Summary
Microvilli are actin bundle-supported surface protrusions assembled by diverse cell types to mediate biochemical and physical interactions with the external environment. Found on the surface of some of the earliest animal cells, primordial microvilli likely contributed to bacterial entrapment and feeding. Although millions of years of evolution have repurposed these protrusions to fulfill diverse roles such as detection of mechanical or visual stimuli in inner ear hair cells or retinal pigmented epithelial cells, respectively, solute uptake remains a key essential function linked to these structures. In this mini review, we offer a brief overview of the composition and structure of epithelial microvilli, highlight recent discoveries on the growth of these protrusions early in differentiation, and point to fundamental questions surrounding microvilli biogenesis that remain open for future studies.
Keywords: actin, filament, cytoskeleton, membrane, protrusion, apical, epithelial
What’s a microvillus?
Epithelial monolayers lining the lumens of hollow organs consist of polarized cells that build large numbers of microvilli on their apical surface, which maximize the potential for interacting with lumenal contents [1]. In the specific case of nutrient absorbing enterocytes, the dominant cell type in the intestinal monolayer, a single cell builds several thousand microvilli on its apical surface during differentiation. The result is a unique array - referred to as the ‘brush border’ - defined by its striking morphological order and uniformity [2–4]. Given that the vertebrate gut epithelium is an abundant source of brush border material, enterocyte microvilli have been the focus of ultrastructural and biochemical investigations for many years. Indeed, our understanding of the cytoskeletal composition and structure of microvilli is in large part based on decades of investigation in this system, which began with Mooseker and Tilney’s discovery of actin as a highly abundant protein in isolated brush border fractions [5].
Moving forward ~50 years to present day, we now know that microvilli exhibit a simple architecture consisting of a supporting core bundle of 20–40 actin filaments encapsulated by plasma membrane [6, 7]. Filaments in this bundle are packed together by a complement of bundling proteins including VIL1 [8], ESPN [9], PLS1 [10], and MISP [11]. The core bundle is laterally tethered to the enveloping plasma membrane by membrane-actin linking proteins, including ERM family protein EZRIN [12], and class 1 myosins such as MYO1A [13, 14] and MYO1D [15]. The resulting structure is a cylindrical membrane protrusion ~1–2 um in length and ~100 nm in diameter. Importantly, individual actin filaments in a core bundle are polarized, with all barbed ends - the favored site of actin monomer incorporation – at the distal tip [6, 16, 17]. Conversely, all pointed ends extend down into the sub-apical cytoplasm, where they are embedded in a cytoskeletal meshwork referred to as the ‘terminal web’ [18–21]. Most but not all these filaments appear to run continuously from the distal tip down to the proximal end [7]. The two ends of the core bundle are also defined by the enrichment of specific factors; all barbed ends at the distal tip are strongly decorated with EPS8 and BAIAP2L1 [22, 23], which both contain structural motifs with the potential to link plasma membrane to the actin cytoskeleton. The basal ends of core bundles are associated with distinct factors including COBL (a putative actin nucleator)[24–26], TMOD3 (a pointed end capper)[27, 28], and MYH4 (a force generator and potential actin disassembly factor) [29]. Finally, in a mature brush border, microvillar packing is driven and organized by tip-tip adhesion links that consist of a heterophilic complex of two protocadherins, CDHR2 and CDHR5 [30]. These adhesive factors are positioned at microvillar tips by the barbed end directed motor MYO7B [31, 32] via interactions that are mediated by the two scaffolding proteins, ANKS4B and USH1C [30, 33–37]. A similar complex links the distal tips of stereocilia on the surface of mechanosensory hair cells [35–38] and inactivating mutations in those factors or their binding partners result in sensory disorders, including Usher syndrome [39].
When and where do cells build microvilli?
As microvilli are defining features of the functionally mature epithelial cells, they are assembled in large numbers during differentiation. In the context of the intestinal epithelium, this process takes place on the surface of nascent enterocytes as they migrate out of stem cell-containing crypts on to the villus surface. Although the precise timing of this transition remains unclear, in mice the entire ~500 μm voyage from crypt to villus tip takes ~5 days [40]. Differentiation takes place over the course of only a few cell diameters (< 25 μm) along this axis as they pass through the crypt-villus transition. These spatial and temporal points of reference allow us to estimate that the 2000–3000 microvilli that ultimately comprise a mature brush border are assembled over a time scale of hours. Thus, timely control of the factors that regulate the growth of microvilli is essential for completing differentiation.
How are microvilli assembled?
Given that microvilli are actin bundle-supported structures, understanding how constituent actin filaments are polymerized and bundled to create these core cytoskeletal units are key goals. To build a core bundle, actin polymerization must first be initiated at the proper sites, immediately subjacent to the apical plasma membrane. The resulting nascent filaments must also be elongated to the required length, aligned, and bundled in a polarized manner such that all their growing barbed ends are situated directly against the membrane. In this arrangement, they can work together to generate the mechanical force needed to produce the outward membrane curvature that is characteristic of microvilli [41, 42]. Importantly, approximately 30 filaments must be polymerized, such that the collective force generated at the distal end of a bundle will exceed the threshold bending stiffness of the plasma membrane without buckling [43–45]. Moreover, recent live cell imaging studies demonstrate that not all core bundles are assembled in a de novo fashion, as a subset are templated from the bundles in pre-existing microvilli [46]. How microvillar actin polymerization is initiated or templated, and how filaments are elongated, oriented and bundled in parallel are all fundamental open questions, but recent studies are offering exciting mechanistic hints, which are discussed in more detail below.
(1). How are core bundle actin filaments generated?
As an individual immature epithelial cell migrates through the cryptvillus transition, a sparse lawn of nascent microvilli transforms into a densely packed array consisting of thousands of microvilli [47]. Because an individual microvillus contains thousands of actin monomers, building an entire brush border demands 10s of millions of subunits. Interestingly, high levels of G-actin accumulate in the sub-apical region of both immature and differentiated enterocytes [48], and incorporation of these subunits into filaments might be regulated by the monomer binding protein, profilin [49, 50], which is also enriched sub-apically [48]. Recent high resolution live-cell imaging also revealed that ‘clouds’ of G-actin accumulate at the plasma membrane in the minutes preceding protrusion growth [46]. Actin monomer availability, which can impact the growth of specific actin networks [49–51] is limiting during the assembly of microvilli, as increasing the supply of G-actin (e.g. by inhibiting the ubiquitous nucleation activity of ARP2/3) increases the actin content and length of microvilli [48].
Beyond the availability of cytoskeletal building blocks, assembling a microvillus also requires molecular machinery for controlling actin monomer-monomer interactions and forming polymerization competent ‘nuclei’ in space and time. Proteomic studies of brush border enriched fractions have identified some interesting candidates [52, 53]. One such factor is Cordon bleu (COBL), which harbors three actin monomer-binding Wiskott-Aldrich Homology 2 (WH2) domains arranged in tandem near its C-terminus. COBL might promote filament nucleation by stabilizing three monomers configured in a short pitch helix [54, 55]. Although such nucleation activity is predicted to be weak [56], studies have indicated that it may be enhanced by Ca2+ and calmodulin [57], and potentially by PRMT2-driven methylation at the arginine residue within its second WH2 domain, which promotes strong G-actin binding in the context of neuronal branching [58]. In vitro studies of COBL fragments have also revealed other diverse activities including filament severing and monomer sequestration [54, 59]. Importantly, loss- and gain-of-function studies in epithelial cell culture models implicate COBL in the control of microvillar growth and length control, although the details appear model dependent [24, 60]. Mice lacking COBL exhibit structural perturbations in the brush border terminal web, consistent with a role in organizing the cytoskeleton in this compartment [26]. Intestinal tissue in these animals appear grossly normal, but this is expected given the high capacity for functional compensation noted in previous studies of mice lacking key brush border structural components [13, 53, 61]. COBL also contains an N-terminal polyproline rich domain that associates with the SH3 domain of PACSIN2, an F-BAR protein that drives enrichment of this factor on the apical membrane [24, 60, 62]. To date, aside from COBL, no other factors with actin nucleation potential have been identified in microvilli. Moreover, a contribution from the ubiquitous ARP2/3 nucleation complex, which generates branched actin filament networks, has also been ruled out [24, 48]. Thus, despite persistent confusion in the literature, COBL must remain a candidate for generating the actin filaments that comprise the microvillar core bundle.
(2). How are core bundle actin filaments elongated?
Once actin monomers are brought together to form a stable nucleus, subsequent elongation of its barbed end to produce a filament of functional length is likely driven by one or more factors that prevent barbed end capping. Formins hold filament elongation potential [63] and were identified as brush border components in two proteomic studies [52, 53]. Although SMIFH2 inhibition of formins failed to impair microvillar growth in cell culture models [24], those results are difficult to interpret in light of this compound’s off target effects on myosin motors [64], which have also implicated in microvillar length control [29]. Other leading candidate elongation factors are EPS8 and its binding partner, BAIAP2L1, which target to the distal ends of microvilli where filament barbed ends reside. EPS8 contains N-terminal motifs that may support membrane association, a central SH3 domain capable of dimerization, and C-terminal actin binding motifs, and was originally suggested to function in filament capping and bundling [65]. Through polyproline motifs, EPS8 interacts with the SH3 domain found in BAIAP2L1. BAIAP2L1 in turn contains an I-BAR domain that drives localization to regions of outward membrane curvature (like that found at microvillar tips) and may promote dimerization; a poorly conserved C-terminal WH2 domain also provides additional actin binding potential. Indeed, overexpression of BAIAP2L1 is sufficient to rescue the microvillus length defect in cells lacking EPS8, while a mutant of BAIAP2L1 lacking the WH2 domain does not [22]. Based on experiments in intestinal epithelial cell culture models, EPS8 and BAIAP2L1 work together to promote core bundle elongation [22]. Although EPS8 and BAIAP2L1 colocalize, EPS8 in particular demonstrates exquisitely specific enrichment in puncta at the distal ends of microvilli [22], and related structures including the filopodia found at the edge of motile cells and stereocilia that extend from the apex of mechanosensory cells [23, 66]. As one might expect based on its unique localization, EPS8 has been implicated in protrusion length control in these different contexts [22, 23, 66, 67].
Recent live imaging studies revealed that EPS8 puncta target to the apical surface and mark future sites of microvillar growth minutes before core bundle assembly begins, further suggesting that this factor may be involved in initiation as well as elongation [46]. Moreover, after a core bundle forms, the initiating EPS8 punctum persists at its distal end, and loss of this punctum is followed immediately by core bundle disassembly [46]. Notably, the core bundles that support nascent microvilli exhibit robust treadmilling [68, 69], an activity that drives the movement of these protrusion across the apical surface [70]. Such dynamics are also regulated by EPS8 and BAIAP2L1 and are hypothesized to play a key role in organizing microvillar packing as protrusion density increases [70]. Whether the EPS8/BAIAP2L1 complex exerts definitive anti-capping activity, which one might expect for factors that promote elongation, remains to be determined.
In addition to the de novo polymerization of core bundle actin filaments, which would necessarily require an actin nucleator, newer live imaging data also points to a second distinct pathway for growing core bundles, which takes advantage of filaments in pre-existing core bundles, rather than growing new polymers de novo. Indeed, a subset of nascent microvilli emerge from a process whereby a ‘mother’ microvillus gives rise to a ‘daughter’, which typically grow from the base of the pre-existing protrusion [46]. Notably, classic ultrastructural studies attempting to capture microvillus re-growth events in tissue explants perturbed with hydrostatic pressure, were the first to document evidence of ‘forked’ microvilli that were consistent with a mother/daughter growth mechanism [71]. Evidence of daughter growth was also recently captured in vivo in the C. elegans intestine [72], suggesting this mode of microvilli growth may be widespread. How might a daughter microvillus grow from the base of a mother? Although EPS8 and BAIAP2L1 are tip specific factors, they occasionally travel in a retrograde fashion to the base of a treadmilling mother microvillus and eventually appear to template daughter growth laterally from the pre-existing core bundle [46, 70]. One possibility is that retrograde movement of these factors represents a subset of filaments that fall behind in terms of elongation relative to other filaments in the treadmilling core, and then - with the help of EPS8 and BAIAP2L1 - restart elongation near the base of mother microvillus where monomer concentrations are expected to be higher.
(3). How are actin filaments bundled with uniform polarity?
Classic ultrastructural studies used myosin subfragment-1 (S1) labeling to establish that all actin filaments in the core bundle extend their barbed ends toward the distal tip [18]. Such ‘barbed end out’ orientation is a common feature of actin-supported surface protrusions, which makes physical sense considering that barbed end elongation potentially generates pushing force for creating outward membrane curvature. How this orientation is created during microvillar assembly remains unclear, although one possibility is that nascent filaments are oriented as they are brought together during filament bundling, which is driven by multiple enterocyte-expressed factors including VIL1, ESPN and PLS1 [8–10]. Intriguingly, triple KO mice lacking all three of these molecules (VEP KO) still assembled microvilli suggesting the existence of other bundling proteins [53]. Although EPS8 was initially invoked as a bundling protein that might compensate, its specific localization to the distal tips of protrusions argues against a conventional bundling activity throughout the length of the core bundle [46]. More recently, Mitotic Spindle Positioning (MISP) was identified as a new unconventional bundling protein that selectively bundles the rootlet ends of microvilli [11, 73]. Whether MISP or other yet-to-be-identified bundlers compensate bundling activity in VEP KO mice remains an open question.
Are any of these bundling factors sufficient for driving uniform filament polarity? Intriguingly, PLS1-actin bundles assembled in vitro exhibit polarized S1 labeling, suggesting that this factor may be capable driving uniform polarity in cells [74]. In enterocyte brush borders, PLS1 appears to accumulate closer to the pointed ends of core bundle filaments [75]. MISP also exhibits highly preferential localization near the basal/pointed ends of microvillar actin filaments [11], and can recruit PLS1 to bundled actin in cells, suggesting their preferential binding to one end could provide a mechanism for orienting filament polarity. Binding near filaments ends might not be essential, however, as ESPN is generally found along the length of the actin core and its overexpression in cultured cells appears to be sufficient for assembling exaggerated bundles consisting of filaments of uniform polarity [76]. The filopodial bundler, FASCIN, also localizes uniformly along the length of bundles it creates and is sufficient for driving uniform filament polarity in these structures [77, 78].
Whereas actin bundling proteins generally exhibit filament side binding potential, another possible non-mutually exclusive mechanism for orienting actin filaments involves barbed end binding factors such as EPS8, which as alluded to above, marks sites of microvillar growth and remains persistently attached to the core bundle distal end through its lifetime [46]. Given that EPS8 also associates with the apical plasma membrane - either directly or through binding partner, BAIAP2L1 - it is well positioned to orient filament barbed ends against the membrane during core bundle assembly. The proposal that specific membrane-associated protein machinery organize core bundle growth dates back to classic ultrastructural studies, which collectively revealed that the distal ends of core bundles in newly forming and fully formed microvilli make contact with the plasma membrane at electron dense patches [6, 71]. Such sites were hypothesized to be enriched in factors that control actin assembly. Although EPS8 is a reasonable candidate constituent of these electron dense regions, this remains to be confirmed experimentally at the ultrastructural level.
Open questions
Recent discoveries are now starting to illuminate the detailed molecular mechanisms that enable epithelial cells to generate apical specializations such as the brush border. Despite these advances, however, key fundamental questions surrounding this process remain unanswered and should be targeted in future investigations. For example, how do epithelial cells define the dimensions of a microvillus core bundle? The number of actin filaments in a core bundle dictates protrusion width and is highly stereotyped in brush border microvilli, yet it remains unclear how this parameter is controlled during differentiation. Filament numbers will be impacted by actin monomer concentration, actin bundler stoichiometries, and perhaps even by barbed end binding proteins (e.g. EPS8 or BAIAP2L1) that are well positioned to control how many filaments impinge on the apical membrane. Some tip specific factors including EPS8 assemble condensates in vitro [38, 79], which might also play a role in controlling filaments numbers. This question is also pertinent to understanding the formation of functionally diverse apical protrusions including the stereocilia found on sensory hair cells and the giant microvilli assembled by intestinal tuft cells; in both of these cases, the core bundles contain ~10-fold more actin filaments than we find in microvilli. Another intriguing open question relates to the origin of mechanical force that enables cells to overcome the bending stiffness of the apical membrane during microvillus formation. Although the canonical view is that polymerizing actin filaments are the primary force generators in this system, microvilli and other structurally similar protrusions contain high levels of force-generating myosin motors. Yet their contribution to growth promoting force remains unexplored. Related to this point, it would be also worth investigating the contribution of external mechanical force in protrusion growth, as fluid shear has been implicated in the growth of placental microvilli [80]. Finally, as most recent investigations have focused on identifying the protein machinery that is directly involved in core bundle assembly, our understanding of how sites of microvillar growth are specified on the apical membrane during differentiation remains limited. Phosphoinositides such as PI(4,5)P2, PI(4,5)P2, and PI(3,4,5)P3 are strongly implicated in specifying apical vs. basolateral membrane domains [81–83]. Whether specific membrane lipid species serve as spatial cues to organize actin polymerization machinery during core bundle growth remains unclear, but this area would also make an appealing target for future studies.
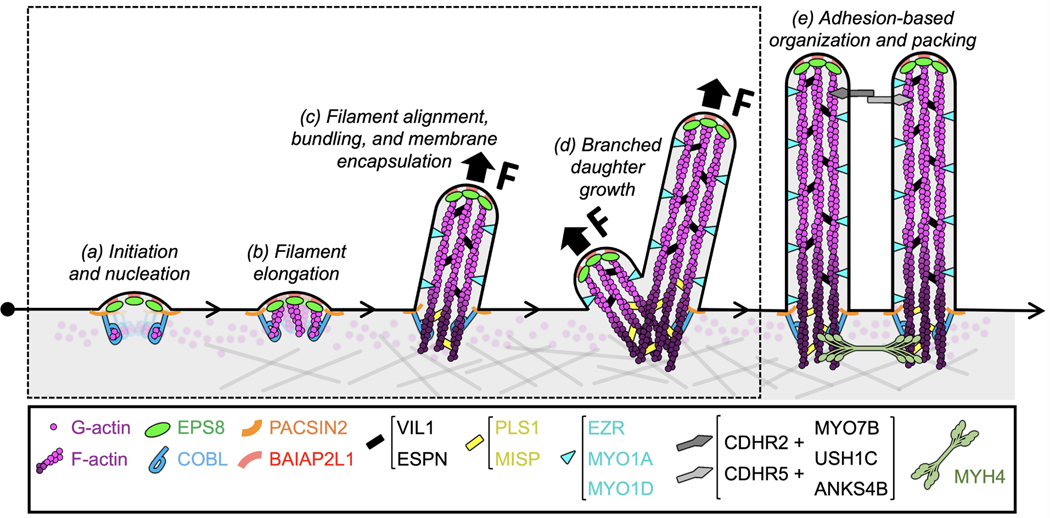
Figure 1 -. Model of microvilli biogenesis.
(a) Sites of microvillus growth are marked by discrete puncta containing EPS8 and BAIAPP2L1. As the only actin nucleator identified in microvilli thus far, COBL might play a role in generating actin filaments de novo. (b) A complex of EPS8/BAIAP2L1 drives the elongation of core bundle filaments. (c) Filament alignment could be promoted by EPS8 together with bundling activities of MISP, PLS1, VIL1, and ESPN. Membrane encapsulation of the core actin bundle is orchestrated by the membrane-cytoskeleton linking factors EZRIN, MYO1A, and MYO1D, and this coincides with core bundle elongation. (d) In addition to de novo growth, a nascent microvillus can also give rise to a daughter microvillus laterally in a process that takes advantage of recycled EPS8 and BAIAP2L1. (e) Microvilli packing is driven by the intermicrovillar adhesion complex, which consists of protocadherins CDHR2 and CDHR5, the actin-based motor, MYO7B and scaffold proteins, USH1C and ANKS4B. Steps highlighted by the dashed box denote events that are focus of discussion in this mini-review.
ACKNOWLEDGEMENTS
The authors would like to thank all members of the Tyska laboratory for discussion and feedback. This work was supported in part by NIH grants F31-DK122692 (IMG), R01-DK111949, R01-DK095811, and R01-DK125546 (MJT).
Abbreviations used herein and alternate designations found in the literature
- ANKS4B
Ankyrin repeat and SAM domain-containing protein 4B, a.k.a. harmonin-interacting ankyrin repeat-containing protein, HARP
- BAIAP2L1
Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, a.k.a. Insulin receptor tyrosine kinase substrate, IRTKS
- CDHR2
Cadherin-related family member 2, a.k.a. protocadherin-24, protocadherin LKC
- CDHR5
Cadherin-related family member 5, a.k.a. m-protocadherin, MUPCDH, MLPCDH
- COBL
Cordon bleu, a.k.a. KIAA0633
- DIAPH1
Diaphanous homolog 1, a.k.a. diaphanous-related formin-1, DRF1
- ESPN
Ectoplasmic specialization protein, a.k.a. espin
- EPS8
Epidermal growth factor receptor kinase substrate 8
- EZR
Ezrin, a.k.a p81, villin-2
- MISP
Mitotic interactor and substrate of PLK1, a.k.a. mitotic spindle positioning, C19ORF21
- MYO1A
Myosin-1a
- MYO1D
Myosin-1d
- MYO7B
Myosin-7B, unconventional myosin-VIIb
- MYH4
Non-muscle myosin-2c, a.k.a. non-muscle myosin heavy chain IIc, NM2C
- PACSIN2
Protein kinase C and casein kinase substrate in neurons protein 2, a.k.a. syndapin-2, syndapin-II
- PLS1
Plastin-1, a.k.a. I (intestine specific)-plastin, fimbrin
- TMOD
Tropomodulin-3
- USH1C
Usher syndrome type-1C, a.k.a. harmonin, antigen NY-CO-38/37, PDZ-73, renal carcinoma antigen NY-REN-3
- VIL1
Villin-1
- PRMT2
Protein arginine N-methyltransferase 2, a.k.a. Histone-arginine N-methyltransferase, HMT1
- ARP2/3
Actin-related protein 2/3 complex
Footnotes
DECLARATION OF INTEREST
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Helander HF and Fandriks L, Surface area of the digestive tract - revisited. Scand J Gastroenterol, 2014. 49(6): p. 681–9. [DOI] [PubMed] [Google Scholar]
- 2.Delacour D, et al. , Plasticity of the brush border - the yin and yang of intestinal homeostasis. Nat Rev Gastroenterol Hepatol, 2016. 13(3): p. 161–74. [DOI] [PubMed] [Google Scholar]
- 3.Crawley SW, Mooseker MS, and Tyska MJ, Shaping the intestinal brush border. J Cell Biol, 2014. 207(4): p. 441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauvanet C, et al. , Structure, regulation, and functional diversity of microvilli on the apical domain of epithelial cells. Annu Rev Cell Dev Biol, 2015. 31: p. 593–621. [DOI] [PubMed] [Google Scholar]
- 5.Tilney LG and Mooseker M, Actin in the brush-border of epithelial cells of the chicken intestine. Proc Natl Acad Sci U S A, 1971. 68(10): p. 2611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mooseker MS and Tilney LG, Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J Cell Biol, 1975. 67(3): p. 725–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohta K, et al. , Helical arrangement of filaments in microvillar actin bundles. J Struct Biol, 2012. 177(2): p. 513–9. [DOI] [PubMed] [Google Scholar]
- 8.Bretscher A. and Weber K, Villin: the major microfilament-associated protein of the intestinal microvillus. Proc Natl Acad Sci U S A, 1979. 76(5): p. 2321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartles JR, et al. , Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol, 1998. 143(1): p. 107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretscher A. and Weber K, Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol, 1980. 86(1): p. 335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morales EA, et al. , Mitotic Spindle Positioning (MISP) is an actin bundler that selectively stabilizes the rootlets of epithelial microvilli. Cell Rep, 2022. 39(3): p. 110692. * This study reports MISP as a fourth actin bundler in brush border microvilli, which protects the core bundle rootlets from ezrin-driven membrane wrapping.
- 12.Gould KL, et al. , The protein-tyrosine kinase substrate, p81, is homologous to a chicken microvillar core protein. J Cell Biol, 1986. 102(2): p. 660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyska MJ, et al. , Myosin-1a is critical for normal brush border structure and composition. Mol Biol Cell, 2005. 16(5): p. 2443–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conzelman KA. and Mooseker MS, The 110-kD protein-calmodulin complex of the intestinal microvillus is an actin-activated MgATPase. J Cell Biol, 1987. 105(1): p. 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benesh AE, et al. , Differential localization and dynamics of class I myosins in the enterocyte microvillus. Mol Biol Cell, 2010. 21(6): p. 970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mooseker MS, Pollard TD, and Wharton KA, Nucleated polymerization of actin from the membrane-associated ends of microvillar filaments in the intestinal brush border. J Cell Biol, 1982. 95(1): p. 223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard TD and Mooseker MS, Direct measurement of actin polymerization rate constants by electron microscopy of actin filaments nucleated by isolated microvillus cores. J Cell Biol, 1981. 88(3): p. 654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirokawa N. and Heuser JE, Quick-freeze, deep-etch visualization of the cytoskeleton beneath surface differentiations of intestinal epithelial cells. J Cell Biol, 1981. 91(2 Pt 1): p. 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirokawa N, et al. , Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J Cell Biol, 1982. 94(2): p. 425–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palay SL and Karlin LJ, An electron microscopic study of the intestinal villus. II. The pathway of fat absorption. J Biophys Biochem Cytol, 1959. 5(3): p. 373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palay SL and Karlin LJ, An electron microscopic study of the intestinal villus. I. The fasting animal. J Biophys Biochem Cytol, 1959. 5(3): p. 363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postema MM, et al. , IRTKS (BAIAP2L1) Elongates Epithelial Microvilli Using EPS8-Dependent and Independent Mechanisms. Curr Biol, 2018. 28(18): p. 2876–2888 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croce A, et al. , A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol, 2004. 6(12): p. 1173–9. [DOI] [PubMed] [Google Scholar]
- 24.Grega-Larson NE, et al. , Cordon bleu promotes the assembly of brush border microvilli. Mol Biol Cell, 2015. 26(21): p. 3803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahuja R, et al. , Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell, 2007. 131(2): p. 337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beer AJ, et al. , The actin nucleator Cobl organises the terminal web of enterocytes. Sci Rep, 2020. 10(1): p. 11156. * This study provides ultrastructural evidence that COBL KO mice exhibit a disorganized terminal web and perturbations in microvillar length.
- 27.Weber KL, Fischer RS, and Fowler VM, Tmod3 regulates polarized epithelial cell morphology. J Cell Sci, 2007. 120(Pt 20): p. 3625–32. [DOI] [PubMed] [Google Scholar]
- 28.Weber A, et al. , Tropomodulin caps the pointed ends of actin filaments. J Cell Biol, 1994. 127(6 Pt 1): p. 1627–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chinowsky CR, et al. , Nonmuscle myosin-2 contractility-dependent actin turnover limits the length of epithelial microvilli. Mol Biol Cell, 2020. 31(25): p. 2803–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawley SW, et al. , Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell, 2014. 157(2): p. 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weck ML, et al. , Myosin-7b Promotes Distal Tip Localization of the Intermicrovillar Adhesion Complex. Curr Biol, 2016. 26(20): p. 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen ZY, et al. , Myosin-VIIb, a novel unconventional myosin, is a constituent of microvilli in transporting epithelia. Genomics, 2001. 72(3): p. 285–96. [DOI] [PubMed] [Google Scholar]
- 33.Crawley SW, et al. , ANKS4B Is Essential for Intermicrovillar Adhesion Complex Formation. Dev Cell, 2016. 36(2): p. 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weck ML., Crawley SW, and Tyska MJ, A heterologous in-cell assay for investigating intermicrovillar adhesion complex interactions reveals a novel protrusion length-matching mechanism. J Biol Chem, 2020. 295(48): p. 16191–16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu IM, et al. , Myosin 7 and its adaptors link cadherins to actin. Nat Commun, 2017. 8: p. 15864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, et al. , Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex. Dev Cell, 2016. 36(2): p. 179–89. [DOI] [PubMed] [Google Scholar]
- 37.Li J, et al. , Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins. Proc Natl Acad Sci U S A, 2017. 114(19): p. E3776–E3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He Y, Li J, and Zhang M, Myosin VII, USH1C, and ANKS4B or USH1G Together Form Condensed Molecular Assembly via Liquid-Liquid Phase Separation. Cell Rep, 2019. 29(4): p. 974–986 e4. ** This study shows that cytosolic factors comprising the intermicrovillar adhesion complex and their homologues in the stereocilia (lower) tip complex assemble condensates via multivalent interactions in vitro and in cells.
- 39.Geleoc GGS and El-Amraoui A, Disease mechanisms and gene therapy for Usher syndrome. Hear Res, 2020. 394: p. 107932. [DOI] [PubMed] [Google Scholar]
- 40.van der Flier LG and Clevers H, Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol, 2009. 71: p. 241–60. [DOI] [PubMed] [Google Scholar]
- 41.Footer MJ, et al. , Direct measurement of force generation by actin filament polymerization using an optical trap. Proc Natl Acad Sci U S A, 2007. 104(7): p. 2181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theriot JA, The polymerization motor. Traffic, 2000. 1(1): p. 19–28. [DOI] [PubMed] [Google Scholar]
- 43.Mogilner A. and Rubinstein B, The physics of filopodial protrusion. Biophys J, 2005. 89(2): p. 782–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orly G, Naoz M, and Gov NS, Physical model for the geometry of actin-based cellular protrusions. Biophys J, 2014. 107(3): p. 576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atilgan E, Wirtz D, and Sun SX, Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys J, 2006. 90(1): p. 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gaeta IM, et al. , Direct visualization of epithelial microvilli biogenesis. Curr Biol, 2021. 31(12): p. 2561–2575 e6. ** This study shows that EPS8 and BAIAP2L1 enrich at the distal tips of growing microvilli, where they are needed for protrusion survival. This work also revealed that in addition to de novo growth, microvilli can also grow laterally from the sides of pre-existing mother protrusions. This novel branching mechanism of protrusion growth also appears to involve EPS8 and BAIAP2L1.
- 47.Pothier P. and Hugon JS, Characterization of isolated villus and crypt cells from the small intestine of the adult mouse. Cell Tissue Res, 1980. 211(3): p. 405–18. [DOI] [PubMed] [Google Scholar]
- 48.Faust JJ, Millis BA, and Tyska MJ, Profilin-Mediated Actin Allocation Regulates the Growth of Epithelial Microvilli. Curr Biol, 2019. 29(20): p. 3457–3465 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suarez C, et al. , Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell, 2015. 32(1): p. 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotty JD., et al., Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell, 2015. 32(1): p. 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burke TA, et al. , Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol, 2014. 24(5): p. 579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McConnell RE, et al. , Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol, 2011. 300(5): p. G914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Revenu C, et al. , A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol Biol Cell, 2012. 23(2): p. 324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Husson C, et al. , Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell, 2011. 43(3): p. 464–77. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, et al. , Structural basis of actin filament nucleation by tandem W domains. Cell Rep, 2013. 3(6): p. 1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dominguez R, The WH2 Domain and Actin Nucleation: Necessary but Insufficient. Trends Biochem Sci, 2016. 41(6): p. 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou W, et al. , The Actin Nucleator Cobl Is Controlled by Calcium and Calmodulin. PLoS Biol, 2015. 13(9): p. e1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou W, et al. , Arginine Methylation by PRMT2 Controls the Functions of the Actin Nucleator Cobl. Dev Cell, 2018. 45(2): p. 262–275 e8. [DOI] [PubMed] [Google Scholar]
- 59.Jiao Y, et al. , Mutagenetic and electron microscopy analysis of actin filament severing by Cordon-Bleu, a WH2 domain protein. Cytoskeleton (Hoboken), 2014. 71(3): p. 170–83. [DOI] [PubMed] [Google Scholar]
- 60.Wayt J. and Bretscher A, Cordon Bleu serves as a platform at the basal region of microvilli, where it regulates microvillar length through its WH2 domains. Mol Biol Cell, 2014. 25(18): p. 2817–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferrary E, et al. , In vivo, villin is required for Ca(2+)-dependent F-actin disruption in intestinal brush borders. J Cell Biol, 1999. 146(4): p. 819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwintzer L, et al. , The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J, 2011. 30(15): p. 3147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kovar DR and Pollard TD, Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A, 2004. 101(41): p. 14725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishimura Y, et al. , The formin inhibitor SMIFH2 inhibits members of the myosin superfamily. J Cell Sci, 2021. 134(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hertzog M, et al. , Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PLoS Biol, 2010. 8(6): p. e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manor U, et al. , Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr Biol, 2011. 21(2): p. 167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Disanza A, et al. , Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol, 2006. 8(12): p. 1337–47. [DOI] [PubMed] [Google Scholar]
- 68.Loomis PA, et al. , Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J Cell Biol, 2003. 163(5): p. 1045–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tyska MJ and Mooseker MS, MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys J, 2002. 82(4): p. 1869–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meenderink LM, et al. , Actin Dynamics Drive Microvillar Motility and Clustering during Brush Border Assembly. Dev Cell, 2019. 50(5): p. 545–556 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tilney LG. and Cardell RR, Factors controlling the reassembly of the microvillous border of the small intestine of the salamander. J Cell Biol, 1970. 47(2): p. 408–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu H, et al. , In situ structure of intestinal apical surface reveals nanobristles on microvilli. Proc Natl Acad Sci U S A, 2022. 119(24): p. e2122249119. * This study features ultrastructural images of radial nanobristles surrounding single intestinal microvillus in C. elegans, which promote the formation of two identical daughter microvilli from a mother microvillus.
- 73.Kumeta M, et al. , Caprice/MISP is a novel F-actin bundling protein critical for actin-based cytoskeletal reorganizations. Genes Cells, 2014. 19(4): p. 338–49. [DOI] [PubMed] [Google Scholar]
- 74.Glenney JR Jr., et al. , F-actin binding and bundling properties of fimbrin, a major cytoskeletal protein of microvillus core filaments. J Biol Chem, 1981. 256(17): p. 9283–8. [PubMed] [Google Scholar]
- 75.Grimm-Gunter EM, et al. , Plastin 1 binds to keratin and is required for terminal web assembly in the intestinal epithelium. Mol Biol Cell, 2009. 20(10): p. 2549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loomis PA, et al. , Targeted wild-type and jerker espins reveal a novel, WH2-domain-dependent way to make actin bundles in cells. J Cell Sci, 2006. 119(Pt 8): p. 1655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jansen S, et al. , Mechanism of actin filament bundling by fascin. J Biol Chem, 2011. 286(34): p. 30087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vignjevic D, et al. , Role of fascin in filopodial protrusion. J Cell Biol, 2006. 174(6): p. 863–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin L, et al. , Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex. Cell Rep, 2021. 34(8): p. 108770. * This study shows that factors comprising the stereocilia (upper) tip complex, including EPS8, assemble condensates via multivalent interactions, and promote actin bundling in vitro.
- 80.Miura S, et al. , Fluid shear triggers microvilli formation via mechanosensitive activation of TRPV6. Nat Commun, 2015. 6: p. 8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roman-Fernandez A, et al. , The phospholipid PI(3,4)P2 is an apical identity determinant. Nat Commun, 2018. 9(1): p. 5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gassama-Diagne A, et al. , Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat Cell Biol, 2006. 8(9): p. 963–70. [DOI] [PubMed] [Google Scholar]
- 83.Martin-Belmonte F, et al. , PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell, 2007. 128(2): p. 383–97. [DOI] [PMC free article] [PubMed] [Google Scholar]