Abstract
The transcription factor c-Maf has been suggested to regulate the activity of γ-crystallin promoters in lens fibre cells. We here show that the transactivation potential of c-Maf and MafB for the rat γD-crystallin Maf-responsive element (γD MARE) is dependent upon the cellular context and, using chimeric and single domain mutants, that c-Maf is most likely to be the cognate factor for the γD MARE in the lens. Transactivation of the γD MARE by c-Maf in lens cells was not enhanced by c-Fos or c-Jun and was not blocked by dominant negative c-Fos or c-Jun constructs. c-Maf can activate the γD MARE as a homodimer since activation of the γD-crystallin promoter in P19 embryonic carcinoma cells required only c-Maf, but none of a number of c-Fos and c-Jun family members tested. Transactivation by c-Maf was inhibited by activation of protein kinase A (PKA) (by signal transduction agonist forskolin) or of protein kinase C (PKC) (by signal transduction agonist tetradecanoyl phorbol acetate). Site-directed mutagenesis showed that this effect is not mediated by phosphorylation of the consensus PKA/PKC site in the extended DNA-binding domain, but likely involves activation of MAP kinase kinase, as inhibition by PD98059 increased transactivation by c-Maf.
INTRODUCTION
The Maf transcription factors are a subclass of the family of bZIP factors. Maf proteins can be either small or large, the small ones lacking the activation domain present in the large proteins. Maf proteins act as homo- or heterodimers with a variety of factors, including members of the AP1 family (for reviews see 1,2), and bind to an extended tetradecanoyl phorbol acetate (TPA) response element (T-MARE) or extended cAMP response element (CRE) (C-MARE) (3,4). Maf proteins have been implicated in the control of development and differentiation, most recently in that of the lens. In the chicken and quail, a special large Maf (L-Maf or MafA) is found in lens and neural retina (5,6). Ectopic expression of L-Maf induces lentoid body formation (6). No mammalian homologue of L-Maf has as yet been isolated. Of the three Maf proteins detected in mammalian lenses, Nrl, c-Maf and MafB (7), the role of c-Maf has been most firmly established (8–10). The recent showing that c-Maf is up-regulated by Pax-6 ties c-Maf to the regulatory network specifying ocular development and differentiation (11). c-Maf knockout mice fail to develop fully formed lenses (8–10) and do not express most of the crystallin genes, i.e. the genes encoding the abundant water-soluble lens proteins. In agreement with this finding, putative MAREs have been detected in the crystallin promoters. For example, the promoters of the six rodent γ-crystallin genes, the γA–γF-crystallin genes, all contain a MARE ∼20 bp upstream from the TATA box (6,8,12). Hence, it has been suggested that c-Maf directly transactivates the γ-crystallin promoters. However, as the γ-crystallin genes are only active during terminal fibre cell differentiation (13,14), it cannot be excluded that the lack of γ-crystallin gene expression in c-Maf knockout mice is due to incomplete fibre cell differentiation rather than to a direct effect on the activity of the γ-crystallin gene promoter. We show here that the ability of MafB or c-Maf to transactivate the rat γD-crystallin promoter is dependent upon the cellular context and that in the lens fibre cell c-Maf indeed regulates the rat γD-crystallin promoter.
As the rat γD-crystallin promoter is only fully active in lens fibre cells cultured with FGF-2 or insulin (15), we also investigated whether c-Maf was involved in transmission of the growth factor signal to the γD-crystallin promoter. We here show that, unexpectedly, c-Maf activity is negatively regulated by common components of the signal transduction pathways: transactivation of the rat γD promoter decreased upon activation of the cAMP or protein kinase C (PKC) pathway. The mechanism of the latter inhibition does not involve phosphorylation of the consensus protein kinase A (PKA)/PKC site in the DNA-binding domain of c-Maf but appears to be transmitted through MAP kinase.
MATERIALS AND METHODS
Cell culture and transfection
Lens epithelial cell explants were prepared from newborn rat lenses as previously described (12). Explants were cultured in M199 (Life Technologies) with 0.1% BSA and 25 ng/ml FGF-2 (a kind gift from Scios, Mountain View, CA) for 10 days to allow fibre cell differentiation to occur. Explants were transfected using the PDS-1000/He Biolistic Particle Delivery System (Bio-Rad) as described (12) using 1 µg DNA (0.8 µg reporter and expression constructs or empty vector, as indicated, and 0.2 µg CMV–β-gal). Explants were harvested 3 days after transfection, lysed and assayed as described (12). Chloramphenicol acetyltransferase (CAT) activity was assayed using the Quan-T-CAT system (Amersham). Luciferase activity was determined using the Luciferase Assay System from Promega according to the manufacturer’s instructions. P19 cells were cultured in a 1:1 mixture of DMEM and F12 medium with 10% foetal calf serum. One hour before transfection, the medium was refreshed. Cells, in a 35 mm plate, were transfected with 1 µg DNA (0.4 µg reporter construct, 0.4 µg appropriate expression construct or empty vector and 0.2 µg CMV–β-gal) using 3 µl of Fugene 6 under the conditions recommended by the manufacturer (Roche Boehringer). Cells were harvested 48 h after transfection and assayed for β-galactosidase activity and CAT.
CHO-IR800 cells (CHO cells stably expressing the insulin receptor) were cultured in DMEM (Life Technologies) with 10% foetal calf serum. Cells were transfected as described above for P19 cells. The medium was changed to DMEM with 0.5% foetal calf serum 24 h after transfection and 8 h later the signal transduction (ant)agonists were added. After 32 h the cell density was determined by measuring cleavage of the tetrazolium salt WST-1 using the reagent supplied by Roche Boehringer. Cells were then harvested and β-galactosidase and luciferase activities were assayed. All transfections were done in duplicate or triplicate; all data reported are the averages of at least three independent experiments in the case of explants and of at least two independent experiments in the case of P19 or CHO cells. Reporter gene activity was corrected for differences in transfection activity on the basis of the activity of the co-transfected CMV–β-gal. In the case of CHO cells treated with (ant)agonists, transfection efficiencies were assumed to be equal and data were corrected for differences in cell density.
Reporter gene constructs
Most of the γD promoter–CAT fusion genes used in this study have been previously described (12,16,17). The –73/+10 γD promoter–luciferase fusion gene was made by excising the promoter fragment from the corresponding CAT clone and inserting it in the polylinker of pGL3 (Promega). The Sox-binding site was mutated from TTTTGT to TGGATC using the oligo GCGGGCCCCTGGATCCCTGTTCCTGCCAAC and its complement and a QuikChange kit (Stratagene) according to the manufacturer’s instructions.
Trans-acting factor expression clones and mutants thereof
The expression clones for rat Maf-1 (MafB) and Maf-2 (c-Maf) were obtained from Dr M. Sakai. In these clones MafB or c-Maf expression is driven by the β-actin promoter (7). For the c-Maf binding and dimerisation domain expression clone, the β-actin–c-Maf clone was digested with StuI (which cuts the sequence 7 nt upstream of the beginning of the extended DNA-binding domain) and BamHI (which cuts 3′ of the insert). The StuI–BamHI fragment was inserted into SmaI + BamHI-digested vector pAS2-1 (Clontech) to obtain an in-frame 5′ NcoI site, and the NcoI–BamHI fragment was then inserted in the NcoI + BamHI-digested β-actin vector. The MafB single domain expression clones were made in a similar manner except that a SmaI site instead of the StuI site was used. The chimeric c-Maf–MafB clones were made by replacing the NcoI–StuI fragment of c-Maf with the NcoI–SmaI fragment of MafB for the MafB–c-Maf clone or, for the c-Maf–MafB clone, by replacing the StuI–BamHI fragment of c-Maf with the SmaI–BamHI fragment of MafB. In the construction of site-directed mutants, we encountered considerable problems with amplification of the c-Maf coding sequence, presumably because of the CG-rich repeated region encoding the stretch of glycines. Hence, for site-directed mutation of the putative phosphorylation sites, the 3′-part of c-Maf was first subcloned as a NotI–XbaI fragment into pBluescript. The putative phosphorylation sites were then mutated using the QuikChange kit (Stratagene) with the following primers (only the sense sequence given): c-MafT291A, GAAGAGGCGGGCCCTGAAAAACC; c-MafT291D, GAAGAGGCGGGACCTGAAAAACC; c-MafY340F, GAAAGGGACGCCTCCAAGGAGAAATACG. The constructs were sequenced to confirm the presence of the desired mutations. The StuI–XbaI mutated region was reinserted into the StuI + XbaI-digested β-actin–c-Maf expression vector.
EGFP–c-Maf constructs
For the EGFP–cMaf construct, the NcoI–XbaI(blunt) c-Maf fragment was first inserted into NcoI + XhoI-digested vector pJGBR. c-Maf was excised from the recombinant as a BglII–SalI fragment, which was inserted into BglII + XhoI-digested pEGFP-C1 (Clontech). For the EGFP–c-Maf mutant constructs, the NotI–BamHI fragment of the wild-type clone was replaced with corresponding fragments containing the desired mutations. Expression of properly sized fusion products was verified by western blotting of extracts of CHO cells transfected with the various EGFP–c-Maf expression clones, using an anti-GFP antibody (Clontech).
The chicken L-Maf expression construct
The L-Maf coding sequence was amplified from chicken lens RNA using the forward primers ACAGCCCGACCTCACCCCATTGC and CCTCCATGGACTGAGATGGCCTCGGAG, with the reverse primers GAGGGAGGGGGTGGAAGGGGCATTG and GGGCTCACATGAAGAAGTCAGCGGCAG. The PCR product was cloned in pBluescript and sequenced. The cDNA clone obtained contained a D174G mutation, which was corrected by site-directed mutagenesis according to the QuikChange (Stratagene) procedure using the primer GGAGAGGTTTTCCGACGACCAGTTGGTG and its complement. The coding sequence was then cloned as a NcoI–BamHI fragment in the β-actin expression vector.
RESULTS
Transactivation of the rat γD promoter by MafB, c-Maf and L-Maf
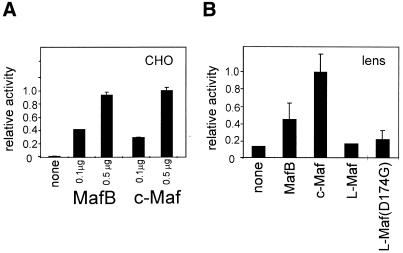
The mouse γF-crystallin promoter has been shown to be activated by c-Maf and L-Maf in non-lens cells (6,8). Similarly, the –73/+10 rat γD promoter is activated equally well in CHO cells by either MafB or c-Maf (Fig. 1A). When in vitro differentiating lens cells, which express the endogenous γ-crystallin genes and in all respects resemble in vivo lens fibre cells, were used as transfection hosts, L-Maf did not activate the –73/+10 γD promoter, while MafB did stimulate activity of the γD promoter, although not as well as c-Maf (Fig. 1B).
Figure 1.
Activation of the γD promoter by different members of the Maf family. CHO cells (A) or lens explants (B) were transfected with the –73/+10 γD–CAT construct and an expression construct of MafB, c-Maf, chicken L-Maf or a chicken L-Maf mutant (see Materials and Methods) as indicated. Results are expressed relative to the activity of the γD construct in cells with added c-Maf, which was set at 1.
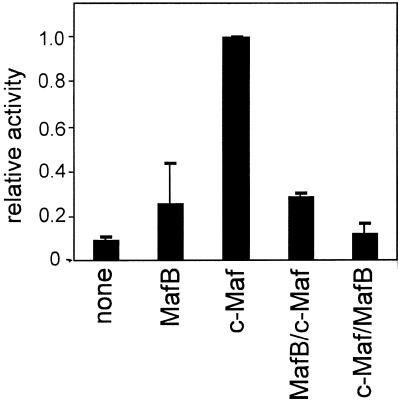
To determine whether the higher activity of c-Maf is due to its DNA-binding and dimerisation domain, to its activation domain or to both, chimeric MafB–c-Maf constructs were made. The construct containing the MafB activation domain coupled to the c-Maf DNA-binding and dimerisation domain had the same low activity as MafB itself; the reverse chimera, with the c-Maf activation domain linked to the MafB DNA-binding and dimerisation domain, was completely inactive (Fig. 2). Hence, for optimal activation both the c-Maf activation domain and the DNA-binding and dimerisation domain are required.
Figure 2.
Transactivation by chimeras between MafB and c-Maf. Explants were co-transfected with the –73/+10 γD–CAT construct and an expression construct for MafB, c-Maf, the N-terminal domain of MafB linked to the C-terminal domain of c-Maf (MafB/c-Maf) or the N-terminal domain of c-Maf linked to the C-terminal domain of MafB (c-Maf/MafB), as indicated. Results are expressed relative to the activity of the γD construct in cells with added c-Maf, which was set at 1.
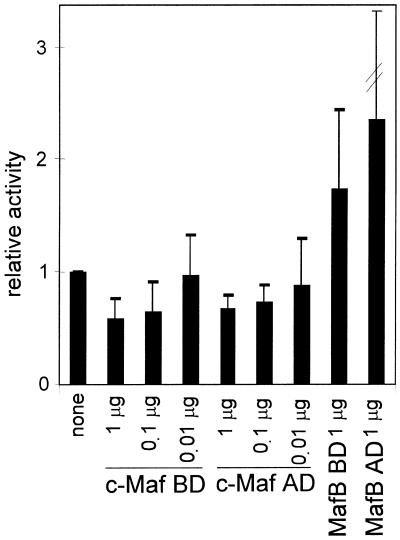
These data show that c-Maf is an efficient activator of the γD promoter in the context of the lens fibre cell, however, they do not prove that c-Maf is the endogenous γD-activating factor. We therefore used a c-Maf mutant that inhibits activation by c-Maf (18). This mutant (c-Maf BD) contains only the DNA-binding and dimerisation domain and should compete with endogenous c-Maf for the MARE but cannot activate the γD promoter. Hence, if c-Maf is the endogenous cognate factor, the c-Maf BD mutant should inhibit activity. As shown in Figure 3, it does. As a control the comparable mutant of MafB (MafB BD) was also tested. This mutant does not inhibit activity of the –73/+10 γD–CAT construct, in agreement with the finding that MafB is a poor activator of the γD promoter (see Fig. 1B). We tested the effect of a c-Maf mutant containing only an activation domain (c-Maf AD) as well. This mutant also inhibited the activity of the –73/+10 γD–CAT construct (Fig. 3). The N-terminal domain could capture co-activating factors, thus quenching activity. As expected, co-transfection of the MafB activation domain had no effect on γD promoter activity (Fig. 3).
Figure 3.
The effect of single domains of c-Maf or MafB on activity of the γD promoter. Explants were co-transfected with the –73/+10 γD–CAT construct and an expression construct for the C-terminal domain of c-Maf (the DNA-binding and dimerisation domain; c-Maf BD) or of MafB (MafB BD) or an expression construct for the N-terminal domain of c-Maf (the activation domain; c-Maf AD) or of MafB (MafB AD), as indicated. The c-Maf constructs were transfected at increasing ratios to γD–CAT DNA, as indicated; for the MafB constructs a 1:1 ratio with γD–CAT DNA was used (see also Materials and Methods). Results are expressed relative to the activity of the γD construct in cells without added Maf, which was set at 1.
The role of AP1 factors in activation of the γD promoter
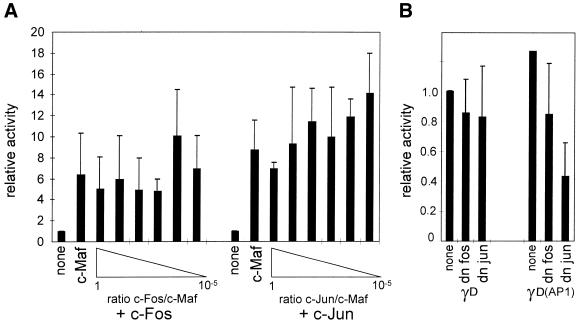
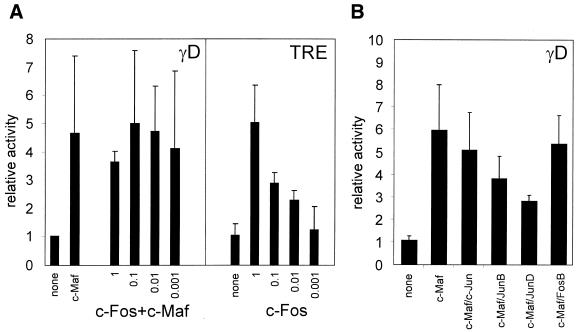
c-Maf can act as a homodimer and can also heterodimerise with other Maf factors or with Fos family proteins, but not with Jun family proteins (19). We therefore tested the effect of co-transfection of both c-Maf and c-Fos on the activity of the γD promoter. As a control, similar experiments were performed using a c-Jun expression clone. No significant effect of co-expression of either c-Fos or c-Jun was found, showing that neither c-Fos nor c-Jun participates in activation of the γD promoter (Fig. 4A). To confirm these data, we co-transfected dominant negative c-Fos or c-Jun constructs (20). Neither of these constructs inhibited activity of the γD–CAT construct (Fig. 4B). That these dominant negative constructs do inhibit the activity of a promoter with an AP1 site in the context of lens fibre cells was shown by testing the effect of the dominant negative expression constructs on the activity of the –73/+10 γD(AP1) mutant promoter. In this mutant an AP1 site was created directly downstream of the MARE (such an apposition of a MARE and an AP1 site is found in the γB-crystallin gene promoter; 12). The γD(AP1) promoter was inhibited by ∼25% by the dominant negative c-Fos construct and by ∼50% by the dominant negative c-Jun construct.
Figure 4.
(A) The effect of added c-Fos or c-Jun on transactivation of the γD promoter by c-Maf. Explants were co-transfected with the –73/+10 γD–CAT construct and an expression construct for c-Maf (at 0.2 µg/transfection) and an expression construct for c-Fos or c-Jun using a decreasing DNA ratio to c-Maf DNA, as indicated. Results are expressed relative to the activity of the γD construct in cells without added Maf, which was set at 1. (B) The effect of dominant negative c-Fos or c-Jun on activity of the γD promoter. Explants were co-transfected with the –73/+10 γD–CAT construct and an expression construct for a dominant negative c-Fos (dn c-Fos) or c-Jun (dn c-Jun), as indicated. As a control, the same experiment was performed using a –73/+10 γD construct containing an AP1 site directly downstream of the MARE [γD(AP1); this mutant is described as γD4 in Klok et al. (12)]. Results are expressed relative to the activity of the γD construct in cells without added factors, which was set at 1.
The data presented in Figure 4 argue against an involvement of AP1 factors in MARE-directed activation of the γD promoter. However, recognition of the MARE by Maf–AP1 heterodimers is sequence dependent (4,19) and the possibility that the γD MARE requires an AP1-like factor other than c-Fos cannot be excluded. We therefore turned to P19 embryocarcinoma cells, which lack AP1 factors. The –73/+10 γD–CAT construct is inactive in P19 cells. Activity in these cells can be restored by co-transfection with the c-Maf expression vector (Fig. 5A). Co-transfection with c-Fos did not increase activity of the γD promoter further. The activity of a known target of c-Fos, a TRE, was stimulated under these conditions, as shown by increased activity of a TRE–CAT reporter. Hence, in the context of P19 cells, c-Maf does not cooperate with c-Fos in activation of the γD promoter. A number of other AP1 family members were tested (Fig. 5B): none stimulated activity and, indeed, the Jun family factors tended to inhibit activity of the –73/+10 γD–CAT construct when co-transfected with c-Maf. Why the Jun factors tend to inhibit c-Maf activity is not clear, since full-length c-Maf has been shown not to interact with Jun proteins (19), although the C-terminal domain of c-Maf and full-length v-Maf do interact with Jun (4,19).
Figure 5.
(A) The effect of added c-Fos on transactivation of the γD promoter by c-Maf in P19 cells. (Panel γD) P19 cells were co-transfected with the –73/+10 γD–CAT construct and an expression construct for c-Maf (at 0.2 µg/transfection) and an expression construct for c-Fos using a decreasing DNA ratio to c-Maf DNA, as indicated. Results are expressed relative to the activity of the γD construct in cells without added Maf, which was set at 1. (Panel TRE) P19 cells were co-transfected with a TRE–CAT reporter construct and decreasing amounts of c-Fos, where 1 indicates 0.2 µg/transfection, 0.1 indicates 0.02 µg/transfection, etc. Results are expressed relative to the activity of the TRE–CAT reporter gene without added c-Fos, which was set at 1. (B) The effect of added c-Fos or c-Jun family members on transactivation of the γD promoter by c-Maf in P19 cells. P19 cells were co-transfected with the –73/+10 γD–CAT construct and an expression construct for c-Maf (at 0.2 µg/transfection) and an expression construct for c-Fos or c-Jun family members (also at 0.2 µg/transfection), as indicated. Results are expressed relative to the activity of the γD construct in cells without added Maf, which was set at 1.
The MARE suffices for growth factor response of the γD promoter
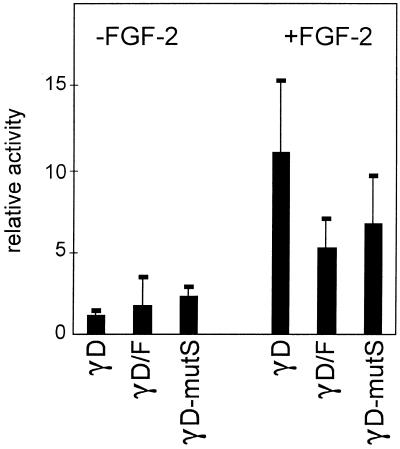
Progression through fibre cell differentiation and sequential activation of the crystallin promoters requires continuous growth factor signalling, which can be provided by FGF-2 or insulin (21,22). The –73/+10 γD promoter is also activated in the presence of FGF-2 (Fig. 6; note that promoter activity in the absence of FGF-2 is close to background and was not corrected for background). Previous mapping experiments have delineated three elements in the –73/+10 region: a Sox target site at –70/–67 (23), the MARE at –57/–46 and a positive acting element around –10 (12,17,24). Mutation of the Sox target site decreased promoter activity by 50% but did not affect the growth factor response (Fig. 6). To test the effect of the –10 element, we used a γD/F chimeric promoter, in which the region downstream of the γD TATA box is replaced with the corresponding region of the γF promoter, which lacks the –10 element (12). This γD/F chimeric promoter is less active than the γD promoter, as expected, but is still activated by FGF-2 (Fig. 6). Hence, the MARE must suffice for FGF-2 stimulation. As this element is absolutely required for promoter activity (12,17,25), the effect of mutating this element on FGF-2 stimulation cannot be tested.
Figure 6.
Mapping of the growth factor-responsive region of the γD promoter. Lens explants were precultured for 10 days with FGF-2 to induce fibre cell differentiation, the medium was then changed to medium without FGF-2 and the explants were transfected with the wild-type –73/+10 γD–CAT construct (γD), with a –73/+10 γD–CAT construct in which the Sox-binding site was mutated (γD-mutS) or a construct in which the region downstream of the TATA box was replaced by the corresponding region of the γF promoter (γD/F). The medium was then supplemented with FGF-2 (25 ng/ml) where indicated. Cells were harvested after 72 h and assayed for activity. Results are expressed relative to the activity of the –73/+10 γD construct in cells without FGF-2, which was set at 1. Note that the activity of the –73/+10 γD–CAT (mutant) construct in the absence of FGF-2 is close to background and that the background was not substracted. Exact activation ratios cannot therefore be calculated from these data.
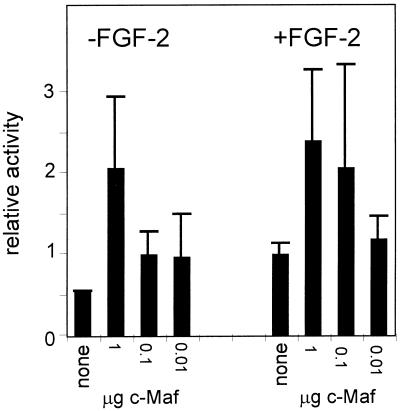
In the absence of FGF-2, added c-Maf still stimulated the –73/+10 γD promoter. The promoter activity reached in the absence of FGF-2 was the same as that reached in the presence of FGF-2 (Fig. 7), showing that excess c-Maf can compensate for the lack of growth factor signalling.
Figure 7.
The effect of co-transfection of c-Maf on activity of the γD promoter in the presence or absence of growth factors. Lens explants were treated and transfected as described in the legend to Figure 6 except that the c-Maf expression construct was co-transfected at the amount indicated as the ratio to γD–CAT DNA (the amount of which was held constant) in the Figure. Results are expressed relative to the activity of the γD construct in cells with FGF-2 but without added c-Maf, which was set at 1. Note that the activity of the –73/+10 γD–CAT construct in the absence of FGF-2 is close to background and that the background was not substracted. Exact activation ratios cannot therefore be calculated from these data.
c-Maf and signal transduction
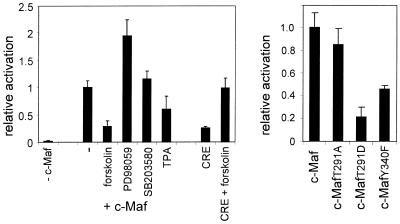
The data reported above strongly support the hypothesis that c-Maf activates the γD promoter as a homodimer, not a c-Maf–AP1 heterodimer. As the data in Figure 6 show that the c-Maf target sequence, the MARE, suffices for FGF-2 stimulation, this would imply that c-Maf is a target of FGF-2 signalling. We tested this hypothesis by determining whether the activity of c-Maf is affected by (ant)agonists of signal transduction pathways. For ease of manipulation, CHO cells were used in these experiments. CHO cells co-transfected with the γD–CAT reporter construct and the c-Maf expression construct were treated either with the signal transduction agonists forskolin and TPA, which activate PKA and PKC, respectively, or the antagonists PD98059, an inhibitor of MAPK/ERK, and SB203580, an inhibitor of p38 MAPK and, at the concentration used here, of protein kinase B kinase (26). Treatment with forskolin strongly inhibited activation of the γD promoter by c-Maf, while treatment with TPA was also inhibitory, but to a lesser extent (Fig. 8). In contrast, inhibition of MAPK/ERK by PD98059 markedly increased the extent of activation by c-Maf; inhibition of p38 MAPK by SB203580 had no effect. In the absence of c-Maf, activity of the γD promoter was not affected by the (ant)agonists, but remained at background level.
Figure 8.
(Left) The effect of signal transduction (ant)agonists on transactivation of the γD promoter by c-Maf in CHO cells. CHO cells were transfected with the –73/+10 γD–luciferase construct and an expression construct for c-Maf as described in Materials and Methods. Signal transduction (ant)agonists were added as indicated at the following concentrations: forskolin, 100 µM; PD98059, 20 µM; SB203580, 10 µM; TPA, 1 µg/ml. The relative activation was calculated from the increase in activity during the time that the signal transduction (ant)agonists were present, where the increase in activity in the absence of (ant)agonists but in the presence of c-Maf was set at 1. As a control, activation of a CRE–luciferase reporter gene by forskolin was also determined. To avoid problems with the scale, the activity of the CRE reporter gene in the presence of forskolin was set at 1. (Right) The behaviour of c-Maf mutants. CHO cells were co-transfected with a –73/+10 γD–luciferase construct and an expression construct for c-Maf or mutants thereof, as indicated. The activity of the mutants is expressed relative to that of wild-type c-Maf, which was set at 1.
The data presented in Figure 8 suggest that c-Maf activity is regulated negatively by the PKA, PKC and MAPK/ERK signalling cascades. A prosite scan of the c-Maf amino acid sequence identified several possible phosphorylation sites: six putative casein kinase II sites, three possible PKC sites, one of which overlapped with the single PKA site, and one tyrosine phosphorylation site. We decided to focus on two sites phosphorylation of which is likely to be inhibitory: the joint PKA/PKC site, which is located in the extended DNA-binding domain, and the tyrosine phosphorylation site, which is at the end of the dimerisation domain. The sequence of the putative PKA/PKC target site is KRRTLK (amino acids 288–293). We mutated T291 either to alanine (c-MafT291A), thus preventing phosphorylation, or to aspartate (c-MafT291D), thereby mimicking phosphorylation. The putative tyrosine phosphorylation site of c-Maf is RLAKERDDLY (amino acids 332–340). In the mutant c-MafY340F the tyrosine was mutated to phenylalanine. This mutant was consistently less active than the wild-type protein. The c-MafT291A mutant was almost as active as the wild-type protein, while the c-MafT291D mutant was essentially inactive (Fig. 8). To ensure that the mutations did not affect nuclear localisation of c-Maf, the mutant c-Maf coding sequences, as well as the wild-type c-Maf coding sequence, were fused to EGFP. The activities of the EGFP–c-Maf fusion proteins were the same as that of the parental c-Maf (mutant) coding sequences (data not shown). Cellular localisation of the EGFP–c-Maf fusion proteins was determined by confocal microscopy. All EGFP–c-Maf fusion proteins accumulated in the nucleus. The EGFP–c-MafY340F protein gave a more particulate pattern and frequently showed large aggregates, suggesting that the lower activity caused by this mutation may be due to aberrant protein–protein interaction (data not shown). Possibly the leucine zipper is disturbed, even though the mutation is C-terminal to the region shown to be required in v-Maf for dimer formation (3,27).
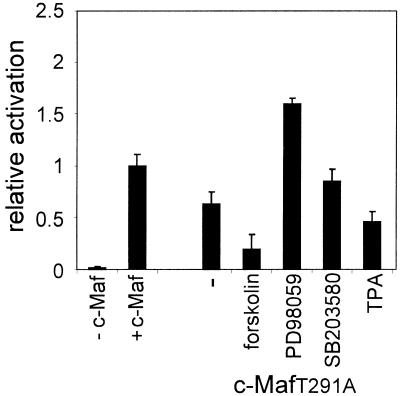
The almost wild-type activity of the c-MafT291A mutant and the lack of activity of the c-MafT291D mutant is consistent with an inhibitory effect of PKA and/or PKC phosphorylation of T291. If so, the c-MafT291A mutant should be refractory to inhibition. To test this, CHO cells were co-transfected with a –73/+10 γD–luciferase construct and the c-MafT291A expression construct and treated with forskolin, TPA, PD98059 or SB203580. The effect of these agents on activation of the γD promoter by c-MafT291A was essentially the same as that found for wild -type c-Maf: forskolin still caused a strong inhibition and TPA was also still inhibitory, while PD98059 still activated and SB203580 had no effect (Fig. 9). Hence, T291 is not a target of PKC or PKA. Similar experiments were performed with the c-MafY340F mutant: this mutant also responded as the wild-type protein to the (ant)agonists used (data not shown). These data suggest that the inhibitory phosphorylation sites could be located in the N-terminal domain. Attempts to mutate the N-terminal domain failed because we were unable to carry out PCR across the glycine repeat region of c-Maf, a problem also encountered by others (9).
Figure 9.
The effect of signal transduction (ant)agonists on transactivation of the γD promoter by the c-MafT291A mutant in CHO cells. CHO cells were transfected with a –73/+10 γD–luciferase construct and an expression construct for the c-MafT291A mutant as described in Materials and Methods. Signal transduction (ant)agonists were added as indicated in the legend to Figure 8. The relative activation was calculated from the increase in activity during the time that the signal transduction (ant)agonists were present, where the increase in activity in the absence of (ant)agonists but in the presence of wild-type c-Maf was set at 1.
DISCUSSION
The three large Maf proteins, MafB, c-Maf and L-Maf (or MafA), are closely related and all are capable of binding to and activating a rodent γ-crystallin MARE in non-lens cells (Fig. 1; 6,8). However, as we show here, the transactivation potential of these proteins is severely constrained by the cellular context. In in vitro differentiating lens fibre cells, in which the endogenous γ-crystallin genes are highly active and which contain all transcription factors involved in regulating activity of the γ-crystallin promoters, c-Maf is a better transactivator than MafB, whereas L-Maf is virtually inactive. Hence, not only the sequence context but also other transcription factors binding in the vicinity, such as Sox in the case of the γD promoter, determine the specificity of a MARE. The constraints imposed by neighbouring transcription factors must also affect the binding affinity, as only the C-terminal domain of c-Maf and not that of MafB inhibited the endogenous transcription factor. Our data also show that the endogenous cognate transcription factor of the γD MARE is most likely c-Maf, in agreement with previous studies showing that c-Maf can activate the mouse γF promoter and that in transgenic mice lacking c-Maf the γ-crystallins are absent (6,8,9).
The ability of c-Maf to transactivate the γD promoter independent of the cellular context and in the absence of AP1 factors suggests that c-Maf can transactivate the γD promoter as a homodimer, at least in non-lens cells. This suggestion is in agreement with the recognition site specificity of the c-Maf homodimer, of the c-Maf–c-Fos heterodimer and of the c-Fos–c-Jun heterodimer: the C found at the fourth position of the γD MARE (instead of the optimal T) abolishes recognition by c-Maf–c-Fos and c-Fos–c-Jun heterodimers, but not by a c-Maf homodimer (3,19).
Recently it was shown that MafA, the quail orthologue of chicken L-Maf, is only active in the phosphorylated form. One of the kinases responsible was found to be ERK2 (28). In contrast, we find that, under our experimental conditions, c-Maf is negatively regulated by stimulation of growth factor signalling pathways. In particular, blocking MEK1/2, and thus indirectly also ERK1/2, activates rather than silences c-Maf transactivation of the γD MARE. The strongly negative effect of forskolin, as well as the more modest effect of TPA, could also be mediated by MEK1/2, as MEK1/2 is downstream from PKA as well as PKC. Thus, our data suggest that c-Maf transmits a negative rather than a positive signal from a growth factor stimulus to the transcriptional apparatus. Our results do not rigorously exclude the possibility that the inhibitory effect of the activation of growth factor signalling pathways on the activity of c-Maf is due to competition for the MARE by other growth factors activated by the growth factor stimulus. However, we think this unlikely, as co-transfection of obvious candidates, the AP1 factors, does not inhibit activation by c-Maf, with the exception of JunD. One would then have to assume that the γD MARE is bound non-productively by a JunD complex, yet the slight deviation of the γD MARE sequence from the TRE consensus abolishes Fos–Jun and Jun–Jun recognition (3).
Our conclusion that the γD MARE is required for positive regulation of the γD promoter by FGF-2 seems completely at odds with our finding that c-Maf is negatively regulated by stimulation of growth factor signalling pathways. However, the effects of growth factor signalling on c-Maf were determined in non-lens cells, under conditions in which we suggest that c-Maf acts as a homodimer and activates the γD promoter in the absence of other factors. There is indirect evidence that c-Maf as part of a heterodimer can transmit a positive PKC or ras signal. c-Maf is known to be a partner of the transcription factor NF-ATc. Activation of NF-ATc-dependent transcription requires both a Ca2+ and a PKC or ras signal, with the latter signal being transmitted by the NF-ATc partner (29). A similar situation could exist in the lens, where other transcription factors cooperate or constrain c-Maf. Our previous mapping studies of the γ-crystallin promoters have shown that in lens fibre cells the MARE is required but not sufficient for promoter activity (12,17; see also for example Fig. 6) and a consensus MARE does not activate the minimal β-globin promoter in lens fibre cells (unpublished data). Hence, we think it likely that c-Maf acts as a heterodimer in the lens in vivo, with the heterodimeric partner modulating the growth factor response of c-Maf. Our data show that the putative c-Maf lens partner is not one of the common AP1 factors. However, other partners of the Maf transcription factors are known, such as members of the CREB family (1,2). CREB-2 is required for proper lens development (30,31), but the phenotype of CREB-2 knockout mice argues against a role for CREB-2 in γ-crystallin gene expression. We favour the hypothesis that c-Maf has a lens-specific partner. A lens-specific partner for c-Maf could also explain why a pentamer of the mouse γF MARE directs expression of a hsp68–lacZ reporter gene only in lens fibre cells and a subregion of the hindbrain in transgenic mice (25), even though c-Maf is widely expressed and is found, for example, in liver, spleen and kidney (7). Unfortunately, direct purification of a c-Maf complex from rat lenses is precluded by the excessive number of lenses required. The larger calf lenses are no alternative, as we have never been able to extract a factor binding to the γD MARE from calf lenses, possibly because of the age of the calves. Finally, a first attempt to select lens-specific partners of c-Maf by a two-hybrid screen failed due to the high intrinsic transcription activation by c-Maf in yeast. Further experimentation is required to overcome this problem.
Acknowledgments
ACKNOWLEDGEMENTS
The dominant negative c-Fos and c-Jun clones were given to us by Dr R. de Groot. The expression clones for the other Jun family members were originally made by Dr P. Angel and were obtained together with the clones for the other Fos family members from Dr A. Zantema. The CHO-IR800 cells were obtained from Dr J. A. Maassen. The financial support of the Netherlands Diabetes Foundation is gratefully acknowledged.
REFERENCES
- 1.Blank V. and Andrews,N.C. (1997) The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci., 22, 437–441. [DOI] [PubMed] [Google Scholar]
- 2.Motohashi H., Shavit,J.A., Igarashi,K., Yamamoto,M. and Engel,J.D. (1997) The world according to Maf. Nucleic Acids Res., 25, 2953–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kataoka K., Noda,M. and Nishizawa,M. (1994) Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol., 14, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerppola T.K. and Curran,T. (1994) Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene, 9, 675–684. [PubMed] [Google Scholar]
- 5.Benkhelifa S., Provot,S., Lecoq,O., Pouponnot,C., Calothy,G. and Felder-Schmittbuhl,M.-P. (1998) MafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene, 17, 247–254. [DOI] [PubMed] [Google Scholar]
- 6.Ogino H. and Yasuda,K. (1998) Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science, 280, 115–118. [DOI] [PubMed] [Google Scholar]
- 7.Sakai M., Imaki,J., Yoshida,K., Ogata,A., Matsushima-Hibaya,Y., Kuboki,Y., Nishizawa,M. and Nishi,S. (1997) Rat maf related genes: specific expression in chondrocytes, lens and spinal cord. Oncogene, 14, 745–750. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.I., Li,T., Ho,I.C., Grusby,M.J. and Glimcher,L.H. (1999) Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc. Natl Acad. Sci. USA, 96, 3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawauchi S., Takahashi,S., Nakajima,O., Ogino,H., Morita,M., Nishizawa,M., Yasuda,K. and Yamamoto,M. (1999) Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem., 274, 19254–19260. [DOI] [PubMed] [Google Scholar]
- 10.Ring B.Z., Cordes,S.P., Overbeek,P.A. and Barsh,G.S. (2000) Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development, 127, 307–317. [DOI] [PubMed] [Google Scholar]
- 11.Sakai M., Serria,M.S., Ikeda,H., Yoshida,K., Imaki,J. and Nishi,S. (2001) Regulation of c-maf gene expression by Pax6 in cultured cells. Nucleic Acids Res., 29, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klok E.J., van Genesen,S.T., Civil,A., Schoenmakers,J.G.G. and Lubsen,N.H. (1998) Regulation of expression within a gene family. The case of the rat γB- and γD-crystallin promoters. J. Biol. Chem., 273, 17206–17215. [DOI] [PubMed] [Google Scholar]
- 13.McAvoy J.W. (1978) Cell division, cell elongation and distribution of α-, β- and γ-crystallins in the rat lens. J. Embryol. Exp. Morphol., 44, 149–165. [PubMed] [Google Scholar]
- 14.Van Leen R.W., Breuer,M.L., Lubsen,N.H. and Schoenmakers,J.G.G. (1987) Developmental expression of crystallin genes: in situ hybridization reveals a differential localization of specific mRNAs. Dev. Biol., 123, 338–345. [DOI] [PubMed] [Google Scholar]
- 15.Peek R., McAvoy,J.W., Lubsen,N.H. and Schoenmakers,J.G.G. (1992) Rise and fall of crystallin gene messenger levels during fibroblast growth factor induced terminal differentiation of lens cells. Dev. Biol., 152, 152–160. [DOI] [PubMed] [Google Scholar]
- 16.Peek R., van der Logt,P., Lubsen,N.H. and Schoenmakers,J.G.G. (1990) Tissue- and species-specific promoter elements of rat γ-crystallin genes. Nucleic Acids Res., 18, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirks R.P.H., Klok,E.J., van Genesen,S.T., Schoenmakers,J.G.G. and Lubsen,N.H. (1996) The sequence of regulatory events controlling the expression of the γD-crystallin gene during fibroblast growth factor mediated rat lens fibre cell differentiation. Dev. Biol., 172, 14–25. [Google Scholar]
- 18.Doerwald L., Nijveen,H., Civil,A., van Genesen,S.T. and Lubsen,N.H. (2001) Regulatory elements in the rat βB2-crystallin promoter. Exp. Eye Res., 73, 703–710. [DOI] [PubMed] [Google Scholar]
- 19.Matsushima-Hibiya Y., Nishi,S. and Sakai,M. (1998) Rat maf-related factors: the specificities of DNA binding and heterodimer formation. Biochem. Biophys. Res. Commun., 245, 412–418. [DOI] [PubMed] [Google Scholar]
- 20.Ransone L.J., Visvader,J., Wamsley,P. and Verma,I.M. (1990) Trans-dominant negative mutants of Fos and Jun. Proc. Natl Acad. Sci. USA, 87, 3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamberlain C.G., McAvoy,J.W. and Richardson,N.A. (1991) The effects of insulin and basic fibroblast growth factor on fibre differentiation in rat lens epithelial explants. Growth Factors, 4, 183–188. [DOI] [PubMed] [Google Scholar]
- 22.Leenders W.P., van Genesen,S.T., Schoenmakers,J.G.G., van Zoelen,E.J.J. and Lubsen,N.H. (1997) Synergism between temporally distinct growth factors: bFGF, insulin and lens cell differentiation. Mech. Dev., 67, 193–201. [DOI] [PubMed] [Google Scholar]
- 23.Kamachi Y., Sockanathan,S., Liu,Q., Breitman,M., Lovell-Badge,R. and Kondoh,H. (1995) Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J., 14, 3510–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peek R., Kraft,H.J., Klok,E.J., Lubsen,N.H. and Schoenmakers,J.G.G. (1992) Activation and repression sequences determine the lens-specific expression of the rat γD-crystallin gene. Nucleic Acids Res., 20, 4865–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goring D.R., Bryce,D.M., Tsui,L.-C., Breitman,M.L. and Liu,Q. (1993) Developmental regulation and cell type-specific expression of the murine γF-crystallin gene is mediated through a lens-specific element containing the γF-1 binding site. Dev. Dyn., 196, 143–152. [DOI] [PubMed] [Google Scholar]
- 26.Lali F.V., Hunt,A.E., Turner,S.J. and Foxwell,B.M. (2000) The pyridinyl imidazole inhibitor SB203580 blocks phosphoinositide-dependent protein kinase activity, protein kinase B phosphorylation and retinoblastoma hyperphosphorylation in interleukin-2-stimulated T cells independently of p38 mitogen-activated protein kinase. J. Biol. Chem., 275, 7395–7402. [DOI] [PubMed] [Google Scholar]
- 27.Kataoka K., Nishizawa,M. and Kawai,S. (1993) Structure–function analysis of the maf oncogene product, a member of the b-Zip protein family. J. Virol., 67, 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benkhelifa S., Provot,S., Nabais,E., Eychene,A., Calothy,G. and Felder-Schmittbuhl,M.-P. (2001) Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol. Cell. Biol., 21, 4441–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crabtree G.R. (1999) Generic signals and specific outcomes: signaling through Ca2+, Calcineurin and NF-AT. Cell, 96, 611–614. [DOI] [PubMed] [Google Scholar]
- 30.Hettmann T., Barton,K. and Leiden,J.M. (2000) Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev. Biol., 222, 110–123. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T., Tsujimura,T., Takeda,K., Sugihara,A., Maekawa,A., Terada,N., Yoshida,N. and Akira,S. (1998) Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells, 3, 801–810. [DOI] [PubMed] [Google Scholar]