Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common B-cell non-Hodgkin lymphoma (B-NHL) in adults. These lymphomas are classified according to gene expression profiling (GEP) into germinal center B-cell (GCB) and activated B-cell type (ABC). Recent studies have suggested new subtypes of large B-cell lymphoma, based on genetic and molecular alterations, among them is large B-cell lymphoma with IRF4-rearrangement (LBCL-IRF4). We used fluorescence in situ hybridization (FISH), GEP (using the DLBCL COO assay by HTG Molecular Inc), and next generation sequencing (NGS) to comprehensively characterize 30 cases of LBCLs located in Waldeyer’s ring in adult patients and to identify LBCL-IRF4. FISH revealed breaks of IRF4 in 2/30 cases (6.7%), BCL2 breaks in 6/30 cases (20.0%), and IGH breaks in 13/29 cases (44.8%). GEP classified 14 cases each as GCB or ABC subtype, and 2 cases remained unclassified; this was concordant with the immunohistochemistry (IHC) in 25/30 cases (83.3%). A subgrouping, based on GEP, was performed: group 1 included 14 GCB cases with the most frequent mutations in BCL2 and EZH2 in 6/14 cases (42.8%). The two cases with IRF4 rearrangement were assigned to this group by GEP and showed IRF4 mutations, supporting the diagnosis of LBCL-IRF4. Group 2 included 14 ABC cases; the most frequent mutations were CD79B and MYD88 identified in 5/14 patients (35.7%). Group 3 included 2 unclassifiable cases in which no molecular patterns were detected. Overall, LBCLs of Waldeyer’s ring in adult patients are a heterogeneous group, including LBCL-IRF4, which shares several features with cases in the pediatric population.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00428-023-03516-7.
Keywords: Diffuse large B-cell lymphoma, Waldeyer’s ring, IRF4 rearrangement, IRF4 mutations
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common B-cell non-Hodgkin lymphoma (B-NHL) in adults. DLBCL are classified based on gene expression profiling (GEP) into germinal center B-cell-type (GCB), activated B-cell-type (ABC), and an unclassifiable subgroup, according to the putative cell of origin (COO). As a surrogate marker, immunohistochemistry (IHC) is currently used in routine diagnosis. Although there are different immunohistochemical algorithms, the Hans algorithm (HA) is the most widely used [1–3]. HA divides DLBCL into GCB and non-GCB based on three antibodies, namely CD10, BCL6, and MUM1 [2]. However, the prognostic impact of IHC algorithms is limited. Trials in DLBCL using COO based on IHC analysis has given disappointing results [4, 5]. More recently, a genetic classification based on structural variants, mutational profile, and somatic copy-number alterations (CNA) was proposed. This genetic classification can further dissect and stratify the GEP-based COO classification into seven genetic subtypes with putative clinical relevance [6, 7]. Hans algorithm remains an important diagnostic tool in routine diagnosis; however, mutational profile of DLBCL contributes to understand the pathogenesis of DLBCL and has the potential to direct targeted therapy in the future [8]. Nevertheless, these mutational subgroups need to be validated and have still only limited relevance for routine diagnostics and treatment. The distinction between ABC and GCB, although it does not capture the complexity of DLBCL, reflects a basic biological distinction, and moving forward, a combination of COO and molecular subgroups may provide a better stratification of the disease [9, 10].
The 2017 World Health Organization (WHO) classification of lymphoid neoplasms introduced for the first time the large B-cell lymphoma with IRF4 rearrangement (LBCL-IRF4) as a provisional entity [11]. This disease is now recognized as definitive entity in both the International Consensus Classification [9] and in the 5th edition of the WHO [12]. LBCL-IRF4 predominates in children and young adults, shows frequent involvement of the Waldeyer’s ring (WR), and has an excellent outcome with chemotherapy. Morphologically, it is composed of large cells, and the growth pattern might be diffuse, follicular/diffuse, or purely follicular [13]. LBCL-IRF4 in children and young adults has a distinct molecular profile characterized by frequent mutations of IRF4, as a result of aberrant somatic hypermutation (aSHM), and NF-κB-related genes despite a GCB-transcriptional program. LBCL-IRF4 has been reported also in adults. However, adult cases show higher genetic complexity, higher mutational burden with frequent MYD88 and KMT2D mutations, and more often an ABC-GEP [14].
Although LBCL-IRF4 in children and young adults are frequently reported in the Waldeyer’s ring, this association has not been thoroughly investigated in adults. Interestingly, a previous study in an adult population suggested that DLBCL of Waldeyer’s ring has distinctive clinicopathological features with high rate of GCB phenotype (61%) and a variable follicular pattern, observed in 44% of the cases [15]. The aim of this study was to comprehensively analyze the genetic landscape of DLBCL and follicular lymphoma (FL) grade 3B of the Waldeyer’s ring in adult patients and to investigate the frequency of LBCL-IRF4 by fluorescence in situ hybridization (FISH), NGS, and GEP.
Material and methods
Patient samples
Thirty samples of adult patients, diagnosed as DLBCL or FL grade 3B of the Waldeyer’s ring (includes lymphoid tissues in the nasopharynx, palatine tonsils, tongue base, soft palate, and oropharyngeal wall) between 2002 and 2020, were retrieved from the files of the Institute of Pathology, University Hospital Tübingen. The cases were classified following the criteria of the 2017 WHO classification [16]. The immunohistochemical staining was performed as part of the diagnostic workup. Expression of the immunohistochemical marker in more than 30% of the neoplastic cells was considered positive. The study was conducted in accordance with the Declaration of Helsinki and was approved by the local Ethics Review Committee and the IRB review panels of the contributing institutions (UKT 199/2020/BO2).
Fluorescence in situ hybridization (FISH)
FISH analyses were performed using 3.5 µm thick FFPE sections and the following probes: IRF4/DUSP22 Dual Color Break Apart Probe (ZytoVision GmbH, Bremerhaven, Germany), IGH Dual Color Break Apart Probe (ZytoVision GmbH), BCL2 Dual Color Break Apart Rearrangement Probe (Abbott Molecular, Des Plaines, IL, USA), and MYC Break Apart Rearrangement Probe (Abbott Molecular). The analyses were carried out according to the manufacturer’s protocols. Results were considered positive if the percentage of positive cells was greater than or equal to 10%.
Gene expression profiling (GEP)
GEP analysis was performed using the EdgeSeq system (HTG Molecular Diagnostics Inc., Tucson, AZ, USA). All samples were analyzed using the HTG EdgeSeq DLBCL Cell of Origin assay for Ion Torrent platforms (HTG Molecular Diagnostics Inc.), according to the manufacturer’s protocol [14]. The COO classification was performed using the HTG EdgeSeq Parser. For the statistical analyses, a quantile normalization, according to Bolstad et al. [17], was performed (see also section statistical analysis). Further details on the GEP methodology can be found in the supplemental methods, gene expression profiling section.
Next-generation sequencing (NGS)
NGS panel sequencing was performed using the Ion GeneStudio S5 prime (Thermo Fisher Scientific, Waltham, MA, USA) with an AmpliSeq Custom DLBCL Panel, designed using the Ion AmpliSeq Designer (Thermo Fisher Scientific). All cases were screened for mutations with a customized panel, encompassing BCL2, BCL6, CARD11, CD79B, EZH2, IRF4, MYD88, PIM1, PRDM1, and TNFAIP3. For CD79B, EZH2 and MYD88 hotspot regions are covered (supplemental table 1). In addition, single amplicons were created for some gene regions, including IRF4 (see also supplemental table 2). The analyses performed for each case are shown in supplemental table 3. Detailed information on the next-generation sequencing is shown in the supplemental methods, mutational analysis section.
Cases with mutations in CD79B and/or MYD88 genes were considered to belong most probably to MCD/C5 subtype, whereas EZH2 mutations and/or BCL2 translocations were considered to belong most probably to EZB/C3 subtype [6, 7].
Statistical analysis
All statistical tests were two-sided, and statistical significance was concluded for values p < 0.05. GEP data was quantile normalized. Significance tests were performed using the Mann–Whitney U test and the Kruskal–Wallis H test. Statistical analyses were carried out using RStudio, version 1.2.1335, and the IBM SPSS Statistics software, version 28 for Microsoft Windows.
Results
Clinicopathological features
Clinical information is summarized in Table 1. A total of 30 patients were included in the study, of whom 12 were male and 18 were female patients, representing an M:F ratio of 1:1.5 with a median age at initial presentation of 69.5 years (range 37–87 years). Information about the clinical stage was available in 23/30 cases, of which 18 had low clinical stage (I or II) at presentation (18/23, 78%), whereas five patients (5/23, 22%) had high-stage disease (III or IV). IPI score was available for 20/30 patients and was low in most cases (14/20, 47%), low-intermediate in three cases (3/20, 10%), high-intermediate in two cases (2/20, 7%), and high in one case (1/20, 3%). Nineteen out of 23 patients (83%) received therapies containing R-CHOP, including 2 patients with a reduced dose of CHOP, whereas three patients (3/23, 13%) received other therapies. One patient (1/23) received no therapy. Clinical follow-up was obtained in 22/30 patients with a mean of 55.0 months (range 1–125 months). Complete remission (CR) was achieved in 17/18 patients, including one patient (case no. 21) who had a relapse (REL) at 69 months of follow-up, but this patient achieved CR with renewed therapy, leading to sustained CR after a total of 109 months of follow-up. One patient achieved partial remission (PR).
Table 1.
Clinical data of 30 large B-cell lymphomas of Waldeyer’s ring in adults
| Case # | Sex | Age | Diagnosis | Clinical stage | IPI | Treatment | R/R | Follow-up |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 44 | DLBCL | NA | NA | NA | NA | NA |
| 2 | F | 45 | DLBCL | IIA | Low | 6 × R-CHOP | CR |
125 mo NED |
| 5 | F | 81 | FL 3B | NA | NA | NA | NA | NA |
| 6 | F | 64 | DLBCL | II | Low-inter |
MAIN 6 × R-CHOP 6 × R |
CR |
61 mo NED |
| 7 | F | 79 | DLBCL | NA | NA | NA | NA | NA |
| 8 | F | 57 | DLBCL | IIA/E | Low | 6 × R-CHOP | CR |
120 mo NED |
| 9 | F | 70 | FL 3B | IIE | Low | 6 × R-CHOP | CR |
98 mo NED |
| 10 | F | 69 | DLBCL | IIA/E | Low | 1 × R-CHOP | NA | NA |
| 11 | M | 52 | DLBCL | II | Low | 6 × R-CHOP | CR |
84 mo NED |
| 13 § | F | 78 | DLBCL | II | Low | 4 × R-CHOP | CR |
97 mo NED |
| 15 | M | 47 | DLBCL | IA | Low |
4 × R-CHOP 2 × R |
CR |
72 mo NED |
| 16 | M | 78 | DLBCL | IIA | Low |
4 × R-CHOP 4 × R |
CR |
53 mo NED |
| 17 | F | 79 | DLBCL | IIA | Low |
6 × R-CHOP 2 × R |
CR |
58 mo NED |
| 19 | F | 87 | DLBCL | IV | NA | No treatment | NA |
1 mo DOD |
| 20 | F | 78 | DLBCL | NA | NA | NA | NA | NA |
| 21 | M | 52 | DLBCL | III | Low-inter |
8 × R-CHOP 8 × R R-BEAM SCT |
REL CR |
REL after 69 mo, NED after a total of 109 mo |
| 22 | F | 47 | DLBCL | IIA | Low |
4 × R-CHOP RTX 39,6 Gy |
CR |
45 mo NED |
| 23 | F | 74 | DLBCL | IVB/E | High | 4 × R-B | PR |
5 mo LFU |
| 24 | M | 74 | DLBCL | IIA/E | Low | 6 × R-CHOP | CR |
65 mo NED |
| 25 | F | 63 | DLBCL | IA | High-inter |
6 × R-CHOP 2 × R MTX |
CR |
48 mo NED |
| 26 | F | 70 | DLBCL | NA | NA | NA | NA | NA |
| 27 | M | 62 |
DLBCL/ FL 3B |
IIA | NA |
4 × R-CHLiP 4 × R |
CR |
48 mo NED |
| 29 | M | 81 | DLBCL | IIIA | Low-inter | 1 × R-miniCHOP | NA | NA |
| 30 | M | 71 | DLBCL | NA | NA | NA | NA |
2 mo LFU |
| 31 | F | 82 | DLBCL | IA | NA | 3 × R-miniCHOP | NA |
2 mo LFU |
| 32 | M | 61 | DLBCL | IVA | High-inter |
TEAM SCT |
NA |
18 mo LFU |
| 33 | M | 58 | DLBCL | I | Low |
4 × R-CHOP 2 × R |
CR |
17 mo NED |
| 34 | M | 37 | DLBCL | IIA | Low |
6 × R-CHOP 2 × R |
CR |
51 mo NED |
| 35§ | F | 37 | DLBCL | IA | Low |
6 × R-CHOP 2 × R |
CR |
30 mo NED |
| 36 | M | 77 | DLBCL | NA | NA | NA | NA | NA |
M, male; F, female; IPI, International Prognostic Index; NA, not available; mo, months; LFU, lost to follow-up; R/R, relapse/refractory; NED, no evidence of disease; DOD, died of disease; PR, partial remission; CR, complete remission; REL, relapse. Therapy: MAIN: study treatment with R-CHOP with/without Bevacizumab (NCT00486759); MTX, methotrexate; R, rituximab; R-B, rituximab, bendamustine; R-BEAM, rituximab, carmustine, etoposide, cytarabine, melphalan; R-CHLiP, rituximab, cyclophosphamide, liposomal vincristine, prednisone; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone; R-miniCHOP, rituximab with dose-reduced CHOP (see above); RTX, radiotherapy; SCT, stem cell transplantation; TEAM, thiotepa, etoposide, cytarabine, melphalan
§Cases with IRF4 translocation/mutation
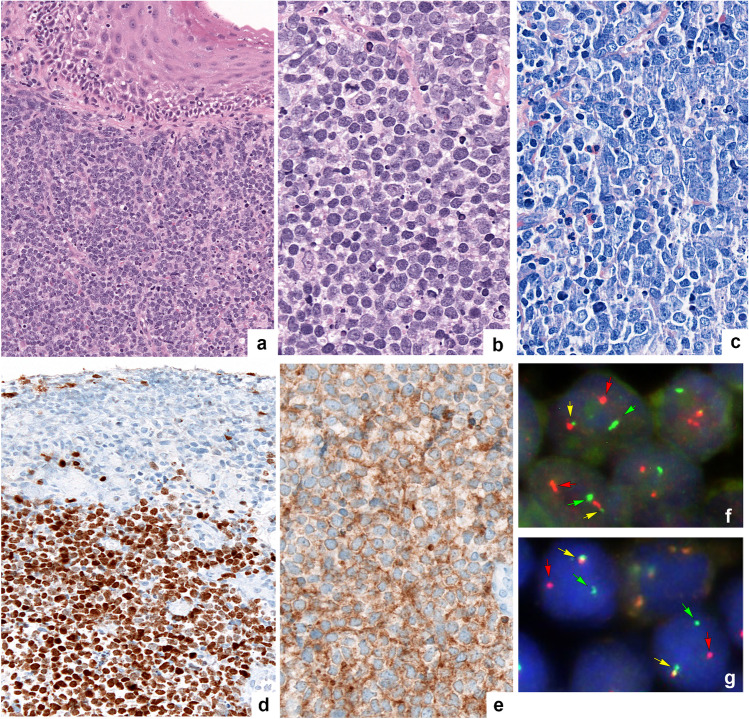
Morphologically, 28 cases were diagnosed as DLBCL, NOS and two as FL grade 3B. One case showed a blastoid morphology with medium-sized cells, inconspicuous nucleoli, and scant cytoplasm (Fig. 1a–c). The rest of the cases showed a characteristic centroblastic morphology. According to the Hans algorithm, 18/30 (60%) cases were classified as GCB type, including five cases, which were CD10 + , BCL6 + , MUM1 + (Fig. 1d, e), whereas 12/30 cases (40%) were classified as non-GCB type. CD10 was positive in 16/30 (53%) cases, BCL6 in 28/30 (93%), and MUM1 in 17/30 (57%). BCL2 expression was observed in 23/30 (77%) cases, whereas MYC was expressed in only 4/27 (15%) cases. The proliferation rate, measured by MIB1, was high in nearly all cases (> 60% in 27/30 cases).
Fig. 1.
Morphological, immunohistochemical, and genetic features of LBCL-IRF4 (case no. 35). a Large B-cell lymphoma with blastoid morphology with medium-sized cells, inconspicuous nucleoli, and scant cytoplasm (original magnification × 200; hematoxylin and eosin stain); b with original magnification × 400: hematoxylin and eosin stain; c original magnification × 400: Giemsa stain. d MUM1. e CD10. f Fluorescence in situ hybridization (FISH) analysis using a break apart probe (BAP) demonstrates an IRF4 break with 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows) consistent with gene rearrangement. g FISH analysis with a BAP demonstrates an IGH break with 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows) consistent with gene rearrangement (Fig. 1 was created using Adobe Photoshop Version CS6)
No significant age difference could be shown between DLBCL ABC (mean age 68.8 years/median age 74 years, SD = 13.6, range 45 to 87 years) and DLBCL GCB (mean age 63.2 years/median age 66.5 years, SD = 13.3, range 37 to 79 years) (p = 0.194).
Fluorescence in situ hybridization
FISH analyses for IRF4, BCL2, and IGH were performed in all cases. MYC analysis was done in cases with MYC expression ≥ 40% of the tumor cells. FISH analysis demonstrated a break of IRF4, indicative of a translocation, in 2/30 cases (6.7%), a BCL2 break in 6/30 cases (20%), and an IGH break in 13/29 cases (44.8%). No MYC break was identified in the 4 cases analyzed. Both cases with an IRF4 break also showed an IGH break (Fig. 1f, g), whereas only in four cases (4/6; 66%) with a BCL2 break, a concordant IGH break was demonstrated. In seven cases, a matching partner for the IGH break was not identified, but BCL6 FISH was not performed. Additionally, IRF4 gains were detected in seven cases (23.3%), BCL2 gains in two cases (6.7%), and IGH gains in one case (3.4%), whereas no MYC gains were detected.
Mutational analyses
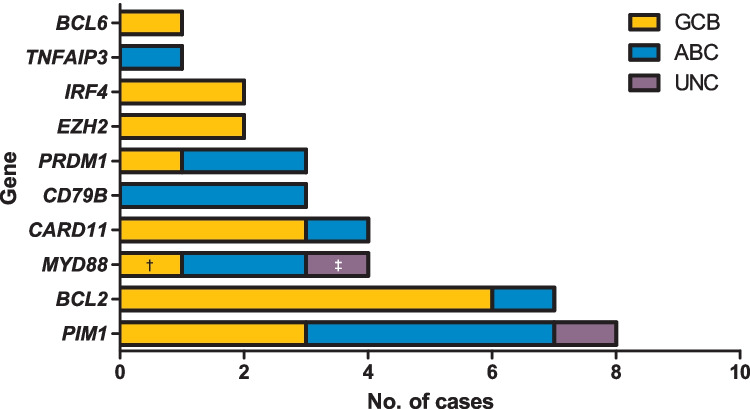
All cases were analyzed by NGS. The results are summarized in Fig. 2. Sixty-five variants were identified in 20/30 (67%) cases (supplemental table 4). Ten cases (33%) were wild type for all analyzed genes. The mean mutational burden was 2.2 variants per case. Overall, the allelic frequency of the mutations ranged from 7 to 89% with a mean of 33.6% (SD = 19.1). Among the mutated cases, 10 of 30 cases showed only one mutated gene (33%), five cases demonstrated two mutated genes (5/30, 25%), and five cases revealed three mutated genes (5/30, 25%). Overall, 58 missense, 3 nonsense, 2 frameshift, and 2 splice site mutations were found. The most frequent genetic alterations were found in PIM1 with 8 mutated cases (8/30, 27%), followed by BCL2 with seven mutated cases (6 of them with BCL2-rearrangement) (7/30, 23%), CARD11 and MYD88 with four mutated cases each (4/30, 13%), CD79B and PRDM1 with three mutated cases each (3/30, 10%), IRF4 and EZH2 with two mutations each (2/30, 6.7%), and BCL6 and TNFAIP3 with one mutated case each (1/30, 3%).
Fig. 2.
Mutation frequencies across GEP subtypes. Total number of cases (x-axis) with a mutation in the gene depicted on the y-axis. †Case no. 23 was classified as GCB by GEP and showed a non-canonical mutation of MYD88 in position p.S219C. ‡Case no. 10 was unclassifiable by GEP and showed a non-canonical mutation of MYD88 in position p.S243N (Fig. 2 was created using GraphPad Prism version 5.00)
Gene expression profiling
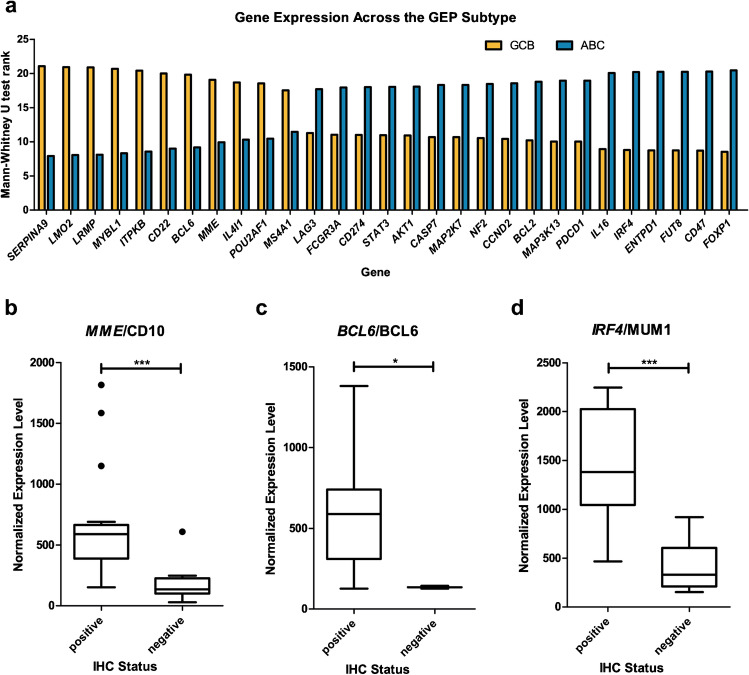
COO classification, using the HTG EdgeSeq Parser, classified 14 cases each as GCB and ABC, whereas two cases remained unclassified (UNC). Significant differences in gene expression among the subtypes were detected in 29 of the gene products examined; 11 of them were highly expressed in the GCB subtype, whereas 18 were highly expressed in the ABC subtype (Fig. 3a and Supplemental Fig. 1). The correlation between the protein and mRNA expression for CD10/MME, BCL6/BCL6, and MUM1/IRF4 was excellent (Fig. 3b–d).
Fig. 3.
Results of the gene expression profiling. a Genes with significant (p < 0.05) differences in the expression levels across GEP subtypes. In total, 11 genes were highly expressed in the GCB subgroup and 18 ABC subgroup respectively. b–d Correlation of the immunohistochemical status (x-axis) with the normalized expression level of the GEP (y-axis) for MME/CD10 (b), BCL6/BCL6 (c), and IRF4/MUM1 (d). The differences were significant with p < 0.001 for MME/CD10, p = 0.018 for BCL6/BCL6, and p < 0.001 for IRF4/MUM1 (Fig. 3 was created using GraphPad Prism version 5.00)
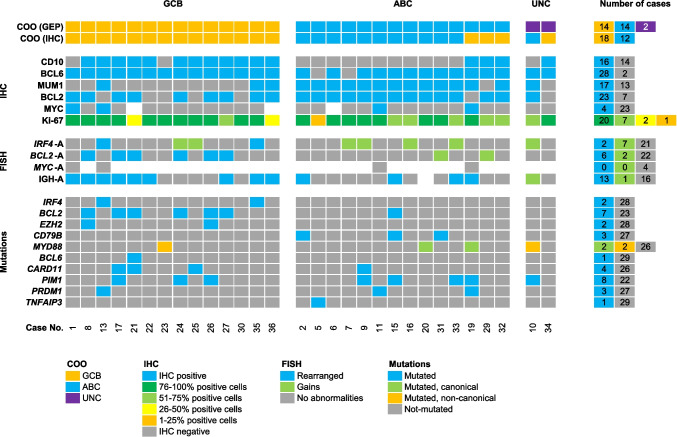
Correlation of morphological features, FISH, GEP, and mutational analysis
The correlation of all parameters is depicted in Fig. 4. The agreement between the COO classification using GEP and IHC was 83% (25/30 cases). From the five discordant cases using GEP as the gold standard, three were in the ABC subtype and two in the unclassifiable cases. The 30 cases were assigned according to GEP into three different groups:
Group 1: 14 cases were classified as GCB type by GEP and IHC. All cases expressed CD10 (12/14 cases, 86%) and/or BCL6 (14/14 cases, 100%) with 3 cases expressing MUM1/IRF4 (3/14 cases, 21%). BCL2-R was demonstrated in 6 cases (49%) and IRF4-R in 2 cases (14%). All of the BCL2 rearranged cases showed BCL2 mutations. Mutational analysis showed that the most frequent mutations were BCL2/EZH2 found in 6 cases (43%). Other mutations frequently found in this group were CARD11 and PIM1 (21.4%, each). This group includes the two IRF4 rearranged cases that were the only cases with IRF4 mutations. One case showed an uncommon immunohistochemical and genetic signature (case no. 23). This case was classified as GCB by GEP and IHC while lacking the expression of CD10 and carrying a non-canonical MYD88 mutation (p.S219C).
Group 2: 14 cases were classified as ABC type by GEP and 11 of these cases by IHC. The three discrepant cases were assigned into the GCB group according to the Hans algorithm because of CD10 expression. By FISH analysis, no BCL2, MYC, or IRF4 translocations were identified. The most frequent mutations were CD79B and MYD88 identified in 5 patients (36%). Other mutations identified were PIM1 (26%) and PRDM1 (14%). One case of this group (case no. 15) was classified as ABC by GEP and IHC and showed mutations of BCL2, CD79B, and PIM1. Furthermore, this case showed a rearrangement of IGH while lacking demonstrable rearrangements in other genes. Because of the BCL2 mutations and an IGH rearrangement without identifiable partner, the possibility of a cryptic BCL2 translocation cannot be excluded.
Group 3: 2 cases were unclassifiable by GEP; however, by Hans algorithm, one case each was assigned to the GCB and the non-GCB group. The case with CD10 and BCL6 expression (no. 34) showed no translocations or mutations. The case with MUM1/IRF4 expression (no. 10) had an MYD88 and PIM1 mutation supporting the non-GCB-type suggested by the Hans algorithm, although the MYD88 mutation was not in the hotspot region.
Fig. 4.
Overview of 30 cases of LBCL-IRF4 clustered in groups according to the COO (GEP). This overview includes the COO results by GEP and IHC, IHC results, and FISH status and mutations. While each column represents one case, each line represents a specific analysis. On the right side of the figure, the raw numbers of specific analyses are shown (Fig. 4 was created using Microsoft Excel version 2210)
Discussion
We investigated the morphological and genetic features of DLBCLs and FLs grade 3B of Waldeyer’s ring in 30 adult patients and screened these cases by FISH analysis for IRF4 translocations to identify cases of the novel entity LBCL-IRF4. Classification of these cases according to their COO using IHC and GEP revealed equal numbers of DLBCL of GCB-type (group 1: 47%) and ABC-type (group 2: 47%). Mutational analysis supported the GCB-type (BCL2/EZH2 mutations) in 43% of the cases and the ABC-type (MYD88/CD79B mutations) in 36% of the cases. Our study demonstrated that LBCL-IRF4 in adults occurs in the WR in 7% of the cases.
In this study, DLBCL of GCB-type was slightly below the rate reported in the WR in a previous study (46.7 vs 62%) [15]. However other groups have found even lower frequency of GCB-type DLBCL (20.4%) [18]. In general, unselected studies of extranodal and nodal DLBCL have reported GCB rates between 40 and 47% [2, 19]. COO assignment was possible in all examined cases in this cohort using the HA by IHC. GEP was able to assign 93% of all cases examined. The agreement between IHC and GEP was over 80%. An important source of disagreement between IHC and GEP is the aberrant expression of CD10 + BCL6 + MUM1 + demonstrated in five cases. These cases per default are classified as GCB type due to CD10 expression. Two of these latter cases were assigned by GEP into the ABC group with gene mutation characteristic of the ABC type. These results support the conclusion that cases with CD10 + , BCL6 + , and MUM1 + should not be classified by HA [14].
DLBCL assigned to COO GCB-type by GEP expressed either CD10 and/or BCL6 (group 1). Only 2 cases did not express CD10. Importantly, three cases in this group coexpressed MUM1/IRF4, thus showing an aberrant phenotype, two of them with IRF4-rearrangement. This finding indicates that cases with aberrant expression of CD10 and MUM1/IRF4 should be investigated for IRF4-rearrangements. Both cases showed a mutation in IRF4, most probably due to aberrant somatic hypermutation. BCL2 rearrangements were demonstrated in half of the cases, sometimes with coexisting BCL2-mutations. The rate of BCL2 translocations in our cohort was more than twice as high as in other studies investigating WR DLBCLs, depending on the comparison reference source, but matched very well with results from studies that included both nodal and extranodal LBCLs [15, 18, 20]. In the majority of the BCL2-rearranged cases, an additional IGH translocation was demonstrated, suggesting the characteristic BCL2:IGH translocation. In two BCL2-rearranged cases, IGH break was not documented indicating the possibility of alternative translocation partners with the light immunoglobulin chain genes κ and λ [21].
According to GEP, 14 DLBCL were assigned to the ABC group (group 2). Eleven of 14 cases showed concordance between HA and GEP with CD10-negativity. In contrast, 3/14 cases, despite the expression of CD10, were assigned to the ABC group based on the GEP and mutational landscape. Two of these latter cases showed aberrant coexpression of CD10, BCL6, and MUM1. No IRF4 translocations were identified in this group. The most frequent mutations (7/14 cases, 50.0%) involved genes of the NF-κB pathway (CD79B, MYD88, CARD11, TNFAIP3) consistent with the MCD/CD5 molecular group [6, 7]. In the study of Chapuy et al., cohort mutations in CD79B and MYD88 were identified in 48% and 50%, respectively [7]. The aberrant activation of NF-κB is well described in the ABC-type and contributes to the pathogenesis of DLBCL, opening up possibilities for targeted anti-NF-κB therapy. Interestingly, two recent studies of LBCL-IRF4 in pediatric and adult populations also demonstrated enrichment in mutations affecting the NF-κB pathway (CARD11, CD79B, MYD88, TNFAIP3) [14, 22].
Finally, one of the aims of this study was to investigate the prevalence of LBCL-IRF4 in the WR in adult patients. Our cohort shows similar results to other adult patients cohorts in and outside Waldeyer’s ring, with a frequency of 8% [15] and in a mixed cohort of children and adults with 13% [13]. Most of the IRF4-rearranged cases in the study by Salaverria et al. [13] affected children and young adults under the age of 18 (14/23 cases). However, studies including only children and young adults have reported higher frequencies, between 21 to 32% of IRF4 rearranged cases, mainly involving the WR [23, 24]. Only one study in adults has reported a relative high frequency (24.5%) of LBCL-IRF4 in the WR in a Chinese population [18]. The mean age of the Chinese cohort was lower when compared to our cohort (58 years vs 69 years); however, this does not explain the difference. The two identified cases with IRF4 rearrangement in our cohort showed both IGH rearrangements, a triple positive phenotype (CD10 + , BCL6 + , MUM1 +), and GCB-type by GEP. The patients were 78 and 37 years old at time of diagnosis, and both showed limited disease (clinical stage II and IA, IPI low); both were treated with R-CHOP and showed no evidence of disease with complete remission at 97 and 30 months. As has been previously reported, mutations in IRF4 were detected only in the two cases with IRF4-translocations and showed the pattern of aberrant somatic hypermutation [14, 22]. Thus, reinforcing the contention that the presence of IRF4 mutations, in the correct context, is supportive of the diagnosis of LBCL-IRF4. Although this lymphoma is more frequently observed in children and young adults, cases in adults share the GCB-type, IRF4 mutations, and frequent coexpression of CD10, BCL6, and MUM1 [22].
In conclusion, large B cell lymphomas involving the Waldeyer’s ring were equally divided in GCB and ABC type. The overall concordance in this study between GEP and HA was excellent except for those cases expressing aberrantly CD10, BCL6, and MUM1. LBCL-IRF4 in adult patients in this study share clinical, morphologic, and genetic features with LBCL-IRF4 in pediatric populations. This relatively small cohort in adults suggests a lower prevalence of LBCL-IRF4 in the Waldeyer’s ring in adults compared to pediatric population. This finding needs to be confirmed in a larger cohort.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Rebecca Braun, Claudia Hermann, Esther Kohler, and Franziska Mihalik for their excellent technical assistance.
Author contribution
L.Q.-M. designed and supervised the study; S.S., L.F., and B.M. performed the FISH analysis; S.S. and F.O. performed the GEP analysis; S.S., F.O., and I.B. performed the NGS mutational analysis; L.Q.-M., L.F., F.F., and S.S. reviewed the cases; S.S., L.F., and L.Q.-M. wrote the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. S.S. received funding from the IZKF fellowship program of the Medical Faculty of the Eberhard Karls University Tübingen (grant number IZKF 2019.2).
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval
The study was approved before by the local ethics committee of the University Hospital Tübingen (199/2020/BO2).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sebastian Streich and Leonie Frauenfeld contributed equally to this work.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, López-Guillermo A, Grogan TM, Miller TP, Leblanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/nejmoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Leonard JP, Kolibaba KS, Reeves JA, Tulpule A, Flinn IW, Kolevska T, Robles R, Flowers CR, Collins R, Dibella NJ, Papish SW, Venugopal P, Horodner A, Tabatabai A, Hajdenberg J, Park J, Neuwirth R, Mulligan G, Suryanarayan K, Esseltine D-L, De Vos S. Randomized phase II study of R-CHOP with or without bortezomib in previously untreated patients with non–germinal center B-Cell–like diffuse large B-cell lymphoma. J Clin Oncol. 2017;35:3538–3546. doi: 10.1200/jco.2017.73.2784. [DOI] [PubMed] [Google Scholar]
- 5.Nowakowski GS, Chiappella A, Gascoyne RD, Scott DW, Zhang Q, Jurczak W, Özcan M, Hong X, Zhu J, Jin J, Belada D, Bergua JM, Piazza F, Mócikova H, Molinari AL, Yoon DH, Cavallo F, Tani M, Yamamoto K, Izutsu K, Kato K, Czuczman M, Hersey S, Kilcoyne A, Russo J, Hudak K, Zhang J, Wade S, Witzig TE, Vitolo U. ROBUST: a phase III study of lenalidomide plus R-CHOP versus placebo plus R-CHOP in previously untreated patients with ABC-type diffuse large B-cell lymphoma. J Clin Oncol. 2021;39:1317–1328. doi: 10.1200/jco.20.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, Roulland S, Kasbekar M, Young RM, Shaffer AL, Hodson DJ, Xiao W, Yu X, Yang Y, Zhao H, Xu W, Liu X, Zhou B, Du W, Chan WC, Jaffe ES, Gascoyne RD, Connors JM, Campo E, Lopez-Guillermo A, Rosenwald A, Ott G, Delabie J, Rimsza LM, Tay Kuang Wei K, Zelenetz AD, Leonard JP, Bartlett NL, Tran B, Shetty J, Zhao Y, Soppet DR, Pittaluga S, Wilson WH, Staudt LM. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378:1396–1407. doi: 10.1056/nejmoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, Staiger AM, Wala JA, Ducar MD, Leshchiner I, Rheinbay E, Taylor-Weiner A, Coughlin CA, Hess JM, Pedamallu CS, Livitz D, Rosebrock D, Rosenberg M, Tracy AA, Horn H, Van Hummelen P, Feldman AL, Link BK, Novak AJ, Cerhan JR, Habermann TM, Siebert R, Rosenwald A, Thorner AR, Meyerson ML, Golub TR, Beroukhim R, Wulf GG, Ott G, Rodig SJ, Monti S, Neuberg DS, Loeffler M, Pfreundschuh M, Trümper L, Getz G, Shipp MA. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, Leppa S, Pasanen A, Meriranta L, Karjalainen-Lindsberg M-L, Nørgaard P, Pedersen M, Gang AO, Høgdall E, Heavican TB, Lone W, Iqbal J, Qin Q, Li G, Kim SY, Healy J, Richards KL, Fedoriw Y, Bernal-Mizrachi L, Koff JL, Staton AD, Flowers CR, Paltiel O, Goldschmidt N, Calaminici M, Clear A, Gribben J, Nguyen E, Czader MB, Ondrejka SL, Collie A, Hsi ED, Tse E, Au-Yeung RKH, Kwong Y-L, Srivastava G, Choi WWL, Evens AM, Pilichowska M, Sengar M, Reddy N, Li S, Chadburn A, Gordon LI, Jaffe ES, Levy S, Rempel R, Tzeng T, Happ LE, Dave T, Rajagopalan D, Datta J, Dunson DB, Dave SS. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494.e415. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campo E, Jaffe ES, Cook JR, Quintanilla-Martinez L, Swerdlow SH, Anderson KC, Brousset P, Cerroni L, de Leval L, Dirnhofer S, Dogan A, Feldman AL, Fend F, Friedberg JW, Gaulard P, Ghia P, Horwitz SM, King RL, Salles G, San-Miguel J, Seymour JF, Treon SP, Vose JM, Zucca E, Advani R, Ansell S, Au W-Y, Barrionuevo C, Bergsagel L, Chan WC, Cohen JI, d’Amore F, Davies A, Falini B, Ghobrial IM, Goodlad JR, Gribben JG, Hsi ED, Kahl BS, Kim W-S, Kumar S, LaCasce AS, Laurent C, Lenz G, Leonard JP, Link MP, Lopez-Guillermo A, Mateos MV, Macintyre E, Melnick AM, Morschhauser F, Nakamura S, Narbaitz M, Pavlovsky A, Pileri SA, Piris M, Pro B, Rajkumar V, Rosen ST, Sander B, Sehn L, Shipp MA, Smith SM, Staudt LM, Thieblemont C, Tousseyn T, Wilson WH, Yoshino T, Zinzani P-L, Dreyling M, Scott DW, Winter JN, Zelenetz AD. The International Consensus Classification of mature lymphoid neoplasms: a report from the Clinical Advisory Committee. Blood. 2022;140:1229–1253. doi: 10.1182/blood.2022015851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song JY, Dirnhofer S, Piris MA, Quintanilla-Martínez L, Pileri S, Campo E. Diffuse large B-cell lymphomas, not otherwise specified, and emerging entities. Virchows Arch. 2022 doi: 10.1007/s00428-022-03466-6. [DOI] [PubMed] [Google Scholar]
- 11.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBDO, Berti E, Bhagat G, Borges AM, Boyer D, Calaminici M, Chadburn A, Chan JKC, Cheuk W, Chng W-J, Choi JK, Chuang S-S, Coupland SE, Czader M, Dave SS, De Jong D, Du M-Q, Elenitoba-Johnson KS, Ferry J, Geyer J, Gratzinger D, Guitart J, Gujral S, Harris M, Harrison CJ, Hartmann S, Hochhaus A, Jansen PM, Karube K, Kempf W, Khoury J, Kimura H, Klapper W, Kovach AE, Kumar S, Lazar AJ, Lazzi S, Leoncini L, Leung N, Leventaki V, Li X-Q, Lim MS, Liu W-P, Louissaint A, Marcogliese A, Medeiros LJ, Michal M, Miranda RN, Mitteldorf C, Montes-Moreno S, Morice W, Nardi V, Naresh KN, Natkunam Y, Ng S-B, Oschlies I, Ott G, Parrens M, Pulitzer M, Rajkumar SV, Rawstron AC, Rech K, Rosenwald A, Said J, Sarkozy C, Sayed S, Saygin C, Schuh A, Sewell W, Siebert R, Sohani AR, Tooze R, Traverse-Glehen A, Vega F, Vergier B, Wechalekar AD, Wood B, Xerri L, Xiao W (2022) The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: lymphoid neoplasms. Leukemia 36:1720–1748. 10.1038/s41375-022-01620-2 [DOI] [PMC free article] [PubMed]
- 13.Salaverria I, Philipp C, Oschlies I, Kohler CW, Kreuz M, Szczepanowski M, Burkhardt B, Trautmann H, Gesk S, Andrusiewicz M, Berger H, Fey M, Harder L, Hasenclever D, Hummel M, Loeffler M, Mahn F, Martin-Guerrero I, Pellissery S, Pott C, Pfreundschuh M, Reiter A, Richter J, Rosolowski M, Schwaenen C, Stein H, Trumper L, Wessendorf S, Spang R, Kuppers R, Klapper W, Siebert R. Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood. 2011;118:139–147. doi: 10.1182/blood-2011-01-330795. [DOI] [PubMed] [Google Scholar]
- 14.Frauenfeld L, Castrejon-de-Anta N, Ramis-Zaldivar JE, Streich S, Salmerón-Villalobos J, Otto F, Mayer AK, Steinhilber J, Pinyol M, Mankel B, Ramsower C, Bonzheim I, Fend F, Rimsza LM, Salaverria I, Campo E, Balagué O, Quintanilla-Martinez L. Diffuse large B-cell lymphomas in adults with aberrant coexpression of CD10, BCL6, and MUM1 are enriched in IRF4 rearrangements. Blood Adv. 2022;6:2361–2372. doi: 10.1182/bloodadvances.2021006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Leval L, Bonnet C, Copie-Bergman C, Seidel L, Baia M, Brière J, Molina TJ, Fabiani B, Petrella T, Bosq J, Gisselbrecht C, Siebert R, Tilly H, Haioun C, Fillet G, Gaulard P. Diffuse large B-cell lymphoma of Waldeyer's ring has distinct clinicopathologic features: a GELA study. Ann Oncol. 2012;23:3143–3151. doi: 10.1093/annonc/mds150. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: International Agency for Research on Cancer; 2017. [Google Scholar]
- 17.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Ma Z, Shi Y, Pang X, Li X, Cui W, Zhang W. Clinicopathologic features and prognostic analysis of Waldeyer ring B-cell lymphoma. Medicine. 2020;99:e18670. doi: 10.1097/md.0000000000018670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott G, Ziepert M, Klapper W, Horn H, Szczepanowski M, Bernd H-W, Thorns C, Feller AC, Lenze D, Hummel M, Stein H, Müller-Hermelink H-K, Frank M, Hansmann M-L, Barth TFE, Möller P, Cogliatti S, Pfreundschuh M, Schmitz N, Trümper L, Loeffler M, Rosenwald A. Immunoblastic morphology but not the immunohistochemical GCB/nonGCB classifier predicts outcome in diffuse large B-cell lymphoma in the RICOVER-60 trial of the DSHNHL. Blood. 2010;116:4916–4925. doi: 10.1182/blood-2010-03-276766. [DOI] [PubMed] [Google Scholar]
- 20.Copie-Bergman C, Gaulard P, Leroy K, Briere J, Baia M, Jais J-P, Salles GA, Berger F, Haioun C, Tilly H, Emile J-F, Banham AH, Mounier N, Gisselbrecht C, Feugier P, Coiffier B, Molina TJ. Immuno–fluorescence in situ hybridization index predicts survival in patients with diffuse large B-cell lymphoma treated with R-CHOP: a GELA study. J Clin Oncol. 2009;27:5573–5579. doi: 10.1200/jco.2009.22.7058. [DOI] [PubMed] [Google Scholar]
- 21.Martín-Subero JI, Harder L, Gesk S, Schlegelberger B, Grote W, Martinez-Climent JA, Dyer MJS, Novo FJ, Calasanz MJ, Siebert R. Interphase FISH assays for the detection of translocations with breakpoints in immunoglobulin light chain loci. Int J Cancer. 2002;98:470–474. doi: 10.1002/ijc.10169. [DOI] [PubMed] [Google Scholar]
- 22.Ramis-Zaldivar JE, Gonzalez-Farré B, Balagué O, Celis V, Nadeu F, Salmerón-Villalobos J, Andrés M, Martin-Guerrero I, Garrido-Pontnou M, Gaafar A, Suñol M, Bárcena C, Garcia-Bragado F, Andión M, Azorín D, Astigarraga I, Sagaseta De Ilurdoz M, Sábado C, Gallego S, Verdú-Amorós J, Fernandez-Delgado R, Perez V, Tapia G, Mozos A, Torrent M, Solano-Páez P, Rivas-Delgado A, Dlouhy I, Clot G, Enjuanes A, López-Guillermo A, Galera P, Oberley MJ, Maguire A, Ramsower C, Rimsza LM, Quintanilla-Martinez L, Jaffe ES, Campo E, Salaverria I. Distinct molecular profile of IRF4-rearranged large B-cell lymphoma. Blood. 2020;135:274–286. doi: 10.1182/blood.2019002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Au-Yeung RKH, Arias Padilla L, Zimmermann M, Oschlies I, Siebert R, Woessmann W, Burkhardt B, Klapper W. Experience with provisional WHO-entities large B-cell lymphoma with IRF4-rearrangement and Burkitt-like lymphoma with 11q aberration in paediatric patients of the NHL-BFM group. Br J Haematol. 2020;190:753–763. doi: 10.1111/bjh.16578. [DOI] [PubMed] [Google Scholar]
- 24.Ramis-Zaldivar JE, Gonzalez-Farre B, Balagué O, Celis V, Nadeu F, Salmeron-Villalobos J, Andres M, Martin-Guerrero I, Garrido-Pontnou M, Gaafar A, Suñol M, Barcena C, Garcia-Bragado F, Andión M, Azorín D, Astigarraga I, Sagaseta De Ilurdoz M, Sábado C, Gallego S, Verdu-Amorós J, Fernandez-Delgado R, Perez V, Tapia G, Mozos A, Torrent M, Solano-Páez P, Rivas-Delgado A, Dlouhy I, Clot G, Enjuanes A, López-Guillermo A, Galera PK, Oberley MJ, Maguire A, Ramsower C, Rimsza LM, Quintanilla-Martinez L, Jaffe ES, Campo E, Salaverria I. IRF4-rearranged large B-cell lymphoma (LBCL) has a genomic profile distinct to other LBCL in children and young adults. Blood. 2019 doi: 10.1182/blood.2019002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.