Abstract
In vivo experimental analysis of human brain tissue poses substantial challenges and ethical concerns. To address this problem, we developed a computational method called the Brain Gene Expression and Network-Imputation Engine (BrainGENIE) that leverages peripheral-blood transcriptomes to predict brain tissue-specific gene-expression levels. Paired blood–brain transcriptomic data collected by the Genotype-Tissue Expression (GTEx) Project was used to train BrainGENIE models to predict gene-expression levels in ten distinct brain regions using whole-blood gene-expression profiles. The performance of BrainGENIE was compared to PrediXcan, a popular method for imputing gene expression levels from genotypes. BrainGENIE significantly predicted brain tissue-specific expression levels for 2947–11,816 genes (false-discovery rate-adjusted p < 0.05), including many transcripts that cannot be predicted significantly by a transcriptome-imputation method such as PrediXcan. BrainGENIE recapitulated measured diagnosis-related gene-expression changes in the brain for autism, bipolar disorder, and schizophrenia better than direct correlations from blood and predictions from PrediXcan. We developed a convenient software toolset for deploying BrainGENIE, and provide recommendations for how best to implement models. BrainGENIE complements and, in some ways, outperforms existing transcriptome-imputation tools, providing biologically meaningful predictions and opening new research avenues.
Subject terms: Genomics, Psychiatric disorders
Introduction
Brain disorders cause considerable disability worldwide [1]. Typically, in vivo molecular assessment of human disease centers on the primarily affected tissue(s) or the site of pathogenesis, but that is not possible for brain disorders unless neurosurgical intervention is required. Collecting ex vivo human brain tissue in an experimental setting for neuropsychiatric research is infeasible, given the considerable risks associated with brain biopsy. There are numerous research questions that would be answered best by studying living human brain tissue, but which therefore remain unaddressed. Transcriptome imputation offers a non-invasive alternative to brain biopsy by allowing investigators to infer tissue-specific gene expression without directly assaying gene-expression levels.
FUSION and PrediXcan are two software tools that model tissue-specific effects of expression quantitative trait loci (eQTLs) on the expression of proximal genes (cis-eQTLs) in order to impute transcriptome profiles. These methods have been successful in prioritizing genome-wide association study (GWAS) hits and have helped reveal putative mechanisms underlying complex disorders [2–6]. With both methods, there is a striking disparity between the number of genes imputable in the brain versus tissues outside of the central nervous system (CNS): to wit, FUSION imputes an average of 3158 genes in the brain (range = 1604–5855 across the 12 brain tissues (including a pair of re-sampled tissues from frontal cortex [BA9] and cerebellum) compared with 5592 in non-CNS tissues; similarly, PrediXcan imputes an average of 4337 genes in the brain (range = 2559–6794) compared with 6262 genes (range = 1642–10,012) outside the CNS. Furthermore, the majority of genes in the brain transcriptome are not significantly predicted by either FUSION or PrediXcan, suggesting that a large amount of variance in transcriptome profiles cannot be captured by eQTLs alone. A recent addition to the suite of genotype-based transcriptome-imputation methods is TIGAR, which uses a Bayesian modeling framework for predicting gene expression from eQTL data. Using a data-driven nonparametric model of cis-eQTL signals, TIGAR further increases the number of imputable genes by 57.8% compared to PrediXcan, but PrediXcan was deemed the preferred method for imputing genes that have few eQTLs influencing expression heritability [7]. A Bayesian hierarchical model called EpiXcan builds upon PrediXcan by applying epigenetic annotations to optimize the weights assigned to cis-eQTLs and increase the predictability of gene-expression levels [8]. EpiXcan increased the number of genes that can be significantly predicted by 94% compared to PrediXcan. Among the 2894 genes for which expression levels can be significantly predicted by both methods, EpiXcan showed better average prediction accuracy (mean cross-validation R2 = 0.19) compared to PrediXcan (mean R2 = 0.16). Despite the methodological improvements that have been made by derivatives of PrediXcan, a common limitation with existing cis-eQTL-based toolsets is that they do not allow for predictions for temporal changes in tissue-specific gene expression.
Tissue-specific and tissue-dependent gene expression help differentiate between brain and peripheral tissues, but compelling evidence also shows that brain and blood exhibit comparable transcriptome profiles [9–12]. Our group systematically reviewed relevant literature on this topic, and found that gene expression profiles in blood and brain are moderately correlated (Pearson’s r of 0.24–0.64), with 35–80% of genes expressed in both tissues [9]. In a later study, we found empirical evidence that ~90% of weighted gene–gene interaction networks identified in prefrontal cortex transcriptomes are preserved in peripheral blood [10]. Brain and blood also show significant overlap with respect to eQTLs [11, 12], signifying that shared genetic effects (albeit with small effect-sizes) may, in part, explain the comparability of gene expression in blood and the brain. Another advantage of capitalizing on human blood transcriptomes for brain gene expression imputation is that such data are widely available in public repositories and also can be generated de novo with relative ease and cost-effectiveness. Unlike DNA variants, transcriptome profiles in blood fluctuate over time, and they may reflect valuable information about corresponding temporal changes in the brain throughout development or over the course of an exposure or intervention.
Based on this evidence and logic, we sought to capitalize on the transcriptomic similarity between the brain and blood (and the easy accessibility of blood) to make predictions about gene expression in the brain solely based on observed expression in the periphery. Simultaneously, we aimed to develop an expression-based transcriptome-imputation method that complements existing cis-eQTL-based transcriptome-imputation methods. We achieved these goals with the Brain Gene-Expression and Network-Imputation Engine (BrainGENIE), which imputes brain tissue-specific gene-expression profiles based on gene-expression profiles assayed from peripheral blood. BrainGENIE is implemented in the R statistical environment and is distributed as freely available software (https://github.com/hessJ/BrainGENIE).
BrainGENIE is not the first or only cross-tissue transcriptome-imputation method, but it has unique strengths that compare quite favorably with other approaches. Tissue Expression Estimation using Blood Transcriptome (TEEBoT), like BrainGENIE, uses principal components (PCs) of peripheral blood transcriptomes to predict transcriptomes of other tissues [13]. TEEBoT was developed using an earlier release of GTEx (v.6) data; hence, its modeling of brain tissue-specific transcriptomes was limited to cerebellum and caudate due to sample-size restrictions. In contrast, BrainGENIE, which was built on the larger and more recent release of GTEx (v.8), enabling transcriptome imputation for 12 brain tissues (ten different brain regions). A second related method is B-GEX, which also modeled brain regional transcriptomes using an older version of GTEx that included fewer donors with paired blood and brain data [14]. Moreover, B-GEX utilizes individual gene transcripts from blood to predict brain gene expression, which captures less variance than PCs and limits predictive power. In short, because BrainGENIE was built on a better training dataset, uses PCs of whole-blood gene expression to optimize predictive power, and imputes transcriptomes for more brain tissues, it matches the strengths of competing methods and overcomes some of their limitations. Since there is no equivalent blood-based transcriptome-imputation method available that has modeled all regions of the brain like BrainGENIE, we benchmarked BrainGENIE against the most popular transcriptome-imputation method, PrediXcan. The two methods are conceptually similar, and there are data available from PrediXcan for all 12 brain tissues modeled by BrainGENIE, enabling direct comparisons. As such, we used the methodology of PrediXcan as a basis for developing BrainGENIE so that the results from the two methods could be directly compared. Comparing these two methods helped us to understand the differences (and points of convergence) between the use of blood-based gene-expression profiles versus eQTLs to impute region-specific gene expression in the brain, and illuminated the relative strengths and weaknesses of each approach. In addition to delivering a convenient software toolkit for BrainGENIE, we describe the application of the BrainGENIE method to real-world peripheral blood transcriptomic data to demonstrate the convergence of BrainGENIE-imputed data with disease-related gene expression changes directly measured in postmortem brain. Lastly, we lay the groundwork for future integrations of both blood gene expression and eQTLs to maximize the prediction of the brain transcriptome.
Methods
Training and evaluation of BrainGENIE
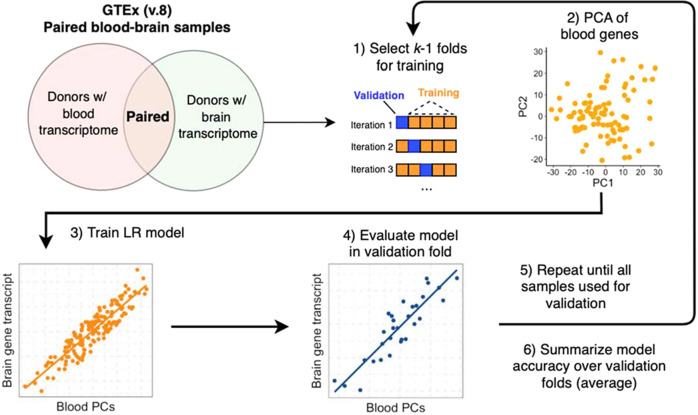
Procedures for normalizing the RNA-sequencing (RNAseq) data from GTEx are described in the Supplement (Supplementary Methods and Supplementary Fig. 1). The process used to train BrainGENIE is illustrated in Fig. 1. We performed a single fivefold cross-validation to estimate the predictive performance of BrainGENIE separately for each brain region. Paired blood–brain transcriptome profiles from GTEx donors were randomly assigned to the fivefolds. For each training subset, a principal component analysis (PCA) was performed on normalized blood transcriptome profiles, and linear regression was trained to predict brain tissue-specific expression levels per-gene using the top k = 5 (11% variance explained), k = 10 (41% variance explained), k = 20 (58% variance explained), and k = 40 PCs (80% variance explained) of whole-blood gene expression (resulting in fold = {1…5} by k = {5, 10, 20, 40} by gene = {1…ngenes} linear models). The normalized transcriptome profiles in the validation subsets were projected onto the PCs of the training subsets. The linear regression model used to train BrainGENIE was formulated as follows: Yi ~ β0+βi Xi+…+ ε, where Yi represents the expression level of a gene in the brain, β0 represents the intercept, βi Xi represents the product of the estimated regression coefficient and value of the ith PC, and ε represents the error term. Our initial work uncovered that prediction accuracies achieved by linear regression were as good as or better than elastic net regression (the model used by PrediXcan); linear regression is also computationally faster to train, thus was the chosen model for BrainGENIE. The trained models were then deployed in the validation set to estimate the predictive performance on unseen data. The metric for prediction performance was the coefficient of determination for observed and predicted per-gene expression levels (R2) in the hold-out fold. This process was repeated until each fold was used as the validation set, and per-gene prediction performance was averaged over the validation sets. In order to have a reasonable side-by-side comparison between BrainGENIE and PrediXcan, we adopted the same criterion for “significantly predicted” as adopted by PrediXcan; i.e., genes that could be predicted with a cross-validation (CV) R2 ≥ 0.01 and with Benjamini–Hochberg false-discovery rate-adjusted p value (FDR) <0.05. When comparing all models, the 40-PC BrainGENIE model exhibited the best performance in the training data in terms of average R2 and number of genes with significantly predicted gene expression levels, and was selected as the final model to deploy on the external test set (described below).
Fig. 1. Schematic illustrating the process for training BrainGENIE using paired blood–brain transcriptome data from the GTEx dataset.
BrainGENIE is trained using paired blood and brain transcriptome profiles collected by GTEx (v8) from adult donors. A single 5-fold cross-validation is performed to estimate the predictive performance of BrainGENIE separately for each brain region. BrainGENIE uses top principal components of transcriptome-wide blood-based gene expression as features to predict brain-regional gene expression levels. The metric used for prediction performance was the coefficinent of determination (R2) to measure how well predicted per-gene expression levels captured observed gene expression levels in the validation folds. Model performance was summarized over the 5 validation folds to obtain an estimate of prediction performance for BrainGENIE.
Accuracy of BrainGENIE versus PrediXcan
We used two-tailed t-tests (alpha = 0.05) to compare the prediction accuracy of BrainGENIE and PrediXcan as indexed by Pearson’s r coefficients for genes that met the criterion for being “significantly predicted”. Tests were performed separately for each of the 12 brain tissues, and Benjamini–Hochberg FDR corrections were applied to resultant p values to adjust for multiple testing. In addition, we used Pearson’s correlation tests to assess the similarity of prediction accuracies between BrainGENIE and PrediXcan for genes that both methods significantly predicted.
Enrichment of cross-disorder pleiotropic gene-sets
The goal of this analysis was to employ gene-set enrichment to determine if BrainGENIE and PrediXcan differed in their ability to significantly predict gene-sets that show significant association with major neuropsychiatric disorders by GWAS. Gene Ontology (GO) identifiers were obtained for 45 gene-sets identified by GWAS meta-analysis as having a shared association across eight neuropsychiatric disorders [15]. GO identifiers were annotated with HGNC gene symbols using the Molecular Signatures Database (v.6.2) [16]. We counted the number of genes in each pleiotropic gene set that were significantly predicted by either method and then performed one-tailed Fisher’s exact tests of enrichment. A significance threshold of FDRp < 0.05 was used to adjust for multiple comparisons.
Concordance with neuropsychiatric-related transcriptomic signatures
To determine how well BrainGENIE captures brain-relevant signatures for neuropsychiatric diagnosis identified in postmortem brain, we performed a Pearson’s correlation analysis to determine the concordance of imputed differential-expression signatures for schizophrenia (SCZ), bipolar disorder (BD), and autism spectrum disorder (ASD) derived by BrainGENIE with “ground-truth” differential-expression signatures from postmortem brain published by the PsychENCODE Consortium and the CommonMind Consortium [17, 18]. For this analysis, we deployed BrainGENIE models on completely independent blood-based transcriptome datasets for SCZ [19–24] (k studies = 7, n cases = 258, n controls = 241), BD [24–30] (k studies = 8, n cases = 335, n controls = 349), and ASD [31–38] (k studies = 5, n cases = 584, n controls = 431). Descriptions for each dataset are provided in Supplementary Table 1. Pre-processing and normalization steps used to prepare blood-based transcriptome profiles for the SCZ, BD, and ASD datasets are described in our previously published studies [10, 39, 40]. Details of our normalization procedure are available in the supplement (Supplementary Methods). The combined set of peripheral blood transcriptome data for each disorder was then supplied to BrainGENIE in order to impute transcriptome profiles for the frontal cortex using the 5-, 10-, 20-, and 40-PC models.
We estimated differential gene expression (DGE) in blood between affected cases and unaffected comparison individuals via combined-samples mega-analyses using linear regression models that covaried for study, age, sex, and abundance of circulating leukocytes inferred using CIBERSORT [41]. Similarly, we estimated DGE using predicted gene-expression profiles for the frontal cortex obtained from BrainGENIE using the same mega-analysis approach. We applied the PrediXcan tool for GWAS summary statistics (S-PrediXcan) to obtain genetically predicted DGE effect-sizes for SCZ, BD, and ASD using the latest GWAS summary statistics for each disorder [42–44]. Transcriptome-wide DGE effect-sizes for each disorder obtained from peripheral blood mega-analyses (t-values), BrainGENIE mega-analyses (t-values), and S-PrediXcan (z-scores) were then compared with DGE effect-sizes directly measured from postmortem brain using Pearson’s correlation test, which was chosen in order to assess the linear monotonic relationship between DGE signals derived from different methods.
Results
BrainGENIE prediction performance
Here, we summarize the performance of BrainGENIE represented by averages per brain region for the 12 brain regions being predicted. We found that BrainGENIE models trained with the top 40 PCs of blood-based transcriptome-wide gene expression yielded a higher average number of imputable genes per brain region relative to 5-, 10-, or 20-PC models. Thus, our summary focuses on the results derived using our 40-PC model. The prediction performance of BrainGENIE, measured by the average cross-validation R2, ranged from 0.03–0.56 for genes that met the criteria of significantly predicted in cross-validation (average CV R2 ≥ 0.01, FDRp < 0.05) (Table 1). The proportion of genes significantly predicted in the brain by BrainGENIE ranged from 16–59% of the total number of genes with detected expression in each brain tissue based on the GENCODE version 26 genome assembly (GRCh38) (mean number of genes = 8151; range: 2947–11,816 genes). The maximum average cross-validation prediction accuracy of BrainGENIE across all brain tissues ranged from R2 = 0.47–0.70. An average of 81% (range: 70–89%) of genes whose expression levels were significantly predicted by BrainGENIE were not significantly predicted by PrediXcan (Table 2). In contrast, an average of 65% of genes significantly predicted by PrediXcan were not significantly predicted by BrainGENIE (range: 50–88%) (Table 2). On average, 1672 genes were found to be significantly predicted by both BrainGENIE and PrediXcan across 12 brain tissues (range: 311 [substantia nigra]–3019 genes [cerebellum]) (Table 2). We found that expression levels of genes in the brain significantly correlated with the prediction accuracy of BrainGENIE (Supplementary Table 2A). Furthermore, higher expression levels of genes in the blood (Supplementary Table 2B) were significantly correlated with higher prediction accuracy of BrainGENIE, indicating that genes that are more abundant in the brain and/or blood are able to be more accurately predicted by BrainGENIE. The mean CV accuracy for BrainGENIE averaged across all brain regions for genes that were significantly predicted was R2 = 0.10, whereas the average CV value for PrediXcan was R2 = 0.15.
Table 1.
Prediction accuracy of BrainGENIE (40-PC model) computed via fivefold cross-validation of the GTEx Project (v.8) release across the 12 brain tissues.
| All predicted genes | Significantly predicted genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain tissue | n donors (paired blood–brain transcriptomes) | # of measured genes | % of genes measured in tissue compared to all annotated genes (GRCh38) | Mean training R2 | Mean CV R2 | Max CV R2 | # of genes | % of measured genes | % of genes measured in tissue compared to all annotated genes (GRCh38) | Mean training R2 | Mean CV R2 | Mean CV Pearson’s r |
| Amygdala | 88 | 18,957 | 34 | 0.65 | 0.05 | 0.39 | 4265 | 22 | 8 | 0.70 | 0.13 | 0.36 |
| Anterior cingulate cortex BA24 | 99 | 19,236 | 34 | 0.56 | 0.05 | 0.43 | 6799 | 35 | 12 | 0.59 | 0.11 | 0.33 |
| Caudate basal ganglia | 137 | 20,524 | 37 | 0.46 | 0.06 | 0.38 | 10,772 | 52 | 19 | 0.50 | 0.09 | 0.30 |
| Cerebellum (Fresh frozen) | 131 | 20,540 | 37 | 0.48 | 0.06 | 0.5 | 9573 | 47 | 17 | 0.53 | 0.11 | 0.33 |
| Cerebellum (PAXgene preserved) | 154 | 21,494 | 38 | 0.42 | 0.05 | 0.39 | 10,098 | 47 | 18 | 0.47 | 0.09 | 0.30 |
| Frontal cortex (PAXgene preserved) | 141 | 20,340 | 36 | 0.42 | 0.06 | 0.5 | 10,186 | 50 | 18 | 0.46 | 0.10 | 0.32 |
| Frontal Cortex (Fresh frozen) | 125 | 19,983 | 36 | 0.49 | 0.07 | 0.5 | 11,816 | 59 | 21 | 0.53 | 0.11 | 0.33 |
| Hippocampus | 122 | 20,189 | 36 | 0.47 | 0.03 | 0.46 | 4008 | 20 | 7 | 0.53 | 0.10 | 0.32 |
| Hypothalamus | 114 | 20,839 | 37 | 0.50 | 0.04 | 0.56 | 5749 | 28 | 10 | 0.56 | 0.11 | 0.33 |
| Nucleus accumbens basal ganglia | 148 | 20,884 | 37 | 0.43 | 0.05 | 0.46 | 11,252 | 54 | 20 | 0.47 | 0.09 | 0.30 |
| Putamen basal ganglia | 125 | 19,330 | 34 | 0.49 | 0.06 | 0.54 | 10,350 | 54 | 18 | 0.53 | 0.11 | 0.33 |
| Substantia nigra | 86 | 18,882 | 34 | 0.62 | 0.04 | 0.51 | 2947 | 16 | 5 | 0.69 | 0.14 | 0.37 |
The proportion of genes expressed in the whole blood accounts for 31% of all genes annotated in the GRCh38 genome assembly. The criteria for declaring genes “significantly predicted” is as follows: cross-validation [CV] R2 ≥ 0.01, CV FDRp ≤ 0.05. The total number of genes detected in each brain tissue for which BrainGENIE models were trained appears in the third column (“# of genes”). The proportion of genes that were significantly predicted by BrainGENIE from the total number of detected genes per brain tissue is presented in the ninth column (“% of genes”).
Table 2.
The number of genes for which brain tissue-specific expression levels can be reliably predicted by BrainGENIE (40-PC model), by PrediXcan, or by both methods.
| Brain tissue | BrainGENIE | PrediXcan | Overlap |
|---|---|---|---|
| Amygdala | 4265 | 2787 | 504 |
| Anterior cingulate cortex BA24 | 6799 | 3544 | 992 |
| Caudate basal ganglia | 10,772 | 5004 | 2225 |
| Cerebellum (Fresh frozen) | 9573 | 5753 | 2530 |
| Cerebellum (PAXGene preserved) | 10,098 | 6794 | 3019 |
| Frontal cortex (PAXGene preserved) | 10,186 | 5500 | 2408 |
| Frontal Cortex (Fresh frozen) | 11,816 | 4563 | 2289 |
| Hippocampus | 4008 | 3688 | 588 |
| Hypothalamus | 5749 | 3652 | 873 |
| Nucleus accumbens basal ganglia | 11,252 | 4851 | 2252 |
| Putamen basal ganglia | 10,350 | 4436 | 2067 |
| Substantia nigra | 2947 | 2559 | 311 |
We found overlap between brain regions in terms of the number of genes that were significantly predicted by BrainGENIE, which was lowest between substantia nigra and amygdala (912 genes) and highest between nucleus accumbens and frontal cortex [frozen] (7604 genes) (Supplementary Fig. 2A). The pattern of inter-regional similarity with respect to the number of genes significantly predicted by BrainGENIE mirrored the spatial pattern of similarity that exists between brain regions with respect to commonly expressed genes (Pearson’s r = 0.55, p = 2 × 10−6) (Supplementary Fig. 2B), indicating that BrainGENIE preserves and recapitulates the spatial relationship between areas of the brain.
The distributions of cross-validation R2 values produced by BrainGENIE and PrediXcan for all significantly predicted genes are shown in Supplementary Fig. 3. The shapes of the distributions found using BrainGENIE were similar to those for PrediXcan; however, two distinctions were consistently noted across brain tissues. First, PrediXcan featured heavier right-tails compared to BrainGENIE (Supplementary Fig. 3), indicative of PrediXcan having more genes with higher prediction accuracies. In contrast, BrainGENIE produced curves whose maxima were consistently shifted to the right relative to those of PrediXcan, indicative of greater average predictability with BrainGENIE. We found a significant association between the cross-validation performance of BrainGENIE in GTEx and the average RNA qualities of brain tissues (Supplementary Fig. 4A), wherein brain tissues that had better RNA quality of the brain tissues exhibited a larger number of imputable genes by BrainGENIE (Pearson’s r = 0.64, p = 0.024, Supplementary Fig. 4B).
Among those genes that are significantly predicted by both methods, PrediXcan showed significantly better overall prediction accuracy for gene expression levels in ten brain tissues (Supplementary Table 3). Prediction accuracies were statistically indistinguishable between BrainGENIE and PrediXcan for the remaining two brain tissues: the amygdala and substantia nigra (FDRp > 0.05, Supplementary Table 3). When evaluating the similarity of prediction accuracy among genes that are significantly predicted by both methods, BrainGENIE showed a small but significantly negative correlation with PrediXcan for genes in the amygdala, caudate, cerebellum (PAXgene preserved), frontal cortex (PAXgene preserved and fresh frozen), putamen, and nucleus accumbens (Supplementary Table 4). This finding is indicative of the methodological designs not converging to achieve consistent predictions, hence lending support to a joint imputation-modeling approach that capitalizes on blood-based gene expression and genotypes to impute brain region-specific gene expression levels.
Cross-disorder gene sets predicted by BrainGENIE versus PrediXcan
Supplementary Fig. 5 shows that 31 of the 45 pleiotropic gene sets for eight neuropsychiatric disorders identified by the Cross-Disorder Group of the Psychiatric Genomics Consortium [15] showed significant enrichment of genes significantly predicted by BrainGENIE. Genes significantly predicted by PrediXcan showed enrichment in 11 pleiotropic gene sets, though all 11 gene sets showed more significant enrichment of genes predicted by BrainGENIE. The null hypothesis of these analyses was that the number of genes significantly imputable by BrainGENIE or by PrediXcan does not relate to the membership of genes to cross-disorder-associated gene-sets. The alternate hypotheses are that either (or both) algorithms allow for significant imputation of more genes that participate in cross-disorder-associated gene ontologies than expected by chance.
Concordance of DGE changes related to neuropsychiatric disorders
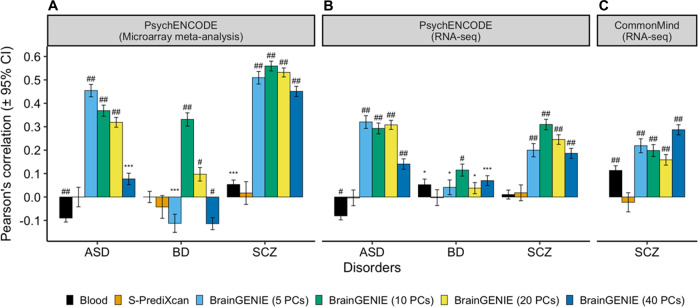
Transcriptome-wide DGE effect-sizes measured in peripheral blood for BD and SCZ demonstrated small but significant correlations with DGE effect-sizes directly measured in postmortem brain (Fig. 2) (Pearson’s r range: 0.05–0.11). The total number of genes that were represented in our analysis are provided in Supplementary Table 5. Conversely, DGE effect-sizes for ASD measured in peripheral blood showed significant inverse correlations with DGE effect-sizes directly measured in postmortem brain (Fig. 2) in the PsychENCODE microarray meta-analysis (Pearson’s r = −0.09, p = 1.7 × 10−25) and RNAseq analysis (Pearson’s r = −0.08, p = 3.3 × 10−20), which may reflect age differences between samples considering that individuals in the peripheral blood datasets were predominantly children whereas those in the postmortem brain studies were predominantly adults. DGE effect-sizes found using S-PrediXcan were not significantly correlated with postmortem brain DGE effect-sizes for SCZ, BD, or ASD (Fig. 2). Conversely, DGE effects estimated from predicted genes’ expression profiles in brain using BrainGENIE were significantly correlated with results directly measured in postmortem brain for ASD, BD, and SCZ (Fig. 2). The strongest correlation that emerged was between DGE effect-sizes obtained using BrainGENIE (ten PCs) and DGE effect-sizes directly measured in postmortem brain for SCZ found by the PsychENCODE Consortium’s microarray meta-analysis (Pearson’s r = 0.56, 95% CI: 0.54–0.58, n genes = 4130, p < 1.0 × 10−300, Fig. 2A). The DGE correlations for ASD, BD, and SCZ found between BrainGENIE and postmortem brain showed significant replication in an independent PsychENCODE Consortium cohort profiled via RNAseq (Fig. 2B). Furthermore, the convergence between BrainGENIE-imputed and measured postmortem brain DGE effect-sizes for SCZ was replicated in a second independent cohort from the CommonMind Consortium, with the strongest concordance found for the 40-PC model of BrainGENIE (Pearson’s r = 0.28, 95% CI: 0.26–0.31, n genes = 6933, p = 1.1 × 10−131, Fig. 2C). When we restricted our analysis to genes differentially expressed in postmortem brain (FDRp < 0.05), the DGE agreement between BrainGENIE-imputed brain gene expression and postmortem brain measured gene expression was as good or better (Supplementary Fig. 6). Scatterplots of the DGE agreement for ASD, BD, and SCZ are provided in Supplementary Fig. 7. The DGE agreement between BrainGENIE-imputed brain gene expression and postmortem brain measured gene expression was significantly stronger than between peripheral blood and postmortem brain for ASD, BD, and SCZ for one or more BrainGENIE models (z-test p values <0.05, Supplementary Table 6). Similarly, measured DGE signals from the postmortem brain were significantly more concordant with those predicted from BrainGENIE than from S-PrediXcan for ASD, BD, and SCZ (Supplementary Table 6). We also evaluated results published by the PsychENCODE Consortium from their approach using FUSION/TWAS to prioritize genes associated with ASD, BD, and SCZ in their cohort. Similar to our findings from S-PrediXcan, the DGE effect-sizes imputed using FUSION/TWAS for ASD, BD, and SCZ were not correlated with the DGE effect-sizes directly measured in postmortem brain (ASD: Pearson’s r = 0.003, p = 0.7; BD: Pearson’s r = 0.005, p = 0.6; SCZ: Pearson’s r = 0.013, p = 0.12) [45].
Fig. 2. BrainGENIE recapitulates disorder-related changes in gene expression found in postmortem brain.
Concordance of case-control differential gene expression (DGE) signals obtained by BrainGENIE and S-PrediXcan compared to A DGE signals derived from postmortem cortical microarray meta-analyses for ASD, BD, and SCZ, B DGE signals derived from RNA-sequencing analysis for ASD, BD, and SCZ by the PsychENCODE Consortium, and C DGE signals obtained from postmortem prefrontal cortex RNA-sequencing analysis for SCZ by the CommonMind Consortium. ASD autism spectrum disorder, BD bipolar disorder, and SCZ schizophrenia. Symbols for significance thresholds: p < 0.05 (*), FDRp ≤ 1 × 10−5 (***), FDRp ≤ 1 × 10−10 (#), FDRp ≤ 1 × 10−20 (##).
Discussion
This study introduced and benchmarked a computational method called BrainGENIE, which predicts brain region-specific gene expression levels based on peripheral blood transcriptomes. Over the past decade, there has been rapid growth in the number of blood-based transcriptome studies aimed at identifying biomarkers for neuropsychiatric disorders. This has led to a vast amount of useful data that may hold untapped information about the brain. Much of the raw data from published blood-based transcriptome studies of neuropsychiatric disorders can be readily downloaded from public repositories (i.e., Gene Expression Omnibus [GEO] and ArrayExpress) or made available to investigators with controlled access (i.e., dbGaP, NIMHGR, and Synapse). It is, therefore, possible to further mine those stores of transcriptome data with BrainGENIE, thus generating novel mechanistic hypotheses about the disease and advancing our understanding of brain disorders in a way that is clearly superior to direct correlation of blood and brain measures.
BrainGENIE exploits PCA as an efficient method for dimensionality-reduction while capturing more variability in the blood transcriptome for prediction compared to individual gene transcripts. PCA helps to reduce the potential of overfitting; i.e., imposing a limit on the number of input features steers a model away from “learning” random noise and fails to generalize to external data. However, the risk of overfitting is not eliminated by PCA alone. The components we derived using PCA are loosely related to “eigengenes”, which are clusters of tightly co-expressed genes identified by the data-driven clustering method Weighted Gene Co-expression Network Analysis (WGCNA) [46]. A fundamental difference between PCA and WGNCA lies in the fact that genes can load onto multiple PCs as opposed to eigengenes being derived with non-overlapping sets of genes. Future work will aim to model networks of genes that are preserved across blood and the brain to improve BrainGENIE’s interpretability. We applied a standard classification approach of k-fold cross-validation to estimate the ability of BrainGENIE to generalize its predictions to unseen data. Further validation is warranted to determine the generalizability of our prediction models for other brain tissues and with an external dataset that is closely matched to the demographic and technical parameters observed in our training set, should one become available. In the future, modifications to BrainGENIE may allow for subsets of genes to be best predicted by PCs and others by individual gene transcripts, or collections of a few (or more) closely correlated transcripts. To improve the prediction performance of BrainGENIE, methods that can account for nonlinear mapping between blood and brain transcriptome profiles may be incorporated.
The predictive performance of BrainGENIE was affected by a number of factors, including the number of samples available for training and the quality of the extracted RNAs. Brain tissues that had large sample sizes (i.e., frontal cortex) showed better prediction performance than brain regions with fewer samples (i.e., amygdala and substantia nigra). In addition, imputation performance improved with the RNA quality of the brain tissues. Differences in imputation performance also were seen between pairs of re-sampled tissue collected from the frontal cortex and cerebellum. The pair of tissues collected and preserved in PAXgene fixative exhibited lower RIN values (likely due to degraded RNAs) and lower imputation performance than the pair of brain tissues that were shipped to the University of Miami Endowment Brain Bank for collection and preservation by flash-freezing. Though the differences in imputation performance of flash-frozen and chemically preserved brain tissues were small, we recommend that investigators use the BrainGENIE models derived from flash-frozen brain tissues. Furthermore, there were, on average, 48 fewer donors (range: 28–63) with paired blood–brain transcriptome data in GTEx (the required input for BrainGENIE) than donors with paired genetic and transcriptome data (the required input for PrediXcan). It is challenging to draw strong conclusions about differences in model performance for individual genes between BrainGENIE and PrediXcan as PrediXcan had more power, and yet BrainGENIE outperformed PrediXcan on a number of benchmarks. Thus, global differences between the methods could not be explained by variation in sample size alone, and even as BrainGENIE was limited by a smaller sample size for model-training, we found that BrainGENIE could impute a substantial fraction of genes that were not imputable using PrediXcan. This suggests that non-genetic components of gene expression ignored by PrediXcan models hold significant information for transcriptome imputation.
GTEx’s transcriptomic data were derived from bulk postmortem brain tissue; thus, we did not model gene expression for any specific brain-cell-type. We instead modeled the cross-tissue overlap at the level of cell mixtures in the brain and blood for BrainGENIE. It is possible that commonalities seen between brain and blood gene expression could be driven by a possible shared lineage between macrophages and microglia [47, 48]. Specificity of brain cell-type transcriptome imputation with BrainGENIE may be achieved with single-cell transcriptomics, but this is not feasible at the moment due to the lack of available data.
The current version of BrainGENIE can predict a substantial proportion of variance in expression levels for 2947 to 11,816 genes in the human brain (depending on the brain region), which accounts for about 16 to 59% of the brain transcriptome. Prior iterations of BrainGENIE made continual improvements in the number of significantly predicted genes, and the variance accounted for in brain tissue-specific gene-expression levels, by moving from an approach that used individual gene transcripts in blood to predict brain gene-expression levels to the current approach of using PCs of blood gene expression. This suggests that further refinement of our models will continue to improve predictions until they reach their (unknown) maximum per-gene and per brain region. As one would expect, not all or even most genes are imputable with BrainGENIE, but the number of new genes that can be imputed with BrainGENIE and not by PrediXcan is considerable. The amount of overlap between BrainGENIE and PrediXcan in terms of genes whose expression can be significantly predicted was relatively small. In addition, prediction accuracies were not strongly correlated between BrainGENIE and PrediXcan, indicating that the different modeling approaches achieve partially orthogonal outcomes when predicting brain transcriptomes. This suggests that there is value to integrating BrainGENIE and PrediXcan for a combined and complementary approach to transcriptome imputation wherever genotypes and blood gene-expression data are available from the same individuals. Ideally, the strengths of multiple modeling approaches, like those in BrainGENIE, PrediXcan, and others, would be combined into a unified framework (or through the integration of outputs from multiple disparate models) to deliver a holistic portrait of the landscape of the human brain transcriptome.
We suggest the following tool selection depending on the type of data available for transcriptome imputation: (1) if only transcriptome data were available from blood, use BrainGENIE, (2) if only GWAS data are available, use PrediXcan (or a derivative), (3) if blood transcriptome and GWAS data are available, use BrainGENIE and PrediXcan (or a derivative) to achieve best-predicted expression levels on a per-gene basis depending on the target brain region. For genes that are predictable by both methods, use the method that achieved better accuracy for the specific gene being imputed in the target brain region, which is often PrediXcan.
Our results showed that transcriptome-wide DGE effect-sizes observed directly in postmortem brain were in better agreement with DGE effect-sizes predicted using BrainGENIE than with DGE effect-sizes found in analyses of peripheral blood and those imputed by PrediXcan. This advantage of BrainGENIE over peripheral blood and PrediXcan was most striking for SCZ but was still evident for BD and ASD. The Concordance of DGE effect-sizes between BrainGENIE and postmortem brain varied based on the number of PCs included in the imputation models. This finding may encourage investigators to parameterize the number of PCs for BrainGENIE based on the model that yields the best overall prediction accuracy. However, it is important to consider which genes are included (or lost) or more significantly predicted when adjusting the number of PCs used by BrainGENIE, as this can be relevant for downstream analyses. For example, a study focused on the frontal-cortical expression of the SCZ risk-gene complement component 4 (C4A) would favor the 20-PC model (average CV R2 = 0.20, p = 3.8 × 10−7) as it yielded higher accuracy than the BrainGENIE models with 5-, 10-, and 40 PCs. Alternatively, an example wherein the 40-PC model yields better imputation is the frontal cortex expression of Synaptic Ras GTPase Activating Protein 1 (SYNGAP1), a leading risk gene for autism (CV R2 = 0.17, p = 5.6 × 10−7). Besides recapitulating disease-related effects with BrainGENIE, it would be valuable to know if disease status impacts the prediction performance of our models. To test for disease-related differences in prediction performance, however, would require paired blood–brain transcriptomes from the same affected individuals with characteristics that are well-matched to the distribution of our GTEx training samples; such a sample, to our knowledge, does not yet exist, highlighting a critical priority for future research.
We applied statistical corrections to remove effects of age, sex, and genetic ancestry from the gene-expression data so that those factors would not systematically bias our models. Still, it is possible that the characteristics of the GTEx sample are not fully representative of the entire population. For example, donors in the GTEx Project were predominantly of European ancestry, hence limiting the applicability of transcriptome-imputation across diverse ancestral groups. Amassing large sample sizes that encompass a broader range of characteristics (e.g., environmental exposures, genetic background, and demographics, to name a few) would allow BrainGENIE to make use of more biological (useful) variability that may help increase the number of significantly predicted genes and improve variance accounted for in gene expression levels of target tissues. Increasing sample ascertainment from diverse human populations, coupled with deeper phenotyping, are strategic ways to enable more effective transcriptome-imputation modeling.
In sum, BrainGENIE is a validated approach to investigating brain region-specific gene-expression profiles. We demonstrated that gene-expression changes associated with disease and imputed in the brain by BrainGENIE were in better agreement (relative to cis-eQTL-based predictions of gene expression by PrediXcan and to gene-expression changes detected in peripheral blood) with corresponding gene-expression changes detected in studies of postmortem brain. The main challenge of transcriptome-imputation is identifying a model and set of predictor variables that can efficiently and significantly predict gene-expression levels while ensuring that downstream analyses of predicted expression levels can yield biologically meaningful results. PrediXcan and FUSION, respectively, can significantly predict an average of 18 and 16% of the brain transcriptome (compared with an average of 40% by BrainGENIE). Those methods have been successful in identifying novel tissue-specific dysregulation of gene expression in complex disorders. A strength of BrainGENIE is that it can capture regulatory impacts of genetic and non-genetic factors on gene expression that are not yet modeled by cis-eQTL-based methods. BrainGENIE fills a void in the study of the brain transcriptome by both allowing analyses of genes that were not previously imputable and improving the predictability of disease-relevant gene sets that PrediXcan can only partially impute.
Though we showed that BrainGENIE has advantages over conceptually similar methods, our intention is for it to serve as a complement to genetic-based transcriptome-imputations methods. In practice, our recommendation would be to integrate BrainGENIE with other methods, whenever possible, to boost confidence in gene-disease associations, hence permitting a deeper understanding of complex phenotypes. As such, BrainGENIE offers an important function in systems-level research into the brain and serves as a valuable hypothesis-generating tool for mechanistic studies. Potential applications of BrainGENIE are far-reaching and would be best indicated (relative to PrediXcan and FUSION) to study gene expression longitudinally, including: across developmental timepoints of the brain, pre- and post-exposure (e.g., environmental risks, traumatic life experiences), and modeling the effects of medication or other clinical interventions. BrainGENIE also could be used to impute brain region-specific transcriptomes at any point in a person’s lifetime, opening the possibility that we could find causal and longitudinal mechanisms underlying neuropsychiatric disease. The reach of our toolset can be extended with additional developments to achieve reliable imputations of cell-type specific transcriptomes, and transcriptomes of other inaccessible tissues, as well as models of alternatively spliced mRNAs and short and long noncoding RNAs, all of which are feasible objectives.
Supplementary information
Acknowledgements
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (commonfund.nih.gov/GTEx). Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\Leidos Biomedical Research, Inc. subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through a Leidos Biomedical Research, Inc. subcontract to Van Andel Research Institute (10ST1035). Additional data repository and project management were provided by Leidos Biomedical Research, Inc.(HHSN261200800001E). The Brain Bank has supported supplements to the University of Miami grant DA006227. Statistical Methods development grants were made to the University of Geneva (MH090941 and MH101814), the University of Chicago (MH090951, MH090937, MH101825, and MH101820), the University of North Carolina—Chapel Hill (MH090936), North Carolina State University (MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University (MH101810), and to the University of Pennsylvania (MH101822). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000424.v8.p2. Dr. Hess is supported by grants from the US National Institute of Mental Health (R21MH126494), the US National Institute of Neurological Disorders and Stroke (R01NS128535), and NARSAD: The Brain & Behavior Research Foundation. Dr. Kong is supported by grants R01MH107205 and R24OD024622. Dr. Faraone is supported by the European Union’s Horizon 2020 research and innovation program under grant agreement No 965381; NIMH grants U01AR076092-01A1, 1R21MH1264940, R01MH116037; 1R01NS128535 – 01; Oregon Health and Science University, Otsuka Pharmaceuticals, Noven Pharmaceuticals Incorporated, and Supernus Pharmaceutical Company. Dr. Glatt is supported by grants from the US National Institutes of Health (R01MH101519, R01AG054002, R01AG064955, and R21MH126494), the Sidney R. Baer, Jr. Foundation, and NARSAD: The Brain & Behavior Research Foundation.
Author contributions
JLH and SJG conceived the study and planned the analyses. JLH carried out the analyses and prepared the first draft of the manuscript under the supervision of SJG. All authors contributed to the study design, interpretation of results, and critical revisions. All authors approved the manuscript before submission.
Data availability
Data and source code can be accessed from the following GitHub repository: https://github.com/hessJ/BrainGENIE.
Code availability
Data and source code can be accessed from the following GitHub repository: https://github.com/hessJ/BrainGENIE.
Competing interests
In the past year, Dr. Faraone received income, potential income, travel expenses continuing education support, and/or research support from Aardvark, Aardwolf, Akili, Atentiv, Corium, Genomind, Ironshore, Medice, Noven, Otsuka, Sandoz, Sky Therapeutics, Supernus, Tris, and Vallon. With his institution, he has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. In previous years, he received support from Alcobra, Arbor, Aveksham, Axsome, CogCubed, Eli Lilly, Enzymotec, Impact, Janssen, KemPharm, Lundbeck/Takeda, Shire/Takeda, McNeil, NeuroLifeSciences, Neurovance, Novartis, Pfizer, Rhodes, Shire, and Sunovion. He also receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health; Oxford University Press: Schizophrenia: The Facts; and Elsevier: ADHD: Non-Pharmacologic Interventions. In addition, he is the program director of www.adhdinadults.com. In the past year, Dr. Glatt has received royalties from a book published by Oxford University Press: Schizophrenia: The Facts, and consulting fees from Cohen Veterans Bioscience. Dr. Cairns is supported by NHMRC project grants (1147644 and 1188493) and an NHMRC Senior Research Fellowship (1121474), and a University of Newcastle College of Health Medicine and Wellbeing, Gladys M Brawn Senior Fellowship.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Stephen J. Glatt, Email: stephen.glatt@psychgenelab.com
Neuropsychiatric Consortium for Analysis and Sharing of Transcriptomes:
Natalie Jane Beveridge, Vaughan Carr, Simone de Jong, Erin Gardiner, Brian Kelly, Nishantha Kumarasinghe, Roel Ophoff, Ulrich Schall, Rodney Scott, Boryana Stamova, and Paul Tooney
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-023-02390-w.
References
- 1.James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 Diseases and Injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018. 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed]
- 2.Hall LS, Medway CW, Pain O, Pardiñas AF, Rees EG, Escott-Price V, et al. A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum Mol Genet. 2020. 10.1093/hmg/ddz253. [DOI] [PMC free article] [PubMed]
- 3.Liao C, Laporte AD, Spiegelman D, Akçimen F, Joober R, Dion PA, et al. Transcriptome-wide association study of attention deficit hyperactivity disorder identifies associated genes and phenotypes. Nat Commun. 2019;10:4450. doi: 10.1038/s41467-019-12450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng B, Liang X, Wen Y, Li P, Zhang L, Ma M, et al. Integrative analysis of transcriptome-wide association study data and messenger RNA expression profiles identified candidate genes and pathways for inflammatory bowel disease. J Cell Biochem. 2019. 10.1002/jcb.28744. [DOI] [PubMed]
- 5.Smit DJA, Cath D, Zilhão NR, Ip HF, Denys D, den Braber A, et al. Genetic meta-analysis of obsessive-compulsive disorder and self-report compulsive symptoms. Am J Med Genet B Neuropsychiatr Genet. 2019;183:208–16. [DOI] [PMC free article] [PubMed]
- 6.Hohman TJ, Dumitrescu L, Cox NJ, Jefferson AL. Genetic resilience to amyloid related cognitive decline. Brain Imag Behav. 2017. 10.1007/s11682-016-9615-5. [DOI] [PMC free article] [PubMed]
- 7.Nagpal S, Meng X, Epstein MP, Tsoi LC, Patrick M, Gibson G, et al. TIGAR: an improved Bayesian tool for transcriptomic data imputation enhances gene mapping of complex traits. Am J Hum Genet. 2019 doi: 10.1016/j.ajhg.2019.05.018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Voloudakis G, Rajagopal VM, Readhead B, Dudley JT, Schadt EE, et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat Commun. 2019 doi: 10.1038/s41467-019-11874-7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain ‘-omes’. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 10.Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, et al. Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr Res. 2016;176:114–24. doi: 10.1016/j.schres.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenzie M, Henders AK, Caracella A, Wray NR, Powell JE. Overlap of expression Quantitative Trait Loci (eQTL) in human brain and blood. BMC Med Genomics. 2014;7:31. doi: 10.1186/1755-8794-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018. 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed]
- 13.Basu M, Wang K, Ruppin E, Hannenhalli S. Predicting tissue-specific gene expression from whole blood transcriptome. Sci Adv. 2021. 10.1126/sciadv.abd6991. [DOI] [PMC free article] [PubMed]
- 14.Xu W, Liu X, Leng F, Li W. Blood-based multi-tissue gene expression inference with Bayesian ridge regression. Bioinformatics 2020. 10.1093/bioinformatics/btaa239. [DOI] [PubMed]
- 15.Lee PH, Anttila V, Won H, Feng YCA, Rosenthal J, Zhu Z, et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019. 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed]
- 16.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011. 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed]
- 17.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018. 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed]
- 18.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner EJ, Cairns MJ, Liu B, Beveridge NJ, Carr V, Kelly B, et al. Gene expression analysis reveals schizophrenia-associated dysregulation of immune pathways in peripheral blood mononuclear cells. J Psychiatr Res. 2013;47:425–37. doi: 10.1016/j.jpsychires.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glatt SJ, Cohen OS, Faraone SV, Tsuang MT. Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:382–92. doi: 10.1002/ajmg.b.31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glatt SJ, Chandler SD, Bousman CA, Chana G, Lucero GR, Tatro E, et al. Alternatively spliced genes as biomarkers for schizophrenia, bipolar disorder and psychosis: a blood-based spliceome-profiling exploratory study. Curr Pharmacogenomics Person Med. 2009;7:164–88. doi: 10.2174/1875692110907030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumarasinghe N, Beveridge NJ, Gardiner E, Scott RJ, Yasawardene S, Perera A, et al. Gene expression profiling in treatment-naive schizophrenia patients identifies abnormalities in biological pathways involving AKT1 that are corrected by antipsychotic medication. Int J Neuropsychopharmacol. 2013;16:1483–503. doi: 10.1017/S1461145713000035. [DOI] [PubMed] [Google Scholar]
- 23.de Jong S, Boks MPM, Fuller TTF, Strengman E, Janson E, de Kovel CGF, et al. A gene co-expression network in whole blood of Schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS ONE. 2012;7:e39498. doi: 10.1371/journal.pone.0039498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuang MT, Nossova N, Yager T, Tsuang M-M, Guo S-C, Shyu KG, et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- 25.Krebs CE, Ori APS, Vreeker A, Wu T, Cantor RM, Boks MPM, et al. Whole blood transcriptome analysis in bipolar disorder reveals strong lithium effect. Psychol Med. 2019. 10.1017/S0033291719002745. [DOI] [PubMed]
- 26.Beech RD, Lowthert L, Leffert JJ, Mason PN, Taylor MM, Umlauf S, et al. Increased peripheral blood expression of electron transport chain genes in bipolar depression. Bipolar Disord. 2010;12:813–24. doi: 10.1111/j.1399-5618.2010.00882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E, et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:494–502. doi: 10.1002/ajmg.b.31006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clelland CL, Read LL, Panek LJ, Nadrich RH, Bancroft C, Clelland JD. Utilization of never-medicated bipolar disorder patients towards development and validation of a peripheral biomarker profile. PLoS ONE. 2013;8:e69082. doi: 10.1371/journal.pone.0069082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padmos RC, Hillegers MHJ, Knijff EM, Vonk R, Bouvy A, Staal FJT, et al. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch Gen Psychiatry. 2008;65:395–407. doi: 10.1001/archpsyc.65.4.395. [DOI] [PubMed] [Google Scholar]
- 30.Savitz J, Frank MB, Victor T, Bebak M, Marino JH, Bellgowan PSF, et al. Inflammation and neurological disease-related genes are differentially expressed in depressed patients with mood disorders and correlate with morphometric and functional imaging abnormalities. Brain Behav Immun. 2013;31:161–71. doi: 10.1016/j.bbi.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y, Green PG, Stamova B, Hertz-Picciotto I, Pessah IN, Hansen R, et al. Correlations of gene expression with blood lead levels in children with autism compared to typically developing controls. Neurotox Res. 2011. 10.1007/s12640-009-9126-x. [DOI] [PMC free article] [PubMed]
- 32.Enstrom A, Krakowiak P, Onore C, Pessah IN, Hertz-Picciotto I, Hansen RL, et al. Increased IgG4 levels in children with autism disorder. Brain Behav Immun. 2009. 10.1016/j.bbi.2008.12.005. [DOI] [PMC free article] [PubMed]
- 33.Stamova B, Green PG, Tian Y, Hertz-Picciotto I, Pessah IN, Hansen R, et al. Correlations between gene expression and mercury levels in blood of boys with and without autism. Neurotox Res. 2011. 10.1007/s12640-009-9137-7. [DOI] [PMC free article] [PubMed]
- 34.Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN. The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect. 2006. 10.1289/ehp.8483. [DOI] [PMC free article] [PubMed]
- 35.Glatt SJ, Tsuang MT, Winn M, Chandler SD, Collins M, Lopez L, et al. Blood-based gene expression signatures of infants and toddlers with autism. J Am Acad Child Adolesc Psychiatry. 2012;51:934–44.e2. doi: 10.1016/j.jaac.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong SW, Collins CD, Shimizu-Motohashi Y, Holm IA, Campbell MG, Lee IH, et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PLoS ONE. 2012;7:e49475. doi: 10.1371/journal.pone.0049475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong SW, Shimizu-Motohashi Y, Campbell MG, Lee IH, Collins CD, Brewster SJ, et al. Peripheral blood gene expression signature differentiates children with autism from unaffected siblings. Neurogenetics. 2013. 10.1007/s10048-013-0363-z. [DOI] [PMC free article] [PubMed]
- 38.Alter MD, Kharkar R, Ramsey KE, Craig DW, Melmed RD, Grebe TA, et al. Autism and increased paternal age related changes in global levels of gene expression regulation. PLoS ONE. 2011. 10.1371/journal.pone.0016715. [DOI] [PMC free article] [PubMed]
- 39.Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, et al. Transcriptomic abnormalities in peripheral blood in bipolar disorder, and discrimination of the major psychoses. Schizophr Res. 2019. 10.1016/j.schres.2019.07.036. [DOI] [PMC free article] [PubMed]
- 40.Tylee DS, Hess JL, Quinn TP, Barve R, Huang H, Zhang-James Y, et al. Blood transcriptomic comparison of individuals with and without autism spectrum disorder: A combined-samples mega-analysis. Am J Med Genet Part B Neuropsychiatr Genet. 2017;174:181–201. doi: 10.1002/ajmg.b.32511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015. 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed]
- 42.Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019. 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed]
- 44.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019. 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed]
- 45.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018. 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed]
- 46.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan WY, Kohsaka S, Rezaie P. The origin and cell lineage of microglia-New concepts. Brain Res Rev. 2007. 10.1016/j.brainresrev.2006.11.002. [DOI] [PubMed]
- 48.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014. 10.1038/nrn3722. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and source code can be accessed from the following GitHub repository: https://github.com/hessJ/BrainGENIE.
Data and source code can be accessed from the following GitHub repository: https://github.com/hessJ/BrainGENIE.