Abstract
The Prp19p-associated complex is essential for the yeast pre-mRNA splicing reaction. The complex consists of at least eight protein components, but is not tightly associated with spliceosomal snRNAs. By a combination of genetic and biochemical methods we previously identified four components of this complex, Ntc25p, Ntc85p, Ntc30p and Ntc20p, all of them being novel splicing factors. We have now identified three other components of the complex, Ntc90p, Ntc77p and Ntc31p. These three proteins were also associated with the spliceosome during the splicing reaction in the same manner as Prp19p, concurrently with or immediately after dissociation of U4 snRNA. Two-hybrid analysis revealed that none of these proteins interacted with Prp19p or Ntc25p, but all interacted with Ntc85p. An interaction network between the identified components of the Prp19p-associated complex is demonstrated. Biochemical analysis revealed that Ntc90p, Ntc31p, Ntc30p and Ntc20p form a subcomplex, which, through interacting with Ntc85p and Ntc77p, can associate with Prp19p and Ntc25p to form the Prp19p-associated complex. Genetic analysis suggests that Ntc31p, Ntc30p and Ntc20p may play roles in modulating the function of Ntc90p.
INTRODUCTION
Introns are excised from precursor mRNA via a two-step reaction which occurs in a large ribonucleoprotein complex called the spliceosome. The spliceosome is composed of five small nuclear RNAs (snRNAs), U1, U2, U4/U6 and U5, and a number of protein factors (for reviews see 1–6). Spliceosome assembly is a stepwise process involving sequential binding of snRNAs to the pre-mRNA in the order U1, U2, then U4/U6 and U5 as a pre-formed tri-snRNP particle. A subsequent conformational rearrangement, which results in dissociation of U1 and U4 accompanied by new base pair formation between U2 and U6, and between U6 and the 5′ splice site, activates the spliceosome and allows the catalytic reactions to take place (7–13). It is believed that such structural rearrangements of the spliceosome are mediated by protein factors. Indeed, several splicing factors containing the DEx(D/H) box motif have been shown to have RNA unwindase activity (6,14–16), although no substrate specificity has been demonstrated. Genetic analysis has provided evidence that Prp28p is involved in unwinding the U1–5′ splice site sequence and Brr2p may be responsible for unwinding of the U4/U6 duplex prior to activation of the spliceosome (17–19).
We have previously shown that yeast Prp19p protein is essential for pre-mRNA splicing and is associated with the spliceosome immediately after or concurrently with dissociation of U4 (20,21). Thus, Prp19p is likely to play a role in mediating this structural change in the spliceosome. Prp19p is not tightly associated with any of the spliceosomal snRNAs, but is associated with a protein complex consisting of at least eight protein components (22). The association of Prp19p with this protein complex appears to be essential for its function.
By a combination of genetic and biochemical approaches we have previously identified four components of the Prp19p-associated complex, Ntc85p, Ntc30p, Ntc25p (Snt309p) and Ntc20p (Ntc stands for PRP nineteen complex) (23–27). Ntc85p is essential for pre-mRNA splicing both in vivo and in vitro. It is important for binding of the Prp19p-associated complex to the spliceosome (24). Ntc25p, although not essential for the splicing reaction, plays a role in stabilizing the Prp19p-associated complex by modulating the interaction of Prp19p with its associated components (28). Ntc30p and Ntc20p are also non-essential splicing factors of unknown function (25). All four proteins are associated with the spliceosome immediately after or concomitantly with U4 dissociation in the same manner as Prp19p, suggesting that the entire complex is associated with the spliceosome as an integral complex.
We report here the identification of another three components of the Prp19p-associated complex, Ntc90p, Ntc77p and Ntc31p. Like the previously identified components, these three proteins were also associated with the spliceosome simultaneously with Prp19p, further strengthening the notion that the entire Prp19p-associated complex may bind to the spliceosome and function as a unit. Immunoprecipitation analysis revealed stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p to form a subcomplex in formation of the Prp19p-associated complex. Genetic analysis further suggests that Ntc31p, Ntc30p and Ntc20p may have overlapping functions in modulating the function of Ntc90p.
MATERIALS AND METHODS
Strains
YSCC1, MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA; YSCC3, MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP4-HA PRP19-MYC; YSCC11, MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA ΔNTC25::LEU2; YSCC14, MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA ΔNTC20::LEU2 ΔNTC30::URA3; YSCC20, MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC20-HA; YSCC31, MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC31-HA; YSCC90, MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC90-HA; YSCC77, MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC77-HA; YSCC771, MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC77-HA ΔNTC25::LEU2; YSCC201, MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC20-HA ΔNTC25::LEU2; CC3020, MATa leu2 trp1 ura3 his3 lys2 PRP19-HA ΔNTC20::LEU2 ΔNTC30::HIS3; CC3120, MATa leu2 trp1 ura3 his3 lys2 PRP19-HA ΔNTC20::LEU2 ΔNTC31::TRP1; CC3130, MATa leu2 trp1 ura3 his3 lys2 PRP19-HA ΔNTC30::HIS3 ΔNTC31::TRP1; CCD3, MATa/MATα leu2/leu2 ura3/ura3 his3/his3 trp1/trp1 suc2/suc2 lys2/LYS2 ADE2/ade2 NTC20/ΔNTC20::LEU2 NTC30/ΔNTC30::HIS3 NTC31/ΔNTC31::TRP1; SEY6210.5, MATa/MATα leu2/leu2 ura3/ura3 his3/his3 trp1/trp1 suc2/suc2 lys2/LYS2 ADE2/ade2; EGY48, MATα ura3 his3 trp1 LexAop-leu2 ade2; Y190, MATa gal4 gal80 his3 trp1 ade2 ura3 leu2 URA3::GAL-LacZ LYS::GAL(UAS)-HIS3c cyhr.
Oligonucleotides
77-1, GAGACAGAATCACAAGA; 77-2, GGCCTCTAGAAACGCCACGGTAAA; 77-3, CAGGAACAGGCACTTTACCCATACGACGTCCCAGACTACGCTTGAATAAAGGTAAGG; 90-1, CTCTAAAGCTGTTGAGT; 90-2, AGAAGCAGCAGCTTGTC; 90-3, ATAGAACTAGATATTTACCCATACGACGTCCCAGACTACGCTTGATATAAGCTATGT; 31-1, GGCCGGATCCGTCAATGTTCGCTT; 31-2, GGCCGGATCCTCGAGCTGGAGGAACAAGC; 31-3, TAAAAGAGAAGCTTAAGCGTAGTCTGGGACGTCGTATGGGTATTCTGATCCTTTTGA.
Plasmids
The construction of plasmids was as follows.
pLW7. The DNA fragment retrieved from the yeast genome by PCR with oligonucleotides 77-1 and 77-2 was digested with HindIII and XbaI and ligated into the HindIII and XbaI sites of plasmid pRS406.
pLW9. The DNA fragment retrieved from the yeast genome by PCR with oligonucleotides 90-1 and 90-2 was digested with HindIII and EcoRI and ligated into the HindIII and EcoRI sites of plasmid pRS406.
pLW71. The HA sequence was inserted into the C-terminus of Ntc77p in pLW7 by site-directed mutagenesis using oligonucleotide 77-3.
pLW91. The HA sequence was inserted into the C-terminus of Ntc90p in pLW9 by site-directed mutagenesis using oligonucleotide 90-3.
pCHC3. The DNA fragment retrieved from the yeast genome by PCR with oligonucleotides 31-1 and 31-2 was digested with BamHI and XhoI and ligated into the BamHI and XhoI sites of plasmid pRS406.
pCHC31. The HA sequence was inserted into the C-terminus of Ntc31p in pCHC3 by site-directed mutagenesis using oligonucleotide 31-3.
pTT91. A 3.4 kb EcoRI fragment containing the entire ORF of the NTC90 gene was inserted into the EcoRI site of plasmid pACT2.
pTT92. A 3.4 kb BamHI–XhoI fragment digested from pTT91 was ligated with BamHI + XhoI-digested plasmid pEG202.
pWC71. A 2.7 kb BamHI–SalI fragment containing the entire ORF of the NTC77 gene was ligated with BamHI + XhoI-digested plasmid pACT2.
pWC72. A 2.7 kb BamHI–SalI fragment containing the entire ORF of the NTC77 gene was ligated with BamHI + SalI-digested plasmid pEG202.
pWC73. A 2.7 kb BamHI–SalI fragment containing the entire ORF of the NTC77 gene was ligated with BamHI + SalI-digested plasmid pAS2.
pCHC32. A 1.4 kb BamHI–XhoI fragment containing the entire ORF of the NTC31 gene was ligated with BamHI + XhoI-digested plasmid pACT2.
pCHC33. A 1.4 kb BamHI–XhoI fragment containing the entire ORF of the NTC31 gene was ligated with BamHI + XhoI-digested plasmid pEG202.
Epitope tagging
DNA fragments of 1 and 1.1 kb from the 3′-end of the NTC90 and NTC77 genes, and the complete NTC31 gene, were isolated by PCR using oligonucleotides 77-1 and 77-2 for NTC77, 90-1 and 90-2 for NTC90 and 31-1 and 31-2 for NTC31 and cloned into plasmid vector pRS406. The NTC90 fragment contained 270 bp of the ORF and its downstream sequence and the NTC77 fragment contained 470 bp of the ORF and its downstream sequence. The HA epitope was then inserted into the C-terminus of each protein by site-directed mutagenesis using oligonucleotides 77-3, 90-3 and 31-3, respectively. The resulting plasmids were linearized by digestion with a restriction enzyme within the ORFs and used to transform yeast strain BJ2168 for directed integration. Correct integration was confirmed by Southern blotting.
Production and purification of antibodies
Ntc90p and Ntc31p were expressed in Escherichia coli under control of the T7 promoter. Total lysates prepared from induced cells were fractionated by SDS–PAGE and the Ntc90p and Ntc31p proteins were eluted from gels for immunization of rabbits. Ntc77p was expressed as a GST fusion using plasmid vector pGEX-5x-3 (Gibco BRL) and the protein was purified by chromatography on a glutathione column for immunization of rabbits. To purify the antibodies, the recombinant proteins were conjugated to CNBr-activated Sepharose according to the manufacturer’s manual (Pharmacia).
Splicing reactions and immunoprecipitation
Splicing assays were performed according to Lin et al. (29) using uncapped actin pre-mRNA as the substrate. Immunoprecipitation was carried out as described by Tarn et al. (20) using protein A–Sepharose.
Two-hybrid assays
The NTC90, NTC85, NTC77, NTC31, NTC30, NTC25, NTC20 and PRP19 genes were fused to the LexA DNA-binding domain (BD) in plasmid pEG202 and the GAL4 activation domain (AD) in plasmid pACT2, and each pair of plasmids was transformed into yeast strain EGY48 together with the β-galactosidase reporter plasmid pSH18-34. PRP19 and NTC25 were also fused to the GAL4 BD in plasmid pAS2 to assay for interaction with Ntc77p in strain Y190. Two-hybrid assays were carried out according to procedures described in the manual for the Matchmaker System (Clontech).
RESULTS
Identification of Ntc90p and Ntc77p as Syf1p and Syf3p
To identify the components of the Prp19p-associated complex, we purified the complex by affinity chromatography on an anti-HA antibody–Sepharose column using extracts prepared from a strain in which Prp19p was tagged with the HA epitope (24). Proteins specifically associated with Prp19p were identified by comparison with those isolated from the extract of an untagged strain. Figure 1 shows the components of the complex fractionated by 10% SDS–PAGE and stained with silver. The high molecular weight region is enlarged and shown on the left. To resolve proteins of higher molecular weight for sequencing, components of the purified complex were fractionated by 7.5% SDS–PAGE, then electroblotted to PVDF membranes. In addition to a peptide from Ntc85p, which was previously identified as Cef1p (24), two other peptide sequences from Ntc90p and Ntc77p were also obtained. A Blast search of the yeast genome database revealed that peptide sequence AMKGVITNVDENIRNDED from Ntc90p is in the sequence of Syf1p and AEQILRDVYKK from Ntc77p is in that of Syf3p.
Figure 1.
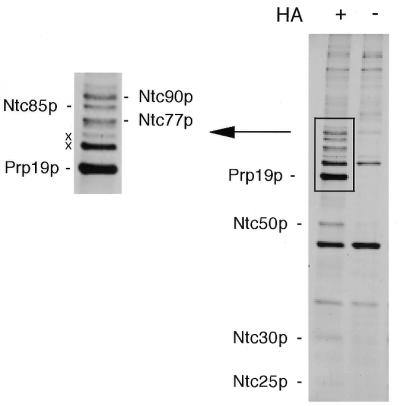
The Prp19p-associated complex was isolated by affinity chromatography on an anti-HA antibody column using the PRP19-HA extract and the components were fractionated by SDS–PAGE on a 10% gel. The extract without a tag was used as the background control (–). The high molecular weight region was enlarged as shown on the left.
To verify that Ntc90p and Ntc77p were indeed encoded by SYF1 and SYF3, respectively, we tagged Syf1p and Syf3p with the HA epitope as described in Materials and Methods for immunoprecipitation analysis to see whether these two proteins are associated with the Prp19p-associated complex. Splicing extracts were prepared from these epitope-tagged strains and subjected to immunoprecipitation with anti-HA antibody, followed by western blotting using antibodies against the identified Prp19p-associated components, Ntc85p, Prp19p, Ntc30p, Ntc25p and Ntc20p. Figure 2 shows that, like Prp19p (lane 2), components of the Prp19p-associated complex were co-precipitated when Syf1p (lane 3) or Syf3p (lane 4) was tagged with HA, but not when no protein (lane 1) or Prp4p (lane 5) was tagged. This indicates that both Syf1p and Syf3p are components of the Prp19p-associated complex and confirms that Ntc90p is Syf1p and Ntc77p is Syf3p.
Figure 2.
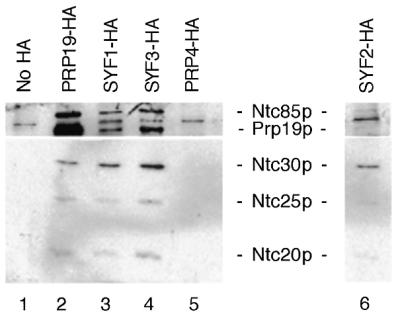
Identification of Syf1p, Syf2p and Syf3p as components of the Prp19p-associated complex. Syf1p, Syf2p and Syf3p were individually tagged with the HA epitope. Immunoprecipitation with the anti-HA antibody of extracts prepared from these strains (lanes 3, 4 and 6) revealed co-precipitation of the identified Prp19p-associated components.
Syf2p is a component of the Prp19p-associated complex
In a screen for mutants synthetic lethal to cdc40/prp17 mutation, SYF1, SYF2 and SYF3 were identified, in addition to several others encoding splicing factors (30). Since both Syf1p and Syf3p were found to be components of the Prp19p-associated complex, we speculated that Syf2p might be another component of the complex. To clarify this, the SYF2 gene was isolated by PCR amplification and tagged with the HA epitope at the C-terminus of the protein. The chromosomal copy of the SYF2 gene in strain BJ2168 was then replaced with SYF2-HA by gene displacement (31). Figure 2, lane 6 shows that immunoprecipitation of splicing extracts prepared from the SYF2-HA strain with the anti-HA antibody indeed precipitated the identified Prp19p-associated components, indicating that Syf2p is also a component of the Prp19p-associated complex. Syf2p was expressed in E.coli and the recombinant protein was used to raise antibodies. Western blot analysis of the Prp19p-associated complex revealed that Syf2p migrated only slightly slower than Ntc30p on SDS–PAGE (data not shown) and was therefore named Ntc31p, which was not detected on the gel possibly due to its poor staining by silver. This makes a total of eight components identified in the Prp19p-associated complex, as listed in Table 1.
Table 1. Components of the Prp19p-associated complex.
| Gene name | Growth requirement | Identification method | |
|---|---|---|---|
| PRP19 | Essential | ||
| NTC90 | SYF1 | Essential | Sequencing |
| NTC85 | CEF1 | Essential | Sequencing |
| NTC77 | CLF1, SYF3 | Essential | Sequencing |
| NTC31 | SYF2 | Non-essential | |
| NTC30 | ISY1 | Non-essential | Genetics |
| NTC25 | SNT309 | Non-essential | Genetics |
| NTC20 | Non-essential | Genetics, sequencing |
Ntc90p, Ntc77p and Ntc31p are associated with the spliceosome in the same manner as Prp19p
We have shown that Prp19p is associated with the spliceosome during the splicing reaction concomitant with or after dissociation of U4 (21,32). The previously identified components of the Prp19p-associated complex are also associated with the spliceosome in the same manner as Prp19p (23–25). It is conceivable that Ntc90p, Ntc77p and Ntc31p are also associated with the spliceosome at the same time as Prp19p during the splicing reaction. Using IgG–agarose to precipitate protein A-fused Syf1p, Syf2p and Syf3p, Russell et al. demonstrated that all three proteins co-precipitated with the spliceosome from the splicing reaction mixture (33). Similarly, spliceosomes formed in NTC90-HA, NTC77-HA or NTC31-HA extracts could be precipitated by anti-HA antibody, as shown in Figure 3A, indicating that these three proteins are associated with the spliceosome during the splicing reaction.
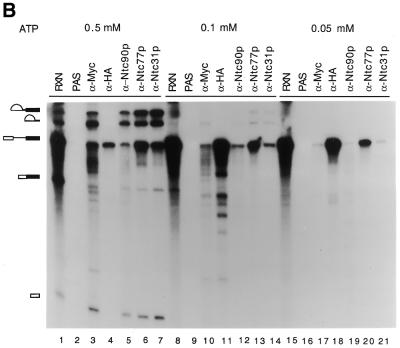
Figure 3.
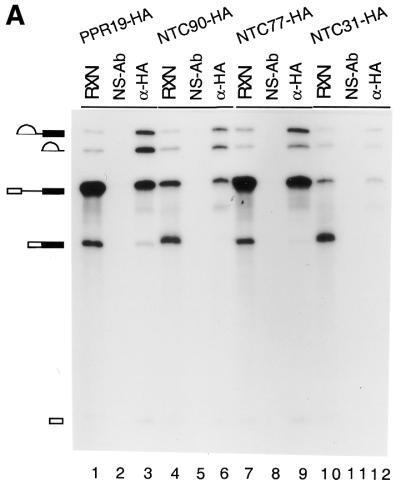
(A) Immunoprecipitation of the spliceosome formed in PRP19-HA (lanes 1–3), NTC90-HA (lanes 4–6), NTC77-HA (lanes 7–9) and NTC31-HA (lanes 10–12) extracts with the anti-HA antibody (lanes 3, 6, 9 and 12). (B) Immunoprecipitation of the spliceosome formed at various ATP concentrations in extracts prepared from a strain in which Prp19p was tagged with the c-Myc epitope and Prp4p tagged with the HA epitope. The splicing reaction was carried out at 0.5 mM (lanes 1–7), 0.1 mM (lanes 8–14) and 0.05 mM (lanes 15–21) ATP and the reaction mixtures (15 µl each) were subjected to immunoprecipitation with protein A–Sepharose alone (lanes 2, 9 and 16), anti-Myc (lanes 3, 10 and 17), anti-HA (lanes 4, 11 and 18), anti-Ntc90p (lanes 5, 12 and 19), anti-Ntc77p (lanes 6, 13 and 20) and anti-Ntc31p antibodies (lanes 7, 14 and 21). RXN, 2 µl of the reaction mixture; NS-Ab, non-specific antibody; PAS, protein A–Sepharose.
To see whether Ntc90p, Ntc77p and Ntc31p also bind to the spliceosome at the same time as Prp19p, we performed immunoprecipitation experiments with splicing reactions carried out in the presence of different concentrations of ATP and compared the pattern of precipitation of the spliceosome by different antibodies. We previously demonstrated that dissociation of U4 from the spliceosome is very sensitive to ATP concentration (21). At higher concentrations of ATP, U4 is rapidly dissociated from the spliceosome after binding of the tri-snRNPs. As a consequence, only a very small amount of U4-containing splicing complex is detected. Dissociation of U4 is blocked at lower concentrations of ATP, resulting in increasing amounts of U4-containing splicing complexes. In contrast, Prp19p is not associated with the spliceosome at lower but only at higher concentrations of ATP (21). We took advantage of this feature to analyze the step of spliceosome assembly in which Ntc90p, Ntc77p and Ntc31p participated. In this experiment, we used a strain in which Prp19p was tagged with the c-Myc epitope and Prp4p was tagged with HA. Prp4p was shown to bind to the 5′-portion of U4 (34) and dissociated from the spliceosome with U4. Therefore, the anti-HA antibody could be used to follow U4 and the anti-Myc antibody to follow Prp19p as controls (25).
Splicing reactions were carried out at different ATP concentrations in extracts prepared from the strain expressing PRP19-Myc and PRP4-HA and the reaction mixtures subjected to immunoprecipitation with anti-Myc, anti-HA, anti-Ntc90p, anti-Ntc77p and anti-Ntc31p antibodies. As shown in Figure 3B, the extract showed a high level of splicing activity at 0.5 mM ATP (lane 1), a low level at 0.1 mM ATP (lane 8) and nearly no splicing activity at 0.05 mM ATP (lane 15). The anti-Myc antibody efficiently precipitated the spliceosome from the reaction mixtures at 0.5 mM ATP (lane 3). At 0.1 mM ATP a small amount of pre-mRNA and lariat intermediate and IVS were precipitated (lane 10) and at 0.05 mM ATP only a negligible amount of pre-mRNA was precipitated by the anti-Myc antibody (lane 17). In contrast, the anti-HA antibody, binding to Prp4p-HA, precipitated little pre-mRNA at 0.5 mM ATP (lane 4), but large amounts of pre-mRNA at 0.1 (lane 11) and 0.05 mM ATP (lane 18). In all cases, no splicing intermediates or products were precipitated, indicating that U4 is dissociated prior to the catalytic steps of the splicing reaction. Precipitation of splicing reaction mixtures with anti-Ntc90p, anti-Ntc77p and anti-Ntc31p antibodies showed similar patterns to that with the anti-Myc antibody. Spliceosome-associated RNAs were precipitated in larger amounts at 0.5 mM ATP (lanes 5–7), less at 0.1 mM ATP (lanes 12–14) and in negligible amounts at 0.05 mM ATP (lanes 19–21). This indicates that Ntc90p, Ntc77p and Ntc31p are associated with the spliceosome immediately after or concurrently with dissociation of U4 in the same way as Prp19p. From these and previous results, all the components of the Prp19p-associated complex bind to the spliceosome at the same time during spliceosome assembly, suggesting that the entire complex may bind to the spliceosome as an integral unit.
Interactions of components of the Prp19p-associated complex
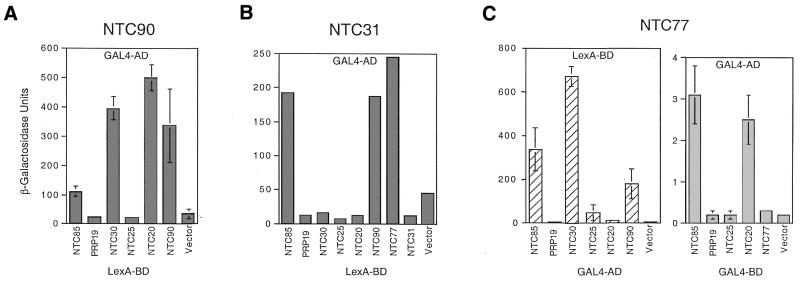
Our previous studies have demonstrated that among the identified components of the Prp19p-associated complex, Ntc85p and Ntc25p interact with Prp19p directly, whereas neither Ntc30p nor Ntc20p interacts with Prp19p. Ntc30p and Ntc20p also show no interaction with Ntc25p or with each other, but both interact with Ntc85p. Ntc85p does not interact with Ntc25p (23–25). To reveal interactions between Ntc90p, Ntc77p, Ntc31p and other components of the complex, two-hybrid analyses were carried out. All of the proteins were fused to the LexA BD and the GAL4 AD as before and interactions between each pair of proteins were assayed by measuring the β-galactosidase activity. Figure 4 shows the results of the two-hybrid assay for Ntc90p (Fig. 4A) and Ntc31p (Fig. 4B) fused to the GAL4 AD. Similar patterns were seen when they were fused to the LexA BD (data not shown) except that an interaction between Ntc90p and Ntc20p was not detected. The interaction of Ntc77p with the other components was primarily based on assays of the Ntc77p–LexA BD fusion with the AD fusion of its counterparts (Fig. 4C, left). The Ntc77p–AD fusion interacted strongly with the LexA BD alone and, therefore could not be used to assay for interaction with LexA BD fusions of other proteins. This may give rise to problems in revealing the interaction of those proteins whose interaction can be detected only when fused to BD, as in the case of Ntc20p. Therefore, we further fused Prp19p, Ntc25p and Ntc20p, whose AD fusions showed weak or no interaction with Ntc77p–BD, to the GAL4 BD and assayed for interaction with Ntc77p–AD. Ntc77p, although it interacted with the LexA BD, did not interact with the GAL4 BD. The result in Figure 4C, right, shows that Ntc77p interacts with Ntc20p, but not with Prp19p or Ntc25p. These results together demonstrate that Ntc90p and Ntc77p have identical interaction patterns in that both proteins interact with Ntc85p, Ntc31p, Ntc30p, Ntc20p and with each other, but not with Prp19p or Ntc25p. Ntc31p, like Ntc30p and Ntc20p, only interacted with Ntc85p, Ntc90p and Ntc77p. Furthermore, Ntc90p, but not Ntc77p nor Ntc31p, could interact with itself. A schematic interaction between all the identified components of the Prp19p-associated complex is shown in Figure 5, with those encoded by essential genes in gray and non-essential genes in black.
Figure 4.
Two-hybrid assays showing interactions between the Prp19p-associated components. (A) Ntc90p was fused to the GAL4 AD and the counterparts fused to the LexA BD. (B) Ntc31p was fused to the GAL4 AD and the counterparts fused to the LexA BD. (C) Left, Ntc77p was fused to the LexA BD and the counterparts fused to the GAL4 AD. Right, Ntc77p was fused to the GAL4 AD and the counterparts fused to the GAL4 BD.
Figure 5.
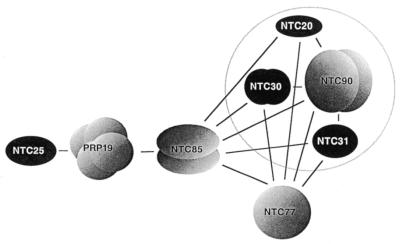
A scheme showing interactions between the components of the Prp19p-associated complex. Prp19p, Ntc90p, Ntc85p and Ntc30p are shown as multi-subunit homo-oligomers due to observed self-interactions in two-hybrid assays. Prp19p was demonstrated to be in tetrameric form. Self-interaction was not detected for Ntc77p, Ntc31p, Ntc25p or Ntc20p. Proteins encoded by essential genes are shown in gray and non-essential genes in black. The circle indicates stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p in a subcomplex.
Genetic interactions between components of the Prp19p-associated complex
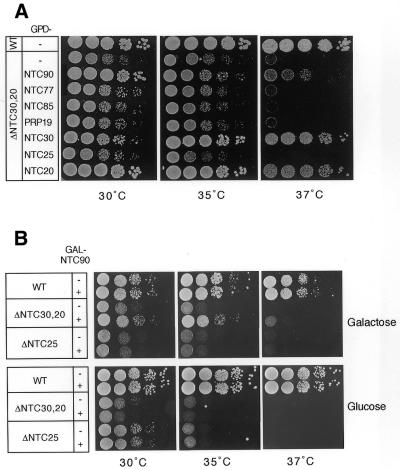
Among the four components encoded by non-essential genes, Ntc25p is distinct from the others in its interaction only with Prp19p and not with the other components. It is interesting that Ntc31p, Ntc30p and Ntc20p show identical patterns of interaction with other associated components in that all of them interacted with Ntc85p, Ntc90p and Ntc77p, but not with Prp19p, Ntc25p or each other. In contrast to deletion of NTC25, which gives a temperature-sensitive phenotype accompanied by accumulation of pre-mRNA (23), deletion of either NTC31, NTC30 or NTC20 alone did not produce an obvious growth defect, as shown in Figure 6A. Deletion of both NTC30 and NTC20 (ΔNTC30/ΔNTC20) has previously been shown to severely impair cell growth (25), suggesting that these two genes may have overlapping functions. We further examined whether NTC31 also functionally overlaps with NTC20 or NTC30 by constructing double deletion mutants of NTC31 and NTC30 (ΔNTC31/ΔNTC30) and of NTC31 and NTC20 (ΔNTC31/ΔNTC20). Figure 6A shows that deletion of NTC31 and NTC30 impairs growth only at higher temperatures, while deletion of NTC31 and NTC20 gave no phenotype. However, deletion of all three genes resulted in cell lethality. As shown in Figure 6B, when the wild-type, double deletion and triple deletion strains carrying plasmid pRS416.NTC30 were streaked on YPD or 5-FOA plates, while the wild-type and double deletion mutants grew well, the triple deletion mutant did not grow on 5-FOA plates. This result, in conjunction with the two-hybrid interaction pattern, indicates that NTC31, NTC30 and NTC20 may perform overlapping functions, although they differ in their degree of functional importance.
Figure 6.
Growth analysis of NTC31, NTC30 and NTC20 single, double and triple deletion mutants. (A) Growth phenotypes of single and double deletion mutants of NTC31, NTC30 and NTC20 were examined by spot assay on YPD plates. (B) Cells of the wild-type, double deletion and triple deletion strains carrying plasmid pRS416.NTC30 were streaked on YPD or 5-FOA plates. WT, wild-type.
We previously showed that Ntc25p plays a role in regulating the interaction of Prp19p with other associated components through its interaction with Prp19p (23). It is possible that Ntc31p, Ntc30p and Ntc20p might play similar roles in regulating their interacting components Ntc90p, Ntc85p and Ntc77p, all of them being essential splicing factors. Taking advantage of the growth defect of the double mutant ΔNTC30/ΔNTC20, we tested functional interactions between NTC30, NTC20 and other genes encoding the Prp19p-associated components by functional complementation. Plasmids carrying individual genes under control of the GPD promoter were transformed into ΔNTC30/ΔNTC20 cells and the resulting transformants were examined for cell growth by spot assay. Figure 7A shows that among all the genes encoding the Prp19p-associated components, only overexpression of NTC90 could rescue cellular growth at 30°C, although it did so less well at higher temperatures. Overexpression of PRP19, NTC85 or NTC77 in ΔNTC30/ΔNTC20 cells did not affect growth significantly. This indicates that overexpression of NTC90 could partially bypass the requirement for NTC30 and NTC20, suggesting functional interactions between NTC30, NTC20 and NTC90.
Figure 7.
The growth defect of a ΔNTC30/ΔNTC20 mutant could be partially rescued by overexpression of NTC90. (A) ΔNTC30/ΔNTC20 cells carrying plasmids containing individual genes encoding components of the Prp19p-associated complex in plasmid vector pG1 were spotted on YPD plates and grown at 30, 35 and 37°C. (B) Wild-type, ΔNTC30/ΔNTC20 or ΔNTC25 cells carrying the NTC90 gene under control of the GAL promoter on plasmid vector pRS414 were spotted on tryptophan drop-out–SMM plates supplemented with galactose or glucose and grown at 30, 35 and 37°C. WT, wild-type.
Like ΔNTC30/ΔNTC20 cells, ΔNTC25 cells also show a slow growing and temperature-sensitive phenotype. To rule out the possibility that overexpression of NTC90 can generally promote growth of cells with a slow growing phenotype, we tested complementation of ΔNTC25 cells by NTC90. Plasmids carrying NTC90 under control of the GAL promoter were transformed into ΔNTC30/ΔNTC20 and ΔNTC25 cells and the resulting transformants were examined for growth on synthetic medium supplemented with galactose or glucose by spot assay. As shown in Figure 7B, while the growth defect of ΔNTC30/ΔNTC20 was partially rescued by NTC90 when grown on galactose plates, growth of ΔNTC25 cells was not improved. This indicates that complementation by NTC90 was specific for ΔNTC30/ΔNTC20, further strengthening the notion of functional interactions between NTC30, NTC20 and NTC90.
Stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p in a subcomplex
We have previously demonstrated that in the Prp19p-associated complex, Ntc25p binds tightly to Prp19p, which interacts with Ntc85p, which then interacts with Ntc30p and Ntc20p (25). In the absence of Ntc25p, Ntc30p and Ntc20p are dissociated from Prp19p and Ntc85p, but remain stably associated with each other (28). Ntc30p and Ntc20p were not detected as interacting with each other by two-hybrid analysis (25) or by far western blotting (data not shown) and therefore are associated through association with unidentified components. Since both Ntc20p and Ntc30p interact with Ntc90p and Ntc77p, it is possible that Ntc90p or Ntc77p is the protein(s) bridging Ntc30p and Ntc20p in ΔNTC25 extracts. Ntc90p is particularly likely to play such a role in view of the fact that NTC90 could functionally complement the ΔNTC30/ΔNTC20 growth defect.
To examine whether Ntc20p and Ntc30p were associated with Ntc90p and/or Ntc77p in ΔNTC25 extracts, Ntc20p was tagged with the HA epitope in a strain in which NTC25 was deleted. Extracts prepared from such a strain were precipitated with anti-HA antibody followed by western blotting to analyze the proteins co-precipitated with Ntc20p. As shown in Figure 8, in the presence of Ntc25p all of the eight identified components of the Prp19p-associated complex were co-precipitated (lane 3). Ntc90p, Ntc31p and Ntc30p, were co-precipitated with Ntc20p in the ΔNTC25 extract (lane 4) as efficiently as in extracts containing Ntc25p (lane 3), indicating stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p independent of Ntc25p. In contrast, Ntc77p and Ntc85p were co-precipitated only in smaller amounts and Prp19p was barely co-precipitated with Ntc20p (lane 4). These results indicate that Ntc90p is the protein bridging Ntc30p and Ntc20p, and these three proteins together with Ntc31p might form a subcomplex, to be called the Ntc90p subcomplex, in formation of the Prp19p-associated complex. Ntc90p, Ntc31p, Ntc30p and Ntc20p are circled together in Figure 5 to indicate this fact. On the other hand, when Prp19p or Ntc77p was tagged with HA for precipitation with anti-HA antibody, none of the associated components co-precipitated efficiently with Prp19p or Ntc77p in the ΔNTC25 extract (Fig. 8, compare lane 2 with lane 1 and lane 5 with lane 6). This suggests that association of Prp19p or Ntc77p with the other components deteriorated in the extract lacking Ntc25p.
Figure 8.
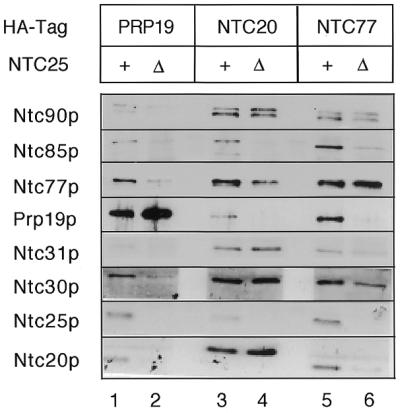
Immunoprecipitation of ΔNTC25 extracts revealed stable association of Ntc90p, Ntc31p, Ntc30p and Ntc20p in a subcomplex. Prp19p, Ntc20p or Ntc77p was tagged with the HA epitope in a strain in which the NTC25 gene was deleted. Extracts prepared from such strains were precipitated with anti-HA antibody followed by western blotting.
DISCUSSION
With the identification of Ntc90p, Ntc77p and Ntc31p, eight components of the Prp19p-associated complex are known. Like previously identified components, these three proteins are associated with the spliceosome in the same way as Prp19p. Thus, all eight known components of the complex bind to the spliceosome at the same time during spliceosome assembly, indicating that the Prp19p-associated complex might be added to the spliceosome as an integral complex. During spliceosome assembly, snRNAs are associated with the spliceosome in a sequential manner and function in splicing in the form of ribonucleoprotein complexes (35–40). Although the RNA components of snRNPs play major roles in the recognition and alignment of splice sites through base pair interactions between the snRNAs and intron sequences, some protein components are suggested to perform important functions in modulating structural rearrangement of RNA (14,41). Aside from these, no protein splicing factors have been shown to function as a complex. The significance of functional association of the Prp19p-associated complex with the spliceosome awaits further study.
Among the identified Prp19p-associated components, the gene encoding Ntc31p, Ntc30p, Ntc25p or Ntc20p is not essential for cell viability. Accordingly, none of these proteins is essential for splicing, despite their association with the spliceosome during the splicing reaction. The function of these proteins in the splicing machinery remains an interesting question. They may play indirect roles in the splicing reaction through regulating the function of other essential components or they may be functionally redundant to each other or to other splicing factors. Indeed, our previous studies showed that Ntc25p interacts strongly with Prp19p and plays a role in stabilizing the Prp19p-associated complex by modulating the interaction of Prp19p with the other components of the complex (28). Yeast cells deleted of NTC25 grow poorly at higher temperatures or in synthetic media, reflecting the requirement for Ntc25p under sub-optimal conditions to stabilize the Prp19p-associated complex. Three lines of evidence suggest that Ntc31p, Ntc30p and Ntc20p might have overlapping functions in regulating the function of Ntc90p. First, Ntc31p, Ntc30p and Ntc20p showed identical patterns on interaction with the other components of the Prp19p-associated complex, including Ntc90p. Secondly, these three proteins together formed a stable complex with Ntc90p, as seen in the extract lacking Ntc25p. Finally, genetic analysis revealed that although individual deletion of the NTC31, NTC30 and NTC20 genes produced no obvious growth defects, deletion of both NTC30 and NTC20 severely impaired cell growth (25) and additional deletion of NTC31 resulted in cell death. The growth defect of ΔNTC30/ΔNTC20 could be partially rescued by overexpression of NTC90. In this context, it is likely that Ntc31p, Ntc30p and Ntc20p play roles in modulating interactions of Ntc90p with other components in a similar way to Ntc25p regulation of Prp19p. These three proteins, being in the same subcomplex with Ntc90p, may share common functions in doing so.
PRP19, NTC85, NTC90 and NTC77 are essential for vegetative yeast growth. Prp19p and Ntc85p have been shown to be required for the in vitro splicing reaction (20,24) and NTC90/SYF1, NTC77/CLF1/SYF3 and NTC85 have been shown to be essential for splicing in vivo (24,33,42). It is interesting that two of these essential splicing factors, Prp19p and Ntc90p, are regulated by auxiliary components in the complex. These proteins function normally without other cofactors under optimal condition, but otherwise require their regulatory components, possibly to ensure their proper function in accommodating environmental change. This may reflect important functions associated with these factors and may also explain why many genes encoding splicing factors are not essential for viability.
Biochemical analysis reveals that Ntc85p may play a role in promoting binding of the Prp19p-associated complex to the spliceosome (24). The functional roles of Prp19p, Ntc90p and Ntc77p remain to be investigated. Chung et al. reported that Clf1p/Ntc77p is required for binding of the tri-snRNP particle to the spliceosome since isolated spliceosomes formed in an in vivo Clf1p/Ntc77p-depleted extract did not contain U4, U5 or U6 (42). This result is inconsistent with our finding that Ntc77p, as a component of the Prp19p-associated complex, became associated with the spliceosome after or concurrently with dissociation of U4. In this case, Ntc77p is not required for tri-snRNP binding. The discrepancy between the results of Chung et al. (42) and ours remains to be resolved.
In an extensive two-hybrid screening, some interactions between components of the Prp19p-associated complex have been identified (30). The reported results are in part consistent with our identified interactions between components of the complex. This type of analysis, although allowing preliminary establishment of protein linkage networks, does not provide complete information on interactions between designated components which are functionally linked. Indeed, interactions between Ntc85p/Cef1p and Prp19p and between Ntc90p/Syf1p and Ntc77p/Syf3p were not identified in their screening. Instead, our systematic two-hybrid analysis between components of the Prp19p-associated complex has revealed a complete interaction map among all the components. This, in conjunction with the immunoprecipitation analysis, enables us to outline the architecture of the Prp19p-associated complex.
In addition to those interactions we observed among the Prp19p-associated components, interactions between Prp22p and Syf1p/Ntc90p and Syf3p/Ntc77p were also identified by Ben-Yehuda et al. (30). Prp22p is a member of the DExH protein family and has been demonstrated to be involved in the second step of the catalytic reaction, as well as release of the intron from the spliceosome after completion of the splicing reaction (43,44). It was demonstrated that in vitro depletion of Prp22p resulted in splicing arrest after the first step of the catalytic reaction (44). In this view, Prp22p is unlikely to be a component of the Prp19p-associated complex. Indeed, western blotting of the purified Prp19p-associated complex using an anti-Prp22p antibody did not detect Prp22p in the complex, although the antibody detected the protein easily in the splicing extract (data not shown). Thus, Prp22p is not tightly associated with the Prp19p-associated complex and interactions between Prp22p and Ntc90p/Syf1p and between Prp22p and Ntc77p/Syf3p might occur only after binding of these components to the spliceosome.
Acknowledgments
ACKNOWLEDGEMENTS
We thank B. Schwer for the anti-Prp22p antibody and M. Tam for synthesis of oligonucleotides and for protein sequencing. We also thank T. Hsieh and P. Lin for critical reading of the manuscript. This work was supported by a grant from the Academia Sinica and by National Science Council (Taiwan) grant NSC89-2321-B-001-008.
REFERENCES
- 1.Beggs J.D. (1993) Yeast protein splicing factors involved in nuclear pre-mRNA splicing. Mol. Biol. Rep., 18, 99–103. [DOI] [PubMed] [Google Scholar]
- 2.Guthrie C. (1991) Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science, 253, 157–163. [DOI] [PubMed] [Google Scholar]
- 3.Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357.
- 4.Rymond B.C. and Rosbash,M. (1992) Yeast pre-mRNA splicing. In Jones,E.W., Pringle,J.R. and Broach,R.J. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 2, pp. 143–192.
- 5.Sharp P.A. (1994) Split genes and RNA splicing. Cell, 77, 805–815. [DOI] [PubMed] [Google Scholar]
- 6.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- 7.Bindereif A. and Green,M.R. (1987) An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J., 6, 2415–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng S.-C. and Abelson,J. (1987) Spliceosome assembly in yeast. Genes Dev., 1, 1014–1027. [DOI] [PubMed] [Google Scholar]
- 9.Lamond A.I., Konarska,M.M., Grabowski,P.J. and Sharp,P.A. (1988) Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc. Natl Acad. Sci. USA, 85, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konarska M.M. and Sharp,P.A. (1986) Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell, 46, 845–855. [DOI] [PubMed] [Google Scholar]
- 11.Konarska M.M. and Sharp,P.A. (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell, 49, 763–774. [DOI] [PubMed] [Google Scholar]
- 12.Pikielny C.W. and Rosbash,M. (1986) Specific small nuclear RNAs are associated with yeast spliceosomes. Cell, 45, 869–877. [DOI] [PubMed] [Google Scholar]
- 13.Pikielny C.W., Rymond,B.C. and Rosbash,M. (1986) Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature, 324, 341–345. [DOI] [PubMed] [Google Scholar]
- 14.Laggerbauer B., Achsel,T. and Lührmann,R. (1998) The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA, 95, 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner J.O., Jankowsky,E., Company,M., Pyle,A.M. and Abelson,J.N. (1998) The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J., 17, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Wagner,J.D.O. and Guthrie,C. (1998) The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol., 8, 441–451. [DOI] [PubMed] [Google Scholar]
- 17.Chen J.Y.-F., Stands,L., Staley,J.P., Jackups,J.R.R., Latus,L.J. and Chang,T.-H. (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell, 7, 227–232. [DOI] [PubMed] [Google Scholar]
- 18.Schwer B. (2001) A new twist on RNA helicases: DExH/D box proteins as RNPases. Nature Struct. Biol., 8, 113–116. [DOI] [PubMed] [Google Scholar]
- 19.Staley J.P. and Guthrie,C. (1999) An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell, 3, 55–64. [DOI] [PubMed] [Google Scholar]
- 20.Tarn W.-Y., Lee,K.-R. and Cheng,S.-C. (1993) The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol., 13, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarn W.-Y., Lee,K.-R. and Cheng,S.-C. (1993) Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl Acad. Sci. USA, 90, 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarn W.-Y., Hsu,C.-H., Huang,K.-T., Chen,H.-R., Kao,H.-Y., Lee,K.-R. and Cheng,S.-C. (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J., 13, 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H.-R., Jan,S.-P., Tsao,T.Y., Sheu,Y.-J., Banroques,J. and Cheng,S.-C. (1998) Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol., 18, 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai W.-Y., Chow,Y.-T., Chen,H.-R., Huang,K.-T., Hong,R.-I., Jan,S.-P., Kuo,N.-Y., Tsao,T.Y., Chen,C.-H. and Cheng,S.-C. (1999) Cef1p is a component of the Prp19p-asociated complex and essential for pre-mRNA splicing. J. Biol. Chem., 274, 9455–9462. [DOI] [PubMed] [Google Scholar]
- 25.Chen C.-H., Tsai,W.-Y., Wang,C.-H., Chen,H.-R. and Cheng,S.-C. (2001) Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem., 276, 488–494. [DOI] [PubMed] [Google Scholar]
- 26.Dix I., Russell,C.S., Ben-Yehuda,S., Kupiec,M. and Beggs,J.D. (1999) The identification and characterization of a novel splicing protein, Isy1p, of Saccharomyces cerevisiae. RNA, 5, 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohi R., Feoktistova,A., McCann,S., Valentine,V., Look,A.T., Lipsick,J. and Gould,K.L. (1998) Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol., 18, 4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H.-R., Tsao,T.Y., Chen,C.-H., Tsai,W.-Y., Her,L.-S., Hsu,M.-T. and Cheng,S.-C. (1999) Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 5406–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin R.-J., Newman,A.J., Cheng,S.-C. and Abelson,J. (1985) Yeast mRNA splicing in vitro. J. Biol. Chem., 260, 14780–14792. [PubMed] [Google Scholar]
- 30.Ben-Yehuda S., Dix,I., Russell,C.S., McGarvey,M., Beggs,J.D. and Kupiec,M. (2000) Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics, 156, 1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winston F., Chumley,F. and Fink,G.R. (1983) Eviction and transplacement of mutant genes in yeast. Methods Enzymol., 101, 211–228. [DOI] [PubMed] [Google Scholar]
- 32.Cheng S.-C., Tarn,W.-Y., Tsao,T.Y. and Abelson,J. (1993) PRP19: a novel spliceosomal component. Mol. Cell. Biol., 13, 1876–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell C.S., Ben-Yehuda,S., Dix,I., Kupiec,M. and Beggs,J.D. (2000) Functional analysis of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA, 6, 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y., Petersen-Bjørn,S. and Friesen,J.D. (1990) The PRP4 (RNA4) protein of Saccharomyces cerevisiae is associated with the 5′ portion of the U4 snRNA. Mol. Cell. Biol., 10, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhani H.D. and Guthrie,C. (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell, 71, 803–818. [DOI] [PubMed] [Google Scholar]
- 36.Newman A. and Norman,C. (1991) Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell, 65, 115–123. [DOI] [PubMed] [Google Scholar]
- 37.Newman A.J. and Norman,C. (1992) U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell, 68, 743–754. [DOI] [PubMed] [Google Scholar]
- 38.Sawa H. and Shimura,Y. (1992) Association of U6 snRNA with the 5′-splice site region of pre-mRNA in the spliceosome. Genes Dev., 6, 244–254. [DOI] [PubMed] [Google Scholar]
- 39.Sawa H. and Abelson,J. (1992) Evidence for a base-pairing interaction between U6 small nuclear RNA and the 5′ splice site during the splicing reaction in yeast. Proc. Natl Acad. Sci. USA, 89, 11269–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassarman D.A. and Steitz,J.A. (1992) Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science, 257, 1918–1925. [DOI] [PubMed] [Google Scholar]
- 41.Fabrizio P., Laggerbauer,B., Lauber,J., Lane,W.S. and Lührmann,R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J., 13, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung S., Mclean,M.R. and Rymond,B.C. (1999) Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA, 5, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Company M., Arenas,J. and Abelson,J. (1991) Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature, 349, 487–493. [DOI] [PubMed] [Google Scholar]
- 44.Schwer B. and Gross,C.H. (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J., 17, 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]