Abstract
The identification of the SARS-CoV-2 Omicron variants BA.4/BA.5, BF.7 and BQ.1.1 immediately raised concerns regarding the efficacy of currently used monoclonal antibody therapies. Here we examined the activity of monoclonal antibody therapies and antiviral drugs against clinical specimens for SARS-CoV-2 Omicron BA.4/BA.5, BF.7 and BQ.1.1 employing an immunofluorescence neutralization assay. Further we explored treatment of BA.4/BA.5 infections with efficient antiviral drugs and monoclonal antibodies in a 3D model of primary human bronchial epithelial cells. We found that the antiviral drugs Molnupiravir, Nirmatrelvir and Remdesivir efficiently inhibit BA.4/BA.5, BF.7 and BQ.1.1 replication. In contrast, only the monoclonal antibody Cilgavimab exerted an inhibitory effect, while Tixagevimab, Regdanvimab and Sotrovimab lost their efficacy against BA.4/BA.5. We found that only the prophylactic treatment with Cilgavimab impacted on tissue inflammation by reducing intracellular complement component 3 (C3) activation following BA.4/BA.5 infection in primary human airway epithelial grown in air-liquid-interphase, which was not the case when using antiviral drugs or Cilgavimab after establishment of infection. Of note, all tested monoclonal antibodies had no neutralizing activity during infection by BF.7 and BQ.1.1 variants. Our results suggest that despite a marked reduction of viral replication, potent antiviral drugs fail to reduce tissue levels of inflammatory compounds such as C3, which can still result in tissue destruction.
In January 2022, the two recent Omicron lineages, BA.4 and BA.5 (BA.4/BA.5), appeared in South Africa and rapidly replaced BA.2 (Tegally et al., 2022). Due to better transmissibility relative to BA.2, Omicron BA.4/BA.5 caused a new COVID-19 wave in Europe in early summer 2022 (Tegally et al., 2022). Omicron BA.4/BA.5 have identical Spike (S) sequences and contain additional mutations in the receptor binding domain (RBD) (Tegally et al., 2022). Novel subvariants defined as SARS-CoV-2 Omicron BF.7 (BF.7) and Omicron BQ.1.1 (BQ.1.1) emerged later in 2022. Several monoclonal antibodies (mAbs) in clinical use target the RBD and, therefore, mutations in that specific region resulted in reduced binding affinity of available mAbs against those recent Omicron variants. Previous studies have shown that SARS-CoV-2 variants exert a minor effect on efficacy of antiviral drugs, while there were huge differences regarding the neutralization capacity of mAbs against Omicron variants (Bojkova et al., 2022). Therefore, we investigated the therapeutic potential of different licensed antiviral drugs as well as the effectiveness of several mAbs in clinical use against these new SARS-CoV-2 variants. For the early treatment of mild to moderate infections in patients at risk as well as for a severe course of the infection, the mAbs Regdanvimab (Regkirona; CT-P59) and Sotrovimab (Xevudy; S309) are in clinical use. Prophylaxis and treatment of mild to moderate infection in subjects with high risk for severe COVID-19, the antibody combination of Cilgavimab and Tixagevimab (Evusheld) has been approved for clinical use. The antiviral drugs Molnupiravir and Remdesivir are both inhibitors of the RNA-dependent RNA polymerase of SARS-CoV-2 (Takashita et al., 2022). Molnupiravir is metabolized into the active compound EIDD-1931, which causes excessive mutations in newly synthesized viral genomes (Bojkova et al., 2022). The combination of Nirmatrelvir, which inhibits the main viral protease, and Ritonavir, which reduces the CYP3A mediated degradation of Nirmatrelvir, is known as Paxlovid. All these small-molecule antiviral drugs are used for the early treatment of mild to moderate infections in patients with an increased risk of the disease becoming severe.
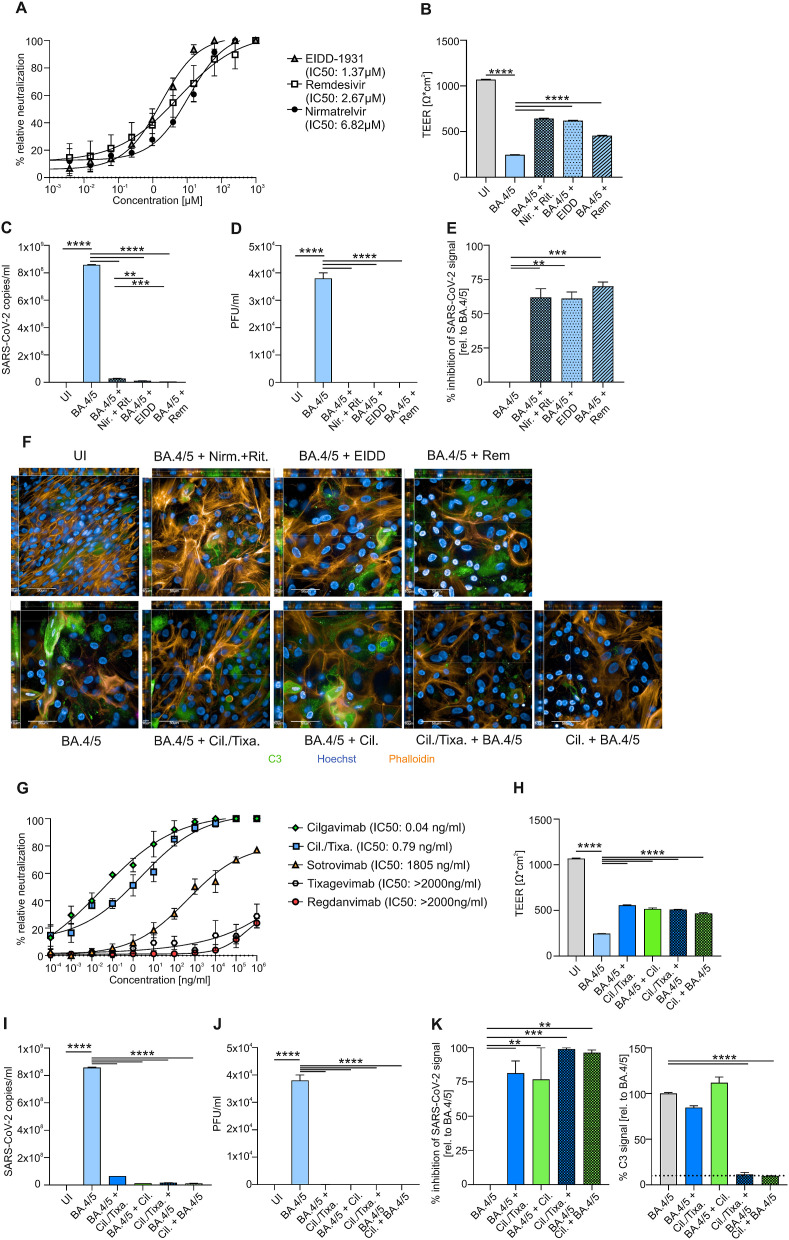
In this efficiency evaluation study, clinical specimen for SARS-CoV-2 Omicron BA.4/BA.5, BF.7 and BQ.1.1 from COVID-19 positive swabs, sequenced by the Austrian Agency for Health and Food Safety, Vienna, Austria (Ethics statement, ECS1166/2020), were propagated and subsequently used in neutralization and infection experiments. For immunofluorescence neutralization assays, VeroE6-TMPRSS2-ACE2 cells were inoculated with SARS-CoV-2 BA.4/BA.5, BF.7 and BQ.1.1 in absence or presence of serial dilutions of antiviral drugs Molnupiravir/EIDD-1931, Nirmatrelvir or Remdesivir (Fig. 1 A, 2A&C). Moreover, cells were treated with Sotrovimab, Regdanvimab, Cilgavimab, Tixagevimab or Cilgavimab/Tixagevimab (Figs. 1G and 2B&D). SARS-CoV-2 infection was evaluated using an anti-SARS-CoV-2-N-mAb and image analysis performed on Operetta CLS™ (PerkinElmer). For detailed analysis primary human airway epithelial (HAE) cells grown in air-liquid-interphase (ALI) for 30 days were treated from the basolateral side using Cilgavimab/Tixagevimab (50 μg/ml) or Cilgavimab (25 μg/ml) 6h prior apical BA.4/BA.5 infection, previously described in (Dichtl et al., 2022). In addition, 50 μg/ml Cilgavimab/Tixagevimab, 25 μg/ml Cilgavimab, 1 μM Nirmatrelvir +0.7 μM Ritonavir (Paxlovid), 10 μM EIDD-1931 or 10 μM Remdesivir were added to the basolateral side 6h after infection. The doses of the mAbs and antiviral drugs used in this study, were previously reported as effective concentrations (Kim et al., 2021).
Fig. 1.
Neutralization curves of antiviral SARS-CoV-2 drugs EIDD-1931, Remdesivir and Nirmatrelvir against SARS-CoV-2 Omicron BA.4/BA.5 variant with corresponding half-maximal inhibitory concentration (IC50) are demonstrated (A). After 72h of infection, TEER (B) and viral RNA (C) was measured from uninfected (UI) and BA.4/BA.5-infected HAE ± Nirmatrelvir + Ritonavir (Nir.+Rit.), EIDD-1931 (EIDD) and Remdesivir (Rem). Plaque assays of supernatants from BA.4/BA.5-infected cultures ± drugs were performed on Vero-TMPRSS2-ACE2 cells (D). Analysis of inhibition of SARS-CoV-2 signal of NHBE cells was analyzed by immunofluorescence (E). Representative pictures of XYZ stacks are shown (F). Scale bars represent 50 μm and 10 μm as indicated. Neutralization curves of injectable SARS-CoV-2 monoclonal antibodies Cilgavimab (Cil.), CIlgavimab/Tixagevimab (Cil./Tixa.), Sotrovimab, Tixagevimab and Regdanvimab against SARS-CoV-2 Omicron BA.4/BA.5 variant with corresponding IC50 are demonstrated (G). TEER (H) and viral RNA (I) was measured from UI and BA.4/BA.5-infected HAE ± antibodies. Cilgavimab/Tixagevimab (BA.4/5 + Cil./Tixa.) or Cilgavimab alone (BA.4/5 +Cil.) was added to BA.4/BA.5-infected cells after 6h of infection or Cilgavimab/Tixagevimab (Cil./Tixa. + BA.4/5) or Cilgavimab alone (Cil. + BA.4/5) was added 6h before infection as prophylaxis. Plaque assays of supernatants from BA.4/BA.5-infected cultures ± drugs were performed (J). Analysis of inhibition of SARS-CoV-2 signal and C3 signal of NHBE cells was analyzed by immunofluorescence (K). The dashed line in the C3 figure demonstrates the background signal of UI samples. At least three experiments were performed. Statistically significant differences were determined by one-way ANOVA with Tukey correction. All values are means ± SEM; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Cells were analyzed using TEER (transepithelial-electrical-resistance), PCR, plaque assay and immunofluorescence for SARS-CoV-2 and C3 expression signal on 3 days post infection (3dpi).
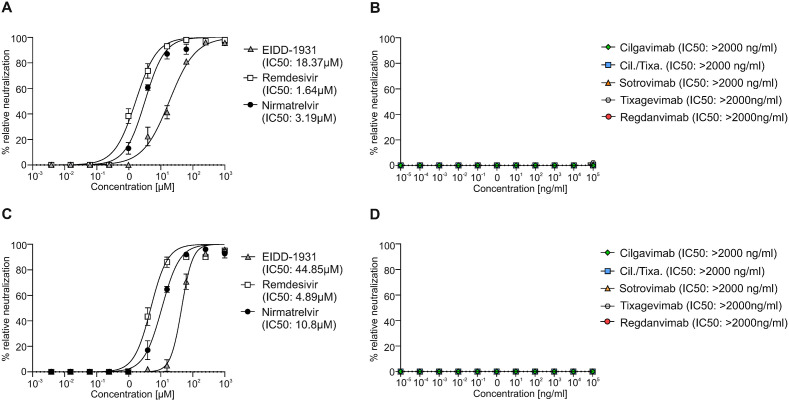
We examined the efficiency of a single dose of antiviral drugs and mAbs toward clinical isolates of BA.4/BA.5, BF.7 and BQ.1.1 in immunofluorescence-based neutralization assay using Vero cells. Additionally, we analyzed their effects in a BA.4/BA.5-infected 3D tissue model consisting of primary normal HAE-cells. ALI cultures of primary HAE-cells display a highly differentiated, pseudostratified, mucus-producing, ciliated respiratory tissue model (Posch et al., 2021). For all experiments primary normal HAE-cells from an early passage (passage 2) were used, which reduces donor specific differences and increases the reliability of the data (Rayner et al., 2019). First, the viral neutralization activity of three approved antiviral drugs, Molnupiravir (EIDD-1931), Remdesivir and Nirmatrelvir demonstrated effective reduction of Omicron BA.4/BA.5 virus replication (Fig. 1A). The immunofluorescence-based neutralization assay also showed that all three antiviral drugs are effective against the novel Omicron subvariants BF.7 and BQ.1.1 (Fig. 2 A&C). Nevertheless, we observed that the IC50 values of all tested antiviral drugs were higher for BQ.1.1 compared to BA.4/BA.5 or BF.7. An effectiveness to neutralize BA.4/BA.5 was not observed for most of the mAbs, here only Cilgavimab exerted an inhibitory effect (Fig. 1G). Tixagevimab and Regdanvimab showed no inhibitory effect against BA.4/BA.5 at all, while Sotrovimab illustrated a drastically reduced IC50 of 1.8 μg/ml against BA.4/BA.5 (Fig. 1G). All tested mAbs were not able to efficiently neutralize BF.7 nor BQ.1.1 (Fig. 2B&D). Neutralization assays were performed in Vero cells that significantly differ from human primary airway cells, which might affect receptor binding affinity and cellular entry of viruses (Bailey and Diamond, 2021). Therefore, we next tested all drugs/mAbs, which showed efficacy in the neutralization assays, in primary BA.4/BA.5-infected HAE cells organized in air-liquid-interphase tissue model. All drugs and components were added 6h post infection, while Cilgavimab alone or in combination with Tixagevimab (Cilgavimab ± Tixagevimab) was also used as prophylaxis (6h prior infection). TEER values, virus quantification, and infectivity assays, analyzed by plaque assay and immunofluorescence SARS-CoV-2 signal as previously shown (Dichtl et al., 2022), indicated that therapeutic treatment with all antiviral drugs (Fig. 1B–F) and prophylactic and therapeutic Cilgavimab-treatment (Fig. 1F, H-K) are effective against BA.4/BA.5. Moreover, prophylactic Cilgavimab-treatment not only blocked BA.4/BA.5 infection, but was able to decrease complement C3 activation following infection, which was evaluated and quantified via immunofluorescence (Fig. 1F&K). This is of interest, since increased intracellular C3 expression, resulting in elevated secreted C3a levels, was also demonstrated in SARS-CoV-2-infected HAE-cells from various donors (Posch et al., 2021). Furthermore, C3 was previously shown to be one of the strongest induced genes in SARS-CoV-2 infections and associated with a highly inflammatory microenvironment, causing severe tissue damage (Posch et al., 2021; Yan et al., 2021). Increased inflammation, detected by e.g. higher TNFα, IL10 and IL6 levels, is associated with the severity of SARS-CoV-2 infections (Dichtl et al., 2022; Guo et al., 2021; Huang et al., 2020). Thus, intracellular C3 activation in lung epithelial cells represents a novel biomarker for inflammatory tissue damage.
Fig. 2.
Neutralization curves of antiviral SARS-CoV-2 drugs EIDD-1931, Remdesivir and Nirmatrelvir (A) and monoclonal antibodies Cilgavimab, CIlgavimab/Tixagevimab (Cil./Tixa.), Sotrovimab, Tixagevimab and Regdanvimab (B) against SARS-CoV-2 Omicron BF.7 variant with corresponding half-maximal inhibitory concentration (IC50) are demonstrated. Neutralization curves of antiviral SARS-CoV-2 drugs EIDD-1931, Remdesivir and Nirmatrelvir (C) and monoclonal antibodies Cilgavimab, CIlgavimab/Tixagevimab (Cil./Tixa.), Sotrovimab, Tixagevimab and Regdanvimab (D) against SARS-CoV-2 Omicron BQ.1.1 variant with corresponding half-maximal inhibitory concentration (IC50) are shown. At least three experiments were performed. All values are means ± SEM.
By using HAE-cells in ALI cultures, we could confirm our findings of the neutralization assay, as the results show that the tested antiviral drugs and the mAb Cilgavimab are effective against BA.4/BA.5 infection. In addition, we further investigated the differences in complement activation between treatment and prophylactic use of Cilgavimab, which was not observed with antiviral drugs. Due to the inefficacy of mABs to neutralize BF.7 or BQ.1.1, these subvariants were only included in an immunofluorescence-based neutralization assay.
Here, we illustrate that the tested antiviral drugs are able to inhibit BA.4/BA.5, BF.7 and BQ.1.1 multiplication, although Molnupiravir (EIDD-1931) and Remdesivir are more effective to treat BA.4/BA.5 infections compared to Nirmatrelvir. For the newly described Omicron subvariants BF.7 and BQ.1.1, the neutralization assays indicate that Nirmatrelvir and Remdesivir are more efficient compared to Molnupiravir. From the group of mAbs, Cilgavimab prophylaxis rather than treatment demonstrated higher inhibitory potential toward BA.4/BA.5 replication. Of note, only Cilgavimab when given as a prophylaxis prior to infection with BA.4/BA.5 reduced the inflammatory response indicated by the decreased C3 expression. Thus, only the prophylactic use of Cilgavimab protected from a highly inflammatory infection side during SARS-CoV-2 BA.4/BA.5 infections and an associated serious tissue damage, while all studied mAbs are ineffective to neutralize BF.7 and BQ.1.1 infections.
Although we used effective concentrations of the mAbs and antiviral drugs (Kim et al., 2021), possible limitations of our study could be the use of one HAE donor as well as single concentrations for all tested drugs. However, we and others could previously demonstrate enhanced intracellular C3 expression upon SARS-CoV-2 infection and that early passages of HAE cells reduce donor-specific variations (Dichtl et al., 2022; Posch et al., 2021; Rayner et al., 2019).
Overall, our data show that despite a significant reduction of viral replication in BA.4/BA.5-infected HAE-cells, potent antiviral drugs or mAbs fail to reduce tissue levels of inflammatory compounds such as C3, which can still result in tissue destruction. Thus, it would be interesting to investigate, if inhibitors of anaphylotoxin receptors in addition to antiviral drugs could improve cell integrity, prevent tissue damage and pathologic inflammation in SARS-CoV-2 infection (Posch et al., 2021). In addition, the use of the combination of Cilgavimab/Tixagevimab should be critically reviewed and replaced with Cilgavimab monotherapy in high-risk patients infected with BA.4/BA.5 infection, since Tixagevimab is not effective in this setting.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: All authors declare no conflicts of interest.
Acknowledgments
We thank our technicians Chiara Ellensohn, Viktoria Kozubowski, Andrea Windisch and Christina Witting (Institute of Hygiene and Medical Microbiology, Medical University of Innsbruck, Innsbruck, Austria) as well as Dr. Lorin Loacker (Central Institute for Medical and Chemical Laboratory Diagnosis, Innsbruck University Hospital, Innsbruck, Austria) and Dr. Lukas Lanser (Department of Internal Medicine II, Medical University of Innsbruck, Innsbruck, Austria) for their valuable help and support regarding this study. The authors were supported by the Austrian Science Fund (FWF; P34070-B13 to W.P., P33510-B13 and DOC82 to D.W.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105581.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Bailey A.L., Diamond M.S. A Crisp(r) new perspective on SARS-CoV-2 Biology. Cell. 2021;184:15–17. doi: 10.1016/j.cell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Widera M., Ciesek S., Wass M.N., Michaelis M., Cinatl J., Jr. Reduced interferon antagonism but similar drug sensitivity in Omicron variant compared to Delta variant of SARS-CoV-2 isolates. Cell Res. 2022;32:319–321. doi: 10.1038/s41422-022-00619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl S., Zaderer V., Kozubowski V., Abd El Halim H., Lafon E., Lanser L., Weiss G., Lass-Florl C., Wilflingseder D., Posch W. Cilgavimab/Tixagevimab as alternative therapeutic approach for BA.2 infections. Front. Med. 2022 doi: 10.3389/fmed.2022.1005589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Wang S., Xia H., Shi D., Chen Y., Zheng S., Chen Y., Gao H., Guo F., Ji Z., Huang C., Luo R., Zhang Y., Zuo J., Chen Y., Xu Y., Xia J., Zhu C., Xu X., Qiu Y., Sheng J., Xu K., Li L. Cytokine signature associated with disease severity in COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.681516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Jang Y.R., Hong J.H., Jung J.G., Park J.H., Streinu-Cercel A., Streinu-Cercel A., Sandulescu O., Lee S.J., Kim S.H., Jung N.H., Lee S.G., Park J.E., Kim M.K., Jeon D.B., Lee Y.J., Kim B.S., Lee Y.M., Kim Y.S. Safety, virologic efficacy, and pharmacokinetics of CT-P59, a neutralizing monoclonal antibody against SARS-CoV-2 Spike receptor-binding protein: two randomized, Placebo-Controlled, phase I studies in healthy individuals and patients with mild SARS-CoV-2 infection. Clin. Therapeut. 2021;43:1706–1727. doi: 10.1016/j.clinthera.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch W., Vosper J., Noureen A., Zaderer V., Witting C., Bertacchi G., Gstir R., Filipek P.A., Bonn G.K., Huber L.A., Bellmann-Weiler R., Lass-Florl C., Wilflingseder D. C5aR inhibition of nonimmune cells suppresses inflammation and maintains epithelial integrity in SARS-CoV-2-infected primary human airway epithelia. J. Allergy Clin. Immunol. 2021;147:2083–2097 e2086. doi: 10.1016/j.jaci.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner R.E., Makena P., Prasad G.L., Cormet-Boyaka E. Optimization of normal human bronchial epithelial (NHBE) cell 3D cultures for in vitro lung model studies. Sci. Rep. 2019;9:500. doi: 10.1038/s41598-018-36735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Yamayoshi S., Simon V., van Bakel H., Sordillo E.M., Pekosz A., Fukushi S., Suzuki T., Maeda K., Halfmann P., Sakai-Tagawa Y., Ito M., Watanabe S., Imai M., Hasegawa H., Kawaoka Y. Efficacy of antibodies and antiviral drugs against Omicron BA.2.12.1, BA.4, and BA.5 subvariants. N. Engl. J. Med. 2022;387:468–470. doi: 10.1056/NEJMc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., Baxter C., Althaus C.L., Anyaneji U.J., Kekana D., Viana R., Giandhari J., Lessells R.J., Maponga T., Maruapula D., Choga W., Matshaba M., Mbulawa M.B., Msomi N., consortium N.-S., Naidoo Y., Pillay S., Sanko T.J., San J.E., Scott L., Singh L., Magini N.A., Smith-Lawrence P., Stevens W., Dor G., Tshiabuila D., Wolter N., Preiser W., Treurnicht F.K., Venter M., Chiloane G., McIntyre C., O'Toole A., Ruis C., Peacock T.P., Roemer C., Pond S.L.K., Williamson C., Pybus O.G., Bhiman J.N., Glass A., Martin D.P., Jackson B., Rambaut A., Laguda-Akingba O., Gaseitsiwe S., von Gottberg A., de Oliveira T. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022 doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Freiwald T., Chauss D., Wang L., West E., Mirabelli C., Zhang C.J., Nichols E.M., Malik N., Gregory R., Bantscheff M., Ghidelli-Disse S., Kolev M., Frum T., Spence J.R., Sexton J.Z., Alysandratos K.D., Kotton D.N., Pittaluga S., Bibby J., Niyonzima N., Olson M.R., Kordasti S., Portilla D., Wobus C.E., Laurence A., Lionakis M.S., Kemper C., Afzali B., Kazemian M. SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.