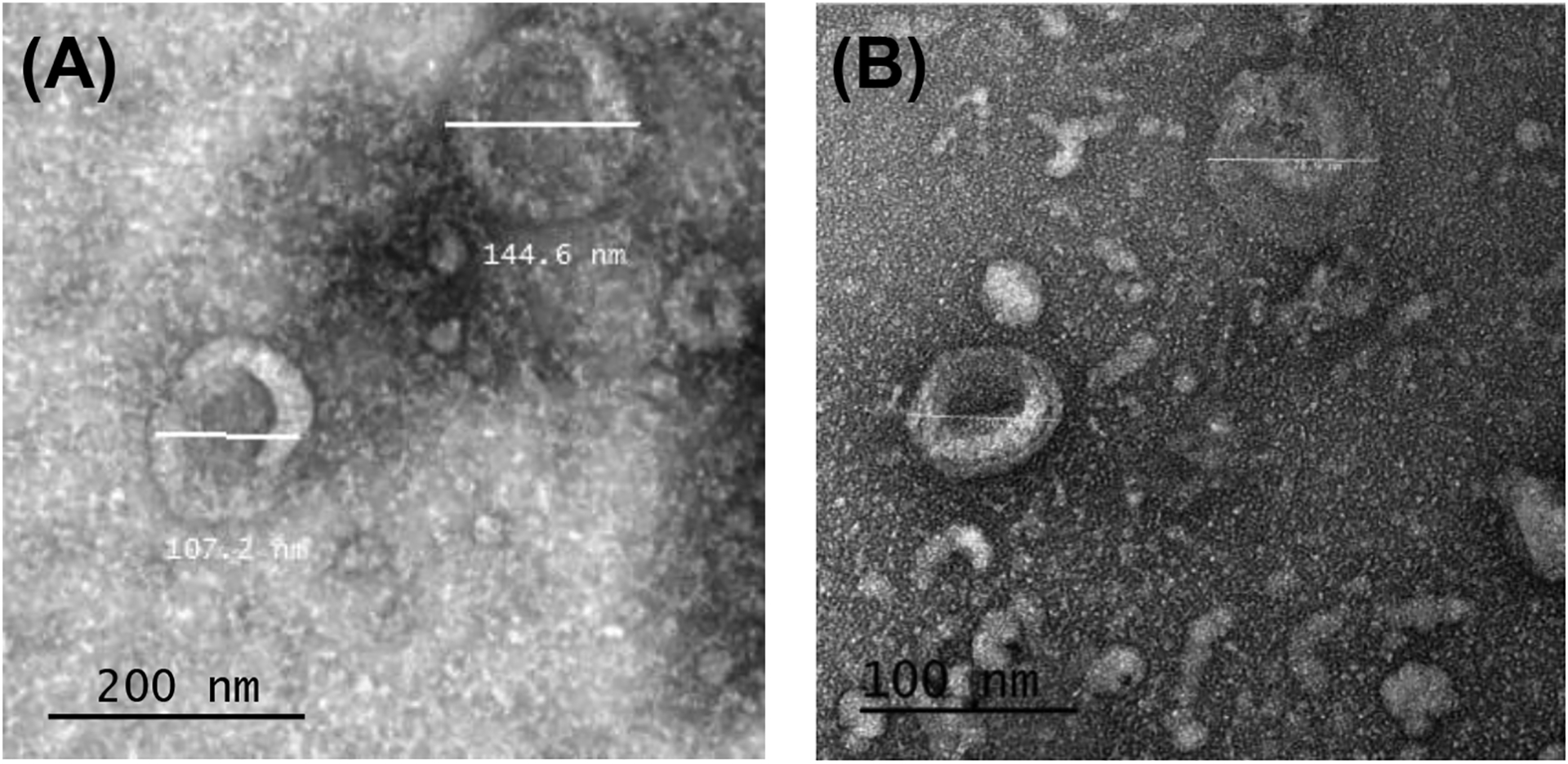
Representative transmission electron microscopy (TEM) images of ultracentrifugation isolates from adipocyte culture with EVs sized 104.2 and 124.7 nm (A) and myotube culture with EVs sized 70.9 and 78.9 nm (B).
Figure 3.
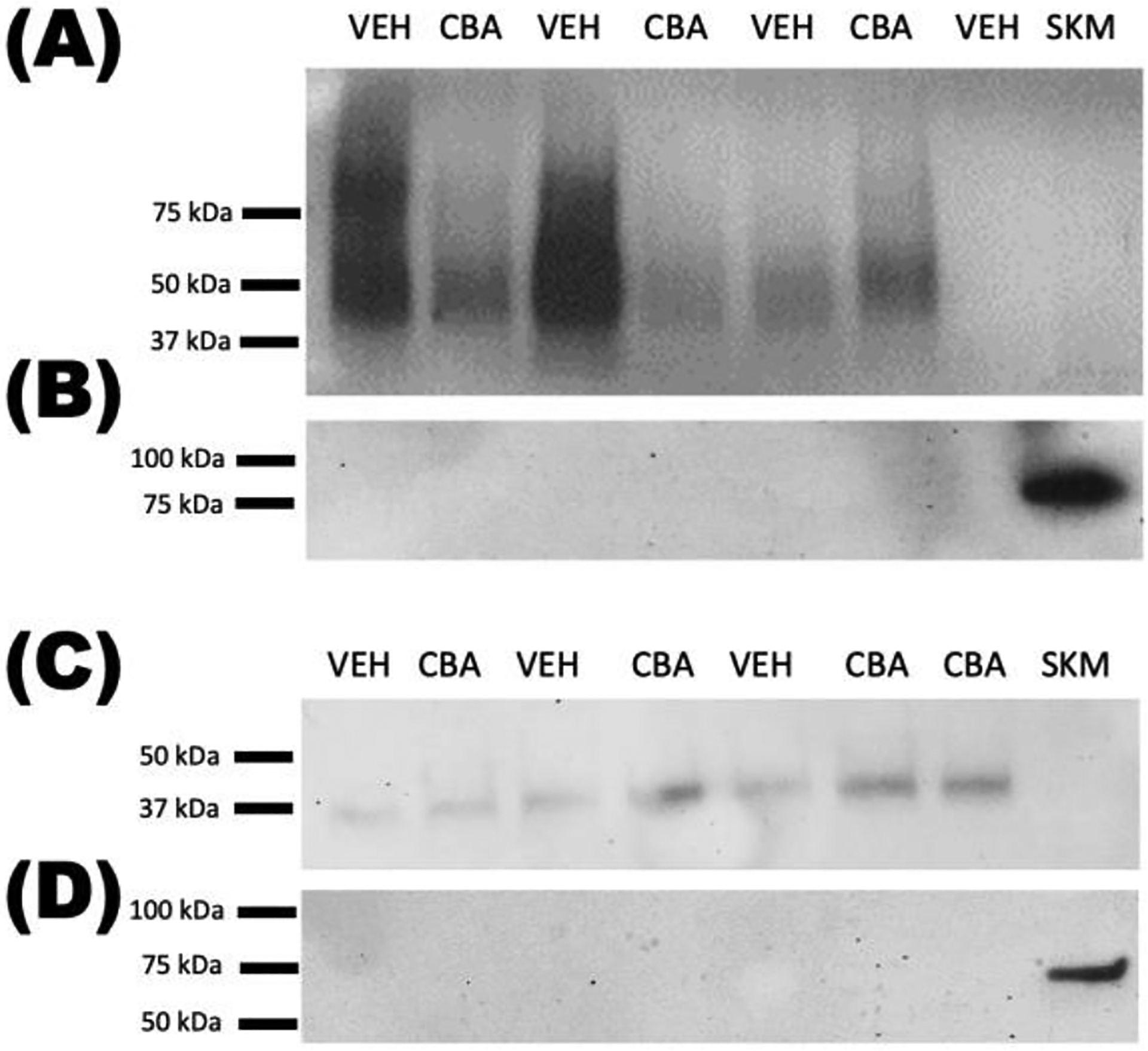
CD63 is expressed in EV isolates from adipocytes (A) and myotubes (C). Calnexin is not expressed in EV isolates from adipocytes (B) or myotubes (D). CBA, EVs from chronic binge alcohol-derived cells; VEH, EVs from vehicle-derived cells; SKM, whole skeletal muscle protein.
EV concentration
EV size distribution and concentrations were analyzed by NTA. There was no significant difference in total particle count (Figure 4) between VEH and CBA groups for adipocyte EV isolates (Figure 4a; 5.0×1011 ± 3.7×1011/ml for VEH, 1.7×1011 ± 0.7×1011/ml for CBA) or myotube EV isolates (Figure 4b; 7.9×1010 ± 8.7×1010 ± 2.4×1010/ml for VEH, 2.1×1011 ± 0.7×1011/ml for CBA). There was no significant difference in the average particle size between VEH and CBA groups for adipocyte EVs (190.8 ± 30.4 nm for VEH, 180.2 ± 17.4 nm for CBA) or myotube EVs (168.5 ± 6.0 nm for VEH, 177.6 ± 5.6 for CBA). Representative size distributions from NTA are shown for one VEH and one CBA isolate for each EV source (Figures 4c–d).
Figure 4.
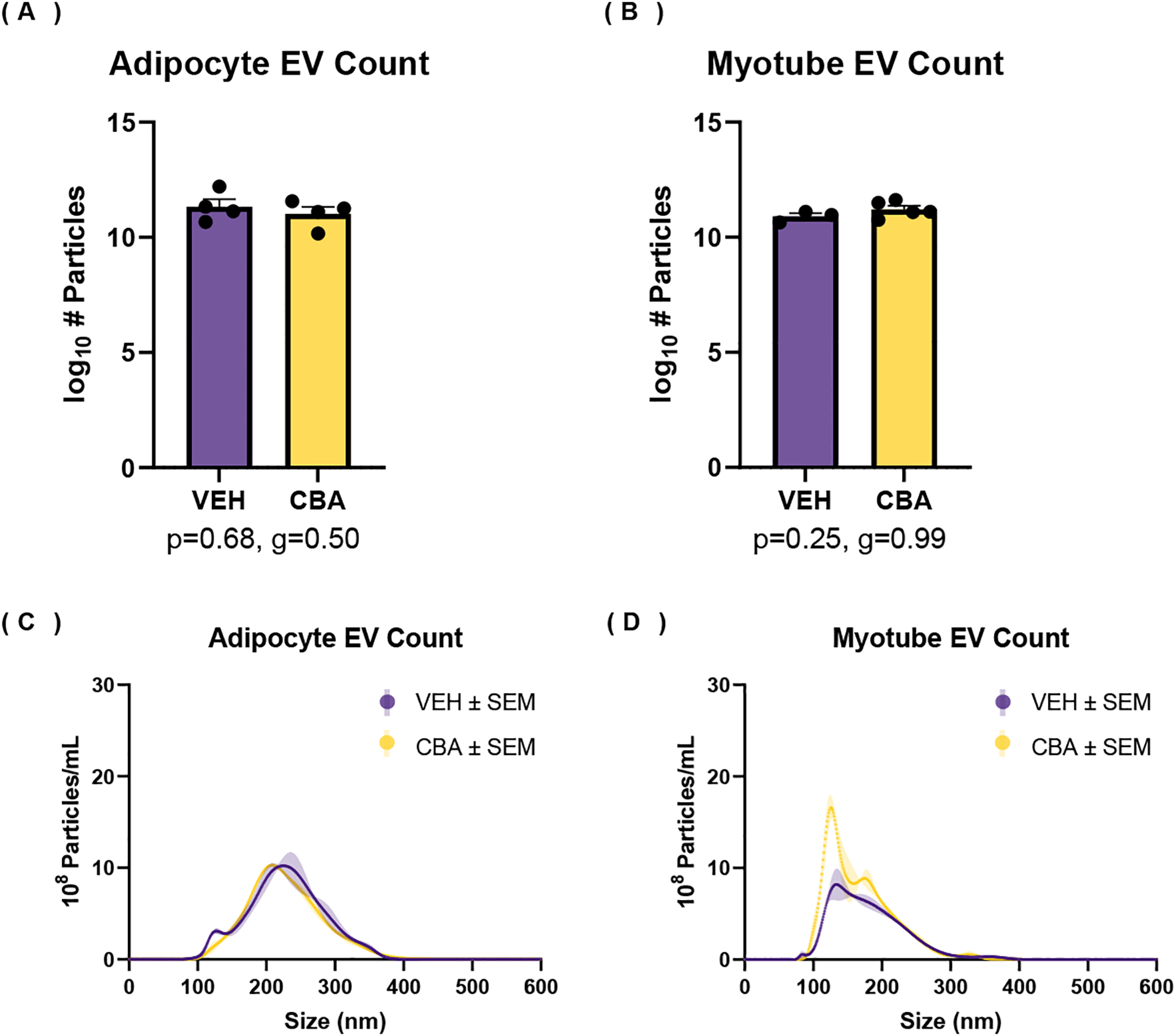
Number of particles from EV isolate measured by nanoparticle tracking analysis. (A-B) Log10 transformed average and standard error of the mean (SEM) of total number of particles isolated from adipocyte culture (A) and myotube culture (B). (C-D) Representative particle size distribution and SEM from isolates of VEH and CBA ASC culture (C) and myotube culture (D). There were no differences in the number of particles isolated from adipocyte or myotube culture between VEH and CBA groups.
Adipocyte Culture EV miRNA Expression
CBA significantly decreased miR-let-7a expression (Hedges’ g=2.08, p<0.05) in EVs isolated from adipocyte cell culture supernatant. There was a large effect size for CBA to increase the expression of miR-16 (g=1.20, p=0.11), miR-125 (g=0.81, p=0.49), and miR-720 (g=1.15, p=0.20) and a large effect size for CBA to decrease the expression of miR-26 (g=2.14, p=0.06) and miR-146a (g=1.54, p=0.11), all which failed to reach statistical significance (Figure 5). There was little to no expression of miR-17 in adipocyte-derived EVs, and no significant differences in expression between VEH and CBA for miRs-9, 103, 181, 142, and 221.
Figure 5.
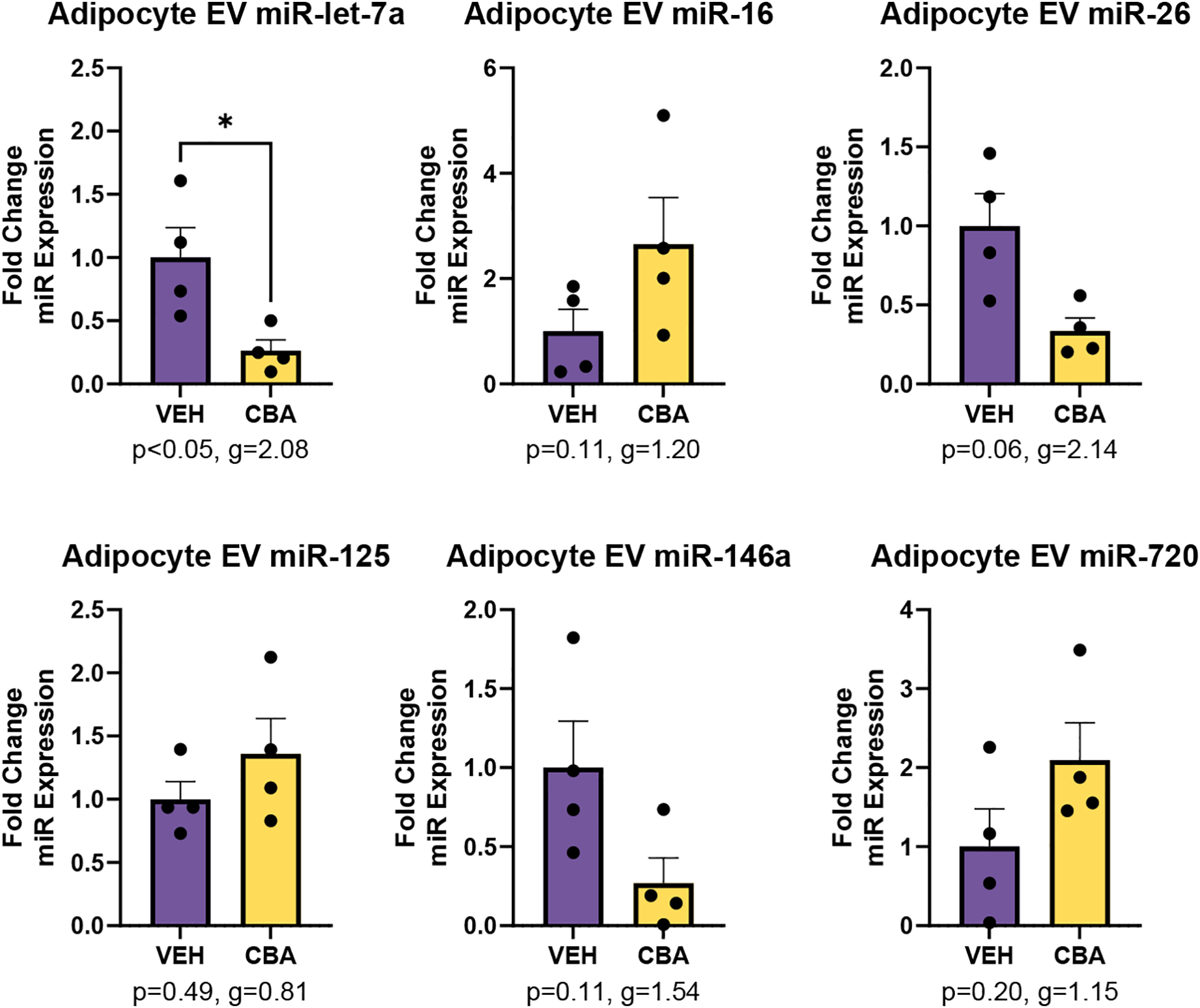
miR expression in EVs isolated from adipocyte culture of VEH (N=4) and CBA (N=4) animals. Values are mean fold change in miR expression ± SEM. There was significant lower expression of miR-let-7a in CBA adipocyte EVs compared to VEH adipocyte EVs. There was a large effect size for CBA to increase miR-16, miR-125, and miR-720 and for CBA to decrease miR-146a. *<0.05 vs. VEH.
Myotube Culture EV miRNA Expression
There was significantly lower miR-16 expression (g=6.03, p<0.05) and significantly higher miR-133a (g=1.05, p<0.05) and miR-133b expression (g=0.99, p<0.05) in EVs isolated from myotube cell culture supernatant of the CBA group. There was a large effect size for CBA to increase the expression of miR-206 (g=1.00, p=0.07) (Figure 6). There was little to no expression of miRs-142, 144, and 203 and no significant difference in the expression of miR-146a between VEH and CBA groups.
Figure 6.
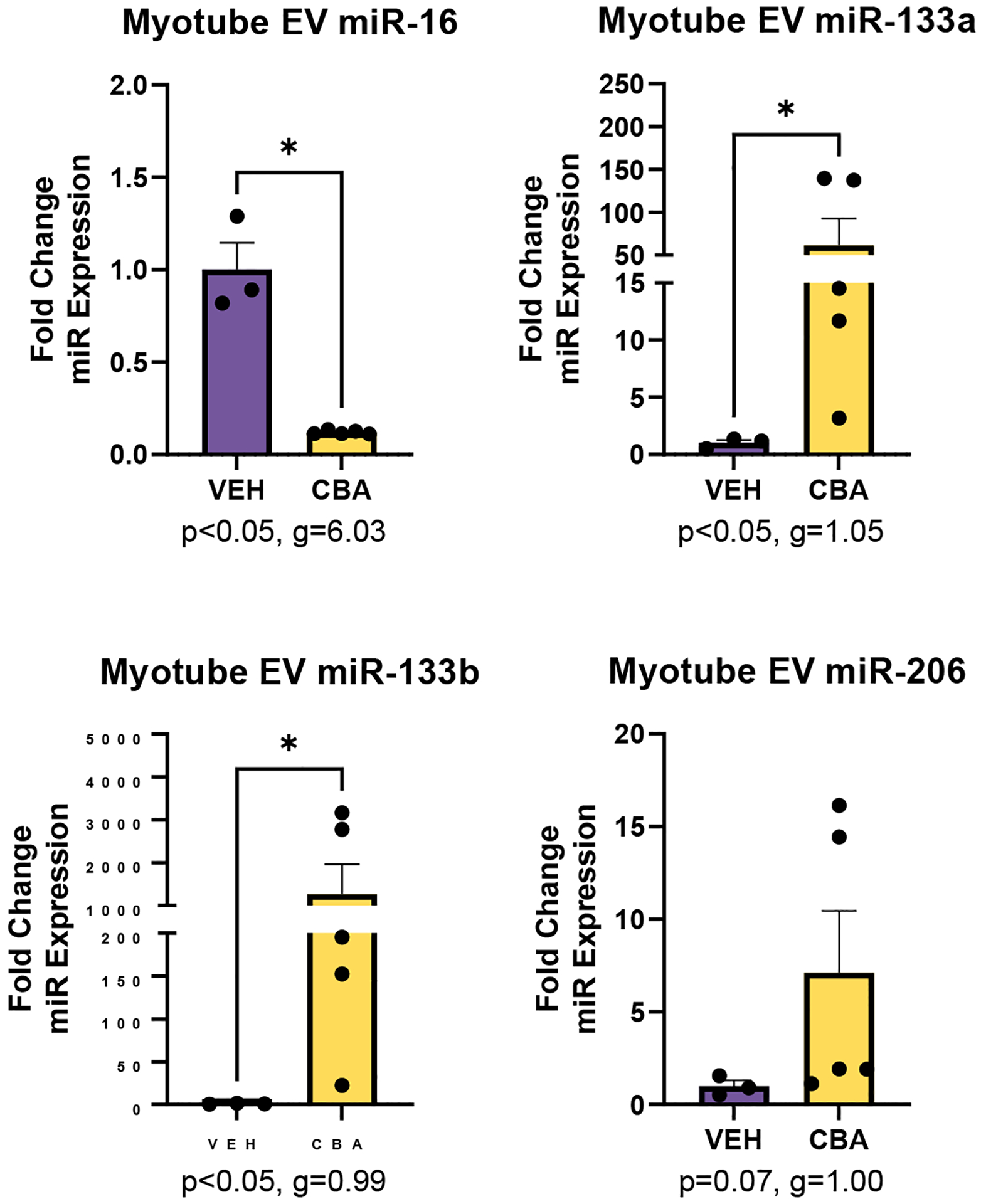
miR expression in EVs isolated from myotube culture of VEH (N=3) and CBA (N=5) animals. Values are mean fold change in miR expression ± SEM. There was significant lower expression of miR-16 and higher expression of miR-133a and 133b in CBA myotube EVs compared to VEH. There was a large effect size for CBA to increase miR-206 expression. *p<0.05 vs. VEH
DISCUSSION
The current study aimed to characterize the effect of CBA on SIV-infected male macaque adipocyte and myotube EV profile. Previous studies using these macaques had shown that CBA decreases disposition index and acute insulin response to glucose regardless of ART treatment (12) suggesting that CBA impairs whole-body glucose/insulin dynamics. At the tissue level, CBA decreases omental adipocyte cell size and increases collagen expression and immune cell infiltration (13). Additionally, CBA impairs skeletal muscle stem cell differentiation (14, 49) and increases skeletal muscle collagen content and profibrotic and inflammatory gene expression (15). Based on these previous findings in our model and the current literature, we hypothesized that CBA alters EV profile, including EV numbers and miR content, of adipocyte and myotube EVs. Based on NTA, our results indicate that CBA does not significantly alter the number and size of EVs released from cultured adipocytes and myotubes isolated from SIV-infected macaques. Whether this would be altered by acute alcohol exposure remains to be determined. However, CBA differentially expressed specific miRs within the EVs released from adipocytes and myotubes in culture.
Once uptaken into cells, miRs within EVs can target specific genes and inhibit their expression (22). Hence, the changes in adipocyte and myotube EV cargo could have functional effects on target cells. The miRs measured in the current study were selected based on their role in targeting genes involved in metabolic dysregulation (28, 29, 21, 30–32) and pathologies associated with alcohol use (14, 33, 34). The miRs that we have identified as being dysregulated with CBA are implicated in metabolic regulation in muscle, adipose, liver, and pancreas.
Previous studies reveal that mice with an alcohol-related liver disease phenotype have a higher amount of circulating EVs compared to control mice (26), and humans with alcohol-induced hepatitis have increased circulating EVs compared to healthy controls (24). We did not detect an increase in the number of EVs released from adipocytes or myotubes from the CBA group. This suggests that chronic alcohol alone without an acute challenge may not be sufficient to increase EV release from adipocytes or myotubes or that the increase in circulating EVs seen with alcohol is specific to conditions with liver pathology. In hepatocytes expressing cytochrome P450 2E1 (Cyp2E1), but not in control or alcohol dehydrogenase (ADH) expressing hepatocytes, in vitro ethanol exposure results in increased EV release (23). This may be due to the production of reactive oxygen species (ROS) caused by Cyp2E1-mediated ethanol metabolism (23) and could also explain why we did not observe an increase in EVs released from adipocytes or myotubes.
Our results show that CBA resulted in alterations of adipocyte and myotube EV miR cargo. Within adipocyte-derived EVs, CBA decreased the expression of miR-let-7a. miR-let-7a has been shown to increase in expression during adipogenesis (42). Additionally, the miR-let-7 family participates in antifibrotic signaling (43). Our previous studies have reported that CBA impairs adipose-derived stem cell differentiation and promotes a pro-fibrotic phenotype in SKM and adipose tissue of SIV-infected macaques (13). Thus, lower expression of miR-let-7a within EVs from adipocytes from the CBA group could be reflective of CBA-induced impairments in adipogenesis and early profibrotic changes, but this remains to be examined.
Within myotube-derived EVs, CBA decreased the expression of miR-16 and increased the expression of miRs-133a and 133b. miR-16 from skeletal muscle EVs modulates beta cell proliferation (21). Additionally, circulating miR-16 is downregulated during insulin resistance in preclinical and clinical studies (29, 44). miR-133a/b are enriched in skeletal muscle and are important for myoblast differentiation (45). Furthermore, miR-133a regulates the browning of adipocytes (30) and inhibits Klf15 mRNA leading to a reduction in GLUT4 expression in cardiomyocytes (31). Additionally, insulin-like growth factor-1 receptor (IGF-1R) expression is inhibited by miR-133a/b (46). Downregulation of miR-16 and upregulation miR-133a/b within myotube EVs further suggests a potential role of EVs in the CBA-induced metabolic dysregulation seen in our model (12). Additionally, an increase in myogenic miRs within the EVs released from CBA-treated macaque myotubes may indicate muscle injury or a compensatory mechanism (47) to restore CBA-mediated impairments in SKM differentiation (14).
This study is not without limitations. One limitation is our small sample size. We have used primary myoblast lines that were cryopreserved from rhesus macaques that were used for a parent study. Because a large number of cells is required to produce measurable amounts of EVs, the current study could only use a subset of primary myoblast lines from the larger parent study. However, the genetic variability of macaques is similar as in humans; therefore, we believe that our statistically significant results indicate biological relevance. Additionally, our study was performed using EVs isolated ex vivo. While this is a limitation, previous studies by our group have found that changes seen in myotube cultures reflect what is seen in the skeletal muscle (48, 49); therefore, we expect changes in myotube-derived EVs to reflect in vivo muscle phenotype. Another limitation is that we used nanoparticle tracking for estimates of EV concentrations. This method detects all particles in solution and is not specific to EVs. Because we utilize macaque samples, a fixative was necessary to avoid contaminating the NTA equipment. This fixative may have caused some clumping of nanoparticles resulting in larger sizes being measured likely explaining why there were particles greater in diameter than the 0.22μm filter pores used prior to ultracentrifugation. Because of the NTA limitations, we have included TEM to confirm our EV isolation methods were successful. An additional limitation is that we did not determine whether CBA-induced EV miR alterations from adipocytes and myotubes reflect what is seen is circulation. Future studies will utilize immunocapture methods (50, 51) to isolate tissue-specific EVs from circulation and compare to those released from cultured cells. Additionally, all of our macaques were SIV-infected and ART treated; therefore, the findings of the current study are limited to CBA’s effects within the context of controlled SIV infection. The current study was performed to characterize whether there are differences in EVs from peripheral metabolic tissues between VEH and CBA groups. Future studies will identify whether there are changes in expression of target genes and whether it translates to alterations in cellular functions.
While alcohol-induced liver pathology is the most commonly studied alcohol-related pathology, our past studies demonstrate that chronic alcohol induces alterations in peripheral metabolic organs including adipose tissue (13), skeletal muscle (14), and pancreas (52). In addition, whole-body dysregulation of glucose/insulin dynamics are seen with at-risk alcohol in PLWH (7) and SIV-infected macaques (12, 52). Characterizing CBA-induced alterations in adipocyte and myotube EV miR cargo is an initial step in determining the role of EVs in alcohol-induced metabolic dyshomeostasis. Our future studies will determine the implications of altered adipocyte and myotube EV miR cargo on cellular functions such as differentiation, hypertrophy, and intercellular or interorgan metabolic crosstalk.
HIGHLIGHTS.
Alcohol impairs metabolic homeostasis in SIV-infected macaques
Chronic alcohol decreases adipocyte miR-let-7a in extracellular vesicles
Chronic alcohol decreases myotube miR-16 in extracellular vesicles
Chronic alcohol increases myotube miR-133 in extracellular vesicles
AKNOWLEDGMENTS
The authors would like to acknowledge Jason Dufour, DVM, DACLAM from the Tulane National Primate Research Center (TNPRC) for his veterinary expertise. The authors are grateful for the excellent technical assistance from Larissa Devlin, Wayne A. Cyprian, and Nancy Dillman at the TNPRC Pathology Laboratory. Members of the LSUHSC Comprehensive Alcohol-HIV/AIDS Research Center (Naveena Chalapati, Curtis Vande Stouwe, Nedra LaCour, Bryant Autin, MS) have been instrumental to the study, and Hui-Yi Lin, PhD provided statistical expertise. We would also like to thank Ying Xiao, TEM specialist at LSU Baton Rouge. We thank the Cellular Immunology and Immune Metabolism Core at the Louisiana Cancer Research Center (Grant 5P30GM114732-05 NIH/NIGMS) for access to the NanoSight N300, and Dorota Wyczechowska, PhD, for her technical support. This work was supported by NIH/NIAAA: P60AA009803 (PEM), K01AA024494 (LS), F30AA029358 (BLB), F32AA027982 (DEL), and T32AA007577 (PEM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Noumegni SRN, Nansseu JR, Ama VJM, Bigna JJ, Assah FK, Guewo-Fokeng M, Leumi S, Katte J-C, Dehayem M, Kengne AP, Sobngwi E. Insulin resistance and associated factors among HIV-infected patients in sub-Saharan Africa: a cross sectional study from Cameroon. Lipids Health Dis 16: 148, 2017. doi: 10.1186/s12944-017-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willig AL, Overton ET. Metabolic Complications and Glucose Metabolism in HIV Infection: A Review of the Evidence. Curr HIV/AIDS Rep 13: 289–296, 2016. doi: 10.1007/s11904-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulte-Hermann K, Schalk H, Haider B, Hutterer J, Gmeinhart B, Pichler K, Brath H, Dorner TE. Impaired lipid profile and insulin resistance in a cohort of Austrian HIV patients. J Infect Chemother 22: 248–253, 2016. doi: 10.1016/j.jiac.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Innes S, Abdullah KL, Haubrich R, Cotton MF, Browne SH. High Prevalence of Dyslipidemia and Insulin Resistance in HIV-infected Prepubertal African Children on Antiretroviral Therapy. Pediatr Infect Dis J 35: e1–7, 2016. doi: 10.1097/INF.0000000000000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galvan FH, Bing EG, Fleishman JA, London AS, Caetano R, Burnam MA, Longshore D, Morton SC, Orlando M, Shapiro M. The prevalence of alcohol consumption and heavy drinking among people with HIV in the United States: results from the HIV Cost and Services Utilization Study. J Stud Alcohol 63: 179–186, 2002. doi: 10.15288/jsa.2002.63.179. [DOI] [PubMed] [Google Scholar]
- 6.Simon L, Ferguson TF, Vande Stouwe C, Brashear MM, Primeaux SD, Theall KP, Welsh DA, Molina PE. Prevalence of Insulin Resistance in Adults Living with HIV: Implications of Alcohol Use. [DOI] [PMC free article] [PubMed]
- 7.Primeaux S, Simon L, Ferguson T, Brashear M, Yeh A, Molina P. Alcohol use and dysglycemia among people living with HIV in the ALIVE-Ex Study.
- 8.Kahn CR, Wang G, Lee KY. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. Journal of Clinical Investigation 129: 3990–4000, 2019. doi: 10.1172/JCI129187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trayhurn P, Drevon CA, Eckel J. Secreted proteins from adipose tissue and skeletal muscle – adipokines, myokines and adipose/muscle cross-talk. Archives of Physiology and Biochemistry 117: 47–56, 2011. doi: 10.3109/13813455.2010.535835. [DOI] [PubMed] [Google Scholar]
- 10.Vechetti IJ, Valentino T, Mobley CB, McCarthy JJ. The role of extracellular vesicles in skeletal muscle and systematic adaptation to exercise. [DOI] [PMC free article] [PubMed]
- 11.Grant PM, Kitch D, McComsey GA, Collier AC, Bartali B, Koletar SL, Erlandson KM, Lake JE, Yin MT, Melbourne K, Ha B, Brown TT. Long-term body composition changes in antiretroviral-treated HIV-infected individuals. AIDS 30: 2805–2813, 2016. doi: 10.1097/QAD.0000000000001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford SM, Simon L, Vande Stouwe C, Allerton T, Mercante DE, Byerley LO, Dufour JP, Bagby GJ, Nelson S, Molina PE. Chronic binge alcohol administration impairs glucose-insulin dynamics and decreases adiponectin in asymptomatic simian immunodeficiency virus-infected macaques. Am J Physiol Regul Integr Comp Physiol 311: R888–R897, 2016. doi: 10.1152/ajpregu.00142.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford SM, Simon Peter L, Berner P, Cook G, Vande Stouwe C, Dufour J, Bagby G, Nelson S, Molina PE. Differential contribution of chronic binge alcohol and antiretroviral therapy to metabolic dysregulation in SIV-infected male macaques. American Journal of Physiology-Endocrinology and Metabolism 315: E892–E903, 2018. doi: 10.1152/ajpendo.00175.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon L, Ford SM, Song K, Berner P, Vande Stouwe C, Nelson S, Bagby GJ, Molina PE. Decreased myoblast differentiation in chronic binge alcohol-administered simian immunodeficiency virus-infected male macaques: role of decreased miR-206. Am J Physiol Regul Integr Comp Physiol 313: R240–R250, 2017. doi: 10.1152/ajpregu.00146.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodd T, Simon L, LeCapitaine NJ, Zabaleta J, Mussell J, Berner P, Ford S, Dufour J, Bagby GJ, Nelson S, Molina PE. Chronic Binge Alcohol Administration Accentuates Expression of Pro-Fibrotic and Inflammatory Genes in the Skeletal Muscle of Simian Immunodeficiency Virus-Infected Macaques. Alcohol Clin Exp Res 38: 2697–2706, 2014. doi: 10.1111/acer.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon L, Hollenbach AD, Zabaleta J, Molina PE. Chronic binge alcohol administration dysregulates global regulatory gene networks associated with skeletal muscle wasting in simian immunodeficiency virus-infected macaques. BMC Genomics 16: 1097, 2015. doi: 10.1186/s12864-015-2329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyczechowska D, Lin H-Y, LaPlante A, Jeansonne D, Lassak A, Parsons CH, Molina PE, Peruzzi F. A miRNA Signature for Cognitive Deficits and Alcohol Use Disorder in Persons Living with HIV/AIDS. Front Mol Neurosci 10: 385, 2017. doi: 10.3389/fnmol.2017.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourgeois BL, Lin H-Y, Yeh AY, Levitt DE, Primeaux SD, Ferguson TF, Molina PE, Simon L. Unique Circulating MicroRNA Associations with Dysglycemia in People Living with HIV and Alcohol Use. [DOI] [PMC free article] [PubMed]
- 19.Ding L, Ai D, Wu R, Zhang T, Jing L, Lu J, Zhong L. Identification of the differential expression of serum microRNA in type 2 diabetes. Bioscience, Biotechnology, and Biochemistry 80: 461–465, 2016. doi: 10.1080/09168451.2015.1107460. [DOI] [PubMed] [Google Scholar]
- 20.Niu Q, Wang T, Wang Z, Wang F, Huang D, Sun H, Liu H. Adipose-derived mesenchymal stem cell-secreted extracellular vesicles alleviate non-alcoholic fatty liver disease via delivering miR-223–3p. Adipocyte 11: 572–587, 2022. doi: 10.1080/21623945.2022.2098583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jalabert A, Vial G, Guay C, Wiklander OPB, Nordin JZ, Aswad H, Forterre A, Meugnier E, Pesenti S, Regazzi R, Danty-Berger E, Ducreux S, Vidal H, El-Andaloussi S, Rieusset J, Rome S. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 59: 1049–1058, 2016. doi: 10.1007/s00125-016-3882-y. [DOI] [PubMed] [Google Scholar]
- 22.Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol 27: 172–188, 2017. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma VK, Li H, Wang R, Hirsova P, Mushref M, Liu Y, Cao S, Contreras PC, Malhi H, Kamath PS, Gores GJ, Shah VH. Alcohol stimulates macrophage activation through caspase-dependent hepatocyte derived release of CD40L containing extracellular vesicles. J Hepatol 64: 651–660, 2016. doi: 10.1016/j.jhep.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med 13: 261, 2015. doi: 10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep 5: 9991, 2015. doi: 10.1038/srep09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha B, Momen-Heravi F, Kodys K, Szabo G. MicroRNA Cargo of Extracellular Vesicles from Alcohol-exposed Monocytes Signals Naive Monocytes to Differentiate into M2 Macrophages. J Biol Chem 291: 149–159, 2016. doi: 10.1074/jbc.M115.694133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng AM, Chung DD, Pinson MR, Salem NA, Eaves SE, Miranda RC. Ethanol Exposure Increases miR-140 in Extracellular Vesicles: Implications for Fetal Neural Stem Cell Proliferation and Maturation. Alcohol Clin Exp Res 43: 1414–1426, 2019. doi: 10.1111/acer.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gesmundo I, Pardini B, Gargantini E, Gamba G, Birolo G, Fanciulli A, Banfi D, Congiusta N, Favaro E, Deregibus MC, Togliatto G, Zocaro G, Brizzi MF, Luque RM, Castaño JP, Bocchiotti MA, Arolfo S, Bruno S, Nano R, Morino M, Piemonti L, Ong H, Matullo G, Falcón-Pérez JM, Ghigo E, Camussi G, Granata R. Adipocyte-derived extracellular vesicles regulate survival and function of pancreatic β cells. JCI Insight 6: 141962, 2021. doi: 10.1172/jci.insight.141962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA, Wiggs MP, Nilsson MI, Crouse SF, Fluckey JD, Washington TA, Greene NP. microRNA-16 Is Downregulated During Insulin Resistance and Controls Skeletal Muscle Protein Accretion: miR-16 A ND P ROTEIN T URNOVER I N M USCLE. J Cell Biochem 117: 1775–1787, 2016. doi: 10.1002/jcb.25476. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Bi P, Shan T, Yang X, Yin H, Wang Y-X, Liu N, Rudnicki MA, Kuang S. miR-133a Regulates Adipocyte Browning In Vivo. PLoS Genet 9: e1003626, 2013. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horie T, Ono K, Nishi H, Iwanaga Y, Nagao K, Kinoshita M, Kuwabara Y, Takanabe R, Hasegawa K, Kita T, Kimura T. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochemical and Biophysical Research Communications 389: 315–320, 2009. doi: 10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- 32.Lu C, Wang D, Feng Y, Feng L, Li Z. miR-720 Regulates Insulin Secretion by Targeting Rab35. Biomed Res Int 2021: 6662612, 2021. doi: 10.1155/2021/6662612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 1779: 682–691, 2008. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovanda A, Režen T, Rogelj B. MicroRNA in skeletal muscle development, growth, atrophy, and disease. Wiley Interdiscip Rev RNA 5: 509–525, 2014. doi: 10.1002/wrna.1227. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut 61: 1600–1609, 2012. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Guo J, Li Y, Jiao K, Zhang Y. let-7a suppresses liver fibrosis via TGFβ/SMAD signaling transduction pathway. [DOI] [PMC free article] [PubMed]
- 37.Molina PE, Amedee AM, Veazey R, Dufour J, Volaufova J, Bagby GJ, Nelson S. Chronic Binge Alcohol Consumption Does Not Diminish Effectiveness of Continuous Antiretroviral Suppression of Viral Load in Simian Immunodeficiency Virus-Infected Macaques. Alcohol Clin Exp Res 38: 2335–2344, 2014. doi: 10.1111/acer.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxi J, Dean M, Zabaleta J, Reiss K, Bagby G, Nelson S, Winsauer P, Peruzzi F, Molina P. Chronic Binge Alcohol Administration Dysregulates Hippocampal Genes Involved in Immunity and Neurogenesis in Simian Immunodeficiency Virus-Infected Macaques. Biomolecules 6: 43, 2016. doi: 10.3390/biom6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poret JM, Molina PE, Simon L. Isolation, Proliferation and Differentiation of Rhesus Macaque Adipose-Derived Stem Cells. JoVE : 61732, 2021. doi: 10.3791/61732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levitt DE, Adler KA, Simon L. HEMA 3 Staining: A Simple Alternative for the Assessment of Myoblast Differentiation. Current Protocols in Stem Cell Biology 51, 2019. doi: 10.1002/cpsc.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenthal R, Cooper H, Hedges L. Parametric measures of effect size. In: The handbook of research synthesis. 1994, p. 231–244. [Google Scholar]
- 42.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 Regulates 3T3-L1 Adipogenesis. Molecular Endocrinology 23: 925–931, 2009. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava SP, Hedayat AF, Kanasaki K, Goodwin JE. microRNA Crosstalk Influences Epithelial-to-Mesenchymal, Endothelial-to-Mesenchymal, and Macrophageto-Mesenchymal Transitions in the Kidney. Front Pharmacol 10: 904, 2019. doi: 10.3389/fphar.2019.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhuri AA, So AY-L, Sinha N, Gibson WSJ, Taganov KD, O’Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol 187: 5062–5068, 2011. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Zhang L, Shu G, Wang B. microRNA-16–5p promotes 3T3-L1 adipocyte differentiation through regulating EPT1. Biochem Biophys Res Commun 514: 1251–1256, 2019. doi: 10.1016/j.bbrc.2019.04.179. [DOI] [PubMed] [Google Scholar]
- 46.Pan Q, Guo C-J, Xu Q-Y, Wang J-Z, Li H, Fang C-H. miR-16 integrates signal pathways in myofibroblasts: determinant of cell fate necessary for fibrosis resolution. Cell Death Dis 11: 639, 2020. doi: 10.1038/s41419-020-02832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karbiener M, Pisani DF, Frontini A, Oberreiter LM, Lang E, Vegiopoulos A, Mössenböck K, Bernhardt GA, Mayr T, Hildner F, Grillari J, Ailhaud G, Herzig S, Cinti S, Amri E-Z, Scheideler M. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 32: 1578–1590, 2014. doi: 10.1002/stem.1603. [DOI] [PubMed] [Google Scholar]
- 48.Runtsch MC, Nelson MC, Lee S-H, Voth W, Alexander M, Hu R, Wallace J, Petersen C, Panic V, Villanueva CJ, Evason KJ, Bauer KM, Mosbruger T, Boudina S, Bronner M, Round JL, Drummond MJ, O’Connell RM. Anti-inflammatory microRNA-146a protects mice from diet-induced metabolic disease. PLoS Genet 15: e1007970, 2019. doi: 10.1371/journal.pgen.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma E, Fu Y, Garvey WT. Relationship of Circulating miRNAs with Insulin Sensitivity and Associated Metabolic Risk Factors in Humans. Metabolic Syndrome and Related Disorders 16: 82–89, 2018. doi: 10.1089/met.2017.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol 11: 34, 2011. doi: 10.1186/1471-213X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang M-B, Xu H, Xie S-J, Zhou H, Qu L-H. Insulin-Like Growth Factor-1 Receptor Is Regulated by microRNA-133 during Skeletal Myogenesis. PLoS ONE 6: e29173, 2011. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandonà M, Consalvi S, Tucciarone L, De Bardi M, Scimeca M, Angelini DF, Buffa V, D’Amico A, Bertini ES, Cazzaniga S, Bettica P, Bouché M, Bongiovanni A, Puri PL, Saccone V. HDAC inhibitors tune miRNAs in extracellular vesicles of dystrophic muscle-resident mesenchymal cells. EMBO Rep 21: e50863, 2020. doi: 10.15252/embr.202050863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duplanty AA, Siggins RW, Allerton T, Simon L, Molina PE. Myoblast mitochondrial respiration is decreased in chronic binge alcohol administered simian immunodeficiency virus-infected antiretroviral-treated rhesus macaques. Physiol Rep 6, 2018. doi: 10.14814/phy2.13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adler K, Molina PE, Simon L. Epigenomic mechanisms of alcohol-induced impaired differentiation of skeletal muscle stem cells; role of Class IIA histone deacetylases. Physiol Genomics 51: 471–479, 2019. doi: 10.1152/physiolgenomics.00043.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guescini M, Canonico B, Lucertini F, Maggio S, Annibalini G, Barbieri E, Luchetti F, Papa S, Stocchi V. Muscle Releases Alpha-Sarcoglycan Positive Extracellular Vesicles Carrying miRNAs in the Bloodstream. PLoS ONE 10: e0125094, 2015. doi: 10.1371/journal.pone.0125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allelein S, Medina-Perez P, Lopes ALH, Rau S, Hause G, Kölsch A, Kuhlmeier D. Potential and challenges of specifically isolating extracellular vesicles from heterogeneous populations. Sci Rep 11: 11585, 2021. doi: 10.1038/s41598-021-91129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon L, Torres D, Saravia A, Levitt DE, Vande Stouwe C, McGarrah H, Coleman L, Dufour JP, Amedee AM, Molina PE. Chronic binge alcohol and ovariectomy-mediated impaired insulin responsiveness in SIV-infected female rhesus macaques. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology 321: R699–R711, 2021. doi: 10.1152/ajpregu.00159.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]


