Abstract
The bulge–helix–bulge (BHB) motif recognised by the archaeal splicing endonuclease is also found in the long processing stems of archaeal rRNA precursors in which it is cleaved to generate pre-16S and pre-23S rRNAs. We show that in two species, Archaeoglobus fulgidus and Sulfolobus solfataricus, representatives from the two major archaeal kingdoms Euryarchaeota and Crenarchaeota, respectively, the pre-rRNA spacers cleaved at the BHB motifs surrounding pre-16S and pre-23S rRNAs subsequently become ligated. In addition, we present evidence that this is accompanied by circularisation of ribosomal pre-16S and pre-23S rRNAs in both species. These data reveal a further link between intron splicing and pre-rRNA processing in Archaea, which might reflect a common evolutionary origin of the two processes. One spliced RNA species designated 16S-D RNA, resulting from religation at the BHB motif of 16S pre-rRNA, is a highly abundant and stable RNA which folds into a three-stem structure interrupted by two single-stranded regions as assessed by chemical probing. It spans a region of the pre-rRNA 5′ external transcribed spacer exhibiting a highly conserved folding pattern in Archaea. Surprisingly, 16S-D RNA contains structural motifs found in archaeal C/D box small RNAs and binds to the L7Ae protein, a core component of archaeal C/D box RNPs. This supports the notion that it might have an important but still unknown role in pre-rRNA biogenesis or might even target RNA molecules other than rRNA.
INTRODUCTION
In Archaea, introns are found either in tRNAs, generally within the anticodon loop and less commonly within the anticodon stem and the variable loop, or at diverse sites within the large rRNA (reviewed in 1,2). Splicing endonucleases involved in the excision of archaeal introns recognise and cleave a conserved secondary structure feature located at exon/intron boundaries in tRNA and rRNA, the bulge–helix–bulge (BHB) motif (2–5). The BHB motif consists of two 3-nt bulges on opposite strands of an RNA, separated by a helix of 4 bp. The splicing endonuclease cleaves at symmetrical positions within each of the 3-nt bulges present on the same minor groove face of the central 4-bp helix of the BHB motif, resulting in 2′,3′-cyclic phosphates and 5′-OH ends. Cleavage is followed by ligation of the ends of the exons and circularisation of the intron. However, up to now, no archaeal RNA ligase catalysing this reaction has been isolated. The solution structure of the BHB motif has been determined by NMR spectroscopy (6) and the crystal structure of an Archaeoglobus fulgidus splicing endonuclease elucidated (7). It was shown that the conformation of the two 3-nt bulges of the BHB motif is stabilised by stacking interactions between bulged nucleotides and bases in the adjacent Watson–Crick helices (6) and that the archaeal splicing endonuclease acts as a symmetrical dimer in the cleavage reaction (7).
The archaeal splicing endonuclease is believed to have a more general processing function, because its hallmark BHB recognition motif is also found at the cleavage sites within the long processing stems surrounding 16S and 23S rRNAs in the archaeal primary rRNA transcript (1,2,8–10). In vivo analysis of rRNA processing intermediates shows that these BHB motifs are used for excision of pre-16S or pre-23S rRNA from the rRNA operon primary transcript (10–12).
We have recently set out to directly identify all small non-mRNA species (snmRNAs) and their genes from various model organisms, an approach for which we have coined the term ‘RNomics’ (13–15). Among Archaea with a completely sequenced genome, we have selected A.fulgidus and Sulfolobus solfataricus as representatives of the major archaeal kingdoms Euryarchaeota and Crenarchaeaota, respectively, and have generated cDNA libraries from these species encoding small, stable RNA molecules sized from 50 to 500 nt. Analysis of these libraries has shed new light on the rRNA processing steps associated with cleavage at the BHB motif, suggesting that these reactions might be more complex than anticipated. From the A.fulgidus library we identified numerous identical cDNA clones encoding a small RNA species, designated 16S-D RNA, which mapped to the 5′-flanking region of the 16S rRNA gene as well as to the 16S–23S intergenic spacer, but lacked the intervening pre-16S rRNA sequence. Its sequence predicts that excision of the pre-16S rRNA from an archaeal primary transcript within the BHB motif is accompanied by religation of the 5′- and 3′-flanking RNA spacers. Interestingly, the 16S-D RNA contains structural motifs found in archaeal C/D box small RNAs and binds to the L7AE protein, a core component of archaeal C/D box RNPs. In addition, we present evidence that the remaining cleavage products at BHB motifs at the boundaries of pre-16S and pre-23S rRNAs from A.fulgidus and S.solfataricus are also ligated. These observations, which imply formation of circular pre-16S and pre-23S rRNA intermediates, reveal a further link between specific steps of archaeal rRNA processing and the splicing of archaeal introns.
MATERIALS AND METHODS
Materials
Dimethylsulphate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulphonate (CMCT) were obtained from Fluka AG (Switzerland) and kethoxal (KE) was purchased from Upjohn (UK); all other reagents and enzymes were purchased from Roche (Germany).
Identification of novel RNA species
We prepared total RNA from A.fulgidus and S.solfataricus by the TRIzol method (Gibco BRL). Total RNA was subsequently fractionated on a denaturing 8% polyacrylamide gel (7 M urea, 1× TBE buffer). RNAs in the size range of ~50–500 nt were excised from the gel, passively eluted and ethanol precipitated. Subsequently, 5 µg RNA was tailed with CTP using poly(A) polymerase as described previously (16). RNAs were reverse transcribed into cDNAs using primer GIBCO1 and cloned into vector pSPORT1 employing the Gibco Superscript system (Gibco BRL). cDNAs were amplified by PCR using primers FSP and RSP. PCR products were spotted by robots in high density arrays on filters (17), performed at the Resource Center of the German Human Genome Project (Berlin, Germany).
DNA sequencing and sequence analysis
We sequenced cDNA clones encoding snmRNAs using appropriate primers and the BigDye terminator cycle sequencing reaction kit (PE Applied Biosystems). We analysed sequences on an ABI Prism 3700 sequencer using the LASERGENE sequence analysis program package.
Northern hybridisation
Total RNA from either A.fulgidus or S.solfataricus was separated on 8% denaturing polyacrylamide gels (7 M urea, 1× TBE buffer) and transferred onto a nylon membrane (Ambion plus) using a Bio-Rad Trans-blot SD semi-dry blotting apparatus. The RNA was immobilised on membranes in a Stratagene crosslinker. Prehybridisation was carried out at 58°C in 1 M sodium phosphate buffer, pH 6.2, with 7% SDS for 1 h. Oligonucleotides complementary to respective RNA sequences were end-labelled with [γ-32P]ATP and T4 polynucleotide kinase for 1 h at 37°C. Hybridisation was carried out at 58°C for 12 h in 1 M sodium phosphate buffer, pH 6.2, 7% SDS. Membranes were washed twice at room temperature for 15 min in wash buffer I (2× SSC, 0.1% SDS), followed by a final wash for 15 min at room temperature in wash buffer II (0.1× SSC, 0.1% SDS). Membranes were exposed to Kodak MS-1 film for 1–3 days.
Determination of 5′-ends of RNAs by primer extension analysis
Detection of the mature ends of RNAs by primer extension was performed according to a standard procedure (18) with minor modifications. Briefly, 10 µg total RNA was hybridised to 0.5 pmol purified 5′-end-labelled primer (see below) in 4.5 µl of 1× buffer containing 50 mM HEPES–KOH, pH 7.0, 100 mM KCl. RNAs were first denatured at 90°C for 2 min and annealed to respective oligonucleotides by slow cooling to 42.5°C. Reverse transcription was performed for 1 h at 42.5°C in a total volume of 8.5 µl containing 0.5 mM dNTP, 10 mM MgCl2, 10 mM DTT, 50 mM Tris–HCl pH 8.5, 60 mM KCl and 2.2 U AMV reverse transcriptase (Roche). Samples were loaded onto 8% (w/v) polyacrylamide, 7 M urea gels. Electrophoresis was performed at 1600 V, 25 mA for 2 h.
Synthesis of DNA templates for in vitro transcription of RNAs
Synthesis of the A.fulgidus 16S-D DNA template. DNA fragments encoding the 16S-D RNA fragment were generated by RT–PCR using 5 µg total RNA from A.fulgidus and oligonucleotides 58.2 and 5′SMALLRNA. The cDNA was cloned into vector pCR2.1 (TA Cloning Kit, Invitrogen); the resulting plasmid is designated pCR2.1-16S-D. For in vitro transcription of 16S-D RNA a PCR product amplified from vector pCR2.1-16S-D with oligonucleotides T7+5′SMALLRNA and 3′SMALLRNA was used as template.
Synthesis of the S.solfataricus 16S-D (stems I–III) DNA template. DNA fragments from 16S-D RNA encoding stems II and III were generated by PCR using plasmid pSS5 E08 (obtained from the cDNA library) and oligonucleotides Sso16SF 5′.1 and Sso23SF1.1. Plasmid pSS5E08 contained stems II and III from the 16S-D RNA as well as stems IV, V and VI from 23S-D RNA (see Fig. 3) of S.solfataricus. A DNA fragment encoding stem I of 16S-D RNA was generated by PCR using a genomic DNA template from S.solfataricus and oligonucleotides Sso16S 5′-2 and 5′ Sso16S low F. For generation of the full-length DNA template encoding stems I, II and III of 16S-D RNA from S.solfataricus, a PCR amplification was performed containing both DNA fragments encoding stems I or II and III (see above) and oligonucleotides Sso16S T7&5′-2 and Sso23SF1.1.
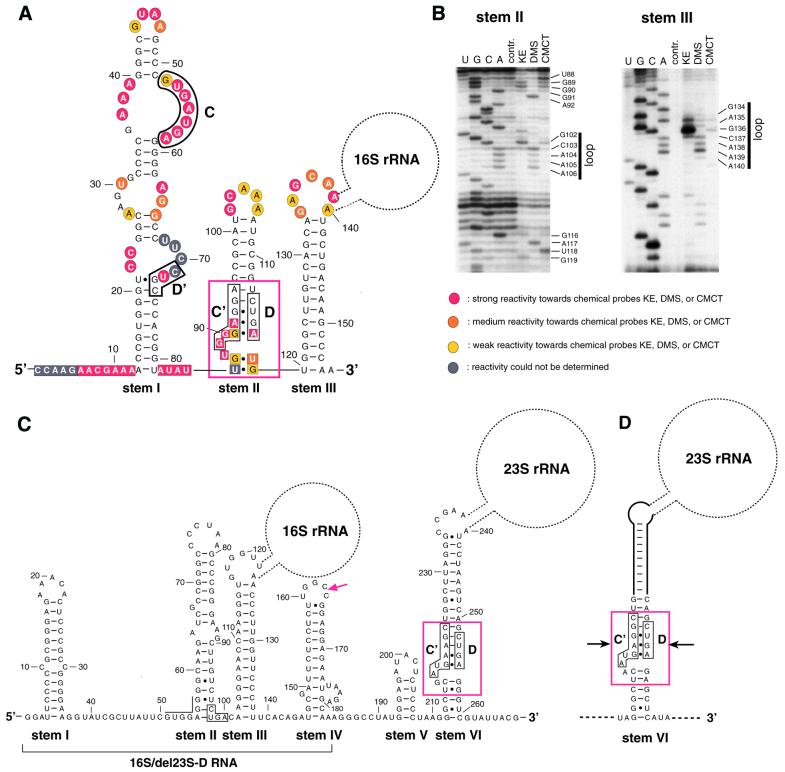
Figure 3.
Structure analysis of 16S-D RNA from A.fulgidus (A and B) and 16S-D/23S-D RNAs from S.solfataricus (C) and S.acidocaldarius (D). (A) Structure prediction of 16S-D RNA by the mfold program and chemical probing data as deduced from (B). Indicated are the C, D′, C′ and D boxes of 16S-D RNA also observed in canonical archaeal C/D box snoRNAs (26,27). The structural hallmark of C/D RNAs, a K-turn motif (29–31) formed by C′/D box apposition, is denoted by a red box. Bases found to be strongly, medium or weakly reactive to chemical probes are shown by red, orange and yellow circles and squares, respectively. Bases whose reactivity could not be determined are indicated by grey circles and squares. The location of the splice site in 16S-D RNA (between positions 139 and 140), including the location of the 16S rRNA, is also shown. (B) Autoradiograms of chemical probing analysis of stems II and III from 16S-D RNA of A.fulgidus. U, C, G, A, sequencing lanes; contr., control lane, no chemical probes added; KE, DMS, CMCT, respective chemical probe added. (Left) The reactive bases in the single-stranded regions in between stems and reactive bases in loop regions of stems are shown. (C) Computer modelling of S.solfataricus 16S/23S-D RNA. The conserved box D motif at the base of stem II is boxed (nucleotide positions corresponding precisely to the box C′ motif present in A.fulgidus are overlined). The red arrowhead denotes the 3′-end of the in vitro synthesised transcript, designated 16S/del23S-D RNA, tested in the Figure 4D gel shift assay. The K-turn at the base of the pre-23S processing stem is delineated by a red box (boxes C′ and D are boxed by black lines). (D) The presumptive L7AE recognition motif (indicated by a red box) at the base of the pre-23S rRNA processing stem of S.acidocaldarius pre-rRNA. Boxes C′ and D are shown by black lines. The previously proposed processing stem (8) extends below the positions denoted by arrows.
Synthesis of the S.solfataricus 16S-D/23S-D (stems I–VI) DNA template. The DNA fragment encoding the full-length 16S/23S-D RNA from S.solfataricus (stems I–VI, see Fig. 3) was generated in two steps. First, a DNA fragment containing stems IV–VI of 23S-D RNA was amplified from plasmid pSS5 E08 (see above) using oligonucleotides Sso16SF 5′.1 and Sso23S.F4. Subsequently, the DNA fragments generated from this reaction and from the 16S-D (stems I–III) DNA template (see above) were used as templates for PCR amplification employing oligonucleotides Sso16S T7&5′-2 and Sso23S.F4. This generated a DNA template containing the full-length 16S/23S-D RNA (stems I–VI).
Synthesis of the sR6 snoRNA template from Pyrococcus abyssi. The DNA fragment containing the coding region for sR6 snoRNA from P.abyssi was generated by PCR amplification using genomic DNA and oligonucleotides P.ab-T7 and P.ab-3′. In all templates a T7 promotor sequence was introduced at the 5′-end by the use of respective oligonucleotides. All PCR products were sequenced prior to in vitro transcription.
T7 in vitro transcription of RNAs
RNAs were in vitro transcribed from DNA templates containing a T7 promoter. Transcription, using T7 RNA polymerase, was performed directly from PCR-amplified DNA fragments (15 µg) in a reaction volume of 400 µl (19). RNAs were purified by electrophoresis on denaturing 8% polyacrylamide, 7 M urea gels and passively eluted from the gels in 0.3 M sodium acetate buffer, pH 5.2, 1 mM EDTA, 0.2% phenol. Subsequently, RNAs were ethanol precipitated and dissolved in 30 µl of H2O.
Chemical probing
Prior to probing, RNAs were heat denatured (at 95°C for 1 min) and renatured by cooling at 4°C for 5 min. Chemical probing of 16S-D RNA from A.fulgidus was performed as reported (18), by addition of 1.5 µl of DMS (1:10 dilution in 95% ethanol), 5 µl of KE (1:5 dilution of a 37 mg/ml stock solution in H2O) or CMCT (84 mg/ml in H2O) to the RNA (10–15 pmol) in a 50 µl reaction volume and incubation at 30°C for 20 min. Reactions were performed in the presence of 100 mM KCl, 7 mM MgCl2 and either 50 mM sodium cacodylate, pH 7.2, (for probing with KE or DMS) or 50 mM sodium borate, pH 8.3, (for probing with CMCT). All reactions were stopped by the addition of 300 µl of ethanol followed by rapid mixing. Prior to addition of ethanol, 25 µl of DMS stop solution (1 M Tris–HCl, pH 7.5, 1 M 2-mercaptoethanol, 0.1 M EDTA) was added to the DMS reactions and KE-modified samples were adjusted to 25 mM sodium borate, pH 7.2. The pellets were resuspended in 200 µl of 0.3 M sodium acetate, pH 5.2, and extracted once with phenol and chloroform. After precipitation, pellets were dissolved in 10 µl of H2O. Primer extension reactions for detection of chemical modifications were performed using 5′-32P-end-labelled primers (18). Samples were loaded onto 8% polyacrylamide, 7 M urea gels. Electrophoresis was performed at 1600 V, 25 mA for 2 h. Probing experiments for 16S-D RNA were repeated five times and shown to be highly reproducible.
Folding prediction of RNA structures
The secondary structures of 16S-D RNA from A.fulgidus and S.solfataricus were folded using mfold v.3.0 by Zuker and Turner (Department of Mathematical Sciences, Rensselaer Polytechnic Institute, Troy, NY).
Purification of L7AE protein from P.abyssi
A DNA fragment containing the coding region of L7AE was generated by PCR amplification using genomic DNA from P.abyssi strain GE5 (kindly provided by Hannu Myllykallio, IGM, Orsay, France) and primers L7AE-5′ and L7AE-3′. The PCR primers introduced a BamHI site at the 5′-end and a XhoI site at the 3′-end of the amplified fragment. The fragment was introduced between the BamHI and XhoI sites of plasmid pGEX-6P-1 (Pharmacia), a GST fusion expression vector. The recombinant GST–L7AE protein was purified from Escherichia coli BL21 cells using glutathione–Sepharose 4B as instructed by the manufacturer (Pharmacia). Cleavage in solution of the fusion protein from GST was performed overnight at 4°C, using 80 U Precission protease (Pharmacia) per ml glutathione–Sepharose bed volume from which the fusion protein was eluted. L7AE has five additional amino acids beyond the GST cleavage site. The mixture was then placed for 10 min at 65°C and microcentrifuged for 15 min at 13 000 r.p.m. After dialysis of the supernatant in buffer D (20 mM HEPES pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol) the samples were concentrated using ultrafiltration (Microcon YM-3) and stored at –20°C.
Electrophoretic mobility shift analysis
All in vitro transcripts, labelled by incorporation of [α-32P]CTP (800 Ci/mmol), were heat denatured (1 min at 90°C) and cooled at room temperature for 20 min before incubation with the recombinant L7AE protein. Aliquots (0.1 pmol) of each transcript were incubated (at 4°C for 20 min) with increasing concentrations of L7AE protein as reported (20), in a final volume of 10 µl of buffer A (20 mM HEPES–KOH, pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol) in the presence of 20 µg E.coli tRNA (Sigma) and 20 U RNase inhibitor (Promega). RNA and RNA–protein complexes were subsequently separated on a native 8% (19:1) polyacrylamide gel containing 0.5× TBE and 5% glycerol. Electrophoresis was performed at 4°C for 5.5 h at 300 V in 0.5× TBE running buffer containing 5% glycerol and the results visualised by autoradiography.
In vitro mobility shift assays were performed in the presence of a large excess of protein as compared to input RNA. Under these conditions, the apparent Kd of the protein–RNA complex corresponds to the protein concentration for which 50% of the input RNA is shifted to an RNP complex. This concentration was estimated by comparison of the shifted/unshifted RNA ratios for the two closest protein concentrations tested around this value.
Oligonucleotides
All oligonucleotides were synthesised by MWG Biotech (Ebersberg, Germany). Oligonucleotides used for generation of cDNA libraries by RT–PCR and amplification of vector inserts: GIBCO1, 5′-GACTAGTTCTAGATCGCGAGC(G15)-3′; FSP, 5′-GCTATTACGCCAGCTGGCGAAAGGGGGATGTG-3′; RSP, 5′-CCCCAGGCTTTACACTTTATGCTTCCGGCTCG-3′. Oligonucleotides used for generation of DNA templates for T7 transcription, chemical probing and primer extension analysis: T7+5′SMALLRNA, 5′-TAATACGACTCACTATAGGCCAAGAACGAAAACCGTGGGTC-3′; 5′SMALLRNA, 5′-CCAAGAACGAAAACCGTGGGTC-3′; 3′SMALLRNA, 5′-TTGGGCTTGTCAGCATATGAGCCC-3′; Ar16sPriExt, 5′-GGGCTTGTCAGCATTTGCTCTGCTGACA-3′; SSo16S-Down, 5′-ACCAAGGGTTAACCACACCC-3′; Sso16SF 5′.1, 5′-GCTTATTCGTGGAGGGGCAAGATCGCCGGGCCCCTAAGC-3′; SSo23SF1.1, 5′-GCCAAGAGGAGAATCGCATC-3′; Sso16S 5′-2, 5′-GGATTCCCCCGCGGGAGAAACAC-3′; 5′ Sso16S low F, 5′-GGTTCCCTTGTCAGAGACAAGCTTCC-3′; Sso16S T7&5′-2, 5′-TAATACGACTCACTATAGGATTCCCCCGCGGGAGAAACAC-3′; SSo23SF1.1, 5′-GCCAAGAGGAGAATCGCATC-3′; SSo23S.F4, 5′-CGTAATACGACCCTCAGCTGA-3′; P.ab-T7, 5′-TCTAATACGACTCACTATAGGGTGAGGAATGATGAGATTTGC-3′; P.ab-3′, 5′-AAAATAGTAAAAGGTCAGTAAC-3′. Oligonucleotides used for northern blot analysis of ligated RNAs species: AF23S-Down, 5′-CGTCCCCAATTTGCATTTGG-3′; Afu 16S Down, 5′-TGTCAGCATTTGCTCTGCTG-3′; tRNAAla 1, 5′-TGGACTCGGCGGATTTGAACC-3′; SSo16SD/F20-5, 5′-GGGTTAACCACACCCTTGGT-3′; SSo23SD/F20-5, 5′-AGGATTTCGGCCCTAAGCCA-3′. Oligonucleotides used for PCR amplification of the P.abyssi L7AE coding region: L7AE-5′, 5′-GGATCCATGGAGGGATGGATGATGG-3′; L7AE-3′, 5′-CTCGAGTCACTTCATGAGCTCCCTA-3′.
RESULTS
Identification of cDNAs reflecting RNA ligation at the BHB motif of pre-16S and pre-23S rRNAs in A.fulgidus
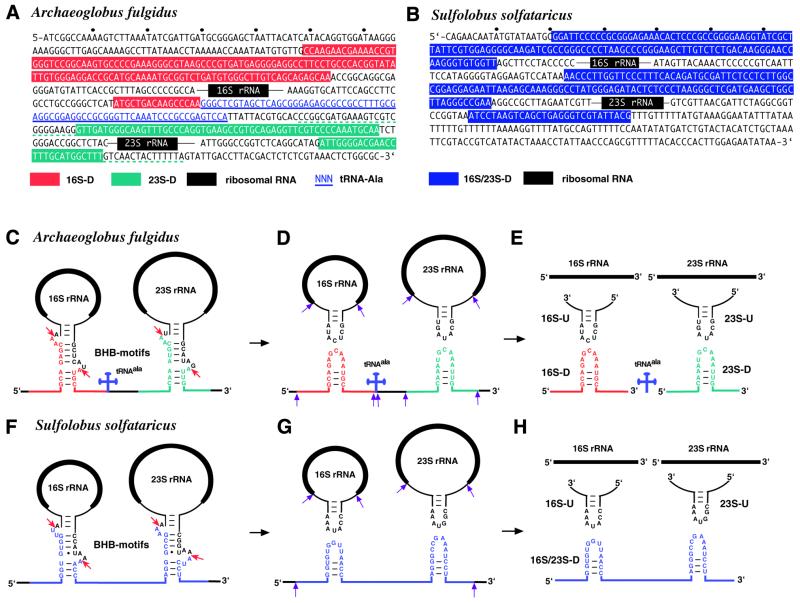
From a cDNA library encoding snmRNA species from the archaeon A.fulgidus we performed sequence analysis of about 2000 cDNA clones, pre-selected as representing novel snmRNAs (15; T.-H.Tang J.-P.Bachellerie, H.Huber, M.Drungowski, T.Elge, J.Brosius and A.Hüttenhofer, in preparation). Subsequently, sequences were analysed by a BLASTN database search for novel unannotated RNA species. Thereby, the most abundant cDNA molecule in A.fulgidus (150 cDNA clones, i.e. 7% of all candidates for snmRNA clones analysed) was derived from a bipartite RNA species mapping to the 5′ external transcribed spacer region (5′-ETS) as well as the 3′ internal transcribed spacer (3′-ITS) of the ribosomal 16S/23S rRNA operon (Fig. 1A and C–E). The corresponding 155-nt-long RNA species (as detailed below) was designated 16S-D. Surprisingly, this RNA lacked the entire intervening 16S pre-rRNA sequence, indicative of a splicing event. Examination of the long stem structures surrounding the 16S rRNA in the precursor transcript indicates that the two RNA fragments fused together correspond exactly to the leader/spacer products predicted to result from pre-16S rRNA excision by the BHB endonuclease (Fig. 1A and C–E). The presence of the 16S-D RNA species can therefore be explained by post-transcriptional religation of the 5′ leader and 3′ spacer following cleavage within the BHB motif.
Figure 1.
Ligated RNA species from processing stems of the ribosomal operon of A.fulgidus (A and C–E) and S.solfataricus (B and F–H). (A) Sequence of the A.fulgidus rRNA operon. Black boxes indicate mature rRNAs and thin black lines depict spacer sequences flanking mature rRNAs in pre-16S and pre-23S rRNAs. tRNAAla is in blue letters and underlined. Bipartite sequences of the ligated 16S-D RNA and 23S-D RNA species, delineated by analysis of the cDNA clones and northern blot and primer extension assays, are highlighted in red and green, respectively, with the terminal heterogeneity observed for 23S-D cDNA clones denoted by dotted green underlining. (B) Sequence of the S.solfataricus rRNA operon. The sequence of the ligated 16S/23S-D RNA species is highlighted in blue; the other symbols are as in (A). (C–H) Proposed pre-rRNA processing pathway in A.fulgidus (C–E) and S.solfataricus (F–H). Mature rRNAs are represented by thick black lines, precursor sequences by thin lines. BHB motifs are shown within processing stems of rRNAs; the cleavage sites within BHB motifs are indicated by a red arrow. Further processing steps of ligated RNA species are shown by purple arrows. Other symbols are as in (A) and (B).
Religation of pre-rRNA fragments generated by endonucleolytic cleavage at BHB motifs reveals an additional resemblance to the splicing of archaeal introns. In the archaeal splicing reaction, endonucleolytic cleavage is followed by two ligations within the BHB motif, one resulting in ligation of the two exons and the other leading to circularisation of the intron (for a review see 2). Therefore, we examined whether the two remaining newly formed RNA ends located in the upper part of the BHB motif would also be subject to religation. In fact, we isolated seven cDNA clones from the A.fulgidus cDNA library encoding such a ligated RNA species, designated 16S-U RNA. This RNA species is likely to be derived from further processing of a presumably circular 16S pre-rRNA precursor species extending from the upper part of the BHB motif (Fig. 1C–E). Unlike those for 16S-D RNA, cDNA clones for 16S-U RNA species vary in size (41–65 nt).
The 23S rRNA precursor stem of A.fulgidus also contains a BHB motif cleaved by archaeal BHB endonucleases. By computer aided analysis, we screened clones from our cDNA library for the presence of ligated RNA species from the 23S pre-rRNA BHB motif. In fact, we identified five cDNA clones encoding ligated RNAs from the lower part of the BHB motif (23S-D RNA, Fig. 1A and C–E). Unlike the 16S-D species, cDNAs encoding the 23S-D RNA were heterogeneous in size, ranging from 68 to 116 nt. More importantly, they do not exhibit a defined 3′-end as observed for 16S-D RNA (Fig. 1A). It should be noted that the mature 5′-ends of RNAs from our cDNA library could not be determined by sequencing due to the library construction strategy by reverse transcription (see Materials and Methods; 15) and therefore had to be analysed by primer extension (see below).
As observed for the 16S BHB motif, we were also able to identify seven cDNA clones encoding a fusion product corresponding to the upper part of the BHB motif, designated 23S-U RNA. As noted for 16S-U RNA and the 23S-D species, the various 23S-U RNA clones differ considerably in size (29–212 nt) and do not exhibit defined 5′- or 3′-ends. We therefore consider it likely that they reflect the presence of relatively unstable rRNA processing intermediates resulting from further cleavage of a circular 23S pre-rRNA intermediate (see above).
Homologous cDNAs reflecting RNA ligation at the BHB motifs of S.solfataricus pre-rRNAs
Next, we wanted to investigate whether cDNA clones presumably reflecting religation in the processing stems of rRNAs following endonucleolytic cleavage at the BHB motifs could also be detected in another distantly related archaeon. Since A.fulgidus belongs to the archaeal kingdom Euryarchaeota, we set out to analyse a cDNA library from a representative of the other major archaeal kingdom Crenarchaeota, S.solfataricus. The canonical BHB motif in the S.solfataricus 16S pre-rRNA processing stem does correspond to a specific site of endonucleolytic cleavage in vitro (21). The S.solfataricus rRNA operon differs from that of A.fulgidus with respect to two features. First, its 16S and 23S rRNA processing stems are not interrupted by a tRNA gene. Secondly, S.solfataricus contains a non-canonical BHB motif within the 23S rRNA processing stem, with the presence of a 4 (instead of 3) nt lower bulge (Fig. 1B and F–H). Despite these differences, cDNA clones reflecting religation of all four BHB fragments from the processing stems of S.solfataricus 16S and 23S pre-rRNAs could be readily identified in our library (Fig. 1H). However, in contrast to A.fulgidus, no separate 16S-D and 23S-D clones were observed, but rather a continuous cDNA clone spanning both ligation sites (designated 16S/23S-D; Fig. 1H). Detection of clones spanning the ligation site at the BHB of the pre-23S processing stem strongly suggests that the S.solfataricus BHB endonuclease can tolerate an extra base in the lower bulge and still cleave within this loop (Fig. 1F).
Detection of the predicted ligated RNAs in A.fulgidus and S.solfataricus
To rule out the possibility that the fused cDNA clones could result from artefactual religation of two unlinked cDNA fragments during the construction of our library, we set out to detect the predicted ligated RNAs by northern blot analysis of total cellular RNA from both archaeal strains, using oligonucleotide probes bridging the presumptive fusion sites.
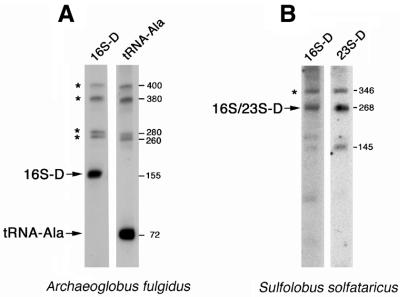
For A.fulgidus we could readily detect the predicted ligated RNA species 16S-D (Fig. 2A). However, we obtained only very weak hybridisation signals from the 23S-D, 16S-U and 23S-U RNA species (data not shown), which probably reflects a rapid turnover and low abundance of these RNA species, consistent with the small number of corresponding cDNA clones isolated in our library. The size of 16S-D RNA as deduced by northern blot analysis corresponds well with the size of the corresponding cDNA clones.
Figure 2.
Northern blot analysis showing expression of 16S-D RNA from A.fulgidus (A) and 16S/23S-D RNA from S.solfataricus (B). (A) Northern blot analysis using an oligonucleotide bridging the proposed fusion site in 16S-D RNA (16S-D) and an oligonucleotide probe specific for tRNAAla, located 3′ to the 16S-D RNA (tRNA-Ala). Mature RNAs (16S-D, tRNAAla) are indicated by arrows, precursor-RNAs by stars. (B) Northern blot analysis of 16S/23S-D RNA from S.solfataricus. Oligonucleotides bridging the 16S (left) or 23S (right) fusion sites were used as hybridisation probes. The mature 16S/23S-D RNA with a size of 268 nt is indicated by an arrow, the precursor RNA by a star. (Right) Length of RNAs as measured by an RNA length marker.
Due to the library construction strategy by reverse transcription followed by second stand synthesis using DNA polymerase I, the very 5′-ends of RNAs could not be cloned (15; T.-H.Tang J.-P.Bachellerie, H.Huber, M.Drungowski, T.Elge, J.Brosius and A.Hüttenhofer, in preparation). We therefore set out to determine the mature 5′-end of the 16S-D RNA species from A.fulgidus by primer extension with the bridging oligonucleotide spanning the fusion site. It was determined to be located at position –177 (+1 indicating the mature 5′-end of 16S rRNA) (Fig. 1A). The 16S-D RNA appears to be processed from a precursor transcript (Fig. 2A). To investigate its processing pathway we also used an oligonucleotide probe specific for tRNAAla which maps immediately downstream from the 16S-D RNA. Analysis of the hybridisation patterns indicates that 16S-D RNA and tRNAAla share a series of common precursors, of ∼400, 380, 280 and 260 nt (Fig. 2A). Since the 3′-end of 16S-D RNA immediately abuts the 5′-end of mature tRNAAla (Fig. 1A and C–E), it is tempting to speculate that separation of both RNA species results from cleavage due to RNase P (22).
For S.solfataricus, in agreement with the identification of cDNA clones corresponding to a continuous RNA species containing both 16S and 23S fusion sites (Fig. 1H), an identical band with the expected size of 268 nt was detected in the northern blot analysis using oligonucleotides bridging either the 16S-D or 23S-D fusion sites. As observed with A.fulgidus, the RNA appears to be processed from a larger precursor species of 346 nt (Fig. 2B). In the case of oligonucleotides derived from the 23S-D portion of the RNA (Fig. 1H), an additional band of 145 nt was observed, possibly reflecting a processing or degradation product from the larger 16S/23S-D RNA.
By primer extension (data not shown) the mature 5′-end of the 16S/23S-D RNA was found to be located at position –135 (+1 indicating the mature 5′-end of 16S rRNA) (Fig. 1B). As observed in A.fulgidus, only very weak hybridisation signals were detected for RNA species 16S-U and 23S-U (data not shown), indicative of a rapid turnover/degradation of these RNA species (see above).
Structure analysis of 16S-D RNA from A.fulgidus and 16S/23S-D RNA from S.solfataricus
On a thermodynamic basis (23), the very abundant A.fulgidus 16S-D RNA species is predicted to fold into a highly stable secondary structure consisting of three stems interrupted by two single-stranded regions (Fig. 3A). Strikingly, the 5′-portion of the 16S/23S-D RNA from S.solfataricus (Fig. 1H) exhibits a very similar three-stem structure, although the sequence is extensively divergent from its A.fulgidus counterpart (Fig. 3C).
We experimentally tested the folding prediction in the case of A.fulgidus by chemical probing of an in vitro synthesised RNA transcript using the base-specific probes KE, DMS and CMCT. These probes modify bases not involved in Watson–Crick interactions, e.g. bases located in single-stranded regions (18). In general, the chemical probing data strongly confirm the proposed secondary structure (Fig. 3A and B, data not shown for stem I). Bases predicted to be located in single-stranded regions appeared accessible, specifically bases in the loop regions and in between stems, whereas bases in the proposed stem regions were mostly not accessible to chemical probes (Fig. 3B). Putative secondary structures of archaeal rRNA operon primary transcripts previously derived on a comparative basis include two partially conserved stem–loop structures, termed helices A and B, defined by their location upstream from the pre-16S processing stem, within the portion of the 5′ external transcribed region spanned by A.fulgidus 16S-D RNA (1,24). While stem III in A.fulgidus 16S-D RNA precisely corresponds to the lower part of the pre-16S processing stem, stems I and II clearly represent equivalents of helices A and B in the A.fulgidus primary transcript.
Intriguingly, further examination of the A.fulgidus 16S-D RNA sequence revealed the presence of hallmark motifs, boxes C, D′, C′ and D in that order (Fig. 3A), also present in archaeal homologues of antisense box C/D snoRNAs which guide rRNA 2′-O-ribose methylation (25–28). However, we could not identify an antisense element matching a potential rRNA target site or other, non-ribosomal RNA targets at the appropriate location within A.fulgidus 16S-D RNA, i.e. over the ∼10 nt immediately upstream from box D or/and box D′. Moreover, the A.fulgidus 16S-D RNA departs from known archaeal box C/D methylation guides by the presence of long 5′ leader and 3′ trailer extensions, upstream from box C and downstream from box D, respectively, and by unusually long D′–C′ and C′–D intervals. Interestingly, in the experimentally derived secondary structure of A.fulgidus 16S-D RNA boxes C′ and D are brought into close proximity to each other at the base of stem II (boxed in Fig. 3A), so as to form the novel RNA fold termed a kink-turn (K-turn), which includes an asymmetrical, internal 3-nt loop flanked on one side by two tandem sheared G·A base pairs (29–32).
Unlike A.fulgidus 16S-D RNA, the 5′-portion of S.solfataricus 16S/23S-D RNA does not exhibit box C and D′ motifs within stem I. Although the box D motif is conserved at an identical location at the base of stem II (Fig. 3C), the matching sequence on the opposite RNA strand substantially departs from a box C/C′ consensus (Fig. 3C, indicated by overlining). As a consequence, neither the two sheared G·A base pairs of the K-turn motif nor the other stem structure delineating an adjacent, asymmetrical 3-nt loop can form at this site in S.solfataricus. Finally, the 10-nt sequence tract upstream from this box D motif in S.solfataricus 16S/23S-D RNA seems unrelated to its counterpart in A.fulgidus 16S-D RNA, ruling out the possibility that it corresponds to an antisense element targeting a common conserved RNA in both species.
Analysis of the interaction of the ligated RNAs with L7AE protein by gel shift analysis
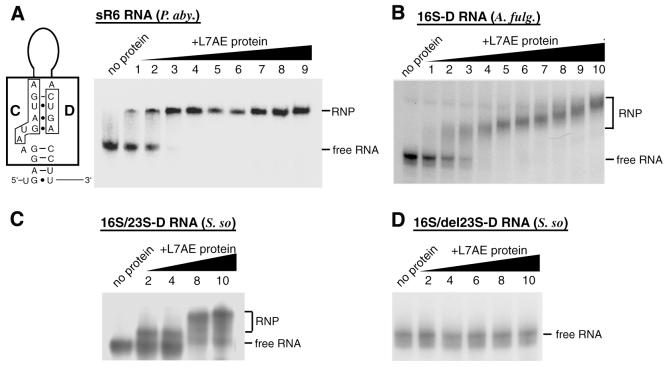
Human box C/D snoRNAs interact directly and specifically with the human 15.5 kDa protein (30). To further assess the biological significance of the presumptive C, C′, D′ and D motifs in A.fulgidus 16S-D RNA and test the notion that this RNA belongs to the family of C/D RNAs, we investigated whether the in vitro transcribed ligated RNA would bind in vitro an archaeal homologue of the human 15.5 kDa protein. Ribosomal protein L7AE is the archaeal protein exhibiting the most significant homology with human 15.5 kDa protein (35% identity between the human protein and P.abyssi L7AE). The archaeal protein interacts with a K-turn in mature 23S rRNA, as revealed by analysis of the Haloarcula marismortui large ribosomal subunit (29).
First, we set out to test the ability of the archaeal protein to selectively recognise a bona fide archaeal C/D RNA guide for rRNA 2′-O-methylation. We selected previously reported P.abyssi sR6 as a reference (26) and examined whether, in the presence of a vast excess of non-specific E.coli tRNA (molar ratio 7400:1), selective binding between the recombinant P.abyssi L7AE protein and in vitro transcribed P.abyssi sR6 could be detected in a gel retardation assay. A complex was indeed formed, with an apparent Kd of 150 nM (Fig. 4A). We next tested whether in vitro transcribed A.fulgidus 16S-D RNA in the presence of a vast excess of non-specific E.coli tRNA (see above) would bind to P.abyssi L7AE protein. As expected, we observed a gel shift reflecting formation of a specific complex, with an apparent Kd value of 400 nM (Fig. 4B). Notably, a decreased mobility of the L7AE–16S-D RNA complex with increasing protein concentration was observed (Fig. 4B). Interestingly, in a previously reported gel shift experiment involving eukaryotic box C/D snoRNAs and the human 15.5 kDa protein (the eukaryotic homologue of L7AE protein), a very similar decrease in RNP complex mobility with increasing protein concentration was observed (30). This might reflect a conformational change in the RNA induced by the protein rather than binding of several protein monomers to the RNA.
Figure 4.
Interaction of the ligated RNA species with L7AE protein detected by electrophoretic mobility shift assay and structure of the C/D box motif within the sR6 snoRNA from P.abyssi. In vitro transcribed, 32P-labelled RNAs were incubated with increasing concentrations of recombinant P.abyssi L7AE protein and the resulting complexes resolved on a native 8% polyacrylamide gel. Left lanes, no protein. (A) (Left) Structure of the C/D box motif located within sR6 snoRNA from P.abyssi; (right) control mobility shift assay with P.abyssi sR6 snoRNA. In lanes 1–9, the sR6 snoRNA was incubated with L7AE protein at concentrations of 0.05, 0.1, 0.25, 0.50, 0.75, 1, 1.25, 1.5 and 2 µM, respectively. (B) Interaction of A.fulgidus 16S-D RNA with the L7AE protein. (C) Interaction of S.solfataricus 16S/23S-D RNA with the L7AE protein. (D) Interaction of the 16S-D domain (as delineated in Fig. 3C, red arrow) of S.solfataricus designated 16S/del23S-D RNA with the L7AE protein. (B–D) In lanes 1–10, incubations were performed with L7AE protein concentrations of 0.21, 0.28, 0.42, 0.56, 0.84, 1.125, 1.7, 2.25, 3.375 and 4.5 µM, respectively.
When the S.solfataricus 16S/23S-D RNA was assayed under the same conditions, a gel shift was also observed, corresponding to an apparent Kd value of ∼500 nM (Fig. 4C). This result was unexpected since the box D motif at the base of stem II in S.solfataricus 16S/23S-D RNA cannot be part of a K-turn motif as mentioned above, unlike its counterpart in A.fulgidus 16S-D RNA. We observed, however, that S.solfataricus 16S/23S-D RNA does indeed contain a canonical K-turn, but the motif is located within its 23S-D domain, instead of the 16S-D domain, at the base of the pre-23S processing stem (Fig. 3C, stem VI). In line with these observations, an in vitro synthesised transcript restricted to the 16S-D domain of S.solfataricus, designated 16S/del23S-D RNA, containing stems I–III only (Fig. 3C, indicated by a red arrow), did not the bind the L7AE protein in the gel retardation assay (Fig. 4D). Remarkably, A.fulgidus 23S-D RNA does not contain a presumptive L7AE recognition motif at the base of its pre-23S processing stem. Instead, the corresponding nucleotides form Watson–Crick base pairs which extend the pre-23S processing stem (data not shown). However, the K-turn motif detected in S.solfataricus is conserved in another Sulfolobus representative, Sulfolobus acidocaldarius, based on a previously reported pre-rRNA secondary structure (Fig. 3D) (8).
DISCUSSION
Our data point to an additional link between splicing of archaeal introns and endonucleolytic cleavages within the long processing stems of archaeal pre-rRNA which give rise to pre-16S and pre-23S rRNAs. They also point to an intriguing but still elusive role for a specific portion of the 5′-ETS which participates in a novel RNA ligation process and appears to represent a novel, atypical member of the box C/D family of archaeal small RNAs which usually guide rRNA 2′-O-methylation (26–28).
Like the BHB at archaeal splice sites, the BHB motif at the boundaries of both pre-16S and pre-23S archaeal rRNAs is also endonucleolytically cleaved at symmetrical positions within each of two 3-nt loops, on opposite strands of the motif and separated by a 4-bp helix (1,10,11) (Fig. 1C–E). The novel ligated A.fulgidus RNA species, 16S-D, 16S-U, 23S-D and 23S-U, all precisely reflected ligation products derived from the expected cleavage sites at each pre-rRNA BHB (Fig. 1E). As for S.solfataricus, which exhibits a 4-nt (instead of 3-nt) lower bulge at the pre-23S rRNA boundaries, inspection of the ligated RNA sequences reveals that cleavage within the non-canonical bulge must occur 2 nt from the central 4-bp stem, i.e. as observed for a canonical bulge. This implies that the endonuclease recognising the lower bulge ‘counts’ from the central 4 bp and not from the adjoining lower stem.
In Archaea, tRNA and rRNA introns are ligated following cleavage within the BHB motif, resulting in circularised RNA species (for reviews see 2,9). Religation within cleaved BHB motifs of the pre-rRNA spacers flanking the pre-16S and pre-23S rRNAs, together with transient formation of circular forms of pre-16S and pre-23S rRNA (Fig. 1C–E), reveals an additional link between rRNA processing and RNA splicing in the two major archaeal kingdoms, Euryarchaeota and Crenarchaeota. Intriguingly, the pre-16S and pre-23S rRNA processing intermediates appear under these conditions as formal equivalents of introns spliced from the rRNA operon transcript. While the archaeal BHB ligase involved in splicing has not yet been identified, our data suggest that the same enzyme could participate in archaeal pre-rRNA processing. It is tempting to speculate that the tight linkage of cleavage and ligation in vivo, in both splicing and pre-rRNA processing, reflects a close physical association of the endonuclease and RNA ligase. In addition, the heterogeneous short length and low number of clones for both the 16S-U and 23S-U RNAs suggests that formation of circular pre-16S and pre-23S rRNA intermediates is rapidly followed by endonucleolytic cleavage within the rRNA spacer sequences and subsequent exonucleolytic trimming (Fig. 1C–E). In line with this notion and consistent with the well-known rapid degradation of all other types of spacer segments processed from the rRNA operon transcript, not only in Archaea but also in Eukarya and Bacteria, only weak hybridisation signals were detected in northern blot analyses for the corresponding processing intermediates in both A.fulgidus and S.solfataricus (data not shown). This is in marked contrast to accumulation of the 16S-D RNA species in A.fulgidus (see below).
We find the A.fulgidus 16S-D species is processed from a larger ligated RNA precursor, which also includes tRNAAla located between the pre-16S and pre-23S rRNAs in the rRNA operon primary transcript (Fig. 2A). Consistent with the notion that processing of the ligated precursor largely depends on RNase P cleavage (22) at the 5′-terminus of the tRNAAla sequence, a single continous RNA harbouring both the 16S-D and the 23S-D ligation sites could be identified in S.solfataricus, which lacks a tRNA gene in the 16S–23S pre-rRNA spacer (Fig. 1B and F–H). A continuous RNA harbouring both the 16S-D and 23S-D ligation sites also seems likely to form transiently in A.fulgidus (Fig. 1D), but we suspect that its detection is hampered by rapid processing of its 23S-D moiety, in addition to cleavage by RNase P.
We analysed in more detail structural features of the 16S-D RNA species which represented by far the most abundant snmRNA from our cDNA library from A.fulgidus. Particularly intriguing in this ligated RNA is the presence of the hallmarks of archaeal C/D box snoRNA, namely the C, D′, C′ and D boxes in that order (for reviews see 25,28). The 16S-D RNA species is substantially larger than known archaeal C/D small RNAs guiding rRNA methylation (26–28). This enlargement is found in the 5′ leader and 3′ trailer sequences, upstream from box C and downstream from box D, respectively. Moreover, the folding pattern derived for A.fulgidus 16S-D RNA (Fig. 3) by chemical probing analysis seemed to rule out formation of the characteristic structural motif of eukaryotic box C/D snoRNAs involving boxes C and D (30,31). This motif, termed a K-turn (29), in which an asymmetrical, 3-nt internal loop is flanked by a regular stem on one side and two non-Watson–Crick, sheared G·A base pairs on the other, is recognised by the homologue of human 15.5 kDa protein and yeast Snu13p (30,31). However, box C′ can substitute for box C in formation of a canonical K-turn at the base of stem II in A.fulgidus 16S-D RNA.
Consistent with this idea, by gel shift analyses we could demonstrate that A.fulgidus 16S-D RNA binds specifically to the purified P.abyssi L7AE protein, the archaeal homologue of the human 15.5 kDa and yeast Snu13 proteins (30). Preliminary experiments involving immunoprecipitation from partially purified cell extracts indicate that the L7AE protein represents a core component of archaeal C/D small RNPs (Omer and Dennis, Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, Canada, personal communication). Interestingly, the apparent Kd we have measured for the heterologous L7AE/16S-D RNA complex is only slightly higher than that of a homologous complex between L7AE and a bona fide archaeal C/D small RNA in P.abyssi (Fig. 4). Taken together, these data support the notion that 16S-D RNA does correspond to a novel, atypical member of the family of archaeal C/D small RNAs. Remarkably, a L7AE-binding site is conserved in the crenarchaeon S.solfataricus 16S/23S-D RNA, but its location differs dramatically from that of the 16S-D RNA site in the euryarchaeon A.fulgidus (Fig. 3C), indicating that different strategies have been used in the two distantly related Archaea to provide for a L7AE protein-binding site in the religated rRNA spacers.
However, the absence of any bona fide rRNA antisense element and its atypical structure does not provide a hint as to the potential role of 16S-D RNA. In eukaryotes, several C/D snoRNAs devoid of rRNA antisense elements have been reported (14,15,33–37). While some of them are able to guide methylation of RNA targets other than rRNA, others so far remain without any proposed role and a third subset of them, including snoRNAs U3, U8 and U22, which all lack the typical 10–20-nt-long rRNA complementarities, are essential for rRNA processing, although some of the mechanisms involved remain elusive (38).
In this context it is interesting to note that a portion of the 5′-ETS that maps within the structural counterpart of 16S-D has been proposed to represent a functional equivalent of eukaryotic U3 snoRNA in the archaeon Halobacterium cutirubrum (39). It was noted that this portion had potential to base pair with its cognate 16S rRNA resulting in structures that are similar to those proposed between U3 and the small subunit rRNA (40). However, close inspection of the A.fulgidus and S.solfataricus pre-rRNA sequences clearly indicates that the potential for bipartite intramolecular interaction between the 5′-ETS and the mature 16S rRNA sequence which was proposed in H.cutirubrum is not conserved in these two additional Archaea. Moreover, our finding that 16S-D RNA exists as a stable, separate entity after excision from the rRNA operon transcript suggests it might function in trans, rather than in cis. The present work sets the stage for further study of the novel 16S-D RNA, particularly with the aid of combined computer/phylogenetic analyses of completely sequenced archaeal genomes to test the possibility that it might target a non-rRNA RNA species.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the German Human Genome Project through the BMBF (no. 01KW9966) to J.B. and A.H., by an IZKF grant (Teilprojekt IKF G6, Münster) to A.H. and by laboratory funds from the Centre National de la Recherche Scientifique and Université Paul Sabatier, Toulouse, and by a grant from the Ministère de l’Education Nationale, de la Recherche et de la Technologie (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires, 2001–2002) to J.P.B.
REFERENCES
- 1.Garrett R.A., Dalgaard,J., Larsen,N., Kjems,J. and Mankin,A.S. (1991) Archaeal rRNA operons. Trends Biochem. Sci., 16, 22–26. [DOI] [PubMed] [Google Scholar]
- 2.Lykke-Andersen J., Aagaard,C., Semionenkov,M. and Garrett,R. (1997) Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci., 22, 326–331. [DOI] [PubMed] [Google Scholar]
- 3.Kjems J. and Garrett,R.A. (1988) Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell, 26, 693–703. [DOI] [PubMed] [Google Scholar]
- 4.Thompson L.D. and Daniels,C.J. (1988) A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J. Biol. Chem., 263, 17951–17959. [PubMed] [Google Scholar]
- 5.Thompson L.D. and Daniels,C.J. (1990) Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J. Biol. Chem., 265, 18104–18111. [PubMed] [Google Scholar]
- 6.Diener J.L. and Moore,P.B. (1998) Solution structure of a substrate for the archaeal pre-tRNA splicing endonucleases: the bulge-helix-bulge motif. Mol. Cell, 1, 883–894. [PubMed] [Google Scholar]
- 7.Li H. and Abelson,J. (2000) Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol., 302, 639–648. [DOI] [PubMed] [Google Scholar]
- 8.Durovic P. and Dennis,P.P. (1994) Separate pathways for excision and processing of 16S and 23S rRNA from the primary rRNA operon transcript from the hyperthermophilic archaebacterium Sulfolobus acidocaldarius: similarities to eukaryotic rRNA processing. Mol. Microbiol., 13, 229–242. [DOI] [PubMed] [Google Scholar]
- 9.Belfort M. and Weiner,A. (1997) Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell, 89, 1003–1006. [DOI] [PubMed] [Google Scholar]
- 10.Dennis P.P., Ziesche,S. and Mylvaganam S. (1998) Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol., 180, 4804–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chant J. and Dennis,P.P. (1986) Archaebacteria: transcription and processing of ribosomal RNA sequences in the Halobacterium cutirubrum. EMBO J., 5, 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell A.G., Ebhardt,H. and Dennis,P.P. (1999) Substrate requirements for a novel archaeal endonuclease that cleaves within the 5′ external transcribed spacer of Sulfolobus acidocaldarius precursor rRNA. Genetics, 152, 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipowicz W. (2000) Imprinted expression of small nucleolar RNAs in brain: time for RNomics. Proc. Natl Acad. Sci. USA, 97, 14035–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavaillé J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Hüttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hüttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.-P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeChiara T.M. and Brosius,J. (1987) Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc. Natl Acad. Sci. USA, 84, 2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt A.O., Herwig,R., Meier-Ewert,S. and Lehrach,H. (1999) High density cDNA grids for hybridization fingerprinting experiments. In Innis,M.A., Gelfand,D.H. and Sninsky,J.J. (eds), PCR Applications: Protocols for Functional Genomics. Academic Press, San Diego, CA, pp. 457–472.
- 18.Stern S., Moazed,D. and Noller,H.F. (1988) Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol., 164, 481–489. [DOI] [PubMed] [Google Scholar]
- 19.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Lührmann,R. (1999) Functional interaction of a novel 15.5 kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciammaruconi A. and Londei,P. (2001) In vitro processing of the 16S rRNA of the thermophilic archaeon Sufolobus solfataricus. J. Bacteriol., 183, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank D.N. and Pace,N.R. (1998) Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem., 67, 153–180. [DOI] [PubMed] [Google Scholar]
- 23.Zuker M. (2000) Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol., 10, 303–310. [DOI] [PubMed] [Google Scholar]
- 24.Kjems J. and Garrett, RA. (1990) Secondary structural elements exclusive to the sequences flanking ribosomal RNAs lend support to the monophyletic nature of the archaebacteria. J. Mol. Evol., 31, 25–32. [DOI] [PubMed] [Google Scholar]
- 25.Bachellerie J.P. and Cavaillé,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 7, 257–261. [DOI] [PubMed] [Google Scholar]
- 26.Gaspin C., Cavaillé,J., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 27.Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 28.Dennis P.P., Omer,A. and Lowe,T. (2001) A guided tour: small RNA function in Archaea. Mol. Microbiol., 40, 509–519. [DOI] [PubMed] [Google Scholar]
- 29.Klein D.J., Schmeing,T.M., Moore,P.B. and Steitz,T.A. (2001) The kink-turn: a new RNA secondary structure motif. EMBO J., 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watkins N.J., Segault,V., Charpentier,B., Nottrott,S., Fabrizio,P., Bachi,A., Wilm,M., Rosbash,M., Branlant,C. and Lührmann,R. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- 31.Vidovic I., Nottrott,S., Hartmuth,K., Luhrmann,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- 32.Winkler W.C., Grundy,F.J., Murphy,B.A. and Henkin,T.M. (2001) The GA motif: an RNA element common to bacterial antitermination systems, rRNA and eukaryotic RNAs. RNA, 7, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- 34.Ganot P., Jady,B.E., Bortolin,M.L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jady B.E. and Kiss,T. (2000) Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res., 28, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavaillé J., Vitali,P., Basyuk,E., Hüttenhofer,A. and Bachellerie,J.P. (2001) A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem., 276, 26374–26383. [DOI] [PubMed] [Google Scholar]
- 37.Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- 39.Dennis P.P., Russell,A.G. and Moniz de Sa,M. (1997) Formation of the 5′ end pseudoknot in small subunit ribosomal RNA: involvement of U3-like sequences. RNA, 3, 337–343. [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes J.M. (1996) Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]